Abstract
Background
Previous work has identified CD11c+CD1c- dendritic cells (DCs) as the major “inflammatory” dermal DC population in psoriasis vulgaris and CD1c+ DCs as the “resident” cutaneous DC population.
Objective
To further define molecular differences between these two myeloid dermal DC populations.
Methods
Inflammatory and resident DCs were single-cell sorted from psoriasis lesional skin biopsies, and the transcriptome of CD11c+CD1c- versus CD1c+ DCs was determined. Results were confirmed with RT-PCR, flow cytometry, immunohistochemistry, and double label immunofluorescence. Human keratinocytes were cultured for functional studies.
Results
TNF-related apoptosis-inducing ligand (TRAIL), Toll-like receptors (TLRs) 1 and 2, S100A12/EN-RAGE, CD32, and many other inflammatory products were differentially expressed in inflammatory DCs compared to resident DCs. Flow cytometry and immunofluorescence confirmed higher protein expression on CD1c- versus CD1c+ DCs. TRAIL receptors, death receptor 4 (DR4), and decoy receptor 2 (DcR2) were expressed in keratinocytes and dermal cells. In vitro culture of keratinocytes with TRAIL induced CCL20 chemokine.
Conclusions
CD11c+CD1c- inflammatory DCs in psoriatic lesional skin express a wide range of inflammatory molecules compared to skin resident CD1c+ DCs. Some molecules made by inflammatory DCs, including TRAIL, could have direct effects on keratinocytes or other skin cell types to promote disease pathogenesis.
Keywords: TRAIL, psoriasis, dendritic cell, inflammation, CCL20, TLR1, TLR2, S100A12, CD32
Implications.
Inflammatory myeloid dermal DCs in psoriasis expressed TNF-related apoptosis-inducing ligand (TRAIL), TLR1, TLR2, S100A12, CD32, and many other inflammatory cytokines and receptors. These molecules could contribute to many different inflammatory pathways and they form a basis for discriminating two key DC populations in the skin.
Introduction
In normal human skin, CD11c+ myeloid DCs express the MHC-class II-like lipid recognition protein CD1c/blood dendritic cell antigen-1 (BDCA-1)1. However, CD1c is not a sensitive marker for cutaneous myeloid DCs during inflammation. We have recently characterized a population of “inflammatory dermal” CD1c- myeloid DCs that infiltrate cutaneous lesions of psoriasis, which is a prototypic inflammatory disease of the skin2. CD1c- DCs are phenotypically immature and produce cytokines and inflammatory mediators including TNF, inducible nitric oxide synthase (iNOS), IL-203, IL-23p19 and IL-12/IL-23p404, 5. In addition, these inflammatory myeloid DCs induce allogeneic T cell proliferation and polarize T cells towards Th17 and Th1, two types of T cells implicated in psoriasis2, 6, 7.
Myeloid DCs are important bridges between the innate and adaptive immune systems, but their functions in many inflammatory diseases are still not entirely clear. In this present study, these inflammatory CD1c- myeloid DCs were single-cell-sorted from psoriatic lesional dermis and further characterized by microarray analysis in order to elucidate molecules associated with CD1c- versus CD1c+ DCs, and determine what inflammatory mediators these cells might be producing. We are also seeking a more useful “positive” marker to be able to study these cells specifically, rather than relying on a “negative” (CD1c-) definition.
In this study, we identified several differentially expressed gene products, including TNF-related apoptosis-inducing ligand (TRAIL), as selective markers for inflammatory myeloid DCs. TRAIL is a member of the TNF superfamily of cytokines that is involved in different kinds of inflammatory responses8. TRAIL is a type II transmembrane protein, which can be cleaved at its C-terminus to form a soluble ligand, and both the membrane bound and soluble form can induce apoptosis9. Multiple cell types at different stages of activation express TRAIL, including myeloid lineage cells (monocytes, DCs and macrophages)10, plasmacytoid DCs11, NKT12, and T cells13. Five TRAIL receptors have been identified in humans14-16. Although TRAIL is best known for induction of apoptosis in tumor cells while sparing normal tissue12, TRAIL has also been implicated in inflammatory disease processes including allergic asthma17 and atopic dermatitis18. In this manuscript, we demonstrate that inflammatory DCs in psoriasis vulgaris express TRAIL, lesional epidermal and dermal cells co-express TRAIL receptors, and the addition of TRAIL to keratinocytes induces CCL20. Thus TRAIL and other differentially expressed molecules identified in inflammatory DCs may be direct inflammatory mediators in the skin. In addition, these markers provide a means to isolate different DC populations for future functional studies.
Methods
Skin Samples
Skin biopsies were obtained from normal volunteers and psoriasis patients under a Rockefeller University Institutional Review Board approved protocol. Written informed consent was obtained and the study was performed in adherence with the Declaration of Helsinki Principles. Lesional and non-lesional psoriasis punch biopsy skin samples were used for immunohistochemistry and immunofluorescence. Dermal single cell suspensions from psoriasis shave biopsies were obtained after removing the epidermis with dispase, and culturing the dermis for 2-3 days, as previously described2.
FACS
FACS-sorting of dermal single cell suspensions was performed as previously described2 using a FACS Aria (BD Biosciences). Dermal cells were sorted into two populations: CD11c+CD163-HLA-DR+CD1c+ and CD11c+CD163-HLA-DR+CD1c- (n=4), to 99% purity. Seven psoriatic DC samples were obtained: 3 paired CD1c+/- DCs and an additional unpaired CD1c- sample. FACS-sorted cells were processed for microarray as biological replicates. FACS phenotyping of dermal single cell suspensions was performed on an LSR-II (BD Biosciences) (n=3). Antibodies used are outlined in Table E1 (see Online Repository).
