Abstract
Cell-cell signalling mediated by Notch regulates many different developmental and physiological processes and is involved in a variety of human diseases. Activation of Notch impinges directly on gene expression through the Suppressor of Hairless [Su(H)] DNA-binding protein. A major question that remains to be elucidated is how the same Notch signalling pathway can result in different transcriptional responses depending on the cellular context and environment. Here, we have investigated the factors required to confer this specific response in Drosophila adult myogenic progenitor-related cells. Our analysis identifies Twist (Twi) as a crucial co-operating factor. Enhancers from several direct Notch targets require a combination of Twi and Notch activities for expression in vivo; neither alone is sufficient. Twi is bound at target enhancers prior to Notch activation and enhances Su(H) binding to these regulatory regions. To determine the breadth of the combinatorial regulation we mapped Twi occupancy genome-wide in DmD8 myogenic progenitor-related cells by chromatin immunoprecipitation. Comparing the sites bound by Su(H) and by Twi in these cells revealed a strong association, identifying a large spectrum of co-regulated genes. We conclude that Twi is an essential Notch co-regulator in myogenic progenitor cells and has the potential to confer specificity on Notch signalling at over 170 genes, showing that a single factor can have a profound effect on the output of the pathway.
Keywords: Drosophila, Gene regulation, Myogenesis, Notch, Twist
INTRODUCTION
Signalling through the Notch receptor controls a broad spectrum of cell fates and developmental processes in organisms ranging from sponges to human and is associated with various pathologies including different types of cancer (Radtke and Raj, 2003; Lasky and Wu, 2005; van Es et al., 2005). This multitude of roles is associated with different outputs, depending upon the cellular context. For example, in certain contexts Notch activation promotes proliferation and maintenance of progenitor populations [e.g. myogenesis (Kopan et al., 1994; Delfini et al., 2000; Hirsinger et al., 2001)], whereas in others it promotes cell type differentiation [e.g. T-cell formation (Robey, 1999) or neural fate (Gaiano and Fishell, 2002)]. Transduction of the Notch pathway is relatively simple: activation of Notch elicits proteolytic cleavage, releasing the Notch intracellular domain (NICD), which enters the nucleus and collaborates directly with DNA-binding proteins of the CSL family {CBF1 (RBPJ) in mammals, Suppressor of Hairless [Su(H)] in Drosophila and LAG-1 in worms (Bray, 2006; Kopan and Ilagan, 2009)}. Effects on gene expression are thus a primary consequence of activating the pathway, implying that the different outputs of Notch activation require a selective transcriptional response. This is highlighted by the observations that the sites of Su(H) occupancy after Notch activation differ according to cell type (Krejci and Bray, 2007) and that only a fraction (slightly more than 10%) of the potential Notch-regulated targets are shared between two cell types (Krejci et al., 2009). The identification of co-regulators that confer such specificity on the Notch response is of primary importance for understanding the context-specific responses.
To date only a few cell type-specific transcription factors have been shown to combine with the Notch pathway to regulate induction of specific targets. These include GATA-like factors, which act in combination with Notch to regulate ref-1 in the C. elegans endoderm (Neves et al., 2007), and NFκB family members, which co-regulate HES and deltex 1, which are both Notch targets during B-lymphocyte development (Moran et al., 2007). The best-characterised co-regulators are the proneural bHLH transcription factors that combine with Notch to activate targets of the E(spl)/HES gene family during neurogenesis in a wide range of species (Nakao and Campos-Ortega, 1996; Parras et al., 1996; Culi and Modolell, 1998). Proneural co-regulation is proposed to depend on a particular binding site architecture, consisting of paired CSL binding sites and a proneural protein binding site (Cave et al., 2005). As this code has been derived from the evolutionarily related genes of the E(spl) family it is not clear whether such specific binding site architecture extends more broadly to other Notch targets, nor whether other co-regulators would involve similar binding site organisation at their targets. Furthermore, in none of the partnerships analysed, including that involving proneural proteins, is it clear whether the regulatory relationship is specific for the targets analysed or whether it extends to a broad spectrum of genes.
In order to discover the scale and breadth of transcriptional responses to Notch, a combination of genome-wide chromatin immunoprecipitation (ChIP) and expression array analysis was used to identify direct Notch targets in different Drosophila cell lines, including DmD8 cells, related to adult muscle progenitors (AMPs) (Krejci et al., 2009). In this context, Notch is required to maintain the progenitors and prevent their differentiation, a function that appears similar to the role of Notch in vertebrate myogenesis, in which activation of Notch signalling prevents muscle differentiation both in embryonic precursors and in satellite cells (Delfini et al., 2000; Hirsinger et al., 2001; Conboy and Rando, 2002; Kopan and Ilagan, 2009). Among the 235 direct Notch targets identified in DmD8 muscle progenitor-like cells, several have been specifically linked to roles in muscle development. However, many others are involved in fundamental aspects of cell differentiation such as signalling and morphogenesis (Krejci et al., 2009). In addressing how the genome integrates Notch activation differentially according to cell type, one question therefore is whether one or many transcription factors are required to specify this diverse cohort of targets and to confer an output that is distinct for myogenic precursors. A second question is how these factors affect CSL binding.
Several key transcription factors are known to have important roles in regulating myogenesis. These include the bHLH proteins of the MyoD/myogenin family and the MADS box proteins of the Mef2 family (Parker et al., 2003; Tajbakhsh, 2003). The latter are important in promoting muscle differentiation in all species (Black and Olson, 1998). In Drosophila, the bHLH protein Twist (Twi) acts prior to Mef2 in both embryonic and adult myogenesis (Cripps et al., 1998; Baylies and Michelson, 2001; Castanon and Baylies, 2002). Initially, Twi is expressed broadly throughout the embryonic mesoderm and subsequently it regulates whether or not the cells are routed into somatic muscle fates. Its expression is then lost in larval muscles when they differentiate (Baylies and Bate, 1996), but persists in adult muscle precursors (Bate et al., 1991), only declining during pupation when the myoblasts fuse and differentiate into the muscles of the adult (Anant et al., 1998). As both the Notch pathway and Twi are important in maintaining AMPs in an undifferentiated state (Anant et al., 1998), Twi is a potential candidate to co-operate with Notch in muscle precursors.
Taking advantage of the Notch targets identified in DmD8 cells, we have investigated how specificity is conferred on the Notch response in the myogenic cell type. Focussing initially on three targets, we find that their expression in vivo is dependent on Twi acting in combination with Notch. Twi binding to target enhancers precedes Su(H) recruitment, which is detected only after activation. To assess whether Twi is likely to confer specificity on a broader range of targets, we analysed whether Twi binding regions identified genome-wide in the DmD8 myogenic precursor-related cells are found in proximity to the Su(H) binding regions identified in the same cells. This revealed a significant association and identified more than 170 genes that are potentially co-regulated by both factors. These results demonstrate that one co-factor, Twi, is likely to regulate on a large scale the output from the Notch pathway in muscle progenitor cells.
