Abstract
We recently identified 15 genes encoding putative surface proteins with features of MSCRAMMs and/or pili in the Enterococcus faecium TX16 (DO) genome, including four predicted pilus-encoding gene clusters; we also demonstrated that one of these, ebpABCfm, is transcribed as an operon, that its putative major pilus subunit, EbpCfm (also called PilB), is polymerized into high molecular weight complexes, and that it is enriched among clinical E. faecium isolates. Here, we created a deletion of the ebpABCfm operon in an endocarditis-derived E. faecium strain (TX82) and showed, by a combination of whole-cell ELISA, flow cytometry, immunoblot and immunogold electron microscopy, that this deletion abolished EbpCfm expression and eliminated EbpCfm-containing pili from the cell surface. However, transcription of the downstream sortase, bpsfm, was not affected. Importantly, the ebpABCfm deletion resulted in significantly reduced biofilm formation (p < 0.0001) and initial adherence (p < 0.0001) versus the wild-type; both were restored by complementing ebpABCfm in trans, which also restored cell surface expression of EbpCfm and pilus production. Furthermore, the deletion mutant was significantly attenuated in two independent mixed infection mouse urinary tract experiments, i.e., outnumbered by the wild-type in kidneys (p = 0.0003 and <0.0001, respectively) and urinary bladders (p = 0.0003 and = 0.002). In conclusion, we have shown that the ebpABCfm locus encodes pili on the E. faecium TX82 cell surface and provide the first evidence that pili of this emerging pathogen are important for its ability to form biofilm and to cause infection in an ascending UTI model.
Key words: Enterococcus faecium, ebp, pilus, biofilm, virulence, pathogenesis, UTI
Introduction
Enterococci are commensal inhabitants of the human intestinal tract and also long known as an important cause of nosocomial infections, such as catheter-associated urinary tract infections (UTI) (ranking second among all organisms recovered from this site),1–3 surgical site infections, bloodstream infections and endocarditis, among others.4–6 During the last 15 years, a major epidemiological shift has been noted in the incidence of E. faecium in both US and European hospitals, with the ratio of E. faecium to Enterococcus faecalis gradually changing in favor of E. faecium, from 1:9 in the past to approximately 4:6 in a recent CDC study.1,7–9
In addition to attachment to and colonization of host tissue surfaces, the ability to form biofilm is considered potentially important for enterococcal infections.10–13 Supporting this, our previous evaluation of 163 E. faecalis isolates indicated that strains derived from endocarditis form biofilm significantly more often than non-endocarditis isolates.14 We recently demonstrated multimeric proteinaceous surface appendages called pili (also known as fimbriae) on the surface of E. faecalis cells and showed, by mutagenesis, that these pili are involved in its ability to form biofilm on an abiotic surface and cause infection in experimental models of endocarditis and ascending UTI; formation of biofilm is generally considered as an important characteristic for both infections.10–12 Similar to the recently characterized pili of other gram-positive pathogens, such as corynebacteria and streptococci,15–17 pili of E. faecalis are encoded by a three-gene locus (ebp), encoding structural pilus subunit proteins (also known as pilins). An adjacent gene (bps in E. faecalis), encodes a pilus-dedicated class C sortase, which tethers individual pilins to form an elongated pilus.15,17,18 So far, multiple other genes or gene clusters have also been reported to affect biofilm formation of E. faecalis isolates,14,19–26 thus highlighting the complexity and multicomponent nature of this phenotype. However, much less is known about E. faecium. Heikens et al.27 showed that Esp is involved in biofilm formation of a clinical E. faecium blood isolate (E1162) and, recently, sgrA was also implicated as a potential additional factor for biofilm formation of the same isolate.28 However, insertional mutagenesis of the highly homologous esp gene of E. faecalis resulted in variable effects among different strains, ranging from complete loss of biofilm formation to no effect.26 Furthermore, our recent analysis of biofilm formation by E. faecium isolates of diverse origin showed efficient biofilm production in the absence of esp by many of the isolates (Almohamad S, Singh KV, Nallapareddy SR, Murray BE; manuscript in preparation). Hence, we considered it likely that other factor(s) also contribute to this phenotype in E. faecium.
In silico searches of the draft genome sequence of E. faecium TX16 by our group and by Hendrickx et al.29,30 recently led to the identification of 22 putative LPXTG-family cell wall anchored proteins, which we named Fms (E. faecium surface protein). Among these are four gene clusters, each located with an adjacent class C sortase, and predicted to encode four distinct types of pili. We demonstrated that one of these gene clusters, ebpABCfm (fms1-5-9), is transcribed as an operon and that its predicted major pilus subunit protein, EbpCfm (EbpAfm and EbpBfm are putative accessory pilus subunits), forms high molecular weight (HMW) complexes in E. faecium, characteristic of gram-positive pili. Recently, Hendrickx et al.31 showed pilus-like structures stained by an antiserum against PilB (EbpCfm) on cells of an E. faecium wound isolate.
In this study, we have created a deletion of the pilus-encoding ebpABCfm operon in E. faecium TX82, complemented the deletion in trans, and characterized production of EbpCfm and polymerized pili on the E. faecium cell surface by these constructs. We then evaluated the effect of the ebpABCfm deletion on the ability of E. faecium to form biofilm in vitro and to cause UTI in a mouse model.
Results
Creation of a deletion of the pilus-encoding ebpABCfm cluster of E. faecium TX82 and its effect on the activity of the downstream sortase gene, bpsfm.
We selected the endocarditis-derived E. faecium strain TX82 for deletion of all three genes of the pilus-encoding ebpABCfm operon, ebpAfm, ebpBfm and ebpCfm, using homologous recombination with allelic replacement by a cat gene; our previous studies showed that this gene cluster (Fig. 1A) is present in and expressed under in vitro growth conditions by TX82.30 Comparison of growth characteristics of the deletion mutant (TX82ΔebpABCfm) with its isogenic parental strain (TX82) showed no significant differences in their growth kinetics or viability when grown in broth cultures of either BHI or TSBG (Suppl. Fig. 1).
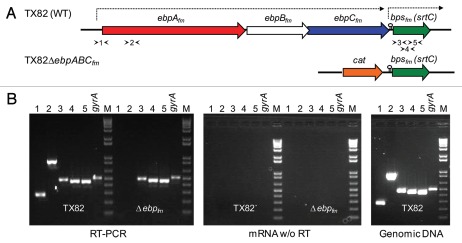
Figure 1.