Cell culture myeloid cells to determine their transcriptome
We have previously published the transcriptomes of the myeloid cells used for principal component analysis19 (see Figure E1 in Online Repository). Macrophages, immature DCs and mature DCs were prepared in a standard manner from monocytes, and RNA hybridized to HGU95 chips. In order to apply principal component analysis to our experimental samples on Affymetrix Hu133A2.0 platform and previously published cell lineage data on Affymetrix HGU95av2 platform19, probe sets were averaged to obtain a unique expression for each gene.
Microarray hybridization
CD1c+ and CD1c- DCs were FACS-sorted directly into Trizol (Invitrogen). RNA was extracted using the RNeasy Mini Kit (Qiagen), GeneChip two-cycle target labeling kit (Affymetrix) was performed, and fragmented cRNA were then hybridized to HGU133A Affymetrix gene chips as previously described20. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE20264.
RT-PCR
RT-PCR was performed using EZ PCR core reagents, primers, and probes (Applied Biosystems) as previously published21. Primer and probe sets used were TLR1 (Hs00413978_m1), TLR2 (Hs00152932_m1), TRAIL (Hs00234355_m1), S100A12 (Hs00194525_m1), CD32 (Hs00234969_m1), and CCL20 (Hs00171125_m1). Custom probe for human acidic ribosomal protein (HARP) was used as a housekeeping gene21.
Immunohistochemistry and immunofluorescence
These techniques were performed in a standard manner, as previously described 4. Each staining was performed on 3-5 patient samples. Antibodies used are outlined in Table E1. All immunofluorescence images are from lesional skin. In all immunofluorescence figures, single stained controls are above the merged image, white line denotes dermo-epidermal junction, dermal collagen fibers gave green autofluorescence, and antibodies conjugated with a fluorochrome often gave background epidermal fluorescence.
CCL20 detection in TRAIL-treated cultured keratinocytes
Primary pooled human keratinocytes (n=4) were obtained from Yale Skin Diseases Research Center core facility. Keratinocytes were cultured in EpiLife medium with defined growth supplement (Cascade Biologics) at 37°C. Once 80% confluent, cytokines were added to keratinocytes and harvested after 24 hours. Cytokines added were rh-TRAIL (Peprotech Inc.) 0.1ng/mL, 1ng/mL, 10ng/mL, 100ng/mL, TNF (R&D) 10ng/ml, Etanercept (Enbrel, Amgen) 10ug/ml. Concentrations of CCL20 in cell-free supernatants of TRAIL-treated keratinocytes were measured using the Quantikine Human CCL20/MIP-3 alpha ELISA (R&D Systems).
Statistics
Two-tail, Wilcoxon signed rank test was used to compare lesional versus non-lesional mRNA expression, and control versus TNF or TRAIL treated keratinocytes. A t-test was also used to compare the effects on CCL20 protein of TRAIL and other mediators. Effect size was calculated by subtracting control CCL20 protein from the mediator added. For all figures p<0.05 (*), p<0.01 (**), p<0.001 (***). To compare microarray data from CD1c- with CD1c+ DCs, a moderated paired t-test was used, available in limma package from R/Bioconductor (http://www.bioconductor.org/). For multiple hypotheses testing, false discovery rate was controlled by using the Benjamini–Hochberg procedure. Genes were considered significant if they had a fold change > 3, and false discovery rate < 0.2 (maximum observed p value =0.03). (using ENTREZ identifiers) and then matched between the two platforms.
Results
CD11c+CD1c- inflammatory DCs were closely related to CD1c+ resident DCs and in vitro-derived immature DCs
We analyzed the transcriptome of these CD1c- DCs from psoriasis lesions together with the transcriptomes of other in vitro-derived myeloid cells19, using principal component analysis (PCA). PCA is a data analysis algorithm to visually assess similarities and differences between samples22. The genomic similarity of these different cell types is represented in a 2D map (see Figure E1 in the Online Repository). While the grouping of similar samples was expected, this analysis showed that there was a clear (right-to-left) progression from in vitro-derived macrophages, in vitro-derived immature DCs (iDC), to psoriatic CD1c- DCs, psoriatic CD1c+ DCs, and in vitro-derived mature DCs (mDC). Monocytes were located far away from this “line” as a negative control. CD1c- DCs were most closely related to CD1c+ DCs followed by iDCs.
The CD1c- DC subset in psoriasis expressed inflammatory genes including TRAIL, TLR1/2, S100A12 and CD32
The gating strategy to obtain pure CD11c+CD163-HLA-DR+CD1c- (n=4) and CD11c+CD163-HLA-DR+CD1c+ (n=3) populations is shown in Figure 1A. Gene array analysis was performed to compare the two DC populations using fold change >3, p<0.03, false discovery rate < 0.2. We found that 555 genes were up-regulated and 131 genes were down-regulated in the CD1c- DC population compared to the CD1c+ DC population (see Table E2 in the Online Repository). From this list, we selected known inflammatory genes of interest, and found that the majority of these genes were upregulated in the CD1c- population (Figure 1B), supporting prior observations that the CD1c- DC subset is inflammatory in function2. Of particular interest were inflammatory mediators TNF-receptor apoptosis-inducing ligand (TRAIL)/TNFSF10 and S100A12/EN-RAGE (fold change 6.1 and 18.1 respectively). Notable receptor genes included Toll-like receptors 1 and 2 (TLR1 and TLR2) and Fc fragment of IgG, low affinity IIa and IIb receptors (FCgR2a/CD32a and FCgR2b/CD32b) (fold change 9.8, 8.4, 8.7, and 12.8, respectively). Although our single-cell sorting strategy gated out CD163 protein expressing macrophages, CD163 mRNA was expressed in CD1c- DCs (fold change 24.3). In addition, CCR2 monocyte chemotactic receptor and intracellular pattern recognition receptor nucleotide-binding oligomerization domain protein 2 (NOD2) were expressed in CD1c- myeloid DCs (fold change 13.7 and 5.6, respectively).