MATERIALS AND METHODS
Drosophila experiments
All alleles and stocks are described in FlyBase unless indicated otherwise. The following reporters were used: m6-GFP (Lai et al., 2000); 4.0Him-GFP (Rebeiz et al., 2002); aosNRE-lacZ, aosNREmutSu(H)-lacZ, edlNRE-lacZ (Krejci et al., 2009); and m8-lacZ (Kramatschek and Campos-Ortega, 1994). In overexpression and RNAi experiments, Gal4 driver stocks [1151:Gal4 (Anant et al., 1998), Ptc:Gal4 Tub:Gal80ts, sca:Gal4] were combined with UAS lines [UAS:twi (Baylies and Bate, 1996), UAS-Ni79.2] and/or RNAi lines [UAS-Notch-RNAi (BL7078), UAS-twi-RNAi (GD37092), UAS-da-RNAi (GD51297) (Dietzl et al., 2007), UAS-twi-RNAi2x (Wong et al., 2008)] and larvae were shifted to 30°C 48 hours before dissections. Immunofluorescence was performed as described previously (Cooper and Bray, 1999) with the following antibodies: mouse anti-NICD (DSHB, 1/20), mouse anti-Cut (DSHB, 1/20), mouse anti-β-gal (DSHB, 1/20), rabbit anti-GFP (Invitrogen, 1/1000), guinea-pig anti-Myc [1:500; gift of F. Martin and G. Morata (Herranz et al., 2008)], rabbit anti-Roughest [1/50; gift of K. Fishbach (Schneider et al., 1995)]. β-galactosidase expression was detected histochemically.
Cell experiments and lacZ reporters
E(spl)m3, NME (–ve control) and aosNRE luciferase reporters have been described (Krejci et al., 2009). A fragment encompassing the region –2098 to +37 of E(spl)m6 (Nellesen et al., 1999) was amplified and cloned into a luciferase vector containing a minimal promoter from Hsp70 (pGL3::Min). Twi sites were mutated using oligonucleotides with three mismatches (introducing T at positions 3, 4 and 8) as described (Krejci et al., 2009). After mutagenesis, aosNRE fragments were cloned into either pGL3::Min or pBlueRabbit (which contains lacZ downstream of a minimal Hsp70 promoter). Cell culture conditions and transfections were as described previously (Nagel et al., 2005; Narasimha et al., 2008). To activate Notch, cells were treated with EDTA as described (Krejci and Bray, 2007). For twi RNAi treatment of DmD8 cells, double-stranded RNA duplexes were produced from a 750 bp fragment of the twi exon with T7 promoter-containing primers and subsequently transcribed using a T7 Transcription Kit (Ambion). Cells were incubated with twi RNAi for 72 hours before Notch activation. Three biological replicates were performed in all experiments.
RNA isolation, quantitative PCR and ChIP experiments were performed as described (Krejci and Bray, 2007). Antibodies for ChIP were goat anti-Su(H) (Santa Cruz Biotechnology, sc15813) and rabbit anti-Twi [from M. Leptin and S. Roth (Roth et al., 1989); or from E. Furlong (Sandmann et al., 2007)].
Bioinformatics and Twi genome-wide ChIP in DmD8 cells
Data sets
ChIP-enriched regions in DmD8 cells were determined de novo using Nimblegen HD2 tiling arrays following the procedures and post-hybridisation analysis described previously for Su(H) (Krejci et al., 2009). The array data have been deposited at NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE22650. Peaks were defined using Tamalpais (Bieda et al., 2006), with P<0.0001. ChIP-enriched regions for Su(H) (292), Twi (2512), Mef2 (1464) or Biniou (292) in embryos were taken from previously published experiments (Sandmann et al., 2006; Jakobsen et al., 2007; Sandmann et al., 2007; Krejci et al., 2009; Zinzen et al., 2009). In the case of Biniou, Twi and Mef2, for which analysis of binding had been performed at multiple time-points (Sandmann et al., 2006; Jakobsen et al., 2007; Sandmann et al., 2007; Zinzen et al., 2009), a non-redundant data set was generated with all the regions that were occupied in at least one stage. Where necessary, the UCSC Genome Browser (Kuhn et al., 2009) Liftover tool was used to transpose all genomic positions to coordinates of release 5 of the D. melanogaster genome.
Overlap
Custom-written Perl scripts were used to determine the overlap between ChIP-enriched regions and their statistical significance. The significance of overlap was determined by 1000 rounds of random simulation. In each round, every Su(H) site was allowed to deviate around its original position (Fig. 1A and see Table S2 in the supplementary material; uniform distribution within a range around the original site). The overlap between random Su(H) and Twi sites was determined in each round and recorded to derive a Z-score [(actual overlap – average random overlap) / standard deviation of random overlap] and empirical P-value (0.001 increment for each random overlap ≥ actual overlap).
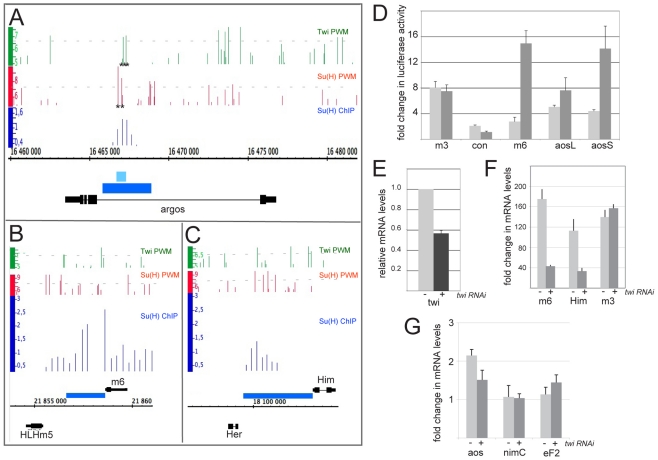
Fig 1.
Requirement for Twist at Notch target enhancers. (A-C) Genomic regions surrounding Drosophila E(spl)m6 (B), Him (C) and aos (A). Black lines and boxes (exons) represent transcribed regions. Graphs show matches to Twi PWM (green bars; Patser score 5-9.3), Su(H) PWM (red bars; Patser score 5-9.79) or the region enriched in ChIP experiments by Su(H) (blue bars; enrichment relative to input; log2 0.3-2.3). Blue rectangles represent regions cloned to create reporters used in subsequent experiments. Asterisks (A) represent binding sites that have been mutated in subsequent experiments. Light-blue rectangle represents fragment aosS, the dark-blue rectangle fragment aosL. Note that aosS contains all Su(H) sites and two of the three Twi sites, aosL=aosNRE and contains additional Su(H) sites. (D) Response of the indicated enhancers to NICD in transient transfection assays in S2 cells (light-grey bars) or DmD8 cells (dark-grey bars). (E-G) twi RNAi treatment of DmD8 cells. (E) Efficiency of twi RNAi treatment on twi levels in DmD8 cells. Values in control untreated cells are normalised to 1. (F,G) Fold change in the expression levels of the indicated genes after Notch activation, as determined by quantitative RT-PCR in the absence (light grey) or presence (dark grey) of twi RNAi. The average of three biological replicates is shown; the data are plotted on separate graphs because of the difference in magnitude of the fold stimulation by Notch. (F) E(spl)m6 and Him, which show 175-fold and 113-fold stimulation by Notch, respectively, are reduced by 76% and 79%, respectively, by twi RNAi, whereas the 140-fold stimulation of E(spl)m3 is not reduced by twi RNAi. (G) The 2-fold stimulation of aos by Notch showed a 56% reduction in the presence of twi RNAi. Control genes (nimC1 and Ef2) were neither induced by Notch nor reduced by twi RNAi. Expression levels in F and G were normalised to that of rp49 (RpL32). Error bars are s.e.m.