Schematic representation of the deleted ebpABCfm locus of TX82 and transcriptional analysis of its effect on bpsfm, the downstream class C sortase gene. (A) A genetic map of the ebp-bpsfm region, showing the segment deleted from TX82ΔebpABCfm by allelic replacement with a cat gene. The previously predicted transcriptional terminator in the intergenic region between ebpCfm and bpsfm is indicated with a lollypop and the lengths of the previously determined mRNA transcripts are marked by arrows above the ebp-bpsfm region.30 Arrowheads indicate locations of each primer pair for RT-PCR (see B below). (B) RT-PCR analysis of bps gene expression of WT TX82 and its isogenic ebpABCfm deletion mutant. Gel on left, RT-PCR of total RNA (20 ng), isolated from mid-exponential cells and treated with DNase; middle gel, control reaction of the same RNA sample amplified without RT; gel on right, control reaction with genomic TX82 DNA as template. An intragenic region of gyrase (gyrA) was included as an internal control. Lane numbers correspond to the primer pairs shown in (A). M, molecular weight marker.
Our previous northern analysis and reverse transcriptase (RT)-PCR data demonstrated that the ∼7 kb ebpABCfm transcript does not include the downstream class C sortase gene, bpsfm, which is separated from the operon by a predicted strong transcriptional terminator and, therefore, likely to be transcribed independently.30 We next performed RT-PCR to confirm that the ebpABCfm deletion did not disrupt the expression of bpsfm. Using a primer pair designed to amplify a 5′ upstream border region of ebpAfm and another primer pair for an internal segment of ebpAfm, no transcription was detected from the ΔebpABCfm deletion mutant, while amplification products of the expected sizes were observed for both primer pairs from the wild-type (WT) mRNA, as anticipated (Fig. 1). However, all three primer pairs spanning different intragenic regions of bpsfm detected its transcription in the ΔebpABCfm deletion mutant at apparently WT levels, indicating lack of a downstream polar effect on bpsfm expression by the deletion of the ΔebpABCfm genes.
Cell surface expression of EbpCfm by WT TX82 and its isogenic ebpABCfm deletion mutant and complementation derivatives.
In an effort to find optimal conditions for the expression of ebpABCfm-encoded pili for subsequent studies (see below), we analyzed the effect of different growth conditions and growth stages on EbpCfm expression by whole-cell ELISA, using a purified EbpCfm-specific antibody.30 With WT TX82 cells harvested from lag (1 h), mid exponential (6 h) and late stationary (16 h) stages (gentamicin was added for pAT392 constructs), no significant differences in surface expression levels of EbpCfm were observed between growth in either BHI or TSBG broth (Fig. 2A). However, more than two-fold higher EbpCfm expression was detected at mid-exponential and late stationary phases versus the lag phase in both media, similar to a previous report with an E. faecium wound isolate, E1165, grown in BHI.31 Comparably high EbpCfm expression was also observed by cells grown on BHI agar. Thus, these results indicate that EbpCfm is readily expressed under multiple in vitro conditions and throughout the growth cycle.
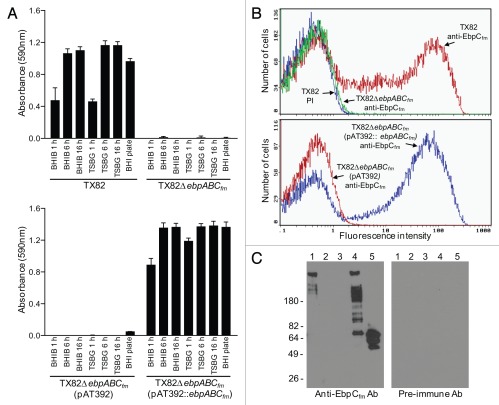
Figure 2.
Characterization of EbpCfm surface expression by TX82 and its ebpABCfm deletion and complementation derivates. (A) Surface expression of EbpCfm in different growth stages and growth media, using whole-cell ELISA. Cells were washed and adjusted to OD600 nm = 0.5 before coating on wells. EbpCfm expression was detected using an affinity-purified anti-EbpCfm antibody.30 Bars represent the means of absorbance at 590 nm ± SD from four independent assays, each performed in triplicate. (B) Flow cytometry analysis. Labeling by the anti-EbpCfm antibody is shown for each strain and by a pre-immune antibody (PI)30 for TX82, and indicated as log fluorescence intensity on the X-axis. Bacteria were analyzed using side scatter as the threshold for detection. Each histogram represents 50,000 events of bacterium-sized particles. (C) Western blot of mutanolysin extracts. Five µg of mutanolysin cell wall extracts were electrophoresed and blotted onto the membrane and probed with affinity-purified anti-EbpCfm antibodies or pre-immune antibodies. Lane 1, TX82; lane 2, TX82ΔebpABCfm; lane 3, TX82ΔebpABCfm (pAT392); lane 4, TX82ΔebpABCfm (pAT392::ebpABCfm), lane 5, rec-EbpCfm.30
As expected, the ΔebpABCfm deletion abolished EbpCfm production. As seen in Figure 2A, complementation of the ebpABCfm mutant led to EbpCfm expression in whole-cell ELISA at a consistently higher level in all conditions tested versus the WT, while no expression was detected by an isogenic control with the empty plasmid. Furthermore, quantification of EbpCfm surface expression by flow cytometry showed a slightly increased percentage of the complemented deletion mutant expressing EbpCfm (grown in the presence of gentamicin) versus WT (47% versus 40%) (Fig. 2B). These cells also showed higher fluorescence intensities (mean fluorescence intensities 41 versus 22, median 51 versus 29, respectively), suggesting that more EbpCfm molecules were expressed on the cell surface. Neither the ΔebpABCfm deletion mutant nor its complementation derivative with the empty vector showed surface expression of EbpCfm. Similarly, the multiple HMW bands seen in a western blot analysis of mutanolysin cell wall extracts of TX82,30 were not produced by the ΔebpABCfm deletion mutant (Fig. 2C). The complemented mutant showed a similar pattern of HMW bands as the WT, although with a higher proportion of lower molecular weight polymers and a band corresponding to the size of monomeric EbpCfm. These results suggest the possibility that the constitutively expressed EbpCfm in the complemented deletion mutant may overload the polymerization capacity of the native sortase, leading to greater amounts of unpolymerized and partially polymerized subunits. Nonetheless, they confirm that plasmid-encoded ebpCfm is polymerized into a ladder-like pattern of HMW complexes characteristic of pilus structures of gram-positive pili.
Analysis of pilus structures on TX82 cells.
