Abstract
The homeodomain protein Pit-1 cooperates with the basic-leucine zipper protein CCAAT/enhancer binding protein α (C/EBPα) to control pituitary-specific prolactin gene transcription. We previously observed that C/EBPα was concentrated in regions of centromeric heterochromatin in pituitary GHFT1–5 cells and that coexpressed Pit-1 redistributed C/EBPα to the subnuclear sites occupied by Pit-1. Here, we used fluorescence resonance energy transfer microscopy to show that when C/EBPα was recruited by Pit-1, the average distance separating the fluorophores labeling the proteins was less than 7 nm. A mutation in the Pit-1 homeodomain, or truncation of the C/EBPα transactivation domain disrupted the redistribution of C/EBPα by Pit-1. Fluorescence resonance energy transfer analysis revealed that the mutant Pit-1 still associated with C/EBPα, and the truncated C/EBPα still associated with Pit-1, but these interactions were preferentially localized in regions of centromeric heterochromatin. In contrast, a truncation in C/EBPα that prevented DNA binding also blocked its association with Pit-1, suggesting that the binding of C/EBPα to DNA is a critical first step in specifying its association with Pit-1. These findings indicated that the protein domains that specify the interaction of Pit-1 and C/EBPα are separable from the protein domains that direct the positioning of the associated proteins within the nucleus. The intimate association of Pit-1 and C/EBPα at certain sites within the living cell nucleus could foster their combinatorial activities in the regulation of pituitary-specific gene expression.
It is the combinatorial interactions between the pituitary-specific homeodomain (HD) protein Pit-1 and other gene-regulatory proteins that controls the transcription of the prolactin (PRL) and GH genes in anterior pituitary cells (1, 2). The pituitary cell-selective programs of gene expression initiated by Pit-1 require the assembly of particular nuclear protein complexes that function to modify chromatin structure and recruit the general transcription apparatus to target genes. Previous observations showed that both Pit-1 and the CCAAT/enhancer binding protein α (C/EBPα) bind to the promoters of the PRL and GH genes where they cooperate to control transcription (3, 4). Recently, the use of the genetically encoded fluorescent proteins (FPs) as in vivo labels has begun to provide insight into how proteins are positioned within the nucleus of living cells (5–8). It is thought that the positioning of proteins at distinct subnuclear sites may function to foster the cooperative protein interactions necessary for the assembly of gene-specific protein complexes (9–17). Here, we use this approach to visualize the relative spatial positioning of C/EBPα and Pit-1 in the nucleus of single living pituitary cells.
In prior studies, we showed that when green fluorescent protein (GFP)-C/EBPα was expressed in the somatolactotrope progenitor GHFT1–5 cell line, it was preferentially positioned to regions of centromeric heterochromatin in the nucleus of the mouse pituitary cells (18–20). This pattern was identical to that of the endogenous C/EBPα protein observed in mouse 3T3-L1 cells (20, 21). Significantly, we found that the coexpression of Pit-1 with C/EBPα resulted in the redistribution of C/EBPα from regions of centromeric heterochromatin to the intranuclear sites occupied by Pit-1. This recruitment activity of Pit-1 for C/EBPα was disrupted by a point mutation in Pit-1 HD that is commonly associated with combined pituitary hormone deficiency (CPHD) syndrome in humans (20). These observations indicated a potential role for Pit-1 in organizing the distribution of C/EBPα in the nucleus of pituitary cells.
It was important to determine how the coexpressed Pit-1 protein affected the redistribution of C/EBPα in the pituitary cell nucleus. The actions of Pit-1 could result from its direct interaction with C/EBPα, or from their mutual association within a nuclear protein complex. Alternatively, the expression of Pit-1 could have global effects on nuclear structure that indirectly alter the positioning of C/EBPα. The investigation of the spatial relationship between proteins using light microscopy is limited by the diffraction of light to approximately 200 nm, and objects that are closer together than this will appear as a single object. Therefore, considerable distances may actually separate proteins that appear to be colocalized by fluorescence microscopy. However, a 50-fold improvement in the spatial resolution of the light microscope can be attained by using the technique of fluorescence resonance energy transfer (FRET). FRET microscopy detects the result of the radiationless transfer of energy from a donor fluorophore to nearby acceptor fluorophores, and color variants of the genetically encoded FPs are suitable as donor and acceptor pairs (22–30). Because the efficiency of energy transfer varies inversely with the sixth power of the distance separating the donor and acceptor fluorophores, the distance over which FRET can occur is limited to less than 7 nm (22–30).
In the present study, we used the approach of acceptor photobleaching FRET microscopy to demonstrate the close physical association of Pit-1 and C/EBPα in the nucleus of living pituitary cells. We observed significant nuclear-localized FRET signals from cells in which the coexpressed Pit-1 redistributed C/EBPα, indicating that the average distance separating the fluorophores labeling the proteins was less than 7 nm. We show that mutations in both Pit-1 and C/EBPα, which disrupt the redistribution activity, still closely associated with their wild-type partners in regions of centromeric heterochromatin. These observations indicated that the Pit-1 HD and conserved regions in the transcriptional activation domain of C/EBPα were required for the recruitment of C/EBPα to the nuclear sites occupied by Pit-1. In contrast, we observed that a truncation of C/EBPα that prevented the binding to DNA failed to associate with Pit-1, suggesting that interactions with DNA are critical for specifying the formation of a complex involving Pit-1 and C/EBPα. These results show the close spatial relationship of Pit-1 and C/EBPα at specific sites within the living cell nucleus that may foster the combinatorial activities of these proteins in the regulation of pituitary gene expression.
Results
The Amino-Terminal Transactivation Regions of C/EBPα Are Required for Interactions with Pit-1
Pit-1 and C/EBPα act cooperatively to induce PRL transcription (4, 20). When coexpressed in pituitary cells, C/EBPα was recruited to the nuclear sites occupied by Pit-1, and disruption of the Pit-1 HD blocked the recruitment activity for C/EBPα (20). Here, we determined the domains of C/EBPα that are necessary for its cooperative actions with Pit-1 and the recruitment from regions of centromeric heterochromatin.
Deletion of the various conserved regions (CR, Ref. 31) of C/EBPα were prepared. C/EBPα lacking CR-1 (amino acids 3–68), CR-2 (amino acids 68–96), and CR-1, CR-2, and CR-3 (amino acids 3–154) were each characterized for their cooperative actions with Pit-1 at the PRL promoter (Fig. 1A). Because mouse GHFT1–5 cells express a low level of Pit-1 (3), we assessed the functional interactions involving C/EBPα and Pit-1 at the PRL promoter in nonpituitary, human HeLa cells. HeLa cells were transfected with the indicated expression plasmids encoding each of the C/EBPα deletion mutants either alone or in combination with Pit-1. Western blot analysis of extracts prepared from the transfected cells showed that each of the deletion mutants was expressed at levels equivalent to C/EBPα (inset, Fig. 1A), indicating that the activity observed for each of the mutant proteins was not a result of altered expression or stability. On average, Pit-1 induced −204 rat PRL promoter activity 20-fold in the HeLa cells, and the combination of Pit-1 and the full-length C/EBPα resulted in approximately 40-fold activation (Fig. 1A). The deletion of the first 68 residues of C/EBPα had no effect on its cooperative activity with the coexpressed Pit-1 (48-fold activation, Fig. 1A), demonstrating that CR-1 was unnecessary for this interaction. Conversely, C/EBP Δ68–96 was impaired in the activation of the PRL promoter when expressed alone (2-fold activation) and was deficient in the cooperative activation with coexpressed Pit-1 (Fig. 1A). The protein lacking CR-1, CR-2, and CR-3 (C/EBPΔ3–154) had similar activity, also failing to interact cooperatively with Pit-1 in inducing PRL transcription (Fig. 1A). These data showed that the CR-2 domain of C/EBPα was critical for the cooperative activation of Pit-1-dependent PRL transcription.
Fig. 1. The CR-2 Domain of C/EBPα Is Required for Induction of Pit-1-Dependent Transcription.