Position weight matrix (PWM) matches
Alignment matrices for Su(H) (Krejci et al., 2009), Twi (Down et al., 2007) and Mef2 were built based on a compilation of previously published binding sites (see Fig. S3 in the supplementary material). The motif scanner nmscan from the NestedMICA package (Down and Hubbard, 2005) was used for genome-wide motif matching, with empirically defined cut-off values for each transcription factor (roughly yielding the same number of genomic binding sites). For sites shown in Fig. 1, matches to PWMs were computed using Patser (Hertz and Stormo, 1999) with a threshold of 5.5 (the approximate cut-off used above is indicated on the resulting graph by a dashed line).
RESULTS
Functional relevance of Twist for Notch target gene expression
We first focussed our investigations on a selection of DmD8 Notch targets with distinct cellular functions: E(spl)m6, an immediate early Notch target involved in regulating Notch pathway activity (Bardin and Schweisguth, 2006); argos (aos), an inhibitor of EGF receptor signalling (Howes et al., 1998); and Him, a repressor that inhibits Mef2 (Liotta et al., 2007). The first question we addressed was whether the cell-context specificity was retained in enhancer fragments isolated from two of these target genes, by testing whether the enhancers responded differently to NICD according to cell type. Fragments encompassing Su(H) ChIP peak regions from E(spl)m6 and aos (Krejci et al., 2009) (see Fig. 1A,B) were tested in comparison to E(spl)m3, which is inducible by Notch in all cells that we have tested (Krejci and Bray, 2007). The resulting reporter genes were co-transfected into DmD8 muscle progenitor type cells and into S2 blood-related cells. E(spl)m6 and aos enhancer fragments both recapitulated the cell specificity, responding to NICD much more robustly in DmD8 cells (Fig. 1D). The magnitude of the response was larger for aosS than for the longer aosL, suggesting that the latter can recruit factors that antagonise NICD or that affect accessibility. Nevertheless, the results suggest that the enhancer fragments are sufficient to confer a context-specific Notch response that is dependent on factors present in DmD8 cells.
Transcription factors that might contribute to the specificity of Notch in DmD8 cells and myogenic precursors include Twi and Mef2 (Castanon and Baylies, 2002; Cripps et al., 2004; Lovato et al., 2005). RNA sequencing profiles have shown that DmD8 cells express high levels of twi mRNA (Celniker et al., 2009) and there is a striking similarity between Notch and twi function in myogenic precursors (Anant et al., 1998), making Twi a good candidate for co-regulatory activity. As a first test of this hypothesis, we treated DmD8 cells with twi RNAi and assayed its effects on a selection of targets (using quantitative RT-PCR to detect transcript levels), under conditions in which the efficacy of twi knockdown was ∼50% to limit any concomitant effects on overall cell fate (Fig. 1E). Even under these limited conditions, E(spl)m6, Him and aos all showed a reduced response to Notch in twi RNAi-treated cells (Fig. 1F,G; 76% reduction for E(spl)m6, 79% for Him and 56% for aos). Neither E(spl)m3, nimC1 (which encodes a phagocytic receptor) nor Elongation factor 2 (Ef2) showed any reduction, arguing that the reduced expression is specific and likely to reveal Twi-dependent targets.
We extended these assays to the muscle progenitors in the wing disc. Enhancers from E(spl)m6, aos and Him all direct expression in AMPs (Lai et al., 2000; Krejci et al., 2009). These cells are apposed beneath the disc epithelium in the notal region (and hence are also known as adepithelial cells) and can be distinguished from the overlying epithelium by the expression of markers such as Cut. All three enhancers directed expression in the Cut-expressing AMPs (Fig. 2A,D,F), although E(spl)m6-GFP expression was restricted to a localised region in the anterior (Fig. 2A). Using several independent RNAi constructs to ablate twi in these cells (expressed using the 1151-Gal4 driver), we consistently detected a reduction of E(spl)m6-GFP, aosNRE-lacZ and 4.0Him-GFP expression (Fig. 2B,C,E,G). Importantly, the muscle progenitors were still present under these conditions and could be detected by expression of markers such as Cut (e.g. Fig. 2B,C). Ablation of Daughterless (Da), a dimerisation partner of Twi in certain contexts (Castanon et al., 2001), had no effect on these targets [although expression of da RNAi in proneural clusters with sca-Gal4 did alter the expression of E(spl)m8 as expected] (see Fig. S1 in the supplementary material). Likewise, no change in E(spl)m6-GFP expression was seen with RNAi targeting nine other bHLH genes (see Table S1 in the supplementary material). Thus, the conditional knockdown of twi, but not da or other bHLH transcription factors, is sufficient to prevent expression from E(spl)m6, aos and Him enhancers.
Fig 2.
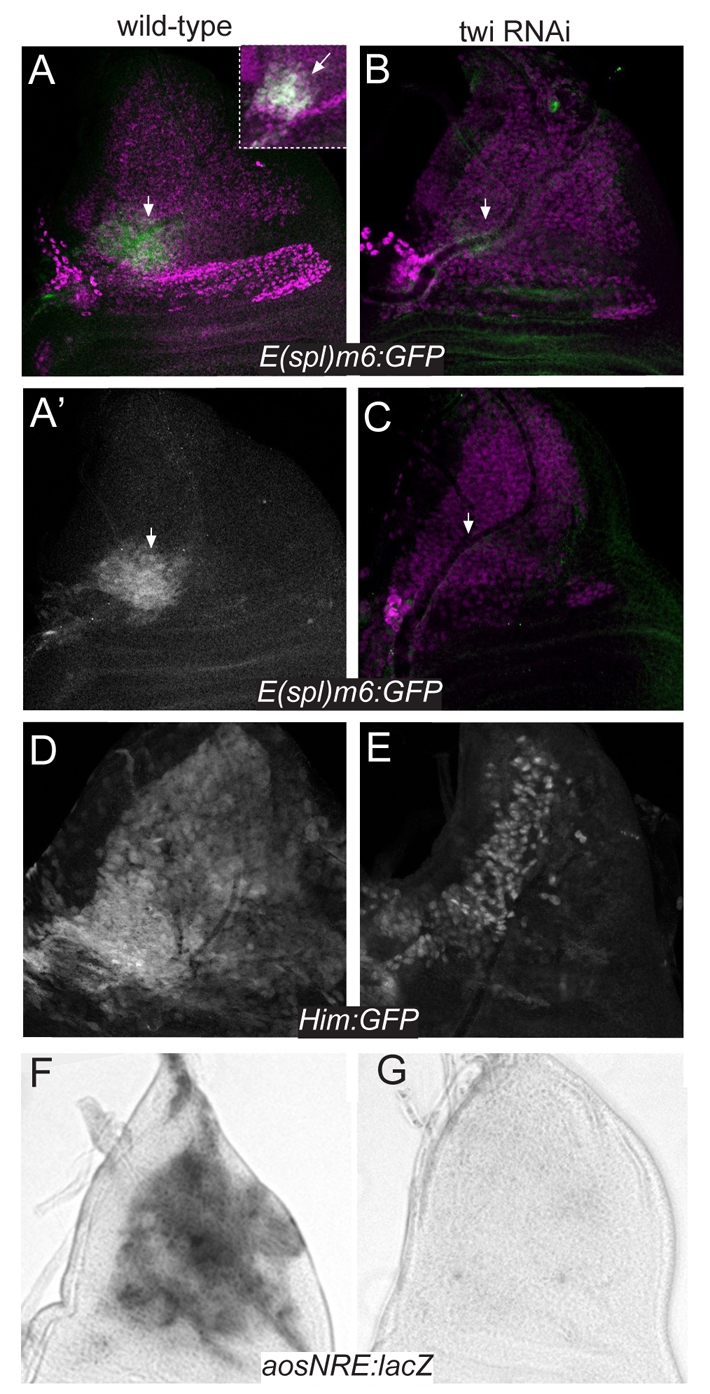
Twist regulation of Notch target enhancers in muscle progenitors. (A-C) Effect of two independent twi RNAi constructs (GD37092, B; twi-RNAi2x, C) on E(spl)m6-GFP expression (green in A,B,C; white in A′). A patch of E(spl)m6-GFP is detectable in wild type in Cut-expressing adult muscle progenitors (AMPs) (A, arrow; inset) but is severely reduced (B, arrow) or abolished (C, arrow) in the presence of twi RNAi. Cut expression (which marks all muscle progenitors; purple, A-C) is unchanged. (D-G) Expression of the indicated Notch target genes in wild type (D,F) and in muscle progenitors expressing twi RNAi (E,G). Expression was monitored using the indicated reporters, detected by antibodies (A-E) or histochemistry (F,G), and colocalisation with AMPs was verified by co-staining with Cut (A-C) or by interference microscopy (F,G).