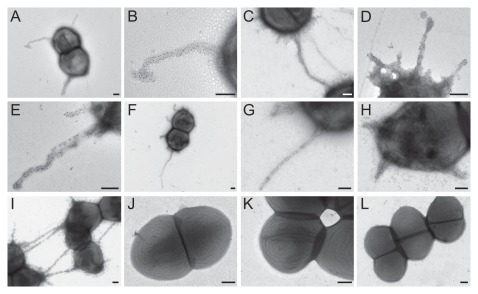
Using high-resolution transmission electron microscopy (TEM) and negative staining, we observed filamentous structures resembling pili attached to the surface of TX82 cells collected from several growth conditions (BHI and TSBG broth, BHI and blood agar plates) (Fig. 3). Furthermore, these structures were specifically recognized by the anti-EbpCfm antibody using immunogold staining, while no staining of pili was seen by purified pre-immune antibodies from the same animal or purified antibodies against Fms13 and Fms16 (unpublished data), two predicted other pilus major subunits of E. faecium TX16.30 Although pleomorphic, these pili morphologically resembled the recently observed pili of another E. faecium isolate, E1165, which represents a wound isolate of European origin.31 The gold particles were found evenly scattered along the shaft of the pilus, supporting the predicted role of EbpCfm as a major pilus subunit. Approximately 40% of TX82 cells grown in BHI broth were seen to express these pili, with no obvious differences observed in the other growth conditions. Of note, some of the Ebp pili appeared to be attached to one or more neighboring cells, suggesting the possibility of involvement in cell-cell interactions (Fig. 3C and I). As anticipated, the ebpCfm-specific antibody did not detect pili on the surface of the ebpABCfm mutant, nor on the mutant with empty vector, while complementation in trans restored pilus production of the ebpABCfm mutant.
Figure 3.
Immuno-EM analysis of pilus production by WT TX82 and its ebpABCfm deletion mutant and complementation derivates. (A) TX82 grown on BHI plate; (B) higher magnification of the pilus seen in (A); (C) TX82 grown in BHI broth, showing two cells possibly connected by a pilus; (D) TX82 grown in TSBG; (E) TX82 grown on a blood agar plate; (F–I) Complemented ebpABCfm deletion mutant (TX82ΔebpABCfm (pAT392::ebpABCfm)) grown in BHI broth. Note the pili apparently linking several cells in (I); (J) TX82ΔebpABCfm grown in BHI broth; (K) TX82ΔebpABCfm (pAT392) grown in BHI broth; (L) TX82 grown in BHI broth. (A–K) were stained with affinity-purified EbpCfm-specific antibodies and (L) with pre-immune antibodies.30 Scale bars 200 nm.
Effect of the ebpABCfm deletion on biofilm formation.
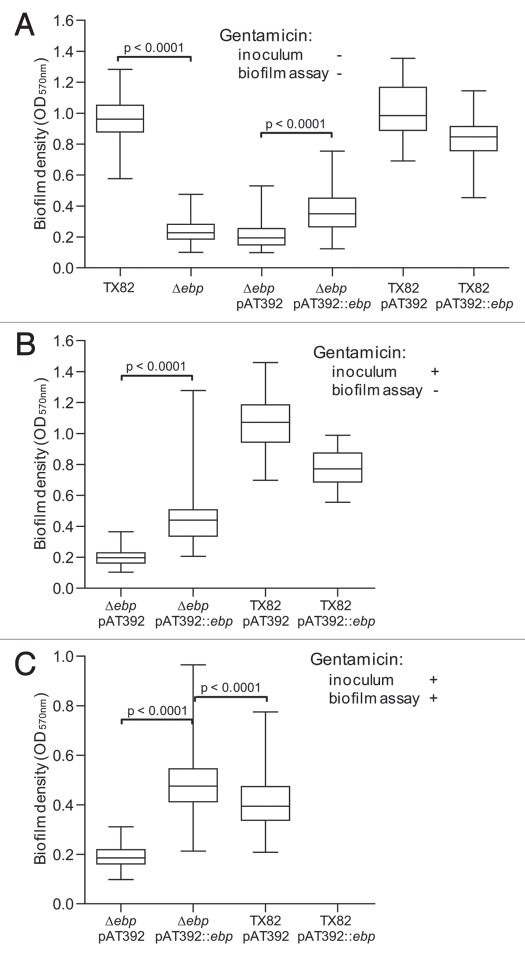
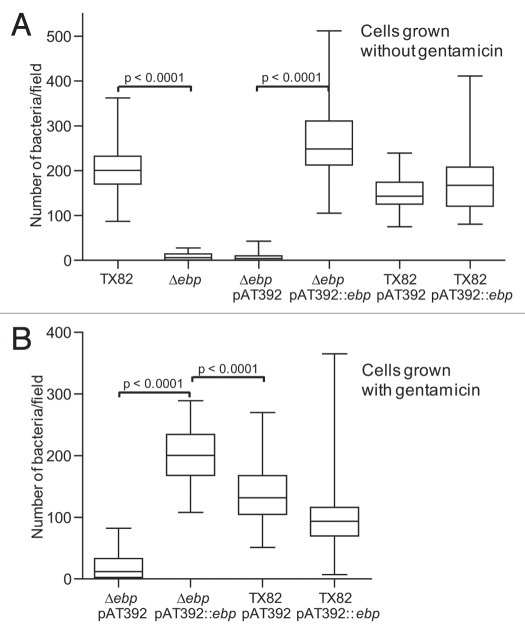
As seen in Figure 4A, deletion of ebpABCfm reduced biofilm formation by 75% versus the WT (p < 0.0001). For testing the complementation constructs, we initially performed the assay using the same conditions, i.e., without the addition of gentamicin, due to the effect some antibiotics may have on biofilm formation.18,32 While a small but significant enhancement was observed by the complemented mutant over the empty vector control (p < 0.0001), its biofilm production was only 38% of WT level. Moreover, WT TX82 supplemented in trans with ebpABCfm [TX82 (pAT392::ebpABCfm)], thus carrying both a chromosomal and plasmid-encoded copy of ebpABCfm, showed reduced biofilm formation compared to TX82 or TX82 with the empty complementation vector [TX82 (pAT392)].
Figure 4.
Comparison of biofilm formation of TX82 (WT) versus its ebpABCfm deletion and complementation derivates. (A) Biofilm formation without added gentamicin. (B) Biofilm formation with overnight inoculum grown in the presence of gentamicin, but cultures in biofilm plate in its absence. (C) Biofilm growth with both inoculum and plate incubated in the presence of gentamicin. Median OD570 nm values and interquartile ranges, with the minimum and maximum values marked by whiskers, represent combined data from at least three independent assays and 48 wells for each construct. Statistical analyses were performed by the Mann-Whitney test and p values are shown in the figure.