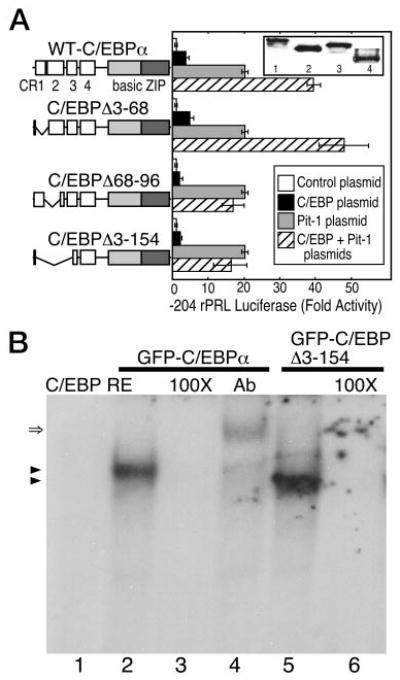
A, HeLa cells were transfected with the −204 rat PRL promoter linked to the Luc reporter gene and the indicated protein expression vectors. The cells were cotransfected with plasmids encoding either the full-length rat C/EBPα or the indicated truncations (10 μg, black bars) or 5 μg of the Pit-1 expression plasmid (gray bars) or the combination of both (hatched bars). Inset, Western blot showing the expressed C/EBPα proteins: lane 1, full-length C/EBPα; lane 2, C/EBPα (Δ3–68); lane 3, C/EBPα (Δ68–96); lane 4, C/EBPα (Δ3–154). Luciferase activity was determined after 24 h and was corrected for total protein. The error is the sem from three independent experiments, each in triplicate and normalized to reporter alone. B, EMSA showing that GFP-C/EBPα (lanes 1–4) and GFP-C/EBPΔ3–154 have similar DNA binding characteristics. Cell extracts were prepared from 3T3-L1 cells expressing the indicated protein, and samples were incubated with a labeled C/EBPα RE as described in Materials and Methods. After gel electrophoresis, a single DNA-protein complex was observed for each protein (arrowheads). Binding specificity was demonstrated by competition with an 100-fold excess of unlabeled oligonucleotide (lanes 3 and 6). The presence of the full-length C/EBPα in the complex was verified by a shift in mobility resulting from the addition of an antibody specific for C/EBPα (double arrow, lane 4).
The C/EBPα deletion mutants were then expressed as fusions to GFP in mouse 3T3-L1 cells, and protein extracts prepared from the transfected cells were subjected to EMSA using a consensus C/EBPα response element (C/EBP RE). Each of the amino-terminal truncations of C/EBPα retained full DNA-binding specificity (not shown), and the results in Fig. 1B compare the DNA-binding activities of the full-length GFP-C/EBPα to the GFP-C/EBPΔ3–154 deletion, which lacked the CR-1, CR-2, and CR-3 domains. Both proteins formed a single complex (arrowheads, lanes 2 and 5), and 100-fold excess of unlabeled oligonucleotide competed completely for binding of the proteins to the labeled probe (lanes 3 and 6). The decreased mobility of the shifted complex upon addition of an antibody recognizing the CR-1 domain demonstrated the presence of C/EBPα. These results are consistent with other studies showing that the carboxy-terminal bZIP domain of C/EBPα is sufficient to direct DNA binding (32–36).
The Amino-Terminal Region of C/EBPα Is Also Required for Pit-1-Mediated Redistribution
The potential role of the amino-terminal domains of C/EBPα in mediating the intranuclear recruitment by Pit-1 was then examined in living pituitary cells. The deletion mutants of C/EBPα were each fused to the blue fluorescent protein (BFP) and coexpressed with GFP-Pit-1 in GHFT1–5 cells. Deletion of the CR-1 (C/EBP Δ3–68), which was fully functional in activation of Pit-1-dependent PRL transcription (Fig. 1A), had no effect on its redistribution by the coexpressed GFP-Pit-1 (Fig. 2A). In striking contrast, the C/EBPα deletion mutant devoid of the CR-1, CR-2, and CR-3 domains (C/EBP Δ3–154) was not recruited to the nuclear sites occupied by GFP-Pit-1 (Fig. 2B). Instead, there was a marked tendency for the GFP-Pit-1 to colocalize in the foci occupied by BFP-C/EBP Δ3–154 (Fig. 2B). Identical to our previous result (20), we observed by immunohistochemical staining that the endogenous Pit-1 in GHFT1–5 cells localized in a reticular pattern throughout the nucleus (Fig. 2C). The endogenous protein was not concentrated in regions of centromeric heterochromatin stained by Hoechst 33342 (H33342). When GHFT1–5 cells expressing the GFP-C/EBPΔ154 protein were stained for Pit-1, however, we observed that some of the endogenous protein was redistributed to the centromeric heterochromatin sites (Fig. 2D). Control experiments showed this was not because of overlap of green fluorescence into the red channel, and there was no nuclear staining observed for cells incubated with secondary antibody alone (data not shown). This result showed that the behavior of the transiently expressed GFP-Pit-1 protein accurately reflected that of the endogenous transcription factor. When combined with our previous observations (20), the results indicated that Pit-1 and C/EBPα act cooperatively to induce PRL transcription, and these actions require both the Pit-1 HD and the amino-terminal activation domains of C/EBPα. These results also imply, but do not conclusively prove, that Pit-1 and C/EBPα associate in the living cell nucleus.
Fig. 2. Both Expressed and Endogenous Pit-1 Colocalized with C/EBPα.
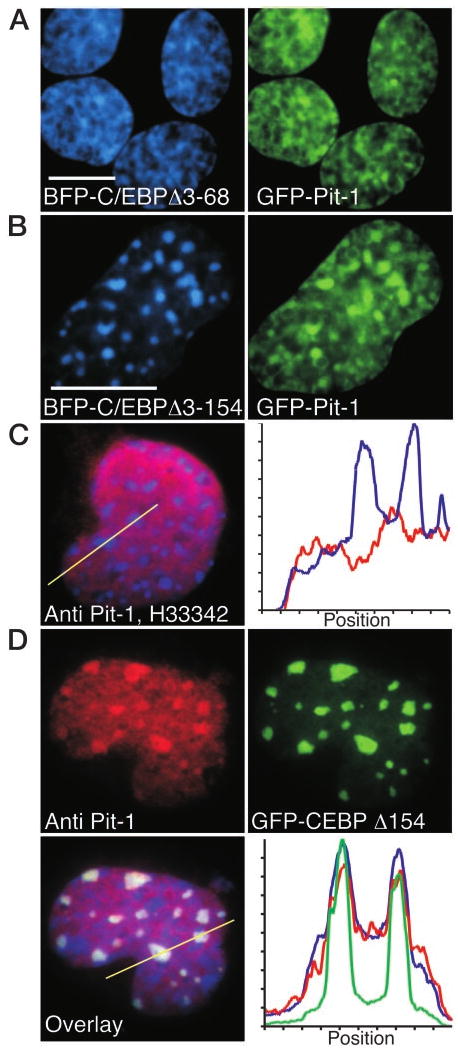
A and B, When coexpressed, GFP-Pit-1 recruited BFP-C/EBPΔ3–68, but not BFP-C/EBPΔ3–154, to the intranuclear domains occupied by Pit-1. Sequential images were acquired of GHFT1–5 cells coexpressing BFP-C/EBPΔ3–68 and GFP-Pit-1 (A) or BFP-C/EBPΔ3–154 and GFP-Pit-1 (B), as described in Materials and Methods. The calibration bars indicate 10 μm. C, Immunohistochemical staining of endogenous Pit-1 in a mouse GHFT1–5 cell. The endogenous Pit-1 was detected with antisera to Pit-1 and a Texas red-linked secondary antibody, and staining with H33342 revealed regions of heterochromatin. The red Pit-1 and blue H33342 fluorescence images were merged, and an intensity profile was obtained for both Texas red emission (red line) and H33342 fluorescence (blue line), and the position indicated by the line was plotted (right panel). D, Immunohistochemical detection of Pit-1 in a mouse GHFT1–5 cell expressing the truncated GFP-C/EBPΔ154. Staining for the endogenous Pit-1 was detected in the red channel, the expressed GFP-C/EBPΔ154 was detected in the green channel, and chromatin stained with H33342 was detected in the blue channel. The different fluorescence images were merged, and an intensity profile obtained for all three colors at the position indicted by the line was plotted (right panel).