Figure 1. Transcriptome of CD1c- DCs versus. resident CD1c+ DCs from psoriatic dermis.
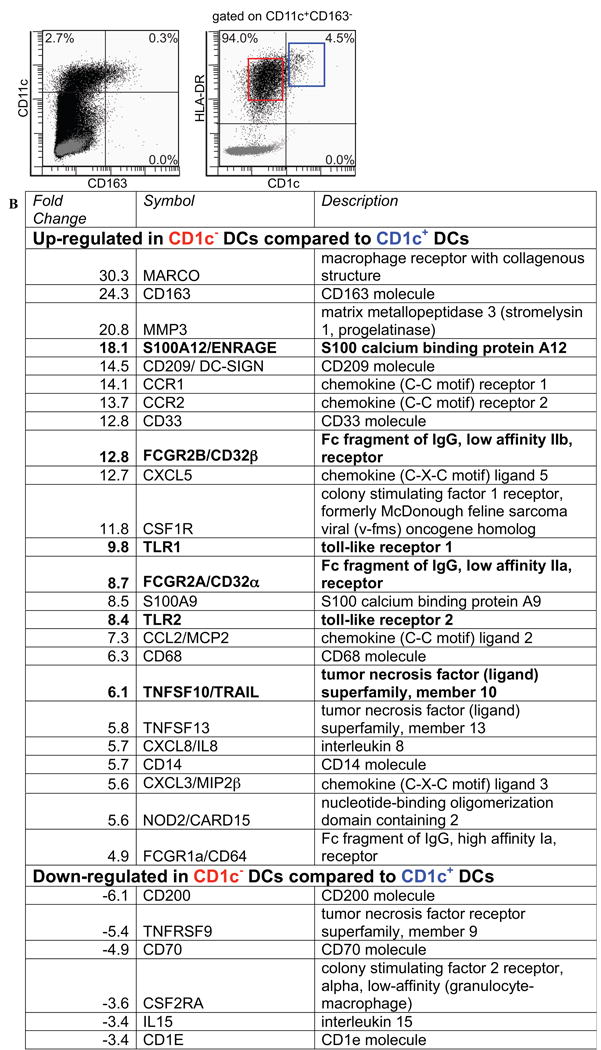
(A) Gating strategy used for cell sorting. HLA-DR+CD11c+CD163- myeloid DCs were sorted into CD1c- (red gate) and CD1c+ (blue gate) populations. (B) Selected up- and down-regulated inflammatory molecules from the transcriptome of CD1c- DC populations. The full list of differentially expressed genes can be found in Table E2.
We verified that these genes were potentially important inflammatory mediators in psoriasis by performing RT-PCR for these genes in additional non-lesional and lesional paired biopsy specimens (n=11 pairs). TLR2, TRAIL, S100A12, and CD32 were significantly up-regulated in lesional psoriatic skin compared to non-lesional (Figure 2). TLR1 expression was not significantly different in lesional versus non-lesional skin (data not shown).
Figure 2. RT-PCR of genes differentially up-regulated in CD1c- DCs in non-lesional and lesional psoriatic skin.
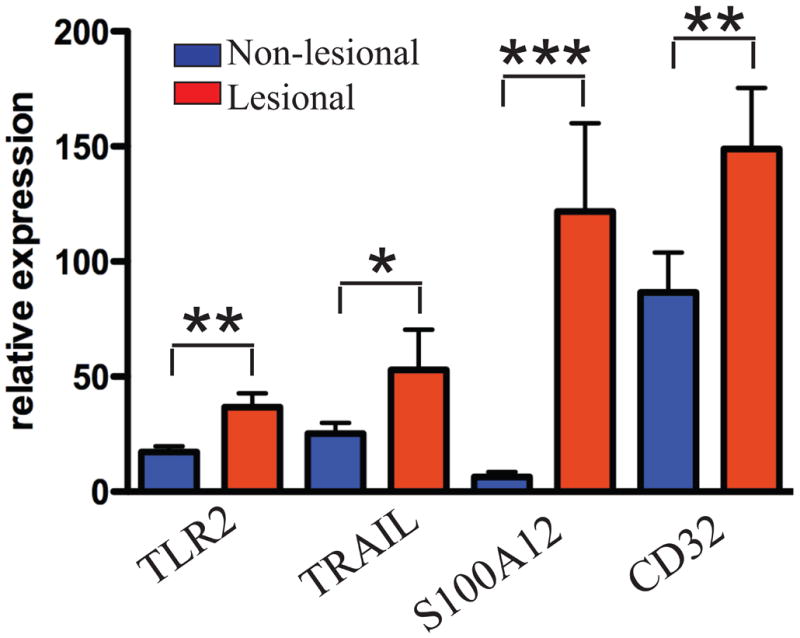
TLR2, TRAIL, S100A12, and CD32 were up-regulated in lesional psoriatic skin (n=11). Error bars indicate SEM. p<0.05 (*), p<0.01 (**), p<0.001(***).