Because only partial restoration of biofilm formation was observed by the complemented mutant, we performed further assays in two additional conditions, (1) by including gentamicin in the overnight culture (16 h) used as the biofilm inoculum and (2) by including gentamicin for both growth of the biofilm inoculum and in the assay incubations (24 h) in microtiter trays. Adding gentamicin to the inoculum significantly increased biofilm activity by the complemented mutant versus the vector control (p < 0.0001, Fig. 4B), and still higher enhancement was observed when gentamicin selective pressure was maintained throughout the experiment (>2.5-fold increase, p < 0.0001, Fig. 4C). The supplemented WT, TX82 (pAT392::ebpABCfm), showed a similar pattern in both conditions as without gentamicin, with the additional copy of ebpABCfm leading to reduced biofilm production versus TX82 (pAT392) (Fig. 4B and C). However, it is notable that the overall level of biofilm formation was reduced when gentamicin was present throughout the biofilm assay. In this condition, the complemented ebpABCfm mutant [TX82ΔebpABCfm (pAT392::ebpABCfm)] showed significantly higher biofilm formation than TX82 (pAT392) (p < 0.0001). The in trans complementation by plasmid-encoded ebpABCfm confirms the role of the ebpABCfm operon in biofilm and shows that this construct leads to overexpression of the biofilm phenotype when antibiotic selection pressure is maintained continuously.
Since the above results pointed to potential plasmid instability, we assessed the amount of plasmid loss in each of the three biofilm conditions. Without gentamicin selection pressure, 90% of cells on average (ranging between 82 and 100% in three independent assays) had lost the complementing plasmid (pAT392::ebpABCfm) at 24 h, while including gentamicin in the inoculum increased its stability slightly (plasmid loss 81%, range 69–94%) and continuous presence of gentamicin resulted in greatly reduced plasmid loss (13%, range 8–16%). In contrast, pAT392 was relatively stable even in the absence of gentamicin throughout the assay (plasmid loss 7%, range 7–8%). Although the specific reason for the more severe instability of pAT392::ebpABCfm versus pAT392 in the absence of gentamicin is not known, it may be related to the observed overexpression of structural pilus components by the complemented deletion mutant (see above) which, by utilizing a portion of the cells metabolic resources, could give an advantage to cells that have lost the plasmid.
Evaluation of primary attachment showed efficient binding of TX82 cells to the polystyrene surface (Fig. 5A). The ebpABCfm deletion nearly abolished (p < 0.0001) this initial adherence, while complementation, either with cells grown in the absence or presence of gentamicin, restored the adherence phenotype (34-fold and 10-fold increase versus the vector control, respectively; p < 0.0001 for both; Fig. 5A and B). The lack of noticeable effect of plasmid instability on the adherence of the complemented mutant may be caused by the shorter incubation of cultures without gentamicin in this assay, oversaturation of the binding surface by the relatively high amount of added bacteria and/or “entrapment” of non-piliated cells by cells retaining the complementing plasmid. Adherence of the complemented deletion mutant was significantly higher than TX82 (pAT392) (p < 0.0001), consistent with the overexpression of EbpCfm by this construct (Fig. 2) and the biofilm formation data above. The additional copy of ebpABCfm in TX82 (pAT392::ebpABCfm) resulted in reduced initial adherence versus TX82 (pAT392) when cells were grown with gentamicin, similar to biofilm formation; the reason for this remains to be determined.
Figure 5.
Effect of the ebpABCfm deletion on primary attachment to polystyrene. (A) Adherence of cells grown without gentamicin. (B) Adherence of cells grown with gentamicin. Median values and interquartile ranges show data combined from four independent assays. Adhering bacteria were counted microscopically from a total of 64 fields (each ∼2,900 µm2) for each construct. Statistical analyses were performed by the Mann-Whitney test and p values are shown in the figure.
Importance of the ebpABCfm locus for UTI in a mouse model.
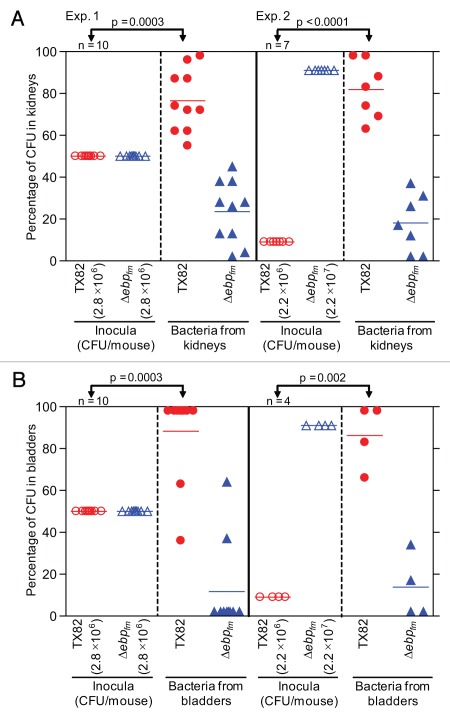
To test whether deleting the ebpABCfm locus has an effect on E. faecium virulence, we used a mixed infection assay and a mouse model of UTI, as previously described.33,34 In brief, an equal suspension of TX82 and the ΔebpABCfm mutant (2.8 × 106 CFU/mouse) was inoculated into 10 mice using catheters and, at 48 h, the mean (geometric) CFUs of total bacteria (WT plus mutant) recovered from kidneys and bladders were 1.2 × 105/gm and 6.7 × 104/gm, respectively. The mean percentages of total CFUs recovered from kidneys were 76.5 for TX82 versus 23.5 for the deletion mutant and, in bladders, 88.3 and 11.7, respectively, showing a clear advantage of TX82 versus the deletion mutant and a significant difference between TX82 in the inoculum and the kidneys and bladders (p = 0.0003 for both). In a second experiment, we used an inoculum 10-fold higher for the deletion mutant (2.2 × 107 CFU/mouse) than the WT (2.2 × 106 CFU/mouse). Bacteria were recovered from the kidneys of all 7 mice inoculated, and the bladders of 4 of these mice, while 3 bladders were not infected. Total CFUs (geometric means of WT + mutant) recovered from kidneys were 1.3 × 104/gm and from infected bladders, 4.9 × 104/gm. Again, significantly higher numbers of WT TX82 were recovered than the mutant (mean percentage 81.9 versus 18.1 in infected kidneys, and 86.2 versus 13.8 in infected bladders, respectively) in both kidneys (p < 0.0001, Fig. 6A) and in bladders (p = 0.002, Fig. 6B). The mean virulence indices18,35,36 of the ebpABCfm mutant versus the WT were 0.307 for kidneys and 0.133 for bladders for the first experiment (with WT to mutant ratio of 1:1) and 0.022 and 0.016 (with WT to mutant ratio of 1:10) for the second experiment, respectively, indicating that the ebpABCfm locus contributes significantly to this infection.
Figure 6.