To determine whether Pit-1 and C/EBPα were in close spatial association with one another, we used the approach of FRET microscopy. FRET microscopy detects the result of the radiationless transfer of excitation energy from a donor fluorophore to a nearby acceptor that can occur only over a distance of less than about 7 nm (22–30). When there is FRET between two fluorophores, the donor signal is quenched and there is sensitized emission from the acceptor (see Fig. 3A). The detection of sensitized FRET emission above the spectral background signal, which is contributed by both the donor and acceptor fluorophores, requires very accurate measurements (24, 28–30). Importantly, because the donor emission is quenched, FRET can also be detected by measuring the increase in donor fluorescence (dequenching) after photobleaching of the acceptor (Fig. 3B). The dequenching of donor emission after acceptor photobleaching provides a direct measure of the FRET efficiency (25–28).
Fig. 3. FRET Microscopy Improves the Optical Resolution.
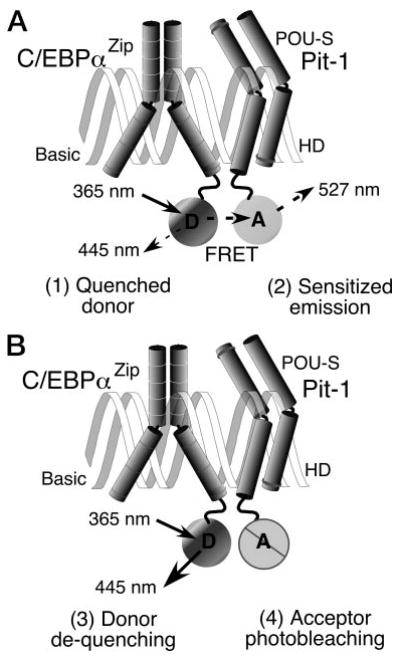
The diffraction of light limits the resolution of the microscope to 200 nm, and objects that are closer together than this will appear as a single object. A, It is possible to realize a 50-fold increase in spatial resolution by using the technique of FRET microscopy. FRET is the radiationless transfer of energy from a donor fluorophore (D) to nearby acceptors (A), resulting in (1) quenching of the donor signal, and (2) sensitized emission from the acceptor. The efficiency of the energy transfer decreases dramatically with distance, limiting FRET to distances less than 7 nm. B, Because the donor emission is quenched with FRET, detecting the (3) dequenching of the donor signal after (4) selective photobleaching of the acceptor provides a direct measurement of FRET efficiency.
In this study, we used the combination of BFP and yellow fluorescent protein (YFP) as donor and acceptor for FRET microscopy. There were three reasons for selecting these particular fluorescent probes. First, in spite of its low quantum yield and sensitivity to photobleaching, the BFP variant used here provides an adequate signal from the nucleus, where the autofluorescence background is low. Because of the high autofluorescence outside the nucleus, this color variant would not be a good choice for studies of cytoplasmic protein interactions. Second, the overlap of the BFP emission and YFP excitation spectra is sufficient for energy transfer, but the spectral background for this pair is much reduced when compared with the cyan FP/YFP combination commonly used for these types of studies (25, 26). Third, the YFP variant is more sensitive to photobleaching than either GFP or the Discosoma sp. red fluorescent protein (25), making it a good choice for acceptor photobleaching FRET. The Förster distance for the BFP and YFP pair, the distance at which energy transfer is 50% efficient, is 3.8 nm (37) and falls to less than 5% efficiency at a distance of 7 nm.
Selectivity of Acceptor Photobleaching
The ability to selectively photobleach the YFP fluorophore in the presence of the coexpressed BFP is essential for acceptor photobleaching FRET. To demonstrate this, we coexpressed nuclear localized, but noninteracting BFP- and YFP-tagged proteins in the same living cells. Fluorescence microscopy was used to identify individual pituitary GHFT1–5 cells coexpressing YFP fused to the corepressor protein silencing mediator of retinoic acid and thyroid hormone receptors (YFP-SMRT), and BFP fused to the basic leucine zipper protein (bZIP) DNA-binding domain of C/EBPα (BFP-C/EBPΔ244). The YFP-SMRT protein localized to discrete foci in the pituitary cell nuclei that were spatially separated from those formed by C/EBPΔ244 (Fig. 4A).
Fig. 4. The Photobleaching of YFP Is Selective.
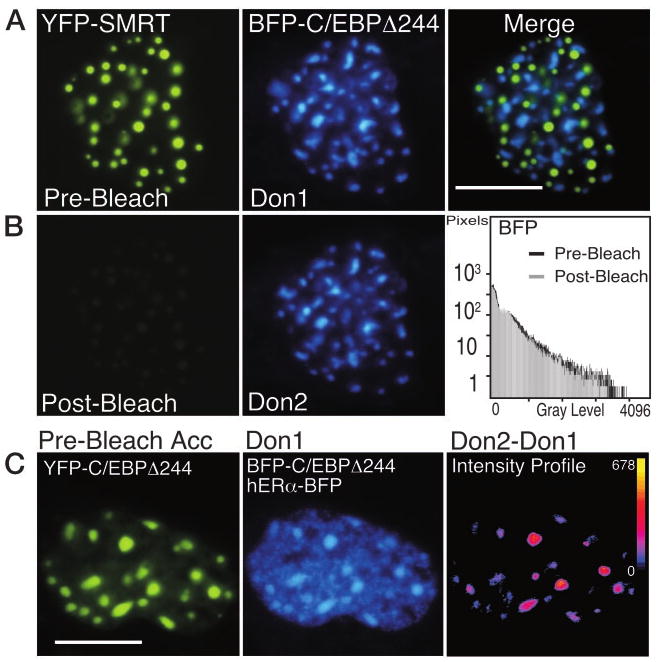
A, Pituitary GHFT1–5 cells were cotransfected with plasmids encoding the corepressor protein SMRT, tagged with YFP, and the C/EBPΔ244 deletion mutant fused to BFP. Sequential images of the nucleus of a cell coexpressing the proteins were acquired at the same focal plane using the filters described in Materials and Methods. The calibration bar indicates 10 μm. The images were merged to show that the proteins were each localized to discrete subnuclear foci. B, The YFP-SMRT was photobleached by 5 min of exposure to 500-nm light. Postbleach images were acquired at the same focal plane and under identical conditions to document the bleaching of YFP and to show that this had no effect on the BFP signal (histogram). C, Acceptor photobleaching FRET measurements detect only specific protein associations. Pituitary GHFT1–5 cells were cotransfected with plasmids encoding both YFP- and BFP-C/EBPΔ244, and hERα-BFP, and cells were identified that expressed all three labeled proteins. The prebleach images of the acceptor, YFP-C/EBPΔ244 (left panel), and the combined fluorescence from the donor, BFP-C/EBP Δ244 (foci) and the nucleoplasmic-localized hERα-BFP (Don1) are shown. The YFP fluorophore was bleached by greater than 90%, and a second donor image (Don2) was acquired at the same focal plane and under identical conditions as the first. The pixel-by-pixel change in gray-level intensity of the donor signal was obtained by digital subtraction of the Don1 image from the Don2 image. The intensity profile (right panel) represents the change in the gray-level intensities in the dequenched donor image (Don2 − Don1). The calibration bar shows the range of gray-level intensities in the dequenched image with black indicating 0 and yellow indicating the maximum gray-level value (shown next to the calibration bar). Note that the increase in donor intensity was limited to the foci.