Twist and Notch co-operate to activate target genes
The reduced expression of several target genes following twi ablation indicates that Twi contributes to their regulation. However, these assays do not distinguish whether Twi alone is sufficient or whether it requires collaboration with Notch. To address this question, we assessed the ability of Twi to confer appropriate Notch responsiveness when expressed in a different cell type in the presence and absence of Notch. Genes such as E(spl)m6 or Him are normally only expressed in AMPs, not elsewhere in the wing disc, despite Notch activity at other locations. Similarly, expression from enhancers identified by Su(H) binding in DmD8 cells is also restricted to muscle progenitors (e.g. aos). We therefore investigated whether Notch and/or Twi were able to activate these enhancers when expressed ectopically in the wing pouch using ptc-Gal4 (Fig. 3T) in combination with thermosensitive Gal80, so that expression was limited to a defined period at the end of larval development.
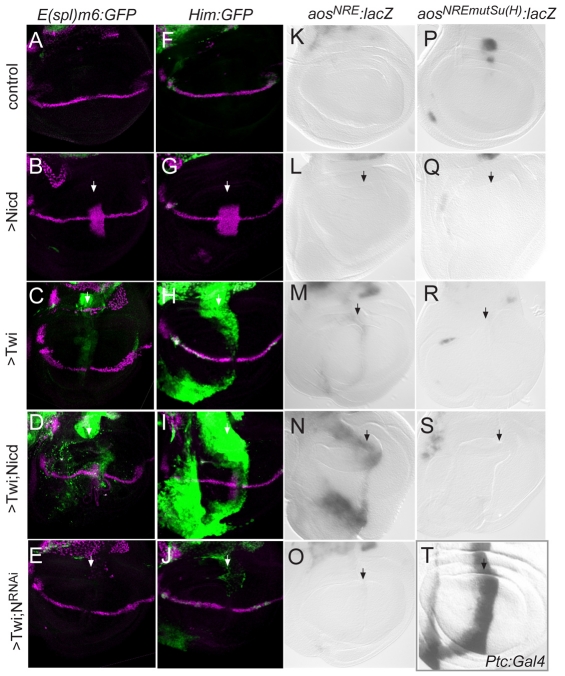
Fig. 3.
Twist acts in combination with Notch to regulate myogenic target enhancers. (A-S) Expression from reporters E(spl)m6-GFP (green, A-E), 4.0Him-GFP (green, F-J), aosNRE-lacZ (grey, K-O) or aosNRESu(H)mut-lacZ (grey, P-S) in wild type (A,F,K,P), and in discs where the levels of Twi and Notch activity were manipulated in a stripe (arrow, ptc-Gal4;Tub-Gal80ts; see T) by NICD expression (>NICD; B,G,L,Q), by Twi overexpression (>Twi; C,H,M,R), by combining NICD and Twi overexpression (>Twi;NICD; D,I,N,S), or by ablating Notch (with Notch RNAi) in cells overexpressing Twi (>Twi;N-RNAi; E,J,O). (T) The expression domain of the Ptc:Gal4 driver, detected with UAS:lacZ. In A-J, Cut expression (magenta) is used as an indicator of Notch activity at the dorsal-ventral boundary; in K-T, β-galactosidase was detected histochemically.
First we assessed whether simply increasing Notch activity in the wing pouch domain would activate the muscle progenitor enhancers. Although this was sufficient to produce ectopic expression of genes that are direct Notch targets at the dorsal-ventral boundary (e.g. Fig. 3B,G), neither E(spl)m6-GFP, 4.0Him-GFP nor aosNRE-lacZ exhibited any ectopic activation (Fig. 3B,G,L). By contrast, expression of Twi using the same driver elicited ectopic expression from all three enhancers (Fig. 3C,H,M), giving strong induction of 4.0Him-GFP and weak induction of E(spl)m6-GFP and aosNRE-lacZ. Their ectopic induction indicates that the enhancers are capable of responding to Twi. This response is highly selective, as increased or ectopic expression of 20 other bHLH proteins, including proneural proteins, failed to elicit any change in E(spl)m6 expression (see Table S1 in the supplementary material).
Combining expression of Twi with NICD yielded much stronger expression from all three enhancers in the wing pouch than expression of Twi alone (Fig. 3D,I,N). These results suggest that Notch and Twi co-operate to induce expression of these genes. To further confirm whether this co-operation was essential, we also tested the consequences of ablating Notch activity when Twi was ectopically expressed. All three enhancers showed substantially reduced expression when Twi was expressed in combination with Notch RNAi (Fig. 3E,J,O), as compared with Twi alone, and two of the three had no residual ectopic expression. This indicates that the expression induced by Twi alone is largely dependent on an underlying activity of Notch [sensitive Notch reporters reveal a low level of Notch activity in most cells of the wing pouch (Furriols and Bray, 2001)]. The dependence on Notch was further confirmed by analysing expression from an aos enhancer in which the Su(H) binding sites had been mutated. This mutated aosNREmutSu(H) enhancer is no longer expressed in muscle progenitors (Krejci et al., 2009) and was not able to respond to ectopic Twi expression or to the combination of Twi and NICD (Fig. 3P-S). Together, these results show that Twi confers the ability on these target enhancers to respond to Notch activity, and vice versa.
Twist binding sites are required for the activity of Notch target gene enhancers
Our results point to a pivotal role for Twi in the context-specific Notch response, but do not distinguish whether this effect is direct. In order to test whether direct binding by Twi is required, we mutated putative Twi binding sites in one of the responsive enhancers. aosNRE contains three matches to a position weight matrix (PWM) for Twi binding sites (see Fig. 1A), with sites 2 and 3 having the best matches. We generated fragments with mutations in site 2 (aosNREΔTwi2) and with mutations in both site 2 and the adjacent site 3 (aosNREΔTwi2,3). When tested in DmD8 cells, both enhancers had lost their ability to respond to Notch activation, suggesting that these sites are essential (Fig. 4A). Similarly, when tested in vivo, the mutated fragments had lost the ability to direct expression in AMPs (Fig. 4B,C). Thus, Twi binding sites are necessary for aosNRE activity in DmD8 cells and in vivo in myogenic precursors.
Fig. 4.