Effect of deletion of ebpABCfm in a murine model of UTI, using a mixed inoculum. (A) Percentage of bacteria recovered from kidneys. (B) Percentage of bacteria recovered from bladders. Two independent experiments are shown in both panels. Exp. 1, mice infected with equal CFUs of TX82 and TX82ΔebpABCfm (2.8 × 106 CFU/mouse); Exp. 2, mice infected with 10-fold overdose of TX82ΔebpABCfm (2.2 × 107 CFU/mouse) versus TX82 (2.2 × 106 CFU/mouse). Horizontal bars represent the means of percentages of total bacteria in kidneys and bladders Statistical analyses were performed by the paired t test and p values are shown in the figure. Empty circles and empty triangles represent percentages of TX82 and TX82ΔebpABCfm in inocula, respectively, while solid circles and solid triangles represent percentages of TX82 and TX82ΔebpABCfm recovered from kidneys and bladders 48 h post infection, respectively.
Discussion
Although usually a harmless resident of the human intestinal tract, some strains of E. faecium, particularly those belonging to the recently emerged CC17 genogroup,9,37 have shown remarkable adaptability to nosocomial environments and have increasingly developed into opportunistic pathogens able to cause a wide variety of healthcare-associated (HA) infections in humans.4–6 In addition to the acquisition of resistance among nosocomial E. faecium isolates to multiple, commonly used antibiotics, such as ampicillin, vancomycin and aminoglycosides, recent epidemiological studies have suggested that these isolates have also evolved or acquired other traits that have contributed to their increased prevalence. For instance, HA isolates more often carry genes e.g., hyl, encoding a hyaluronidase-like glycoside hydrolase,38 esp, encoding an enterococcal surface protein39 or a functional copy of a gene (acm, encoding a collagen-binding adhesin)40,41 associated with virulence. Furthermore, our recent analysis of 433 E. faecium isolates from diverse clinical and non-clinical sources indicated that the presence of most of the 15 fms genes, which encode characterized or putative MSCRAMM and/or pilus proteins including the ebpABCfm gene cluster of this study, are found more often in clinical isolates, including CC17,42,43 and, thus, may have contributed to the emergence of E. faecium isolates with increased survival, colonization and/or ability to cause HA infections.
Here, we have analyzed cell surface expression of EbpCfm, the putative major pilus subunit protein of the ebpABCfm operon, and EbpCfm-containing pili by an endocarditis-derived E. faecium isolate TX82, its isogenic ebpABCfm deletion and complementation derivatives. We found that deletion of ebpABCfm eliminated Ebp pili from the cell surface of TX82 and led to significant attenuation in its ability to adhere to polystyrene and form biofilm, and cause infection in a murine model of ascending UTI. Complementation of the deleted ebpABCfm in trans restored surface expression EbpCfm and its polymerization into HMW complexes and also resulted in pilus production on the cell surface. These pili were variable in length and thickness, and generally similar in appearance to those seen on WT cells, with anti-EbpCfm antibodies evenly scattered along the pilus shaft. Thus, our data unambiguously show that the ebpABCfm locus codes for pili observed on the TX82 cell surface and that complementation of the ebpABCfm deletion in trans leads to efficient pilus synthesis by the native sortase.
The proposed link between biofilm formation and various enterococcal infections, such as UTI, is that bacteria may be less effected by host immune defenses and therapeutic antibiotics and, thus, seems relevant to the pathogenesis of the increasingly antibiotic-resistant enterococci.12,44 Our recent assessment of the prevalence among a large collection of E. faecium isolates of their ability to form biofilm in vitro identified TX82 as an efficient biofilm producer (Almohamad S, Singh KV, Nallapareddy SR, Murray BE; manuscript in preparation). The strong reduction in its biofilm forming ability (75% versus WT) by the ebpABCfm deletion indicates that Ebp pili are an important factor contributing to the biofilm phenotype of this isolate. Our data also showed that this mutant has a clear defect (96% reduction versus WT) in its initial adherence to the polystyrene surface, providing at least one explanation for the attenuation of the biofilm phenotype. TX82 does not carry the esp gene (unpublished data), previously shown to affect the biofilm phenotype of another E. faecium isolate, E1162.27 Although lower biofilm densities by E1162 were observed by Heikens and colleagues,27 in our hands, this strain's biofilm and the reduction by its esp mutant were comparable to what we observed for TX82 (lacking esp) and its ebpfm mutant, respectively (Suppl. Fig. 2); this is consistent with observations for other bacteria that various factors can contribute differently to biofilm formation in different strains. A larger number of isolates needs to be tested to assess the overall impact of these two genes on the biofilm phenotype of E. faecium.
Since antibiotics may affect the biofilm phenotype, we considered it preferable to perform biofilm assays without added antibiotics; indeed, we subsequently observed in this study a decrease in overall biofilm densities in the presence of gentamicin. However, we found the complementing plasmid (pAT392::ebpABCfm) to be highly unstable in the absence of gentamicin (∼10% of cells retained the plasmid), which is likely the cause for only partial, although statistically significant, restoration of biofilm formation in the absence of gentamicin. In contrast, in the presence of gentamicin, pAT392::ebpABCfm was relatively stably maintained (retained by ∼87% of cells), and this led to clearly increased enhancement of biofilm formation, reaching 120% of the level observed with TX82 (pAT392), also grown in gentamicin. Complementation also restored initial adherence and resulted in a similar increase relative to TX82 (pAT392) (135% for cells grown without, 148% with, gentamicin). These observations are consistent with the overproduction of EbpCfm on the cell surface of the complemented deletion mutant observed by both flow cytometry and whole-cell ELISA under various in vitro growth conditions, including TSBG, the biofilm culture medium. While our data demonstrated the importance of the ebpABCfm locus for the initial adherence stage of biofilm formation, its specific function in subsequent biofilm maturation remains unknown. The Ebp pili could mediate physical interactions and adherence between individual cells, thus facilitating the accumulation of a multicellular biofilm network. Supporting this hypothesis, our immuno-EM analysis showed, in some of the fields, two or more neighboring cells apparently connected by Ebp pili, as was also previously seen with the biofilm-associated pili of E. faecalis.18 Another possibility is that the polymerized pilus material could participate in the buildup of the biofilm matrix, surrounding and embedding bacterial cells, as has been proposed for extracellular polysaccharides14,45,46 and DNA.24,25,47–49
Although commonly isolated from nosocomial UTIs, little is known about the factors important for the attachment and colonization of E. faecium at this site. Recently, Leendertse et al.50 showed that Esp is involved in transient aggravation of experimental E. faecium UTI. Since several enterococcal infections in humans, including UTI, are thought to involve biofilm formation in the host, we tested the ebpABCfm deletion mutant in a mouse UTI model using a competition assay, which has been previously established as a UTI model for E. faecalis.33,34,51 With a mixed inoculum containing equal numbers of CFUs of WT TX82 and the ebpABCfm mutant, deletion of ebpABCfm resulted in clear attenuation in the ability of TX82 to cause infection. That is, WT was recovered in significantly higher numbers of CFU's than the deletion mutant in both kidneys (3.3-fold) and bladders (7.5-fold). The severe instability of pAT392::ebpABCfm, similar to the previous observation of another complementation vector in E. faecium,52 prevented its inclusion in the UTI model. Nevertheless, a 10-fold overdose of the mutant relative to WT in the inoculum resulted in a similar outnumbering of WT over the mutant in both kidneys (4.5-fold) and bladders (6.2-fold), thus confirming the significant attenuation of the ebpABCfm mutant. Interestingly, the higher number of colonized kidneys versus bladders in some mice resembles reports with E. faecalis33,53,54 and could reflect similar tropism of both organisms for kidneys.34
In summary, we have shown that the ebpABCfm-encoded pili of E. faecium are important for both the biofilm phenotype and colonization of kidneys and bladders in an experimental UTI model. The recent protective active and/or passive immunization in animal models using pilus subunits from streptococci,55–57 suggests that pili of gram-positive cocci could be useful targets for alternative preventive or therapeutic approaches. In this respect, the presence of the ebpABCfm locus in >95% of clinical E. faecium isolates,42 its similarity to the pilus-encoding genes of E. faecalis pili and their comparable effects on biofilm formation and UTI in both organisms, make this locus particularly attractive for antibacterial strategies against enterococcal infections.
