Abstract
Vaxign is the first web-based vaccine design system that predicts vaccine targets based on genome sequences using the strategy of reverse vaccinology. Predicted features in the Vaxign pipeline include protein subcellular location, transmembrane helices, adhesin probability, conservation to human and/or mouse proteins, sequence exclusion from genome(s) of nonpathogenic strain(s), and epitope binding to MHC class I and class II. The precomputed Vaxign database contains prediction of vaccine targets for >70 genomes. Vaxign also performs dynamic vaccine target prediction based on input sequences. To demonstrate the utility of this program, the vaccine candidates against uropathogenic Escherichia coli (UPEC) were predicted using Vaxign and compared with various experimental studies. Our results indicate that Vaxign is an accurate and efficient vaccine design program.
1. Introduction
Reverse vaccinology is an emerging vaccine development approach that starts with the prediction of vaccine targets by bioinformatics analysis of microbial genome sequences [1]. Predicted proteins are selected based on desirable attributes. Normal wet laboratory experiments are conducted in a later stage to test all or selected vaccine targets. Rino Rappuoli, the pioneer of reverse vaccinology [1, 2], first applied this approach to the development of a vaccine against serogroup B Neisseria meningitidis (MenB), the major cause of sepsis and meningitis in children and young adults [2]. In this study, bioinformatic methods were first used to screen the complete genome of a MenB strain MC58 for genes encoding putative surface-exposed or secreted proteins. These proteins were predicted to be antigenic and therefore may represent the most suitable vaccine candidates. In total, 350 novel vaccine candidates were predicted and expressed in Escherichia coli; 28 were found to elicit protective immunity. It took less than 18 months to identify more vaccine candidates in MenB than had been discovered during the past 40 years by conventional methods [2]. Since then, the concept of reverse vaccinology has also successfully been applied to other pathogens, including Bacillus anthracis [3], Porphyromonas gingivalis [4], Chlamydia pneumoniae [5], Streptococcus pneumoniae [6], Helicobacter pylori [7], and Mycobacterium tuberculosis [8]. Compared to a conventional vaccine development approach that starts from the wet laboratory, reverse vaccinology begins with bioinformatics analysis, which dramatically quickens the process of vaccine development.
Since reverse vaccinology was conceived and applied in a test case ten years ago, this technology has progressed dramatically. Subcellular location is still considered as one main criterion for vaccine target prediction. However, more criteria have been added. For example, since it was found that outer membrane proteins containing more than one transmembrane helix were, in general, difficult to clone and purify [2], the number of transmembrane domains for a vaccine target is often considered in bioinformatics filtering. More and more genomes are now available for each pathogenic species. It is now required to examine all completed genomes and predict vaccine targets that are conserved in all genomes. If genomes from non-pathogenic strains of the species are also available, ideal vaccine targets are those that exist in genomes of virulent pathogen strains but are absent from the avirulent strains. To induce strong immunity and avoid autoimmunity, predicted vaccine targets are required not to have sequence similarity to proteins of hosts (e.g., human). Epitope-based vaccines have been demonstrated to induce protection against many infectious diseases [9]. To optimize epitope vaccines, it has become an essential task to predict immune epitopes from protective antigens.
While reverse vaccinology has been used for a decade, this approach is often not accessible to the general laboratory, due to the lack of software programs that are easy to use and implement. Although many individual software programs are available to aid in vaccine target prediction [10–17], they are individually developed for different purposes and contain disparate data formats and programming settings. This makes tool and data integration difficult. Successful use of these tools often requires local installation, command line execution, and substantial computational power. Many tools are not optimized for high throughput data processing. NERVE, for example, is a new enhanced reverse vaccinology environment that includes several steps of programs for reverse vaccinology [18]. NERVE aims to help save time and money in vaccine design. However, it also requires software download and database setup. In addition, NERVE does not include precomputed data of vaccine target prediction, which makes the prediction time extensive. In addition, NERVE does not perform MHC class I and II epitope predictions.
Many immunoinformatics epitope mapping tools have been developed during the last three decades [19]. For example, DeLisi and Berzofsky developed the earliest computer-driven algorithm for epitope mapping based on empirical observations of amino acid residue periodicity in T-cell epitopes [20]. The anchor-based MHC binding motifs were used for T-cell epitope identification by many researchers, such as Sette et al. in 1989 [21] and Rotzschke et al. in 1991 [22]. Matrix-based approached for T-cell epitope mapping have been developed by a number of research teams such as Sette et al. [23], Davenport et al. [24], De Groot et al. [25], and Reche et al. [15]. Many databases of MHC-binding peptides, starting from MHCPEP developed by Brusic et al. in 1994 [26] to the currently frequently used IEDB [27], have been developed for use with matrices and neural network-based epitope prediction tools.