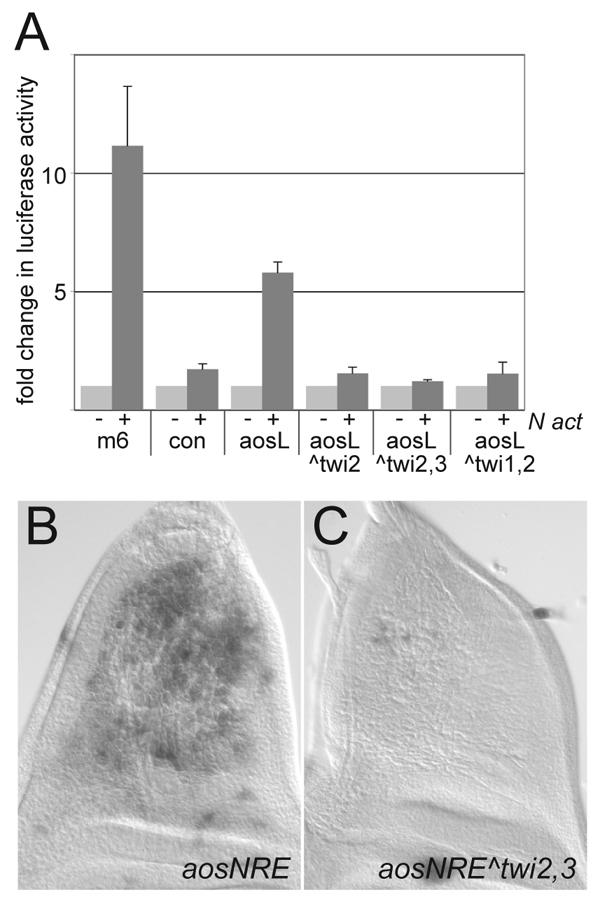
Twist binding sites are required for Notch activation of argosNRE. (A) Response of the indicated enhancers to NICD in transient transfection assays in DmD8 cells, expressed as fold change (dark-grey bars) relative to expression levels in the absence of Notch (light-grey bars). Mutating Twi binding sites as indicated abolishes aosNRE responsiveness. (B,C) In vivo, mutations of the same sites lead to a decrease in expression (C) compared with the control enhancer (B).
Genome-wide association of Twist and Suppressor of Hairless binding
As the tested enhancers all require Twi for their responsiveness to Notch, this regulatory combination might be required for the majority of Notch targets in DmD8 cells and muscle progenitors. If this were the case, there should be a strong association of Su(H) and Twi sites among the DmD8 target loci. To test this possibility, we first used data from previously published genome-wide ChIP experiments performed in embryos (Sandmann et al., 2006; Jakobsen et al., 2007; Sandmann et al., 2007; Zinzen et al., 2009). Using two approaches (Fig. 5A and see Fig. S2 in the supplementary material), we found a significant association between Su(H) and Twi binding regions (based on a null model that constrained the position of the peaks to take account of their non-random distribution across the genome; see Materials and methods and Table S2 in the supplementary material). Furthermore, when the partner combinations between Su(H) and several other transcription factors were calculated on the basis of the embryonic data, there was a strong pair-wise correlation of Su(H) with Twi (see Fig. S2B in the supplementary material, red box) and ChIP peaks of other muscle transcription factors exhibited less association with Su(H)-bound regions (Fig. 5A and see Fig. S2C in the supplementary material), suggesting that the association with Twi is specific.
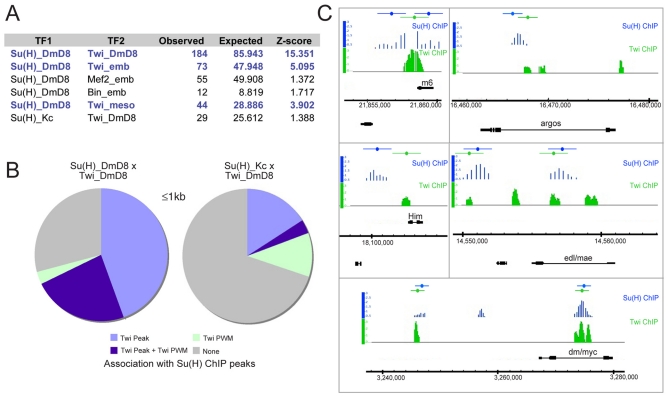
Fig 5.
Genome-wide association of Twist and Su(H) binding. (A) Observed and expected random frequency of overlap between Su(H) peaks (TF1) from DmD8 or Kc cells and the indicated binding regions of other transcription factors (TF2). TF2 binding regions were Twi in DmD8 (this study), Twi, Mef2 and Biniou in embryos (Sandmann et al., 2006; Jakobsen et al., 2007; Sandmann et al., 2007) and a second Twi embryonic data set generated with finer scale peak resolution (Twi_meso) (Zinzen et al., 2009). The Z-score expresses the divergence of the observed result from the random model, with scores above three (blue) indicating significant divergence. In the random model, Su(H) sites were allowed to deviate by five times the average peak width around the actual position, and differences in the number of TF2 peaks are internally controlled for in this randomisation strategy. We note, however, that Su(H)_Kc peaks were narrower than those of Su(H)_DmD8, so the replacement parameters were not directly comparable. (B) Pie charts representing the percentage of Su(H)_DmD8 peaks (left) or Su(H)_Kc peaks (right) that have midpoints within 1 kb of Twi_DmD8 peaks with (dark blue) and without (light blue) matches to the PWM, of Twi PWM matches (light green) or neither association (grey). The midpoint analysis is independent of the peak width; peak numbers in Su(H)_DmD8 (260) and Su(H)_Kc (241) are similar. (C) Examples of genomic regions from representative DmD8 Notch targets showing Su(H) (blue) and Twi (green) ChIP-enriched regions in DmD8 cells. Note that oligonucleotides were spaced at 300 nucleotides in the tiling arrays for Su(H) and at 55 nucleotides for Twi. Gene models are depicted in black. Coloured bars above represent the windows used to calculate peak overlaps using the midpoint method and a 1 kb window.
It is likely that Twi binding differs between cell types and developmental stages. Therefore, although the embryonic data are indicative of regions that have the potential to bind Twi, these might not be identical to the occupied regions in Notch-activated DmD8 cells. We therefore performed ChIP in the DmD8 cells and hybridised the bound DNA to a whole-genome tiling array to determine the sites of Twi occupancy. This identified ∼2500 Twi-enriched regions (Twi peaks called at P<0.0001) in the DmD8 cells. Here, we focus on the relationship of the Twi peaks with Su(H)-bound regions to investigate Notch and Twi co-regulation and identify putative co-regulated genes. First, we found that 184/260 (71%) Su(H) peaks directly overlapped with Twi-bound regions (Z-score>15.35, based on the constrained random model; Fig. 5A and see Table S2 in the supplementary material). To avoid bias due to differences in the average size of the peaks in the two data sets, we also analysed the proximity of Twi and Su(H) peaks using a midpoint-centred method (Fig. 5B,C). This approach identified 181 (70%) Su(H) peaks with midpoints within 1 kb of Twi peak midpoints (Fig. 5B) and 200 (76%) that were within 2 kb of Twi peak midpoints (see Fig. S2 in the supplementary material). Again, these are highly significant when compared with a similar analysis using an Su(H) peak data set from another (Kc) cell type (Fig. 5B; P<<10–16). Thus, our comparisons reveal a striking association between Su(H) and Twi in DmD8 AMP-like cells and we note also that the majority of potential Twi sites (identified by motif-finding with PWM; see Fig. S3 in the supplementary material) in proximity to the Su(H) peaks are occupied by Twi in these cells (Fig. 5B).