TRAIL identified the CD11c+CD1c- inflammatory DC population
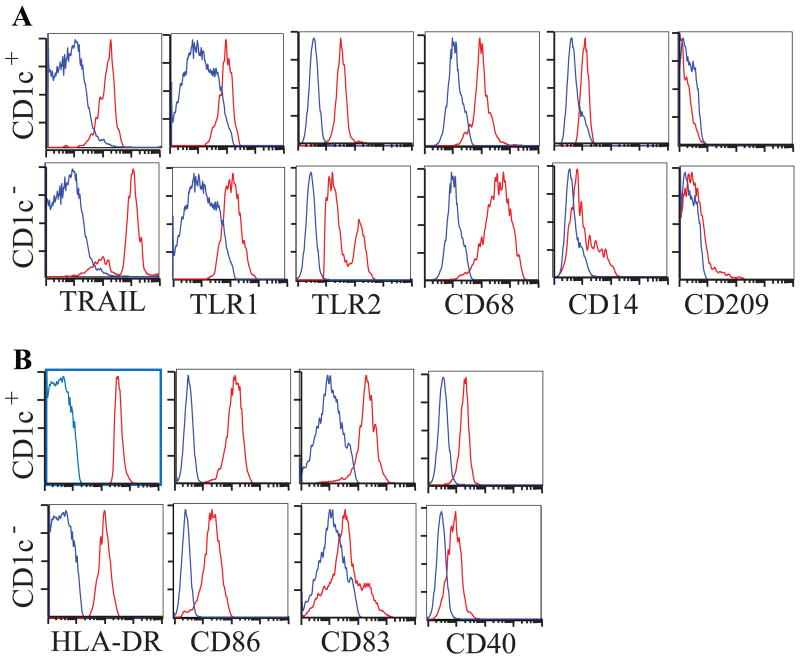
We determined the relative protein expression of TRAIL, TLR1, TLR2, CD68, CD14, and CD209 on CD1c+ versus CD1c- dermal dendritic cells using 8-color FACS analysis (n=3). Representative FACS plots of single cell suspensions from psoriasis lesional biopsies shown in Figure 3. As in the sorting experiments, both CD1c+ and CD1c- DC populations were gated on CD11c+CD163-HLA-DR+ cells. We confirmed the gene array expression data that each of these markers was more highly expressed in CD1c- cells compared to CD1c+ cells. TRAIL was the most consistent marker for CD1c- inflammatory DCs, expressed at a high level on nearly all CD1c- cells. One log difference in level of expression of TRAIL by FACS would allow cell sorting to obtain a pure population of these DCs for further studies. TLR2 was expressed at a high level of CD1c- cells, but was not highly expressed on CD1c+ cells (Figure 3A). CD1c+ DCs expressed higher levels of DC maturation markers HLA-DR, CD86, CD83 and CD40 compared to CD1c- DCs (Figure 3B).
Figure 3. Flow cytometric analysis of psoriatic dermal single cell suspensions.
(A) CD1c- DCs (bottom row) express higher levels of TRAIL, TLR1, TLR2, CD68, CD14 and CD209 compared to CD1c+ DCs (top row), (B) CD1c+ DCs expressed comparatively higher levels of DC maturation antigens HLA-DR, CD86, CD83 and CD40. Red histogram represents antigen expression gated on HLA-DR+CD11c+CD163-CD1c+/- DCs, blue is isotype.
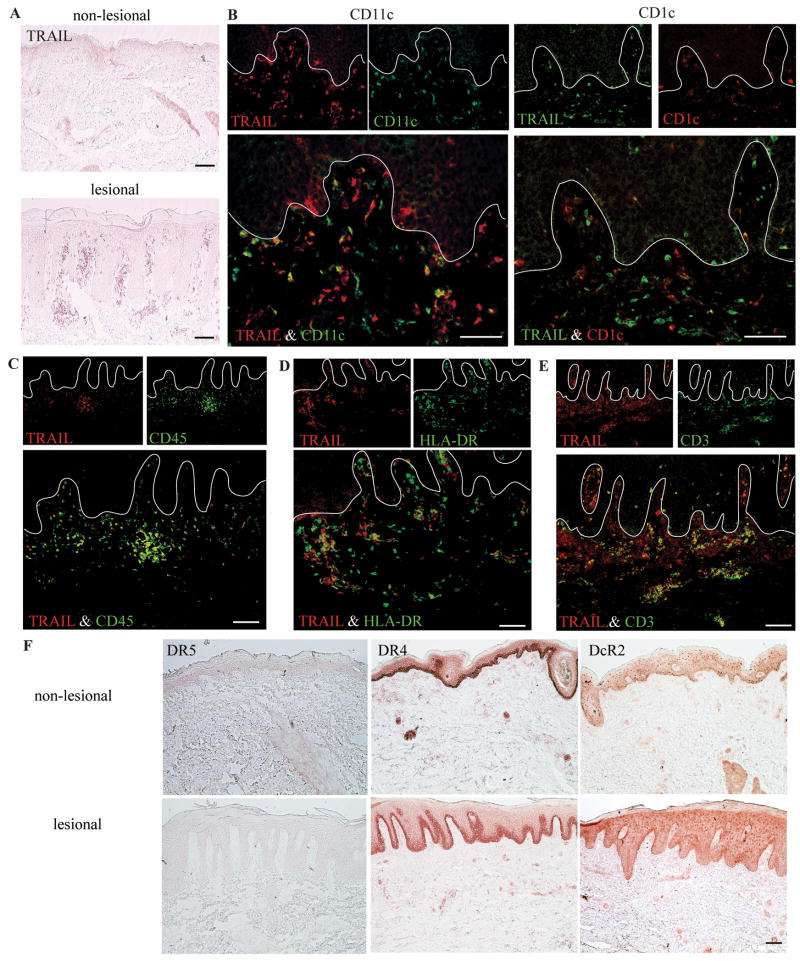
We confirmed that TRAIL+ cells were localized to the papillary dermis and at the dermal-epidermal junction in lesional psoriasis skin, but not in non-lesional skin (Figure 4A), and TRAIL was expressed by many CD11c+ DCs and few CD1c+ cells (Figure 4B). TLR1 (Figure E2A in Online Repository) and TLR2 (Figure E2B) also localized to the papillary dermis and dermo-epidermal junction in psoriatic lesional skin, but not in non-lesional skin by immunohistochemistry. The majority of CD11c+ DCs co-expressed TLR1 and TLR2, but no CD1c+ cells expressed TLR1 or TLR2. S100A12 was expressed in both lesional and non-lesional keratinocytes and in dermal cells (Figure E2C). The majority of S100A12 producing dermal cells expressed CD11c, but very few co-expressed CD1c. CD32 was expressed by papillary dermal cells in both lesional and non-lesional skin (Figure E2D). In addition, CD32 was expressed by cells scattered throughout the epidermis and in the reticular dermis of lesional skin. We found that CD32 was expressed by both CD11c+ and CD1c+ cells.