Materials and Methods
Bacterial strains, plasmids and growth conditions.
E. faecium and Escherichia coli strains as well as plasmids used in this study are listed in Table 1. Plasmid constructs were given pTEX numbers and their host strains corresponding TX numbers. E. faecium strains were routinely grown in/on brain heart infusion (BHI) (Difco Laboratories, Detroit, MI) broth/agar or, for some experiments (see below), in tryptic soy broth supplemented with 0.25% glucose (TSBG) or on trypticase soy agar supplemented with 5% sheep blood (Becton Dickinson, Franklin Lakes, NJ), and E. coli in Luria-Bertani media (Difco Laboratories) at 37°C. With E. faecium TX82, gentamicin (Sigma Chemical Co., St. Louis, MO) was used at 125 µg/mL (150 µg/mL for selection of transformants) and chloramphenicol at 10 µg/mL and, with E. coli, gentamicin was used at 25 µg/mL.
Table 1.
Bacterial strains and plasmids used in this study
| Strains/plasmids | Relevant characteristics* | Reference or source |
| Strains | ||
| E. faecium | ||
| TX82 | Endocarditis isolate. Vanr, Ampr | 61 |
| TX5645 | TX82ΔebpABCfm, ebpABCfm deletion mutant | This study |
| TX5664 | TX5645 (pAT392), deletion mutant with empty complementation vector. Genr | This study |
| TX5665 | TX5645 (pAT392::ebpABCfm), deletion mutant complemented with ebpABCfm. Genr | This study |
| TX5667 | TX82 (pAT392), WT with empty complementation vector. Genr | This study |
| TX5668 | TX82 (pAT392::ebpABCfm), WT supplemented with ebpABCfm. Genr | This study |
| TX2154 | E1162, blood isolate. Ampr | 27 |
| TX2155 | E1162Δesp, deletion mutant. Ampr, Chlr | 27 |
| E. coli | ||
| DH5α | E. coli cloning host | Invitrogen |
| XL1 Blue | E. coli cloning host, Tetr | Stratagene |
| TX5644 | DH5α (pTEX5644) | This study |
| TX5666 | XL1 Blue (pAT392::ebpABCfm) | This study |
| Plasmids | ||
| pTEX5501ts | Temperature sensitive plasmid, used for deletion mutagenesis of E. faecium. Chlr, Genr | 61 |
| pTEX5644 | Plasmid for ebpABCfm deletion; flanking regions of ebpABCfm cloned into pTEX5501ts. Chlr, Genr | This study |
| pAT392 | Complementation vector. Genr | 62 |
| pTEX5665 | pAT392::ebpABCfm; plasmid for complementation of the ebpABCfm mutant. Genr | This study |
Van, vancomycin; Amp, ampicillin; Gen, gentamicin; Tet, tetracycline; Chl, chloramphenicol; superscript “s” designates sensitivity, “r” designates resistance.
General techniques.
Chromosomal DNA from E. faecium TX82 was isolated by the hexadecyltrimethyl ammonium bromide method.58 Pulsed-field gel electrophoresis was performed as described earlier.59,60
Construction of an ebpABCfm deletion mutant and its complementation.
The ebpABCfm region of E. faecium TX82 was deleted by allelic replacement, as described previously.61 Briefly, upstreatm (817 bp) and downstream fragments (803 bp) flanking the ebpABCfm region were amplified using primers listed in Supplemental Table 1, and ligated into pTEX5501ts, on either side of the cat gene, followed by transformation into E. coli DH5α. The resulting plasmid, pTEX5644 (Table 1), was confirmed by sequencing and electroporated into E. faecium TX82. Single-crossover integrants (TX82::pTEX5644) were selected on BHI plates supplemented with gentamicin and chloramphenicol. Transformants were cultured consecutively six times, then plated on non-selective media at 37°C, and resulting colonies scored for sensitivity to gentamicin and resistance to chloramphenicol, to select for plasmid excision (conferring susceptibility to gentamicin) by double-crossover recombination. In the resulting ebpABCfm deletion derivative (TX5645), the deleted region, replaced by the cat gene, begins at and includes the ATG start codon of the ebpAfm coding sequence and extends to 93 bp upstream of the ebpCfm TAG stop codon; the 3′ end of ebpCfm was left intact to avoid disrupting the activity of the downstream sortase gene bpsfm. The deletion site in TX5645 was confirmed by PCR, sequencing and pulsed-field gel electrophoresis.
For complementation, the complete ebpABCfm region, including its own RBS, was amplified from genomic DNA of TX82 (see Suppl. Table 1 for primers), cloned under the constitutive P2 promoter in pAT392,62 and confirmed by sequencing. This plasmid, pTEX5665, was then electroporated into the ebpABCfm deletion mutant (TX5645).
Growth curve.
Pre-warmed BHI or TSBG broth was inoculated with an overnight culture at an initial optical density of 0.05 or 0.1 at 600 nm (OD600 nm). The culture was then grown at 37°C with shaking, and samples were taken at regular intervals to measure their OD600 nm and, at 0, 3, 5, 7 and 24 h, for determining their CFU counts on BHI agar.