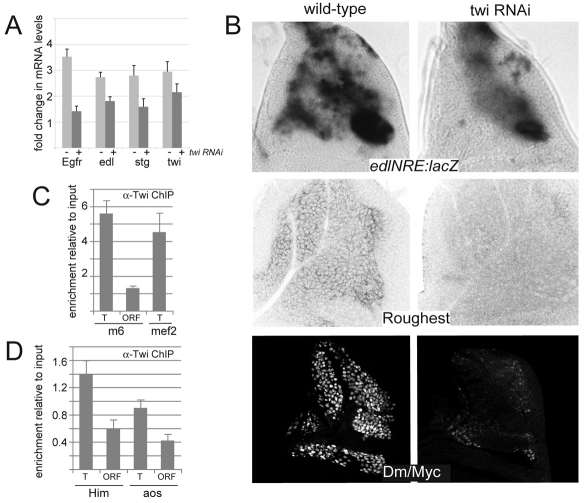
Extrapolating the Twi peaks to the previously assigned direct Notch-regulated target genes in DmD8 cells (Krejci et al., 2009) revealed that 84% (197/235) of the assigned Notch targets had Twi peaks in their vicinity (see Table S3 in the supplementary material). These included E(spl)m6, aos and Him, as well as other targets previously shown to be expressed in muscle progenitors in vivo (Fig. 5C) (Krejci et al., 2009). To further confirm that Twi was required for normal expression of the associated Notch targets, we monitored the expression of four genes in twi RNAi-treated cells (under conditions that reduced Twi by ∼50%, as previously). All showed a decrease in activation (e.g. Fig. 6A). We therefore selected three further target genes and monitored the effects of twi RNAi on their expression in vivo using available reporter genes or antibodies (Fig. 6B). All three genes [edl (mae), roughest and diminutive (myc)] were expressed in the muscle progenitors in wild-type discs and had reduced expression in the presence of twi RNAi, supporting the model that they are co-regulated by Twi.
Fig 6.
Depletion of Twi compromises target expression and Twi is present at target enhancers prior to Notch activation. (A) Fold change in the expression levels of the indicated genes after Notch activation as determined by quantitative RT-PCR in the absence (light grey) or presence (dark grey) of twi RNAi. The average of three biological replicates is shown. (B) Expression of the indicated Notch target genes in wild type and in muscle progenitors expressing twi RNAi. (C,D) Enrichment of fragments from the indicated genes in anti-Twi ChIP from DmD8 cells before Notch activation. T, Twi binding/enhancer region; ORF, open reading frame. Results are the average of three independent experiments; error bars indicate s.e.m.
The genome-wide results suggest, therefore, that Twi has a widespread role in conferring the myogenic-specific response to Notch. We further queried whether the organisation of the Su(H) and Twi sites in the target enhancers had any conserved features such as spacing or topology, as seen for the proneural and Su(H) binding region in E(spl) genes (the A-SPS motif). However, we found no evidence for a consistent motif organisation within the identified regions. We note, however, that in the majority (71%) of the assigned co-regulated targets, the distance between the Su(H) and Twi peak midpoints is less than 500 bp (see Table S3 in the supplementary material).
Twist facilitates Suppressor of Hairless binding to Notch target gene enhancers
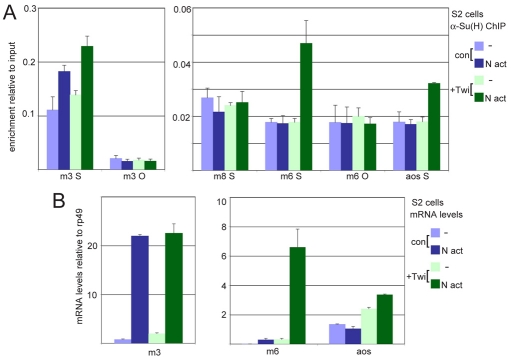
Twi has the characteristics to confer cell context on the Notch response. We therefore first investigated whether it is present on target enhancers prior to Notch activation. Fragments that encompass Twi sites in E(spl)m6, Him and aos were enriched, as compared with the cognate open reading frame, in a Twi ChIP from unactivated DmD8 cells (Fig. 6C,D), suggesting that Twi occupancy precedes recruitment of the Notch-Su(H) complex. Similar results were obtained with an independent anti-Twi antibody (see Fig. S4 in the supplementary material). We therefore asked whether Twi alters the recruitment of Su(H) to target enhancers. For this purpose, we created S2-Notch cells in which twi expression is under the control of an inducible metallothionein (MT) promoter. We then compared Su(H) binding enrichment in the presence and absence of Twi and/or Notch activation (Fig. 7A). As previously, we detected increased binding of Su(H) close to E(spl)m3 after Notch activation in S2 cells. By contrast, no binding was detected at the sites associated with E(spl)m6 or aos. Induction of Twi alone had little effect on the Su(H) binding at these locations in the absence of Notch. However, when we activated Notch in cells in which Twi expression had been induced, we detected a significant enrichment for Su(H) binding (Fig. 7A) and an induction of cognate mRNA expression (Fig. 7B). aos expression was increased above basal levels by Twi alone, supporting the observation that Twi can bind to target enhancers in the absence of Notch activation. As aos is associated with several Twi peaks (Fig. 5C), its increased expression in the absence of Notch might be explained by Twi recruitment at other regulatory modules. As a further control, we examined E(spl)m8, which is a Notch target in the proneural clusters, where it is regulated by proneural bHLH proteins (Achaete and Scute). E(spl)m8 was not activated in the Twi-expressing S2 cells and Su(H) binding was not enriched (Fig. 7A,B). These results suggest, therefore, that Twi enhances Su(H) binding to myogenic Notch target enhancers under conditions in which Notch is also activated.
Fig 7.
Twist enhances Su(H) binding. (A) Enrichment of fragments from the indicated genes in an anti-Su(H) ChIP from control (con, blue) or Twi-expressing (+Twi, green) cells, before (light bars) or after (dark bars) Notch activation (N act). S, Su(H) binding/enhancer region; O, ORF. (B) mRNA levels of the indicated genes. Colours indicate conditions as in A.
DISCUSSION
A major outcome of Notch activation resides in the transcriptional programme elicited in a given cell type. However, little is known about the factors that are required to confer such specificity. This is primarily because most studies have focussed on a small number of HES/E(spl) gene targets. Here, we have taken advantage of novel Notch targets identified in a genome-wide study of muscle progenitor-related cells to uncover a key component in specifying the Notch response in this context: the bHLH transcription factor Twi. Starting initially with a diverse set of Notch targets that are expressed in AMPs, we found that Twi was crucial for their expression in these cells. Furthermore, the enhancers tested in our ectopic assay were only strongly induced when Twi was present in combination with Notch activation. Indeed, an enhancer in which the Su(H) sites were mutated was no longer able to respond to Twi, and vice versa, demonstrating that their combined activities are necessary.
To determine whether the Twi co-regulation could be extrapolated to a broad spectrum of Notch targets in muscle progenitors, we assessed whether there was a significant association between Su(H) and Twi binding in the muscle progenitor-related DmD8 cells. Comparison of the binding regions genome-wide revealed a strong association of Twi and Su(H) among these targets: 71% of Su(H) peaks directly overlapped with Twi peaks. This association was highly significant based on random models that constrained the positions of peaks across the genome to take account of the non-random distribution of transcription factor binding sites and in comparison to several other ChIP data sets. Expression of putative Notch-Twi targets in myogenic precursors was dependent on Twi, as predicted by the association. Together, these data indicate that one transcription factor, Twi, has the potential to co-ordinate the expression of a broad cross-section of Notch targets in muscle progenitors (84% of previously assigned Notch targets are associated with a Twi peak) and thus to confer a specific context on the Notch response.