Figure 4. Lesional CD11c+CD1c- DCs express TRAIL, and keratinocytes expressed TRAIL receptors.
(A) TRAIL antigen was present in lesional dermis. (B) The majority of lesional CD11c+ cells expressed TRAIL, while few cells co-expressed TRAIL and CD1c. (C) Many TRAIL+ cells express CD45, (D) HLA-DR, and (E) some T cells expressed TRAIL. (F) There was no expression of DR5, but abundant DR4 and some expression of DcR2. Bar = 100μm.
Characterization of TRAIL+ cells and TRAIL receptor+ cells
TRAIL+ cells co-expressed blood derived antigen CD45, suggesting that TRAIL was only produced by bone marrow-derived leukocytes (Figure 4C). The majority of TRAIL+ cells co-expressed MHC-II molecule HLA-DR (Figure 4D), and some TRAIL+ cells were also CD3+, indicating they were T cells (Figure 4E). Only occasional CD163+ macrophages expressed TRAIL (see Figure E3 in Online Repository). Thus, the majority of TRAIL expression in psoriatic skin localized to CD11c+CD1c- inflammatory DCs.
To determine the potential site of action of TRAIL in psoriatic skin, we evaluated the expression of its activating receptors DR5, DR4, and DcR2 (Figure 4F). Ligation of DR5 and DR4 may more commonly induce apoptosis but also inflammation, while the DcR2 lacks an apoptosis domain and causes inflammation cells via NFκB14-16. In both non-lesional and lesional skin DR5 was not expressed, but DR4 and DcR2 were localized to basal keratinocytes and dermal cells. Few CD11c+ cells expressed DR4 (Figure E4A in Online Repository), and there was no overlap between DR4 and either CD3 (Figure E4B) or CD163 (see Figure E4C). In the dermis, there was nearly complete overlap between DR4 and blood vessel markers vimentin and CD31, and growth receptor IGF1Rb (Figure E4D, E, F).
TRAIL+ cells produced IL-23p19 and IL-12/IL-23p40 cytokine subunits, as well as iNOS and TNF
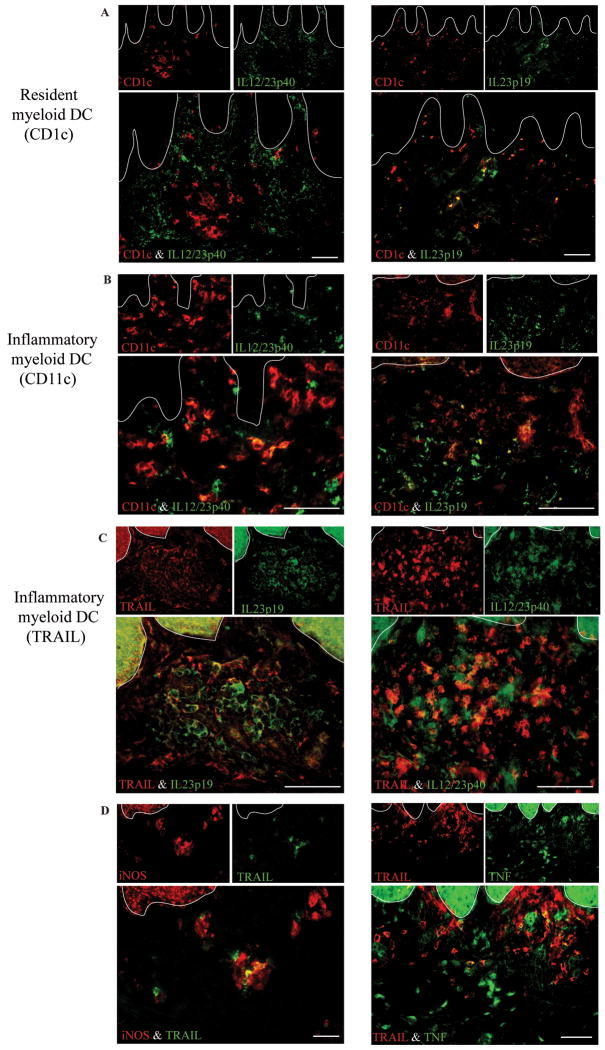
We further characterized the myeloid DC subsets in psoriasis lesions using double label immunofluorescence (Figure 5). The resident myeloid DCs (CD1c+) expressed IL-23p19 but not IL-12/23p40 (Figure 5A), while the inflammatory subset identified by CD11c expressed both IL-23 subunits (Figure 5B)4, 5. Many IL-12/23p40 and IL-23p19 expressing cells also expressed TRAIL (Figure 5C) and some TRAIL+ cells expressed iNOS and TNF (Figure 5D). Results are summarized in Table E3 (see Table E3 in the Online Repository). Thus. TRAIL+ cells are likely a subset of inflammatory myeloid DCs that contributes to pathogenesis in psoriasis by producing these inflammatory mediators.