The distinct intranuclear positioning of these two noninteracting proteins was used to characterize the selectivity of the acceptor photobleaching applied in the FRET experiments described below. Figure 4A shows the prebleach reference images of YFP-SMRT and BFP-C/EBPΔ244 (Don1) in the nucleus of the same cell. Merger of the YFP and BFP images demonstrated that the fluorescence signals from the two differently labeled proteins were localized to multiple, nonoverlapping foci (Fig. 4A, merge). The YFP-SMRT was then selectively bleached by exposure to 500-nm light for 5 min, resulting in a greater than 90% reduction in the YFP signal (Fig. 4B). A second image of BFP-C/EBPΔ244 (Don2) was then acquired in the same focal plane and under identical conditions as the first BFP image. Comparison of the BFP signals before and after the YFP photobleaching showed there was only a slight decrease in the BFP signal (see histogram, Fig. 4B). This result clearly demonstrated the selectivity of the acceptor photobleaching method.
The Specificity of Acceptor Photobleaching FRET
We next evaluated the ability of acceptor photobleaching FRET microscopy to both detect FRET signals and to discriminate the FRET signals from other background signals. We previously used acceptor photobleaching FRET to detect the dimer interactions of the isolated bZIP domain of C/EBPα (C/EBPΔ244) in the nucleus of pituitary GHFT1–5 cells (26). When expressed in GHFT1–5 cells, we observed that, in contrast to full-length C/EBPα, the truncated protein was almost exclusively localized to centromeric heterochromatin in the pituitary GHFT1–5 cells (see Fig. 3 in Ref. 20). Because of this very restricted intranuclear positioning, we were able to distinguish FP-labeled C/EBPΔ244 from the coexpressed, but noncentromeric localized human estrogen receptor-α (hERα) when they were expressed in the same cell. This allowed us to directly compare acceptor photobleaching FRET signals originating from sites containing both donor and acceptor to adjacent regions containing predominantly donor.
Figure 4C shows the nucleus of a GHFT1–5 cell coexpressing YFP-C/EBPΔ244, BFP-C/EBPΔ244, and hERα-BFP. The C/EBPΔ244 fusion proteins were localized to discrete foci, whereas hERα-BFP adopted a granular distribution throughout the nucleoplasm (Fig. 4C, Don1). The YFP-labeled C/EBPΔ244 was selectively photobleached, and a second image of the BFP-C/EBPΔ244 and hERα-BFP was then acquired at the same focal plane and under identical conditions to the first. The change in the BFP signal after YFP photobleaching was quantified at each pixel by digitally subtracting the prebleach BFP image from the postbleach image (Fig. 4C, Don2 − Don1). The pixel-by-pixel changes in the dequenched donor signal were then mapped in the intensity profile (Fig. 4C, right panel). The color-coded look-up table represents the change in donor signal with black indicating no change and yellow indicating the maximum change in gray-level intensity, which is indicated in the figure.
After YFP photobleaching, digital subtraction of the prebleach donor image from the postbleach image revealed that the increase in BFP signal was restricted to the foci where YFP-C/EBPΔ244 and BFP-C/EBPΔ244 were colocalized. These regional changes in fluorescence intensity were also quantified by measuring the signal at ten identically sized regions of interest (ROIs) within the foci or in the surrounding nucleoplasm in both the Don1 and Don2 images (Table 1). Before YFP photobleaching, the average donor signal within the foci was similar to the BFP signal in the nucleoplasm surrounding the foci (651 vs. 378, Table 1). The majority of the acceptor signal, however, originated from the foci (1471 vs. 267, Table 1). After photobleaching of the acceptor, the average BFP signal in the foci was increased by 38%. In contrast, there was a slight decrease in the average signal from hERα-BFP in the nucleoplasm (−0.8%, Table 1), which likely reflects some bleaching of BFP during the acquisition of the two donor images. Thus, despite similar levels of BFP-labeled proteins throughout the entire nucleus, only the donor proteins directly associated with the YFP-C/EBPΔ244 in the foci exhibited dequenching. These results show the highly selective dequenching expected for the dimerized C/EBPΔ244 proteins (26) and demonstrated the ability of acceptor photobleaching FRET to detect these protein interactions in the presence of a high background signal.
Table 1.
Quantification of Regional Changes in Fluorescence Intensity and FRET Efficiency
| Donor | Acceptor | Position | Acca | Don1b | Don2c | E%d |
|---|---|---|---|---|---|---|
| BFP-CEBPΔ244 + hERα-BFP | YFP-CEBPΔ244 | Foci | 1471 ± 400e | 651 ± 74 | 1064 ± 190 | 38.0 |
| NPf | 267 ± 49 | 378 ± 39 | 375 ± 43 | −0.8 | ||
| BFP-Pit-1 | YFP-C/EBPα | Nucg | 1567 ± 452 | 341 ± 83 | 403 ± 106 | 15.4 |
| BFP-Pit-1R271A | YFP-C/EBPα | Foci | 2784 ± 259 | 487 ± 87 | 645 ± 109 | 24.6 |
| NP | 1361 ± 185 | 330 ± 47 | 362 ± 61 | 8.8 | ||
| BFP-Pit-1 | YFP-C/EBPΔ154 | Foci | 3081 ± 340 | 339 ± 29 | 533 ± 28 | 36.3 |
| NP | 1344 ± 185 | 295 ± 41 | 331 ± 49 | 10.9 | ||
| BFP-Pit-1 | YFP-C/EBPΔ318 | Nuc | 1942 ± 298 | 866 ± 174 | 884 ± 181 | 2.0 |
Acceptor before photobleaching.
Donor before acceptor photobleaching.
Donor after acceptor photobleaching.
E% = [1 − (Don1/Don2)] × 100.
Average gray level ± sd.
NP, Nucleoplasm.
Nuc, Entire nucleus.
Detecting the Interactions of Pit-1 and C/EBPα in the Nucleus of the Living Cell
The acceptor photobleaching FRET approach was then used to characterize further the association of Pit-1 and C/EBPα in living GHFT1–5 cells. As demonstrated previously (20), when coexpressed with BFP-Pit-1, the YFP-C/EBPα was redistributed to the nuclear sites occupied by BFP-Pit-1 (Fig. 5A). After selective photobleaching of the YFP fluorophore, acquisition of a second BFP-Pit-1 image revealed an increase in BFP-Pit-1 fluorescence intensity. The pixel-by-pixel change in the donor intensity after acceptor photobleaching was determined by digital subtraction (Don2 − Don1), and this change in signal is shown in the intensity profile (Fig. 5A, right). The average donor signal throughout the entire nucleus was increased by 15.4% (Table 1). The dequenching of the BFP-Pit-1 signal after photobleaching of the YFP linked to C/EBPα provides evidence that the average distance separating the fluorophores was less than 7 nm.
Fig. 5. Acceptor Photobleaching FRET Microscopy Detects the Interactions of Pit-1 and C/EBPα.