Further evidence in support of the instructive role of Twi comes from its ability to confer Notch responsiveness on some muscle precursor targets when expressed in a heterologous cell line. Twi itself was found to occupy sites on the target enhancers prior to Notch activation, and in the heterologous cells it was accompanied by increased Su(H) binding after Notch activation. This suggests that Twi binding precedes Su(H) recruitment. However, the co-regulation does not appear to require the Twi partner Da [the E47 (TCF3) homologue], which was previously reported to contact Notch. Furthermore, there does not appear to be any specific organisation or spacing of Twi and Su(H) motifs among the co-regulated targets, in contrast to the conserved motif, consisting of paired Su(H) sites closely linked to an A-class bHLH binding site, that is thought to underlie Notch-proneural bHLH synergy (Cave et al., 2005). Nevertheless, in most of the co-regulated targets, the Su(H) and Twi mid-peaks are separated by less than 500 bp, suggesting that the interaction operates over a limited range. The Twi-Su(H) co-regulation appears, therefore, more in keeping with models in which binding sites for transcription factors are flexibly disposed and act independently with targets in the basal transcriptional machinery [the so-called `billboard' model (Arnosti and Kulkarni, 2005)]. However, mutation of a single Twi binding motif in the aos enhancer was sufficient to compromise activity, despite the fact that there are several matches to the Twi consensus site, suggesting that only a subset of the possible binding motifs are crucial. In addition, as the characterised Notch-Twi-dependent enhancers do not have identical patterns of expression, it is likely that their activity is further constrained by others factors. This is most evident for E(spl)m6, which is only expressed in a small patch of the AMPs but nevertheless responds very robustly to Twi and NICD.
What are the likely characteristics conferred on cells by the Twi-Notch combination?
The AMPs have the capacity for self-renewal and are not committed to a particular muscle lineage, characteristics similar to those of mammalian muscle satellite cells (Figeac et al., 2007). The genes regulated by the combination of Twi and Notch might therefore be important for maintaining these cells as progenitors with myogenic potential. Normally, Twi expression declines as the muscles differentiate. Interfering with this regulation by persistent expression of Twi or Notch inhibits the development of mature fibres (Anant et al., 1998). Conversely, ablating Notch results in premature differentiation (Krejci et al., 2009). The genes regulated by the combination of Twi and Notch include those with proven roles in myogenic regulation that are relevant to the maintenance of muscle progenitors. These include twi itself, Him [an inhibitor of Mef2 (Liotta et al., 2007)] and zfh1 [a repressor of myogenesis (Postigo et al., 1999)]. As Notch signalling and Twi homologues also inhibit vertebrate myogenic differentiation (Kopan et al., 1994; Rohwedel et al., 1995; Spicer et al., 1996; Delfini et al., 2000; Hirsinger et al., 2001; Schuster-Gossler et al., 2007; Vasyutina et al., 2007), and overexpression of Twi in terminally differentiated myotubes can induce reversal of cell differentiation (Hjiantoniou et al., 2008), it will be interesting to test whether homologues of the identified co-regulated targets of Notch and Twi are similarly regulated.
The Notch-Twi combination might also be required to confer properties on the adult myogenic precursors, such as differential adhesion, migration and proliferation. Besides the genes with proven roles in myogenic regulation, many of the Notch-Twi co-regulated genes are implicated in morphogenesis. These include the Ig-domain proteins Roughest, Kirre and Dscam (Bao and Cagan, 2005; Schmucker and Chen, 2009; Zhuang et al., 2009), the netrin receptor Unc-5 and leucine repeat protein Capricious. Notch and Twi have both been found to contribute to the regulation of the epithelial-mesenchymal transition (EMT), and Twi is proposed to affect malignant progression by inducing EMT and suppressing the senescence response (Ansieau et al., 2008). Therefore, it is possible that the co-regulated genes might also confer specialised behaviours that are required in the adult precursors and in cells undergoing EMT (Huber et al., 2005).
In conclusion, we find that Notch and Twi potentially co-regulate a broad spectrum of genes required for the maintenance of muscle progenitors. This suggests that a single co-regulatory relationship can account for a significant component (>170 genes) of the Notch output in one cell type.
Supplementary Material
Acknowledgments
We are particularly grateful to Maria Leptin for the Twi antibody used for the genome-wide ChIP. We also thank Mary Baylies, Karl Fishbach, Eileen Furlong, Alexis Lalouette, Gines Morata and Joel Silber for generously providing fly stocks or antibodies and Rob White and members of our groups for helpful discussions. This work was supported by programme and project grants to S.B. from the Medical Research Council and by a grant from the Association for International Cancer Research. A.K. and F.B. were European Molecular Biology Organization long-term fellows. B.A. is a Royal Society University Research Fellow and acknowledges the Wellcome Trust for supporting the development of computational methods used in this study. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.053181/-/DC1
References
- Anant S., Roy S., VijayRaghavan K. (1998). Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development 125, 1361-1369 [DOI] [PubMed] [Google Scholar]
- Ansieau S., Bastid J., Doreau A., Morel A. P., Bouchet B. P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., et al. (2008). Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14, 79-89 [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Kulkarni M. M. (2005). Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890-898 [DOI] [PubMed] [Google Scholar]
- Bao S., Cagan R. (2005). Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell 8, 925-935 [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Schweisguth F. (2006). Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell 10, 245-255 [DOI] [PubMed] [Google Scholar]
- Bate M., Rushton E., Currie D. A. (1991). Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 113, 79-89 [DOI] [PubMed] [Google Scholar]
- Baylies M. K., Bate M. (1996). twist: a myogenic switch in Drosophila. Science 272, 1481-1484 [DOI] [PubMed] [Google Scholar]
- Baylies M. K., Michelson A. M. (2001). Invertebrate myogenesis: looking back to the future of muscle development. Curr. Opin. Genet. Dev. 11, 431-439 [DOI] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. (2006). Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16, 595-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. L., Olson E. N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167-196 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Castanon I., Baylies M. K. (2002). A Twist in fate: evolutionary comparison of Twist structure and function. Gene 287, 11-22 [DOI] [PubMed] [Google Scholar]
- Castanon I., Von Stetina S., Kass J., Baylies M. K. (2001). Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128, 3145-3159 [DOI] [PubMed] [Google Scholar]
- Cave J. W., Loh F., Surpris J. W., Xia L., Caudy M. A. (2005). A DNA transcription code for cell-specific gene activation by notch signaling. Curr. Biol. 15, 94-104 [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S., Karpen G. H., Kellis M., Lai E. C., Lieb J. D., MacAlpine D. M., et al. (2009). Unlocking the secrets of the genome. Nature 459, 927-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy I. M., Rando T. A. (2002). The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 3, 397-409 [DOI] [PubMed] [Google Scholar]
- Cooper M. T., Bray S. J. (1999). Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397, 526-530 [DOI] [PubMed] [Google Scholar]
- Cripps R. M., Black B. L., Zhao B., Lien C. L., Schulz R. A., Olson E. N. (1998). The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 12, 422-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps R. M., Lovato T. L., Olson E. N. (2004). Positive autoregulation of the Myocyte enhancer factor-2 myogenic control gene during somatic muscle development in Drosophila. Dev. Biol. 267, 536-547 [DOI] [PubMed] [Google Scholar]