Uropathogenic Escherichia coli (UPEC) is the most common cause of community-acquired urinary tract infection (UTI). Over half (53%) of all women (and 14% of men) experience at least one urinary tract infection (UTI), leading to an estimated 6.8 million annual physician visits in the United States alone, 1.3 million emergency room visits, and 246,000 hospitalizations of women with an annual cost of more than $2.4 billion [28]. Although many groups have attempted to develop vaccines against UPEC [29–33], no preparations are yet in general use in the United States. Complete and annotated genomic sequences have now been determined for four strains of extraintestinal pathogenic E. coli including CFT073, UTI89, 536, and F11; these UPEC strains were isolated from human cases of cystitis, pyelonephritis, and/or bacteremia. These provide a basis for predicting UPEC vaccine targets using these genome sequences based on reverse vaccinology. Recently, we have also performed several high throughput proteomic and genomic studies including in vivo microarray [34], proteomics of urine-grown bacteria [35], and in vivo induced antigen technology (IVIAT) [36]. We hypothesized that vaccine targets predicted based on genome analysis largely correlate with the results obtained from these high throughput data analyses.
Vaxign (http://www.violinet.org/vaxign/), the first web-based, publically available vaccine design system, was first introduced in the second Vaccine Congress meeting in December 2008 in Boston, MA, USA. Vaxign was demonstrated to successfully predict vaccine targets against different pathogens [37]. Since then, Vaxign has significantly been improved in terms of performance and speed. In this report, we systematically introduce the updated Vaxign prediction system, and describe how Vaxign was used to predict vaccine targets against uropathogenic E. coli (UPEC). Many predicted results, based on genome sequence analyses, were also confirmed by wet-lab testing and other studies based on RNA, protein, and antibody analyses.
2. Methods
2.1. Vaxign Software Components for Vaccine Target Prediction
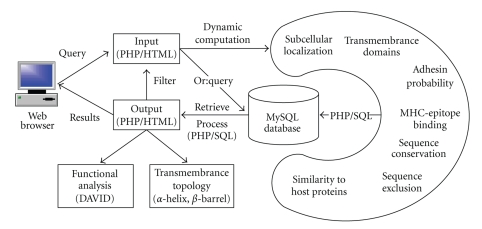
Vaxign integrates open source tools and internally developed programs with user-friendly web interfaces. Input data for Vaxign execution are amino acid sequences from one protein or whole genomes. This Vaxign pipeline includes the following components (Figure 1).
Figure 1.
The Vaxign algorithm pipeline.
(1) Prediction of subcellular localization. Vaxign predicts different subcellular locations using optimized PSORTb 2.0 that has a measured overall precision of 96% [10].
(2) Transmembrane domain prediction. The transmembrane helix topology analysis is performed using optimized HMMTOP based on a general hidden Markov model (HMM) decoding algorithm [11]. A profile-based hidden Markov model implemented in PROFtmb is used in Vaxign for the prediction and discrimination of bacterial transmembrane beta barrels [38]. The resulting PROFtmb method reaches an overall four-state (up-, down-strand, periplasmic-,and outer-loop) accuracy as high as 86% [38]. Since the execution of PROTtmb is very time consuming, not all proteins in all genomes in the Vaxign database were preanalyzed for transmembrane beta barrel analysis.
(3) Calculation of adhesin probability. Adhesin probability is predicted using optimized SPAAN [12]. The SPAAN prediction has a sensitivity of 89% and specificity of 100% based on a defined test set [12]. The probability of being an adhesin has a default cut-off of 0.51.
(4) Protein conservation among different genomes. This program identifies conserved sequences among more than one genome. OrthoMCL is applied to calculate the homology between different sequences [13]. The E-value of 10−5 is set as the default value. An internally developed reciprocal best fit method, based on BLAST, was also developed for result comparison.
(5) Exclusion of sequences present in nonpathogenic strains. OrthoMCL is used to calculate the homology between predicted sequences and all proteins in a specified non-pathogenic strain genome(s) [13].
(6) Comparison of sequence similarity between predicted proteins and host (human and/or mouse) proteome. OrthoMCL is customized for this purpose.