Reverse transcriptase (RT)-PCR.
Total RNA isolation from TX82 and TX5645 (TX82ΔebpABCfm), grown in BHI to mid-exponential phase, and RT-PCR (see Suppl. Table 1 for primers) was performed as described previously.30 A 509-bp fragment of the gyrA (encoding gyrase A) gene was amplified as an internal control of the PCR reaction and reactions without reverse transcriptase were performed to confirm the lack of DNA contamination in the RNA preparation.
Whole-cell ELISA and flow cytometry.
Whole-cell ELISA was performed following a previously described protocol, with minor modifications.30,41 Briefly, E. faecium TX82 and its derivatives were grown in either BHI or TSBG broth for 1, 6 or 16 h or on BHI plates overnight, with cultures containing pAT392 constructs supplemented with gentamicin. Cells were collected from broth cultures by centrifugation or scraped from agar plates into phosphate-buffered saline (PBS), pH 7.4, and washed twice with PBS. High binding microtiter plate wells (4HBX; Thermo Scientific, Wobum, MA) were then coated for 1 h with 100 µl of cells resuspended in 50 mM carbonate-bicarbonate buffer, pH 9.6, and adjusted to OD600 nm = 0.5. EbpCfm expression was detected using affinity-purified polyclonal rEbpC-specific antibodies, as described previously.30 Purified pre-immune antibodies from the same animal were used as a negative control. Antiserum against formalin-killed whole cells of the E. faecium strain TX16, included as a positive control,63 confirmed equal coating of all TX82 constructs included in these assays.
Flow cytometry was used for quantification of surface expression of EbpCfm. Bacteria were grown in BHI (with gentamicin for pAT392) for 12 h from an initial OD600 nm of 0.01 and surface exposed EbpCfm was labeled using the anti-EbpCfm antibody and R-phycoerythrin conjugated secondary antibody, as described previously.30 Samples were analyzed by Coulter EPICSXL AB6064 flow cytometer (Beckman Coulter) and System II software.
Mutanolysin cell wall extracts and western blot.
Cell wall anchored proteins were released by a mutanolysin treatment from cells of E. faecium grown in BHI broth for 12 h (starting OD600 nm ∼0.01), as previously described.30 Five µg of concentrated mutanolysin extracts, with total protein concentrations estimated by the bicinchoninic acid assay (Pierce, Rockford, IL), were analyzed by a western blot, using the anti-EbpCfm antibody and horse radish peroxidase-labeled secondary antibody, as described recently.30
Immunoelectron microscopy.
E. faecium cells were grown in either BHI or TSBG broth for 12 h or on BHI or blood agar plates overnight; cultures containing pAT392 constructs were supplemented with gentamicin. Cells from broth cultures were harvested by centrifugation and from plates by scraping, and washed with 0.1 M NaCl. Immunogold labeling was performed as described previously,64 using anti-EbpCfm antibodies (1:100 dilution) or pre-immune antibodies (1:100 dilution),30 followed by 12-nm gold-donkey anti-goat IgG (1:20 dilution) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Samples were viewed in a Jeol 1400 transmission electron microscope.
Biofilm formation and primary adherence
An assay for detecting in vitro biofilm formation by E. faecium, after incubation for 24 h in TSBG broth, was performed following a previously described method, using 96-well polystyrene plates (Becton Dickinson, Franklin Lakes, NJ).14 For plasmid stability analysis, the total bacterial growth in wells of the biofilm assay plate was resuspended (instead of staining the biofilm by crystal violet), serially diluted and plated on BHI plates. Single colonies (94–96 colonies/assay) were then resuspended in BHI broth in 96-well plates and replica-plated onto BHI plates with and without gentamicin to screen for plasmid loss. For assessing primary attachment to a polystyrene surface, 5 mL of an overnight (16 h) culture in TSB broth, supplemented with 0.5% glucose, was diluted to OD600 = 0.1 and added to six-well plates (Becton Dickinson, Franklin Lakes, NJ), as described previously.14 After 2 h incubation, wells were washed six times with PBS, fixed, gram-stained and counted microscopically.14
Mouse UTI model.
Protocols previously used in our laboratory were followed for the preparation of mice, inoculum volumes, and all other stages of the mixed-infection competition experiment.33 To prepare inocula for infection, BHI-grown 24 h-cultures of E. faecium TX82 and TX82ΔebpABCfm were resuspended in 0.9% saline solution and the suspensions were mixed in equal (1:1) or 1:10 (WT versus mutant) ratios based on their OD600 nm readings; these inocula were also plated for colony counts. At 48 h after infection, CFU counts were enumerated from bladders and kidneys, as previously described.33 Because of prior observations that chloramphenicol resistance is not always expressed by mutants,52,65 the ratio of TX82 and TX82ΔebpABCfm among the recovered bacterial colonies were determined by hybridizing colony lysates with a 488 bp intergenic ebpB-ebpCfm30 probe under high-stringency conditions.60,66 Pre-approved protocols and guidelines of the Animal Welfare Committee of the University of Texas Health Science Center at Houston were followed throughout this study.
Statistical analyses.