- Culi J., Modolell J. (1998). Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12, 2036-2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfini M. C., Hirsinger E., Pourquie O., Duprez D. (2000). Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development 127, 5213-5224 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156 [DOI] [PubMed] [Google Scholar]
- Down T. A., Hubbard T. J. (2005). NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 33, 1445-1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T. A., Bergman C. M., Su J., Hubbard T. J. (2007). Large-scale discovery of promoter motifs in Drosophila melanogaster. PLoS Comput. Biol. 3, e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figeac N., Daczewska M., Marcelle C., Jagla K. (2007). Muscle stem cells and model systems for their investigation. Dev. Dyn. 236, 3332-3342 [DOI] [PubMed] [Google Scholar]
- Furriols M., Bray S. (2001). A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11, 60-64 [DOI] [PubMed] [Google Scholar]
- Gaiano N., Fishell G. (2002). The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci. 25, 471-490 [DOI] [PubMed] [Google Scholar]
- Herranz H., Perez L., Martin F. A., Milan M. (2008). A Wingless and Notch double-repression mechanism regulates G1-S transition in the Drosophila wing. EMBO J. 27, 1633-1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., Stormo G. D. (1999). Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563-577 [DOI] [PubMed] [Google Scholar]
- Hirsinger E., Malapert P., Dubrulle J., Delfini M. C., Duprez D., Henrique D., Ish-Horowicz D., Pourquie O. (2001). Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation. Development 128, 107-116 [DOI] [PubMed] [Google Scholar]
- Hjiantoniou E., Anayasa M., Nicolaou P., Bantounas I., Saito M., Iseki S., Uney J. B., Phylactou L. A. (2008). Twist induces reversal of myotube formation. Differentiation 76, 182-192 [DOI] [PubMed] [Google Scholar]
- Howes R., Wasserman J. D., Freeman M. (1998). In vivo analysis of Argos structure-function. Sequence requirements for inhibition of the Drosophila epidermal growth factor receptor. J. Biol. Chem. 273, 4275-4281 [DOI] [PubMed] [Google Scholar]
- Huber M. A., Kraut N., Beug H. (2005). Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17, 548-558 [DOI] [PubMed] [Google Scholar]
- Jakobsen J. S., Braun M., Astorga J., Gustafson E. H., Sandmann T., Karzynski M., Carlsson P., Furlong E. E. (2007). Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev. 21, 2448-2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Nye J. S., Weintraub H. (1994). The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development 120, 2385-2396 [DOI] [PubMed] [Google Scholar]
- Kramatschek B., Campos-Ortega J. A. (1994). Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development 120, 815-826 [DOI] [PubMed] [Google Scholar]
- Krejci A., Bray S. (2007). Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21, 1322-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A., Bernard F., Housden B. E., Collins S., Bray S. J. (2009). Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2, ra1 [DOI] [PubMed] [Google Scholar]
- Kuhn R. M., Karolchik D., Zweig A. S., Wang T., Smith K. E., Rosenbloom K. R., Rhead B., Raney B. J., Pohl A., Pheasant M., et al. (2009). The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 37, D755-D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., Bodner R., Posakony J. W. (2000). The enhancer of split complex of Drosophila includes four Notch-regulated members of the bearded gene family. Development 127, 3441-3455 [DOI] [PubMed] [Google Scholar]
- Lasky J. L., Wu H. (2005). Notch signaling, brain development, and human disease. Pediatr. Res. 57, 104R-109R [DOI] [PubMed] [Google Scholar]
- Liotta D., Han J., Elgar S., Garvey C., Han Z., Taylor M. V. (2007). The Him gene reveals a balance of inputs controlling muscle differentiation in Drosophila. Curr. Biol. 17, 1409-1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovato T. L., Benjamin A. R., Cripps R. M. (2005). Transcription of Myocyte enhancer factor-2 in adult Drosophila myoblasts is induced by the steroid hormone ecdysone. Dev. Biol. 288, 612-621 [DOI] [PubMed] [Google Scholar]
- Moran S. T., Cariappa A., Liu H., Muir B., Sgroi D., Boboila C., Pillai S. (2007). Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J. Immunol. 179, 195-200 [DOI] [PubMed] [Google Scholar]
- Nagel A. C., Krejci A., Tenin G., Bravo-Patino A., Bray S., Maier D., Preiss A. (2005). Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25, 10433-10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K., Campos-Ortega J. A. (1996). Persistent expression of genes of the enhancer of split complex suppresses neural development in Drosophila. Neuron 16, 275-286 [DOI] [PubMed] [Google Scholar]
- Narasimha M., Uv A., Krejci A., Brown N. H., Bray S. J. (2008). Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell Sci. 121, 747-752 [DOI] [PubMed] [Google Scholar]
- Nellesen D. T., Lai E. C., Posakony J. W. (1999). Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213, 33-53 [DOI] [PubMed] [Google Scholar]
- Neves A., English K., Priess J. R. (2007). Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development 134, 4459-4468 [DOI] [PubMed] [Google Scholar]
- Parker M. H., Seale P., Rudnicki M. A. (2003). Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4, 497-507 [DOI] [PubMed] [Google Scholar]
- Parras C., Garcia-Alonso L. A., Rodriguez I., Jimenez F. (1996). Control of neural precursor specification by proneural proteins in the CNS of Drosophila. EMBO J. 15, 6394-6399 [PMC free article] [PubMed] [Google Scholar]
- Postigo A. A., Ward E., Skeath J. B., Dean D. C. (1999). zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 19, 7255-7263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Raj K. (2003). The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3, 756-767 [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Reeves N. L., Posakony J. W. (2002). SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. USA 99, 9888-9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey E. (1999). Regulation of T cell fate by Notch. Annu. Rev. Immunol. 17, 283-295 [DOI] [PubMed] [Google Scholar]
- Rohwedel J., Horak V., Hebrok M., Fuchtbauer E. M., Wobus A. M. (1995). M-twist expression inhibits mouse embryonic stem cell-derived myogenic differentiation in vitro. Exp. Cell Res. 220, 92-100 [DOI] [PubMed] [Google Scholar]
- Roth S., Stein D., Nusslein-Volhard C. (1989). A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59, 1189-1202 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Jensen L. J., Jakobsen J. S., Karzynski M. M., Eichenlaub M. P., Bork P., Furlong E. E. (2006). A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 10, 797-807 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E. E. (2007). A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D., Chen B. (2009). Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 23, 147-156 [DOI] [PubMed] [Google Scholar]
- Schneider T., Reiter C., Eule E., Bader B., Lichte B., Nie Z., Schimansky T., Ramos R. G., Fischbach K. F. (1995). Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron 15, 259-271 [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K., Cordes R., Gossler A. (2007). Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc. Natl. Acad. Sci. USA 104, 537-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer D. B., Rhee J., Cheung W. L., Lassar A. B. (1996). Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272, 1476-1480 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. (2003). Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 13, 413-422 [DOI] [PubMed] [Google Scholar]
- van Es J. H., Jay P., Gregorieff A., van Gijn M. E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., et al. (2005). Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381-386 [DOI] [PubMed] [Google Scholar]
- Vasyutina E., Lenhard D. C., Wende H., Erdmann B., Epstein J. A., Birchmeier C. (2007). RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc. Natl. Acad. Sci. USA 104, 4443-4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. C., Castanon I., Baylies M. K. (2008). Daughterless dictates Twist activity in a context-dependent manner during somatic myogenesis. Dev. Biol. 317, 417-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang S., Shao H., Guo F., Trimble R., Pearce E., Abmayr S. M. (2009). Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136, 2335-2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R. P., Girardot C., Gagneur J., Braun M., Furlong E. E. (2009). Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65-70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.