(7) Prediction of MHC class I- and class II-binding epitopes. Vaxign uses an internally developed program Vaxitope to predict MHC class I and class II binding epitopes. Vaxitope is developed based on PSSM (Position Specific Scoring Matrix) motif prediction. The PSSMs for the prediction of peptide binders to MHC class I or II are calculated based on a position-based weighting method using the BLK2PSSM utility included in the BLIMPS package [14]. Data for generating the PSSMs came from known epitope data from the IEDB immune epitope database [27]. The P value for the predicted epitope binding to PSSMs is calculated by the MAST sequence homology search algorithm [39]. A receiver operating characteristic (ROC) curve and the values of the area under the ROC Curve (AUC) were used to calculate the accuracy of the Vaxitope prediction [40]. For the AUC analysis, the epitope data from the IEDB immune epitope database [27] were used. A leave-one-out approach was applied to test if a known epitope can be predicted on the condition that this epitope is excluded in initial generation of PSSMs.
(8) Protein functional analysis: Predicted proteins can be selected and automatically exported to the DAVID bioinformatics resources [41] for functional protein analysis.
2.2. Vaxign Server and Web Implementation
Vaxign is implemented using a three-tier architecture built on two Dell Poweredge 2580 servers which run the Redhat Linux operating system (Redhat Enterprise Linux ES 5). Users can submit database or analysis queries through the web. These queries are then processed using PHP/HTML/SQL (middle-tier, application server based on Apache) against a MySQL (version 5.0) relational database (back-end, database server), or executed in runtime based on the Vaxign algorithm pipeline. The result of each query is then presented to the user in the web browser (Figure 1). Two servers are scheduled to regularly backup each others' data.
2.3. Application of Vaxign in Prediction of UPEC Vaccine Targets
To predict vaccine targets against uropathogenic E. coli (UPEC) using Vaxign, four UPEC strains with fully sequenced genomes were used: strains CFT073 (RefSeq ID: NC_004431), 536 (NC_008253), UTI89 (NC_007946), and F11 (NZ_AAJU00000000). Microbial genomes and protein sequences were downloaded from NCBI RefSeq genome database [42]. To determine whether predicted antigens exist in UPEC strains but not in non-pathogenic E. coli, the non-pathogenic E. coli K-12 strain MG1655 (RefSeq ID: NC_000913) [43] was used as a control genome.
2.4. Comparison of Different Methods in UPEC Vaccine Target Prediction
The results of UPEC vaccine targets predicted by Vaxign were manually compared with results from our previous studies using microarray [34], proteomics [35], immunoproteomic analysis [36].
2.5. Verification of UPEC Vaccine Targets Predicted by Vaxign
To experimentally verify the predicted data, UPEC proteins were prepared using recombinant cloning technology. For active immunization, CBA/J mice (N = 10 for each group) were intranasally immunized with individual proteins combined with cholera toxin. As negative control, cholera toxin alone was also used to vaccinate mice. The vaccinated group were boosted at 7 and 14 days. One week after the final boost, control (naïve: Ctx-treated) and vaccinated mice were transurethrally challenged with 5 × 108 CFU E. coli CFT073. After a one-week, the efficacy of protection by individual subunit vaccines was evaluated by measuring the CFU/ml urine and CFU/g bladder or kidney tissue. The vaccine challenge experiments were reported in a recent publication [33].
3. Results
3.1. The Vaxign Algorithm for Vaccine Target Prediction
The workflow of the Vaxign pipeline is shown in Figure 1. The predicted features in Vaxign include protein subcellular location, transmembrane helices, adhesin probability, sequence conservation among pathogen genomes, and sequence similarity to host (human and mouse) proteomes. For those pathogens against which a strong B cell response (for antibody production) is critical, surface-exposed proteins such as secreted proteins and outer membrane proteins (especially adhesins) are ideal targets for vaccine development. For these pathogens, nonsurface proteins such as cytoplasmic or inner membrane proteins, however, may not represent good targets for vaccine development due to lack of close contact with the host cells [1, 2]. However, for the vaccine development against those pathogens where T cell response is critical, subcellular localization is not an issue since a T cell response could be directed to any protein target. It has been reported that 250 out of 600 vaccine candidates from N. meningitidis B failed to be cloned and expressed due to the presence of more than one transmembrane spanning region [2]. Therefore, it might also be prudent to ignore those proteins with multiple transmembrane spanning regions in the first place. The adherence of microbial pathogens to host cells is mediated by adhesins. Adhesins are essential for bacterial colonization and survival and represent possible targets for vaccine development. The conserved vaccine targets among different strains in one pathogen offer protection against these different strains. A vaccine candidate with similar sequence to the host (e.g., human or mouse) is likely to be a poor immunogen due to epitope mimicry, or if an immune response is triggered, cause autoimmunity in the host [44–46]. These aspects are considered in the Vaxign prediction pipeline (Figure 1).