Figure 5. Expression of Th17 polarizing cytokines IL-23p19 and IL-12/IL-23p40 in cutaneous DCs in psoriasis.
(A) CD1c+ resident DCs did not express IL-12/23p40, but did express IL-23p19. (B) Inflammatory myeloid DCs (CD11c+) expressed both IL23 subunits. (C, D) TRAIL+ cells co-expressed some IL-12/IL-23p40 and IL-23p19, as well as iNOS and TNF. Bar = 100μm.
TRAIL induced CCL20 in human cultured keratinocytes
In order to start to evaluate the potential function of TRAIL in the skin, pooled human keratinocytes (n=3) were cultured with recombinant human TRAIL. TRAIL induced dose-dependent CCL20 expression in cultured keratinocytes, although TNF induced 10-fold more CCL20 mRNA expression than 100ng/mL of TRAIL (see Figure E5A in the Online Repository). A second set of experiments (n=3) were performed to analyze the production of CCL20 protein by TRAIL. In Figure E5B, addition of each agent was compared to saline to determine the difference from control. There was some background CCL20 produced (mean 38 pg/ml), most likely due to endogenous TNF production, as there was a reduction in CCL20 production by the addition of etanercept (to mean 22 pg/ml, p=0.1). There was an increase in the production of CCL20 by TRAIL (56 pg/ml) compared to control (p=0.3), but it was greater in magnitude compared to etanercept alone (p=0.12). TRAIL appears to induce some TNF as the addition of etanercept decreased the amount of CCL20 produced (43pg/ml). TNF induced a greater increase in CCL20 (179 pg/ml) compared to control (p=0.13), and this was almost completely abolished by etanercept (14 pg/ml).
Discussion
In this study, we have identified novel molecular markers of different DC subsets in psoriasis, a common human inflammatory skin disease. These markers were initially identified in the transcriptome of FACS-sorted, dermal single cell suspensions from inflamed human skin, and expression was confirmed in situ. This approach has led to identification of new molecules that are associated with infiltrating leukocytes in psoriasis and it has expanded our understanding of how CD11c+CD1c- DCs may contribute to inflammation in human skin.
We first studied TRAIL as an example of how the list of CD1c- differentially expressed genes could be used to dissect pathogenic mechanisms of these inflammatory myeloid DCs. TRAIL is best known as an apoptosis-inducing ligand that selectively “kills” tumor cells12. In BCCs treated with the TLR7/8 agonist imiquimod, TRAIL acted as a cytotoxic molecule11. During the BCC regression phase, imiquimod treated, DR4-expressing tumor islets were surrounded by an inflammatory infiltrate containing TRAIL-expressing CD11c+ myeloid DCs, plasmacytoid DCs, and T cells. In vitiligo, TRAIL was proposed to cause apoptosis of melanocytes, resident skin pigment producing cells. TRAIL+ CD11c+ DCs surrounded depigmented plaques, and both TRAIL and in vitro-derived TRAIL+ DCs were cytotoxic to cultured melanocytes23.
However, recent studies on mouse models of allergic asthma have suggested a potential inflammatory role for TRAIL in immune-mediated diseases. In both human asthmatics and an allergen-specific airway hyperreactivity mouse model, TRAIL was overexpressed in asthmatic sputum and the bronchial epithelium, respectively. In mice deficient for TRAIL, this allergen-specific airway hyperreactivity was abolished, secondary to diminished T cell trafficking to the lungs17. TRAIL and S100A12, both products of inflammatory DCs, potentiate inflammation by upregulating blood vessel leukocyte adhesion molecules and TRAIL promotes vascular proliferation24-26.
There is a third proposed role for TRAIL, as an anti-inflammatory agent. In patients with AD but not in healthy controls, TRAIL was expressed in blood mononuclear cells, T cells, eosinophils, and neutrophils, and in cutaneous T cells. Expression of TRAIL in myeloid DCs was not assessed. The investigators proposed an anti-inflammatory function for TRAIL in human skin, demonstrating that TRAIL induced IL-1R antagonist expression in cultured human keratinocytes18.
While alternate functions of TRAIL in cutaneous diseases have been proposed, the expression in psoriasis is most consistent with its pro-inflammatory or cell-damaging effects in other systems. CCL20, a potent T cell and DC chemokine, was expressed by bronchial epithelial cells in a TRAIL-dependent manner17. Our studies support a similar mechanism of TRAIL-dependent inflammation in psoriatic skin. TRAIL receptors DR4 and DcR2 were expressed on psoriatic basal keratinocytes, and recombinant human TRAIL induced CCL20 in cultured human keratinocytes. TRAIL may function as an inducer of CCL20 and subsequent T cell and DC chemotaxis to sites of inflammation, although the biological relevance of the induction of CCL20 by TRAIL remains to be determined.