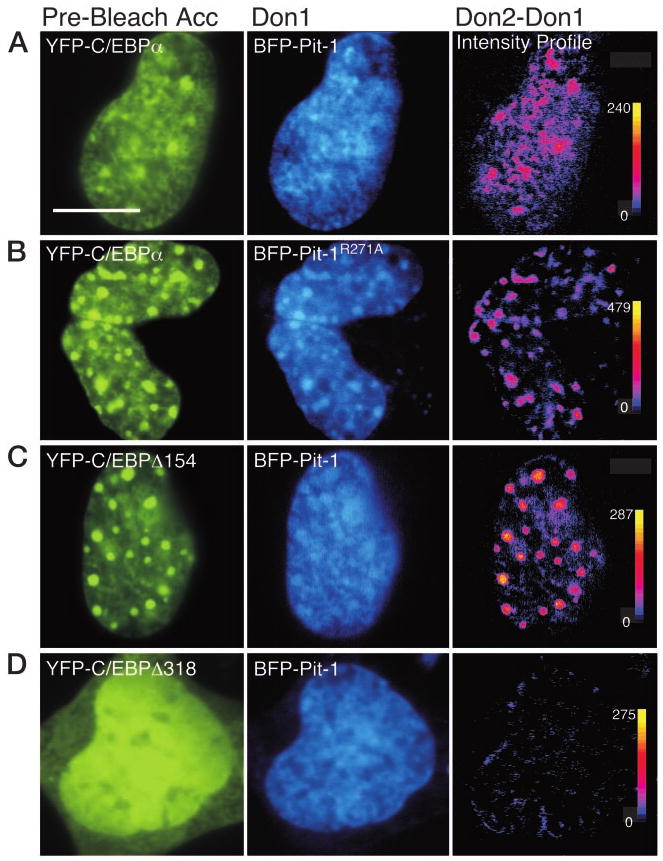
Prebleach acceptor (YFP) and donor (BFP) images are shown for each of the panels A–D. A second donor image (Don2) was acquired at the same focal plane and under identical conditions as the first (Don1) image after acceptor (YFP) photobleaching. The dequenched donor signal (Don2 − Don) was then determined as described in the legend for Fig. 4. For each dequenched donor image, the pixel-by-pixel change in gray-level intensity is shown in the intensity profile (right panels), with black indicating 0 and yellow indicating the maximum gray-level value (shown next to each calibration bar). A, The prebleach acceptor and donor images showing the recruitment of YFP-C/EBPα by BFP-Pit-1 (Don1); the bar indicates 10 μm. The dequenched donor (Don2 − Don1) intensity profile is shown in the right panel. B, The prebleach acceptor and donor images showing subnuclear distribution of YFP-C/EBPα and the mutant Pit-1R271A labeled with BFP (Don1). The dequenched donor (Don2 − Don1) intensity profile is shown in the right panel. C, The prebleach acceptor and donor images showing subnuclear distribution of YFP-C/EBPΔ154 and BFP-Pit-1 (Don1). The dequenched donor (Don2 − Don1) intensity profile is shown in the right panel. D, The prebleach acceptor and donor images showing subnuclear distribution of YFP-C/EBPΔ318 and BFP-Pit-1 (Don1). The dequenched donor (Don2 − Don1) intensity profile is shown in the right panel.
We previously demonstrated that a point mutation in the Pit-1 HD, which resulted in a protein with dominant inhibitory activity, disrupted the ability of Pit-1 to recruit C/EBPα. Instead, we observed that the mutant Pit-1 protein was partially localized to the heterochromatin foci occupied by C/EBPα (20). Here, we used the acceptor photobleaching FRET approach to examine the interaction of the mutant Pit-1 protein with C/EBPα at these subnuclear sites in pituitary GHFT1–5 cells (Fig. 5B). When YFP-C/EBPα and BFP-Pit-1R271A were coexpressed, there was a tendency for the BFP-Pit-1R271A to localize to the intranuclear foci occupied by YFP-C/EBPα (Fig. 5B). After acceptor photobleaching, we observed an increase in the BFP-Pit-1R271A signal throughout the nucleus, with the most prominent change being localized to the foci (Fig. 5B, right). These localized changes in the BFP-Pit-1R271A signal were quantified, and the results are shown in Table 1. The YFP-C/EBPα was enriched 2-fold in the foci (Table 1; 2784 vs. 1361), and BFP-Pit-1was enriched 1.5-fold (Table 1; 487 vs. 330) in the foci relative to the nucleoplasm. The ratio of acceptor to donor was 5.7 and 4.1 in the foci and nucleoplasm, respectively. Although the ratio of acceptor to donor was similar in both regions, the BFP-Pit-1R271A signal associated with YFP-C/EBPα in the foci was increased 24.6%, compared with 8.8% change in signal in the nucleoplasm (Table 1). These results indicate that BFP-Pit-1R271A and YFP-C/EBPα were associated throughout the nucleus. However, the differences in donor dequenching observed for the two subnuclear domains show that either the fraction of Pit-1R271A interacting with C/EBPα, or the spatial relationship between the proteins, were different in the two nuclear locations.
Similar results were obtained from GHFT1–5 cells coexpressing the YFP-C/EBPΔ3–154 deletion and BFP-Pit-1. When coexpressed in the same GHFT1–5 cells, we observed the colocalization of BFP-Pit-1 and YFP-C/EBPΔ3–154 throughout the nucleus, with some accumulation of Pit-1 in the intranuclear foci (Figs. 2 and 5C). After photobleaching of the YFP fluorophore, there was a marked increase in BFP-Pit-1 signal associated with C/EBPΔ3–154 in the foci (Fig. 5C, right). Quantification of the BFP-Pit-1 signal showed that the average BFP-Pit-1 signal associated with YFP-C/EBPΔ3–154 in the foci increased 36%, compared with a 10.9% change in signal in the nucleoplasm (Table 1). The acceptor photobleaching FRET results for both Pit-1R271A and C/EBPΔ3–154 indicate that the fluorophores labeling the proteins are in closer proximity when the proteins are localized to the heterochromatin foci than when the proteins are in the nucleoplasm.
To demonstrate that these FRET signals were not due to the fluorophores being colocalized in the restricted volume of the nucleus, we next examined the coexpression of Pit-1 and a truncated C/EBPα protein defective in DNA binding. In a previous study, we showed that truncation of the C-terminal 40 amino acid residues that form the leucine zipper region of C/EBPα (C/EBPΔ318) disrupts DNA binding (19). When coexpressed with BFP-Pit-1, the YFP-C/EBPΔ318 protein was incompletely localized to the nucleus, but there was substantial spatial overlap with the nuclear localized BFP-Pit-1 (Fig. 5D). Importantly, the levels of donor and acceptor proteins achieved in the nucleus were comparable to that for the other protein pairs tested (Table 1). In stark contrast to the results obtained with C/EBPα and C/EBPΔ154, however, the selective bleaching of YFP-C/EBPΔ318 yielded only a 2% change in the BFP-Pit-1 signal (Fig. 5D, right, and Table 1).
To confirm and extend these observations of single cells, we then analyzed multiple cells expressing each of these protein partners. It is important to note that the vast majority of cells with balanced expression of the indicated protein partners displayed the same subnuclear distributions as those illustrated in Fig. 5. The analysis of 16 different cells expressing YFP-C/EBPΔ318 and BFP-Pit-1 showed that the average change in the donor signal was 1.7% (Fig. 6). By comparison, cells expressing the combination of YFP-C/EBPα and BFP-Pit-1 showed an average increase in donor signal of 10.9% throughout the nuclei (Fig. 6). When Pit-1 and C/EBPα were colocalized in the subnuclear foci, as was the case for Pit-1R271A and C/EBPΔ154, the average increase donor signal measured for 10 different cells was 18% and 29%, respectively (Fig. 6). In contrast, the change in donor signal localized outside the foci was approximately 10% (Fig. 6). Taken together, these results demonstrate the specific associations of Pit-1 and C/EBPα as part of common nuclear protein complexes in the living cell nucleus.
Fig. 6. The FRET Efficiency (E%) for Paired Pit-1 and C/EBPα Protein Variants Measured in Multiple Cells.
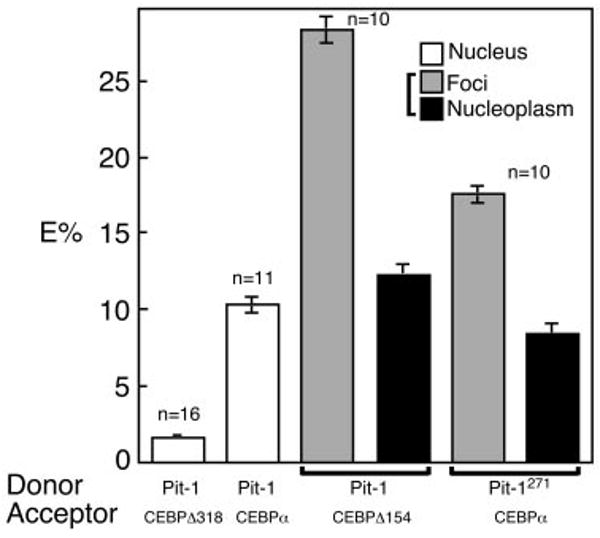
The indicated number (n) of cells, each expressing the donor and acceptor pairs shown below the graph, were analyzed for changes in donor fluorescence after acceptor photobleaching. For protein pairs distributed throughout nucleus (open bars), the average gray-level intensity was determined for the entire nucleus in both the pre- and postbleach donor images. The average E% (±sem) was then determined using the equation shown in the footnotes of Table 1. For protein pairs that were distributed to both foci and nucleoplasm, the average donor gray-level intensity was determined for 10 different ROIs in each region that were identically positioned in both the pre- and postbleach donor images. All ROIs were of identical size. The E% for the foci (gray bars) or nucleoplasm (black bars) in each cell was determined, and the average E% (±sem) for the indicated number of cells is plotted.