Differences in biofilm and primary adherence values between TX82 and its ebpABCfm deletion and complementation derivatives were evaluated by the Mann-Whitney test. The percentages of the ΔebpABCfm mutant in the inoculum versus the percentages of the ΔebpABCfm mutant in the kidneys and bladders of individual mice co-infected with TX82 and TX82ΔebpABCfm in the competition assay were analyzed for significance by the paired t test. The mean virulence index of TX82ΔebpABCfm versus TX82 in the UTI model was calculated using the following equation: Mean virulence index = Σ[(% TX82 in inoculum)/(%ΔebpABCfm in inoculum)]/Σ[(%TX82 in kidneys or bladders%)/(ΔebpABCfm in kidneys or bladders)].18,36,52
Acknowledgements
This work was supported by NIH grants R01 AI067861 to Barbara E. Murray and R01 AI 061381 to Hung Ton-That from the National Institute of Allergy and Infectious Diseases, NIAID. We thank Karen Jacques-Palaz for her technical assistance. We would also like to thank Rob J. Willems and Esther Heikens for providing E. faecium E1162 and its esp deletion mutant.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/11966
Supplementary Material
References
- 1.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 2.Hooton TM, Stamm WE. Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin North Am. 1997;11:551–581. doi: 10.1016/s0891-5520(05)70373-1. [DOI] [PubMed] [Google Scholar]
- 3.Huycke MM, Sahm DF, Gilmore MS. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Murray BE. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther. 2008;6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 5.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 6.Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 7.Treitman AN, Yarnold PR, Warren J, Noskin GA. Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002) J Clin Microbiol. 2005;43:462–463. doi: 10.1128/JCM.43.1.462-463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Top J, Willems R, van der Velden S, Asbroek M, Bonten M. Emergence of clonal complex 17 Enterococcus faecium in The Netherlands. J Clin Microbiol. 2008;46:214–219. doi: 10.1128/JCM.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–828. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Carniol K, Gilmore MS. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol. 2004;186:8161–8163. doi: 10.1128/JB.186.24.8161-8163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donelli G, Guaglianone E. Emerging role of Enterococcus spp in catheter-related infections: biofilm formation and novel mechanisms of antibiotic resistance. J Vasc Access. 2004;5:3–9. doi: 10.1177/112972980400500101. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72:3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 18.Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock LE, Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004;186:5629–5639. doi: 10.1128/JB.186.17.5629-5639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hufnagel M, Koch S, Creti R, Baldassarri L, Huebner J. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J Infect Dis. 2004;189:420–430. doi: 10.1086/381150. [DOI] [PubMed] [Google Scholar]
- 21.Kristich CJ, Li YH, Cvitkovitch DG, Dunny GM. Esp-independent biofilm formation by Enterococcus faecalis. J Bacteriol. 2004;186:154–163. doi: 10.1128/JB.186.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohamed JA, Murray BE. Influence of the fsr locus on biofilm formation by Enterococcus faecalis lacking gelE. J Med Microbiol. 2006;55:1747–1750. doi: 10.1099/jmm.0.46729-0. [DOI] [PubMed] [Google Scholar]
- 23.Tendolkar PM, Baghdayan AS, Shankar N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J Bacteriol. 2006;188:2063–2072. doi: 10.1128/JB.188.6.2063-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72:1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 2008;190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heikens E, Bonten MJ, Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189:8233–8240. doi: 10.1128/JB.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickx AP, van Luit-Asbroek M, Schapendonk CM, van Wamel WJ, Braat JC, Wijnands LM, et al. SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect Immun. 2009;77:5097–5106. doi: 10.1128/IAI.00275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickx AP, van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sillanpaa J, Nallapareddy SR, Prakash VP, Qin X, Hook M, Weinstock GM, et al. Identification and phenotypic characterization of a second collagen adhesin, Scm and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology. 2008;154:3199–3211. doi: 10.1099/mic.0.2008/017319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospitalacquired Enterococcus faecium isolate. Microbiology. 2008;154:3212–3223. doi: 10.1099/mic.0.2008/020891-0. [DOI] [PubMed] [Google Scholar]
- 32.Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun. 2007;75:5399–5404. doi: 10.1128/IAI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195:1671–1677. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun. 2005;73:2461–2468. doi: 10.1128/IAI.73.4.2461-2468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nannini EC, Teng F, Singh KV, Murray BE. Decreased virulence of a gls24 mutant of Enterococcus faecalis OG1RF in an experimental endocarditis model. Infect Immun. 2005;73:7772–7774. doi: 10.1128/IAI.73.11.7772-7774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beuzon CR, Holden DW. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 2001;3:1345–1352. doi: 10.1016/s1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 37.Leavis HL, Bonten MJ, Willems RJ. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006;9:454–460. doi: 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, et al. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187:508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- 39.Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycinresistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357:853–855. doi: 10.1016/S0140-6736(00)04205-7. [DOI] [PubMed] [Google Scholar]
- 40.Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun. 2008;76:4110–4119. doi: 10.1128/IAI.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47:1733–1747. doi: 10.1046/j.1365-2958.2003.03417.x. [DOI] [PubMed] [Google Scholar]
- 42.Sillanpaa J, Prakash VP, Nallapareddy SR, Murray BE. Distribution of genes encoding MSCRAMMs and pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol. 2009;47:896–901. doi: 10.1128/JCM.02283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galloway-Pena JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009;200:1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 45.Sousa C, Teixeira P, Oliveira R. The role of extracellular polymers on Staphylococcus epidermidis biofilm biomass and metabolic activity. J Basic Microbiol. 2009;49:363–370. doi: 10.1002/jobm.200800196. [DOI] [PubMed] [Google Scholar]
- 46.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 47.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 48.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 49.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leendertse M, Heikens E, Wijnands LM, van Luit-Asbroek M, Teske GJ, Roelofs JJ, et al. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis. 2009;200:1162–1165. doi: 10.1086/605609. [DOI] [PubMed] [Google Scholar]
- 51.Singh KV, Lewis RJ, Murray BE. Importance of the epa locus of Enterococcus faecalis OG1RF in a mouse model of ascending urinary tract infection. 2009 doi: 10.1086/600124. (revised manuscript resubmitted to J Infect Dis) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76:4120–4128. doi: 10.1128/IAI.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson JR, Clabots C, Hirt H, Waters C, Dunny G. Enterococcal aggregation substance and binding substance are not major contributors to urinary tract colonization by Enterococcus faecalis in a mouse model of ascending unobstructed urinary tract infection. Infect Immun. 2004;72:2445–2448. doi: 10.1128/IAI.72.4.2445-2448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69:4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gianfaldoni C, Censini S, Hilleringmann M, Moschioni M, Facciotti C, Pansegrau W, et al. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect Immun. 2007;75:1059–1062. doi: 10.1128/IAI.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margarit I, Rinaudo CD, Galeotti CL, Maione D, Ghezzo C, Buttazzoni E, et al. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J Infect Dis. 2009;199:108–115. doi: 10.1086/595564. [DOI] [PubMed] [Google Scholar]
- 57.Garibaldi M, Rodriguez-Ortega MJ, Mandanici F, Cardaci A, Midiri A, Papasergi S, et al. Immunoprotective activities of a Streptococcus suis pilus subunit in murine models of infection. Vaccine. doi: 10.1016/j.vaccine.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, David DM, Scidman JG, Smith JA, et al., editors. Current Protocols in Molecular Biology. Brooklyn, NY: Green Publishing Associates; 1994. pp. 241–242. [Google Scholar]
- 59.Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006;72:334–345. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D,Dcarboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rakita RM, Quan VC, Jacques-Palaz K, Singh KV, Arduino RC, Mee M, et al. Specific antibody promotes opsonization and PMN-mediated killing of phagocytosis-resistant Enterococcus faecium. FEMS Immunol Med Microbiol. 2000;28:291–299. doi: 10.1111/j.1574-695X.2000.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 64.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh KV, Nallapareddy SR, Sillanpaa J, Murray BE. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 6:1000716. doi: 10.1371/journal.ppat.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh KV, Coque TM, Weinstock GM, Murray BE. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Med Microbiol. 1998;21:323–331. doi: 10.1111/j.1574-695X.1998.tb01180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.