Thus, TRAIL may contribute to leukocyte trafficking and transmigration through induction of CCL20 in keratinocytes, endothelial cell integrin and selectin up-regulation, and IGF1R-dependent vascular smooth muscle cell proliferation. In psoriasis skin, there is no evidence for keratinocyte or vessel apoptosis27, however, there is up-regulated blood vessel integrin and selectin expression28, 29. TRAIL-induced vascular hyperproliferation may contribute to the psoriatic phenotype of supra-abundant ectatic blood vessels coursing through the papillary dermis30. Therefore, we propose a unifying hypothesis whereby TRAIL production by inflammatory myeloid DCs acts on TRAIL receptors on keratinocytes, vascular smooth muscle cells, and endothelial cells to induce secretion of CCL20, proliferation of blood vessels, and inducion of E-selectin and ICAM-1 expression (Figure E6 in the Online Repository).
Other molecules in the molecular characterization of inflammatory myeloid DCs that are of great interest include TLR1 and TLR2, S100A12 and CD32. We confirmed these genes were specific to CD11c+ cells, and not CD1c+ cells in psoriasis by immunofluorescence. TLR1/2 are very interesting molecules as they are the receptors for Listeria monocytogenes, and infection in mice with this agent induced TIP-DCs31, which we described in psoriasis, and consider a subset of inflammatory myeloid DCs2, 32. DCs co-express the activating (CD32a) and inhibitory (CD32b) isoforms of IgG FcR II (CD32). The balance between these receptors controls DC activation, maturation and function33. Further studies will be required to determine if and how these molecules function in cell-mediated inflammatory skin diseases.
We have now identified a number of positive surface markers that distinguish “inflammatory” DCs from “resident”, CD1c+ DCs in human skin. These markers need to be studied on DC subsets that accumulate in different inflammatory conditions, such as atopic dermatitis or Crohn's disease to determine if they are restricted to specific disease tissues or different types of cell mediated inflammation (Th1/Th17 versus Th2). Our transcriptome also identified a wide set of molecules in bona-fide inflammatory DCs that should be compared to molecules induced during in vitro manufacture of DCs with various cytokines and ligands. These markers give the potential to more easily sort distinct DC subsets for functional studies. Furthermore, these molecules identify potential therapeutic targets to prevent the development or emigration of pathogenic myeloid DCs into the skin for the prevention or treatment of psoriasis.
Supplementary Material
Acknowledgments
Research was supported by National Institutes of Health (NIH) grant UL1 RR024143 from the National Center for Research Resources (NCRR). LCZ is supported by NIH MSTP grant GM07739; KCP is supported by the Dana Foundation (Human Immunology Consortium Grant); KEN is supported by the Clinical Scholars Program at The Rockefeller University; MSF is partially supported by NIH grant UL1 RR024143; MAL is supported by 1 K23 AR052404-01A1 and The Doris Duke Charitable Foundation. We thank Yale Skin Diseases Research Center core facility for keratinocyte culture.
Abbreviations used
- DC
dendritic cell
- iNOS
inducible nitric oxide synthase
- Tip-DC
TNF-and-iNOS producing dendritic cell
- TRAIL
TNF-related apoptosis-inducing ligand
- DR4
death receptor 4
- DR5
death receptor 5
- DcR2
decoy receptor 2
- IGF1Rb
insulin-like growth factor 1 receptor b
- BDCA-1
blood dendritic cell antigen-1
- VSMC
vascular smooth muscle cell
- KC
keratinocyte
- NOD2
nucleotide-binding oligomerization domain protein 2
- HUVEC
human umbilical vein endothelial cell
- HD-MEC
human dermal microvascular endothelial cell
- ICAM-1
intercellular adhesion molecule 1
- AD
atopic dermatitis
- BCC
basal cell carcinoma
- TLR
Toll-like receptor
- iDC
immature dendritic cell
- mDC
mature dendritic cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zaba LC, Fuentes-Duculan J, Steinman RM, Krueger JG, Lowes MA. Normal human dermis contains distinct populations of CD11cBDCA-1 dendritic cells and CD163FXIIIA macrophages. J Clin Invest. 2007;117:2517–25. doi: 10.1172/JCI32282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC, et al. Psoriasis Is Characterized by Accumulation of Immunostimulatory and Th1/Th17 Cell-Polarizing Myeloid Dendritic Cells. J Invest Dermatol. 2009;129:79–88. doi: 10.1038/jid.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Lee E, Lowes MA, Haider AS, Fuentes-Duculan J, Abello MV, et al. Prominent production of IL-20 by CD68+/CD11c+ myeloid-derived cells in psoriasis: Gene regulation and cellular effects. J Invest Dermatol. 2006;126:1590–9. doi: 10.1038/sj.jid.5700310. [DOI] [PubMed] [Google Scholar]
- 4.Zaba LC, Cardinale I, Gilleaudeau P, Sullivan-Whalen M, Farinas MS, Fuentes-Duculan J, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204:3183–94. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Zaba LC, Cardinale I, Nograles KE, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol. 2008;181:7420–7. doi: 10.4049/jimmunol.181.10.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kryczek I, Bruce AT, Gudjonsson JE, Johnston A, Aphale A, Vatan L, et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: mechanism and pathological relevance in psoriasis. J Immunol. 2008;181:4733–41. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, et al. Psoriasis Vulgaris Lesions Contain Discrete Populations of Th1 and Th17 T Cells. J Invest Dermatol. 2008;128:1207–11. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- 8.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–82. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 9.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161:2833–40. [PubMed] [Google Scholar]