Discussion
In sum, our results have demonstrated the cooperative actions of Pit-1 and C/EBPα that function in the control of PRL transcription. In the previous study, we used the direct visualization of FP-labeled Pit-1 and C/EBPα to demonstrate that, when coexpressed, Pit-1 recruited C/EBPα from regions of centromeric heterochromatin to the intranuclear sites occupied by Pit-1 (20). The recruitment activity of Pit-1 was disrupted by deletion of the HD and, significantly, by a point mutation in the HD earlier identified in humans with CPHD. These results indicated that the Pit-1 HD played a critical role in the recruitment with C/EBPα, either through direct protein-protein interactions, or by association with common protein partners. Further, these results suggest that the dominant inhibitory activity of the CPHD Pit-1 mutant could be a consequence of the disruption of the intranuclear positioning of the protein and, potentially, other interacting protein partners.
The present study extended these observations to show that the amino-terminal activation domain of C/EBPα mediated the transcriptional cooperativity with Pit-1 and was required for the intranuclear recruitment by Pit-1. Based upon sequence alignment across species, Erickson et al. (31) identified four conserved regions, CR-1–4, in the transactivation domain of C/EBPα. We found that deletion of C/EBPα CR-1 had no effect on its cooperative actions with Pit-1 at the PRL promoter or on the ability of Pit-1 to recruit the truncated C/EBPα in living pituitary cells (Fig. 1). This result is consistent with the earlier observation that a C/EBPα CR-1 deletion retained full activity when assayed for its ability to induce differentiation in 3T3-L1 cells (31). In contrast, deletion of the CR-2 and CR-3 domains resulted in the loss of this differentiation function in 3T3-L1 cells (31). We observed here that a C/EBPα deletion removing CR-1–3 (C/EBPΔ3–154) was deficient in the transcriptional cooperativity with Pit-1. Significantly, the truncated C/EBPΔ3–154, which bound DNA with appropriate specificity and localized to the centromeric heterochromatin, was not reorganized by the coexpressed Pit-1. Instead, we observed a tendency for Pit-1 to become colocalized with the C/EBP Δ3–154 in the regions of heterochromatin (Figs. 2 and 5). We also observed that the endogenous Pit-1 protein in GHFT1–5 cells was redistributed to centromeric heterochromatin sites occupied by C/EBP Δ3–154. These results were remarkably similar to our earlier observations with the dominant inhibitory Pit-1R271A mutant, which failed to reorganize C/EBPα and also became colocalized at sites of centromeric heterochromatin (Fig. 5 and Ref. 20). Taken together, these results indicated that an association between Pit-1 and C/EBPα, either directly or as part of a common protein complex, mediates both intranuclear positioning and transcriptional cooperativity.
The Intranuclear Positioning of Pit-1 and C/EBPα
If this association required only the Pit-1 HD and the C/EBPα CR2/CR3 domains, we would expect that deletion of either of these domains should prevent the colocalization of the proteins. The CR deletions, however, did not block the colocalization, but rather changed the locations of these proteins in the nucleus. These results imply that protein domains specifying colocalization are separable from the protein domains that direct the final positioning of the protein complex within the nucleus. In this regard, the DNA binding activities of both Pit-1 and C/EBPα appear critical to specifying their intranuclear positioning. The positioning of both C/EBPα and C/EBPΔ3–154 to regions of centromeric heterochromatin (18–20) probably results from the binding of the conserved carboxy-terminal bZIP domain to multiple repeated sequences in centromeric satellite DNA (21). Disruption of the leucine-zipper dimerization domain prevents the formation of the scissors-grip structure that is necessary for DNA binding (32–34), and we showed that the truncated C/EBPΔ318 does not bind to a consensus C/EBPα DNA element (19). We observed here that YFP-C/EBPΔ318 expressed in GHFT1–5 cells was not localized to regions of centromeric heterochromatin. This mutant also failed to interact with the coexpressed Pit-1 (Figs. 5 and 6), suggesting that DNA binding may be a necessary first step in specifying the association with Pit-1.
Likewise, Pit-1 is also known to interact with different classes of DNA elements. For example, Gaiddon et al. (38) found that Pit-1 could bind specifically to the serum RE in the c-fos gene promoter. Structural studies have shown considerable flexibility in the conformations Pit-1 adopts on different DNA elements (39, 40), and this appears to be a common theme among transcription factors (41). Thus, the binding of Pit-1 or C/EBPα to different classes of DNA elements could specify protein conformations that direct their interactions with particular protein partners, resulting in their colocalization. Because of the limited optical resolution of the light microscope, however, the colocalization of FP-tagged Pit-1 and C/EBPα could not establish whether the proteins were separated by less than approximately 200 nm.
Detecting Protein Interactions Using Acceptor Photobleaching FRET Microscopy
To determine whether Pit-1 and C/EBPα were in close spatial association, we took advantage of the spectral properties of the FP labels and applied FRET microscopy. When using intensity-based imaging, the detection of sensitized FRET emission upon donor excitation is limited by the spectral background contributed by both the donor and acceptor fluorophores (22–24, 28, 30). An alternative approach is to measure the dequenching of donor emission after photobleaching of the acceptor (25–29). This approach requires only the detection of the donor signal, which is less prone to artifacts resulting from the spectral background. Further, each individual cell acts as its own internal standard, allowing small changes in donor signal to be quantified (28). Finally, the increase in the donor signal provides a direct measure of FRET efficiency (25–28). Because FRET efficiency decreases as the sixth power of the distance separating the donor and acceptor, significant donor dequenching will occur only if the fluorophores are separated by less than 7 nm.
We verified the acceptor photobleaching FRET approach for our living cell model in two different ways. First, we demonstrated that the signal from a nuclear localized BFP-fusion protein was not altered by the photobleaching of another nuclear localized, but noninteracting, YFP-fusion protein (Fig. 4). Second, we showed that protein interactions detected by acceptor photobleaching FRET could be readily distinguished from the signals originating from other noninteracting proteins. When coexpressed in the same cell, the YFP- and BFP-C/EBPΔ244 proteins localized to distinct nuclear foci, whereas the hERα-BFP was dispersed in a granular pattern throughout the nucleoplasm (Fig. 4 and Ref. 20). Although the donor signals originating from the foci and nucleoplasm before acceptor photobleaching were similar, only the signal from the BFP-tagged proteins associated with YFP-C/EBPΔ244 in the foci was increased (dequenched) after acceptor photobleaching. The fluorescence from the noninteracting hERα-BFP at adjacent sites in the nucleoplasm was not increased. Together, these results demonstrated the selectivity of acceptor photobleaching and showed that only donor signal associated with acceptor underwent dequenching.
The Intimate Association of Pit-1 and C/EBPα
Using the acceptor photobleaching FRET approach, we determined that when C/EBPα was recruited from the centromeric heterochromatin to the sites occupied by Pit-1, the two proteins were in close physical association. The dequenching of the BFP-Pit-1 signal upon photobleaching of YFP-labeled C/EBPα (Fig. 5) provided evidence that the fluorophores were spatially positioned on average less than 7 nm apart. This intimate association between Pit-1 and C/EBPα was not simply due to their colocalization in the restricted space within the nucleus. When coexpressed, Pit-1 and a leucine-zipper domain deletion of C/EBPα (Δ318) also had substantial spatial overlap in the nucleus, with donor and acceptor protein levels comparable to that for the other protein pairs tested (Fig. 5 and Table 1). In contrast to the results obtained with C/EBPα, the selective bleaching of YFP-C/EBPΔ318 yielded less than 2% change in the BFP-Pit-1 signal when measured in 16 different cells (Fig. 6). We showed earlier that C/EBPΔ318 is defective in DNA binding (19). The failure of this mutant protein to interact with Pit-1 suggests that the binding of C/EBPα to DNA is necessary for their association. The detection of FRET between Pit-1 and C/EBPα indicates that the average distance separating the fluorophores was less than 7 nm but is not proof of a direct protein-protein interaction. The association of Pit-1 and C/EBPα could result from the interactions of these proteins with a common protein partner. Indeed, both proteins interact with CBP (18, 19, 40), and we observed earlier that GFP-CBP was colocalized with C/EBPα when expressed in GHFT1–5 cells (18).
We also observed that the mutant variants of Pit-1 and C/EBPα proteins, which were still able to associate with their wild-type partners, were also in close physical proximity (Fig. 5 and Ref. 20). As was shown previously (20), the mutant BFP-Pit-1R271A had a marked propensity to localize at the sites of centromeric heterochromatin occupied by YFP-C/EBPα (Fig. 5B). Similarly, there was also a tendency for both coexpressed and endogenous Pit-1 to colocalize to these sites with the expressed C/EBPΔ3–154 (Figs. 2 and 5). In both cases, however, this distribution was incomplete, and there was a significant concentration of the proteins colocalized in the nucleoplasm surrounding the subnuclear foci. After acceptor photobleaching, the BFP signal in both the foci and nucleoplasm was increased, but in both cases the change in signal associated with the foci was more robust. For both Pit-1R271A and C/EBPΔ154, we observed that the average change in the nucleoplasm measured in 10 different cells was approximately 10%, which was very similar to the results obtained for the wild-type proteins (Fig. 6). In contrast, the interaction of C/EBPΔ154 or Pit-1R271A with their wild-type partners in the centromeric heterochromatin foci resulted in an average increase in donor signal of 29% and 18%, respectively.
The difference in FRET efficiencies from the foci and the nucleoplasm of the same cell could reflect a difference in the fraction of proteins that were available to form complexes in the two subnuclear regions. For example, the fraction of Pit-1 available to interact with C/EBPα in the nucleoplasm might be reduced by its interactions with other cellular proteins. This would result in an increase in background donor fluorescence, reducing the apparent change in the donor signal upon acceptor photobleaching. Alternatively, the spatial relationship between the fluorophores labeling the Pit-1 and C/EBPα proteins might be significantly different for proteins localized in the two subnuclear regions. For example, both Pit-1 and C/EBPα may be bound to arrays of repeated DNA sites in the centromeric heterochromatin, as was suggested for C/EBPα (21). The arrangement of repeated DNA elements in centromeric heterochromatin could allow several acceptor proteins to fall within the Förster radius of each donor and increase FRET efficiency. A second possibility is that there are changes in the conformation of the protein complex when bound to particular DNA sites. For example, C/EBPα and Pit-1 may adopt a different conformation when associated with centromeric heterochromatin that positions the fluorophores in closer proximity, resulting in an increased FRET efficiency. The fact that the C/EBPΔ154 protein had consistently higher dequenching values suggests that placement of the fluorophore nearer to the bZIP domain allowed it to be in closer proximity to the fluorophore linked to Pit-1.
It should be noted that the FPs based upon the Aequorea GFP can weakly associate when at high concentrations, although GFP was crystallized as a monomer (42). Although it is possible that the colocalization of FP-labeled proteins in a restricted volume, such as the nucleus, could favor this weak interaction, several observations suggest this is not the case. First, we showed that when coexpressed, YFP-SMRT and BFP-C/EBPΔ244 each localized to distinct and spatially separate foci in the pituitary cell nucleus (Fig. 4). If the FPs associated with one another as dimers, one would not expect these discrete patterns of subnuclear localization. Second, we showed that the C/EBPΔ318 deletion, which is defective in DNA binding (19), failed to associate with Pit-1 despite its colocalization within the nucleus (Figs. 5 and 6). Third, there are examples in the literature of different FRET-based indicator proteins in which a donor and acceptor fluorophore are tethered to one another through a short linker peptide (25, 42–45). In most cases, a change in the linker conformation resulting from its modification, or the binding of a substrate, induces a FRET signal. These indicators could not work in this manner if the juxtaposed FPs were already dimerized.
Pit-1 and the Organization of the Pituitary Cell Nucleus
Our results indicate that subtle changes in protein sequence, such as the Pit-1R271A CPHD mutant, may dramatically affect its association with other nuclear proteins, as well as the final intranuclear positioning of these protein complexes. These results could reflect changes in DNA binding affinity or the altered conformation of the mutant Pit-1 protein that favor its interaction with C/EBPα bound to the repeated satellite DNA elements, but not to other chromatin sites. If we are to understand disease processes and design therapeutic strategies, we must establish the rules that govern the organization of the nuclear environment. The distribution of transcription factors in the living cell is dynamic, and their positioning in the nucleus reflects the balance of their interactions with other protein partners and their association with the chromatin. We showed here the steady-state interactions of two transcription factors on the nanometer scale in the intact pituitary cell nucleus. The approach of acceptor photobleaching FRET microscopy, however, is an endpoint assay that requires that the higher-order protein complexes be relatively stable. The detection of the dynamic interactions of these higher-order protein complexes within the three-dimensional volume of the living cell nucleus is difficult to achieve using these intensity-based imaging techniques. Future studies characterizing these dynamic interactions will require both high spatial and temporal resolution. These types of measurements can be achieved using the combination of fluorescence life-time imaging microscopy and FRET (29, 46). The application of these live cell-imaging techniques will be essential to establish how the subnuclear targeting of Pit-1 and its interacting partners contribute to the combinatorial code directing pituitary-specific gene expression.
Materials and Methods
Construction of Expression Vectors, Transfection of Cell Lines, and Reporter Gene Assays
The construction of plasmids and maintenance of cell lines were described in the previous paper (20). GHFT1–5, 3T3-L1, or HeLa cells were transfected with the indicated plasmid DNA(s) by electroporation as described previously (26). The total amount of DNA was kept constant using empty vector DNA.
Western Blotting and EMSA
The Western blot analysis of the expressed proteins was described previously (20, 47). EMSAs were performed on whole-cell extracts prepared from transiently transfected 3T3-L1 cells as described previously (47). A duplex oligonucleotide corresponding to a consensus C/EBP binding site: 5′-GATCGAGCCCCATTGCGCAATCTATATTCG (Geneka Biotechnology, Inc., Montréal, Québec, Canada) was end-labeled using [γ-32P]ATP and T4 polynucleotide kinase and used as probe.
Acceptor Photobleaching FRET Microscopy and Image Analysis
Pituitary GHFT1–5 cells were transfected with the indicated expression plasmid DNA encoding the FP-fusion proteins, inoculated into culture dishes containing 25-mm coverglass, and then subjected to fluorescence microscopy as described previously (18–20, 22, 24, 26). The fluorescence images were acquired using an inverted IX-70 equipped with a ×60 aqueous-immersion objective lens (Olympus Corp., Lake Success, NY). The filter combinations were 500/15-nm excitation, 525-nm dichroic mirror, and 545/25-nm emission for YFP, and 365/15-nm excitation, 390-nm dichroic mirror, and 460/50-nm emission for BFP (Chroma Technology Corp., Brattelboro, VT). Grayscale images with no saturated pixels were obtained using a cooled digital interline camera (Orca-200, Hamamatsu, Bridgewater, NJ). All images were collected at a similar gray-level intensity by controlling the excitation intensity using neutral density filtration and by varying the on-camera integration time. The acceptor photobleaching method used here was described previously (25–29). A reference image of acceptor (YFP) fluorescence was first acquired followed by the acquisition of the first donor image (Don1). For photobleaching of the YFP acceptor, exposure of the specimen for approximately 5 min to unfiltered 500-nm excitation light typically achieved greater than 90% decrease in the YFP signal. After the photobleach period, a second acceptor reference was acquired to document photobleaching. A second donor image (Don2) was then acquired at the same focal plane and under identical conditions as the first, using the BFP filter set. The image of the donor before acceptor photobleaching (Don1) was digitally subtracted from donor after acceptor bleaching (Don2) to demonstrate the change in donor signal. To quantify these changes in donor fluorescence in different regions of the nucleus, the average signal was determined for 10 different ROIs identically positioned in the pre- and postbleach donor images. All ROIs were of identical size. Digitial subtraction of the Don1 image from the Don2 image using ISEE software (Inovision Corp., Raleigh, NC) generated the dequenched donor images. The intensity profile maps using the same look-up table represent the pixel-by-pixel change in the gray-level intensity in each of the dequenched donor images. For the printed images the background was subtracted and the resulting image files were processed for presentation using Canvas 7.0 (Deneba, Inc., Miami, FL) and rendered at 300 dots per inch.