- 10.Kemp TJ, Elzey BD, Griffith TS. Plasmacytoid dendritic cell-derived IFN-alpha induces TNF-related apoptosis-inducing ligand/Apo-2L-mediated antitumor activity by human monocytes following CpG oligodeoxynucleotide stimulation. J Immunol. 2003;171:212–8. doi: 10.4049/jimmunol.171.1.212. [DOI] [PubMed] [Google Scholar]
- 11.Stary G, Bangert C, Tauber M, Strohal R, Kopp T, Stingl G. Tumoricidal activity of TLR7/8-activated inflammatory dendritic cells. J Exp Med. 2007;204:1441–51. doi: 10.1084/jem.20070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Kayagaki N, Yagita H, et al. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J Exp Med. 2002;195:161–9. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang XR, Zhang LY, Devadas S, Li L, Keegan AD, Shi YF. Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ. 2003;10:203–10. doi: 10.1038/sj.cdd.4401138. [DOI] [PubMed] [Google Scholar]
- 14.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–8. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 15.Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–3. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 16.Collison A, Foster PS, Mattes J. Emerging role of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) as a key regulator of inflammatory responses. Clin Exp Pharmacol Physiol. 2009;36:1049–53. doi: 10.1111/j.1440-1681.2009.05258.x. [DOI] [PubMed] [Google Scholar]
- 17.Weckmann M, Collison A, Simpson JL, Kopp MV, Wark PA, Smyth MJ, et al. Critical link between TRAIL and CCL20 for the activation of TH2 cells and the expression of allergic airway disease. Nat Med. 2007;13:1308–15. doi: 10.1038/nm1660. [DOI] [PubMed] [Google Scholar]
- 18.Vassina E, Leverkus M, Yousefi S, Braathen LR, Simon HU, Simon D. Increased expression and a potential anti-inflammatory role of TRAIL in atopic dermatitis. J Invest Dermatol. 2005;125:746–52. doi: 10.1111/j.0022-202X.2005.23878.x. [DOI] [PubMed] [Google Scholar]
- 19.Haider AS, Lowes MA, Suarez-Farinas M, Zaba LC, Cardinale I, Khatcherian A, et al. Identification of Cellular Pathways of “Type 1,” Th17 T Cells, and TNF- and Inducible Nitric Oxide Synthase-Producing Dendritic Cells in Autoimmune Inflammation through Pharmacogenomic Study of Cyclosporine A in Psoriasis. J Immunol. 2008;180:1913–20. doi: 10.4049/jimmunol.180.3.1913. [DOI] [PubMed] [Google Scholar]
- 20.Zaba LC, Suarez-Farinas M, Fuentes-Duculan J, Nograles KE, Guttman-Yassky E, Cardinale I, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol. 2009;124:1022-10 e1-395. doi: 10.1016/j.jaci.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamian F, Lowes MA, Lin SL, Lee E, Kikuchi T, Gilleaudeau P, et al. Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proc Natl Acad Sci U S A. 2005;102:2075–80. doi: 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–4. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 23.Kroll TM, Bommiasamy H, Boissy RE, Hernandez C, Nickoloff BJ, Mestril R, et al. 4-Tertiary butyl phenol exposure sensitizes human melanocytes to dendritic cell-mediated killing: relevance to vitiligo. J Invest Dermatol. 2005;124:798–806. doi: 10.1111/j.0022-202X.2005.23653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li JH, Kirkiles-Smith NC, McNiff JM, Pober JS. TRAIL induces apoptosis and inflammatory gene expression in human endothelial cells. J Immunol. 2003;171:1526–33. doi: 10.4049/jimmunol.171.3.1526. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 26.Kavurma MM, Schoppet M, Bobryshev YV, Khachigian LM, Bennett MR. TRAIL stimulates proliferation of vascular smooth muscle cells via activation of NF-kappaB and induction of insulin-like growth factor-1 receptor. J Biol Chem. 2008;283:7754–62. doi: 10.1074/jbc.M706927200. [DOI] [PubMed] [Google Scholar]
- 27.Gottlieb AB, Chamian F, Masud S, Cardinale I, Abello MV, Lowes MA, et al. TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol. 2005;175:2721–9. doi: 10.4049/jimmunol.175.4.2721. [DOI] [PubMed] [Google Scholar]
- 28.Sigmundsdottir H, Johnston A, Gudjonsson JE, Bjarnason B, Valdimarsson H. Methotrexate markedly reduces the expression of vascular E-selectin, cutaneous lymphocyte-associated antigen and the numbers of mononuclear leucocytes in psoriatic skin. Exp Dermatol. 2004;13:426–34. doi: 10.1111/j.0906-6705.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Wang H, Peng B, Zhang M, Zhang D, Hou S, et al. Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance. Proc Natl Acad Sci U S A. 2009;106:4349–54. doi: 10.1073/pnas.0810844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–73. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 31.Machata S, Tchatalbachev S, Mohamed W, Jansch L, Hain T, Chakraborty T. Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J Immunol. 2008;181:2028–35. doi: 10.4049/jimmunol.181.3.2028. [DOI] [PubMed] [Google Scholar]
- 32.Lowes MA, Chamian F, Abello MV, Fuentes-Duculan J, Lin SL, Nussbaum R, et al. Increase in TNF-alpha and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a) Proc Natl Acad Sci U S A. 2005;102:19057–62. doi: 10.1073/pnas.0509736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boruchov AM, Heller G, Veri MC, Bonvini E, Ravetch JV, Young JW. Activating and inhibitory IgG Fc receptors on human DCs mediate opposing functions. J Clin Invest. 2005;115:2914–23. doi: 10.1172/JCI24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.