Acknowledgments
We thank Ignacio DeMarco and Jesse Bible for technical assistance.
This work was supported by NIH Grants DK-47301 (to R.N.D.), F32 DK-GM10093 (to J.F.E.), and F32 DK-60315-01 (to T.C.V.).
Abbreviations
- BFP
Blue fluorescent protein
- bZIP
basic leucine zipper protein
- C/EBPα
CCAAT/enhancer binding protein α
- CPHD
combined pituitary hormone deficiency
- CR
conserved region
- Don1 and Don2
first and second donor image, respectively
- FPs
fluorescent proteins
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- H33342
Hoechst 33342
- HD
homeodomain
- hERα
human estrogen receptor α
- PRL
prolactin
- RE
response element
- ROIs
regions of interest
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
- YFP
yellow fluorescent protein
References
- 1.Gutierrez-Hartmann A. INSIGHT: Pit-1/GHF-1: a pituitary-specific transcription factor linking general signaling pathways to cell-specific gene expression. Mol Endocrinol. 1994;8:1447–1449. doi: 10.1210/mend.8.11.7877613. [DOI] [PubMed] [Google Scholar]
- 2.Dasen JS, Rosenfeld MG. Combinatorial codes in signaling and synergy: lessons from pituitary development. Curr Opin Genet Dev. 1999;9:566–574. doi: 10.1016/s0959-437x(99)00015-5. [DOI] [PubMed] [Google Scholar]
- 3.Schaufele F. CCAAT/enhancer-binding protein α activation of the rat growth hormone promoter in pituitary progenitor GHFT1–5 cells. J Biol Chem. 1996;271:21484–21489. doi: 10.1074/jbc.271.35.21484. [DOI] [PubMed] [Google Scholar]
- 4.Jacob KK, Stanley FM. CCAAT/enhancer-binding protein α is a physiological regulator of prolactin gene expression. Endocrinology. 1999;140:4542–4550. doi: 10.1210/endo.140.10.7076. [DOI] [PubMed] [Google Scholar]
- 5.Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 7.Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2000;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 8.Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol Endocrinol. 2000;14:518–534. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- 9.van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- 10.Thanos D, Maniatis T. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 11.Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB, Penman S, Hiebert S, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iborra FJ, Jackson DA, Cook PR. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J Cell Sci. 1996;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- 13.Nardozza TA, Quigley MM, Getzenberg RH. Association of transcription factors with the nuclear matrix. J Cell Biochem. 1996;61:467–477. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C467::AID-JCB14%3E3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.Kim TK, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 15.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 16.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMGI(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol Cell. 1998;1:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 17.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–186. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaufele F, Enwright JF, III, Wang X, Teoh C, Srihari R, Erickson R, MacDougald OA, Day RN. CCAAT/enhancer binding protein α assembles essential cooperating factors in common subnuclear domains. Mol Endocrinol. 2001;15:1665–1676. doi: 10.1210/mend.15.10.0716. [DOI] [PubMed] [Google Scholar]
- 19.Liu W, Enwright JF, III, Hyun W, Day RN, Schaufele F. CCAAT/enhancer binding protein α uses distinct domains to prolong pituitary cells in the growth 1 and DNA synthesis phases of the cell cycle. BMC Cell Biol. 2002;3:6. doi: 10.1186/1471-2121-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enwright JF, III, Kawecki-Crook MA, Voss TC, Schaufele F, Day RN. A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAATT/enhancer binding protein α required for prolactin gene transcription. Mol Endocrinol. 2003;17:209–222. doi: 10.1210/me.2001-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day RN. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Mol Endocrinol. 1998;12:1410–1419. doi: 10.1210/mend.12.9.0168. [DOI] [PubMed] [Google Scholar]
- 23.Periasamy A, Day RN. Visualizing protein interactions in living cells using digitized GFP imaging and FRET microscopy. Methods Cell Biol. 1999;58:293–314. doi: 10.1016/s0091-679x(08)61962-7. [DOI] [PubMed] [Google Scholar]
- 24.Weatherman RV, Chang CY, Clegg NJ, Carroll DC, Day RN, Baxter JD, McDonnell DP, Scanlan TS, Schaufele F. Ligand-selective interactions of estrogen receptor detected in living cells by fluorescence resonance energy transfer. Mol Endocrinol. 2002;16:487–496. doi: 10.1210/mend.16.3.0813. [DOI] [PubMed] [Google Scholar]
- 25.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 26.Day RN, Periasamy A, Schaufele F. Fluorescence resonance energy transfer microscopy of localized protein interactions in the living cell nucleus. Methods. 2001;25:4–18. doi: 10.1006/meth.2001.1211. [DOI] [PubMed] [Google Scholar]
- 27.Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 28.Siegel RM, Chan FKM, Zacharias DA, Swofford R, Holmes KL, Tsien RY, Lenardo MJ. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Science's STKE. 2000 doi: 10.1126/stke.2000.38.pl1. http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2000/38/pl1. [DOI] [PubMed]
- 29.Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 30.Mahajan NP, Linder K, Berry G, Gordon GW, Heim R, Herman B. Bcl-2 and Bax interactions in mitochondria probed with green fluorescent protein and fluorescence resonance energy transfer. Nat Biotechnol. 1998;16:547–552. doi: 10.1038/nbt0698-547. [DOI] [PubMed] [Google Scholar]
- 31.Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein. J Biol Chem. 2001;276:16348–16355. doi: 10.1074/jbc.m100128200. [DOI] [PubMed] [Google Scholar]
- 32.Baxevanis AD, Vinson CR. Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Gen Dev. 1993;3:278–285. doi: 10.1016/0959-437x(93)90035-n. [DOI] [PubMed] [Google Scholar]
- 33.Wedel A, Ziegler-Heitbrock HW. The C/EBP family of transcription factors. Immunobiology. 1995;193:171–185. doi: 10.1016/s0171-2985(11)80541-3. [DOI] [PubMed] [Google Scholar]
- 34.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 35.Williams SC, Angerer ND, Johnson PF. C/EBP proteins contain nuclear localization signals imbedded in their basic regions. Gene Expr. 1997;6:371–385. [PMC free article] [PubMed] [Google Scholar]
- 36.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson GH, Piston DW, Barisas BG. Forster distances between green fluorescent protein pairs. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 38.Gaiddon C, de Tapia M, Loeffler JP. The tissue-specific transcription factor Pit-1/GHF-1 binds to the c-fos serum response element and activates c-fos transcription. Mol Endocrinol. 1999;13:742–751. doi: 10.1210/mend.13.5.0275. [DOI] [PubMed] [Google Scholar]
- 39.Jacobson EM, Peng L, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 40.Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 41.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 42.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 43.Nagai Y, Miyazaki M, Aoki R, Zama T, Inouye S, Hirose K, Iino M, Hagiwara M. A fluorescent indicator for visualizing cAMP-induced phosphorylation in vivo. Nat Biotechnol. 2000;18:313–316. doi: 10.1038/73767. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci USA. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elangovan M, Day RN, Periasamy A. Nanosecond fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J Microsc. 2002;205:3–14. doi: 10.1046/j.0022-2720.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- 47.Day RN, Liu J, Sundmark V, Kawecki M, Berry D, Elsholtz HP. Selective inhibition of prolactin gene transcription by the ETS-2 repressor factor. J Biol Chem. 1998;273:31909–31915. doi: 10.1074/jbc.273.48.31909. [DOI] [PubMed] [Google Scholar]