During the past decades, many algorithms and software programs have been developed to address individual processes in the Vaxign vaccine design pipeline. Many software programs have been widely tested and validated. To avoid reinventing the wheel, we have incorporated many existing software programs into Vaxign as described in the Section 2. All open source programs (e.g., BLAST) have been customized. The Vaxitope (vaccine epitope prediction) is a new program that is internally developed and will be described later in this paper in more detail. One focus of the Vaxign development was to seamlessly incorporate different programs with different development styles and even program languages into a comprehensive analysis system. To achieve this goal, MySQL relational database was used to replace plain text input files typically used in original programs. In a typical scenario, output data of one program is stored in MySQL, and SQL query scripts are used to retrieve and process the data as input for another program. Each component program except Vaxitope in the Vaxign pipeline has individually been tested and validated in the literature [10–13]. The testing of Vaxitope is described below.
The Vaxign database contains precomputed prediction results using 76 genomes from 13 pathogens (Table 1). In total, 191,192 proteins have been precomputed. These data can be queried using the Vaxign web interface. A user can also input protein sequence data for dynamic computation and result output.
Table 1.
Pathogens currently analyzed by Vaxign.
| species | # of Genomes | # of proteins |
|---|---|---|
| Bacterial species: | ||
| (1) Bacillus anthracis | 3 | 16182 |
| (2) Brucella | 9 | 28559 |
| (3) Campylobacter | 10 | 17443 |
| (4) Clostridium | 10 | 35130 |
| (5) Corynebacterium diphtheriae | 1 | 2272 |
| (6) Coxiella burnetii | 5 | 9686 |
| (7) Escherichia coli | 7 | 34592 |
| (8) Francisella | 9 | 14771 |
| (9) Haemophilus influenzae | 4 | 6735 |
| (10) Helicobacter pylori | 6 | 9261 |
| (11) Mycobacterium tuberculosis | 2 | 8178 |
| (12) Neisseria meningitidis | 4 | 7909 |
| Virus strains: | ||
| (13) HIV | 2 | 33 |
| (14) Measles virus | 1 | 7 |
| (15) Vaccinia virus | 1 | 223 |
| (16) Variola virus | 1 | 197 |
| (17) Yellow fever virus | 1 | 14 |
| Total # | 76 | 191192 |
3.2. Vaxitope: Prediction of MHC Class I and Class II Binding Epitopes
Vaxign predicts both MHC class I and class II binding epitopes using an internally developed tool Vaxitope. Vaxitope is based on Position Specific Scoring Matrix (PSSM), a type of scoring matrix used in protein similarity searches in which amino acid substitution scores are given separately for each position in a protein multiple sequence alignment. In PSSM, a Tyr-Trp substitution at position A of an alignment may receive a very different score than the same substitution at position B. In contrast, in position-independent matrices such as the PAM and BLOSUM matrices, the Tyr-Trp substitution receives the same score no matter at what position it occurs. The general strategy of using PSSMs for prediction of MHC Class I and II binding has proven effective in RANKPEP [15].
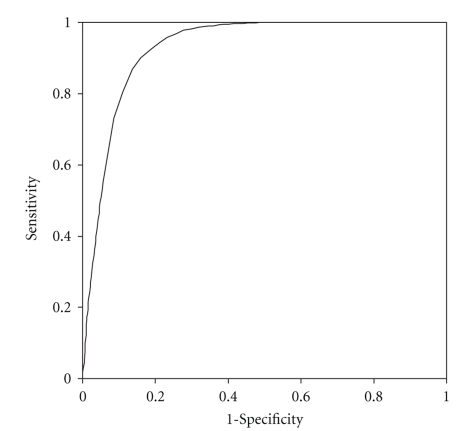
To evaluate the performance of Vaxitope, a receiver operating characteristic (ROC) curve analysis was generated for prediction of epitopes against 40 MHC class I or II alleles (Table 2). The ROC analysis detects the ability of predictions to classify each predicted epitope peptide into MHC class I or II binding based on its comparison with existing epitope database [40]. Plotting the rates of true-positive predictions (sensitivity) as a function of the rate of false-positive predictions (1-specificity) gives an ROC curve. For example, a ROC curve based on Vaxign analysis was generated using HLA A*0201 specific PSSM (Figure 2). HLA A*0201 is one of the most studied HLA MHC Class I allele. According to the IEDB immune epitope database [27], 3216 epitopes are known to positively bind to this allele (as positive testing dataset), and 4826 epitopes cannot bind to this allele (as negative testing dataset). The positive HLA A*0201 alleles were used to calculate the True Positive Rate (Sensitivity). The negative alleles were used to calculate the False Positive Rate (1-Specificity) (Figure 2). The areas under the ROC curve (AUC) provide a way to measure prediction quality. An AUC of 0.5 represents random predictions, and an AUC of 1.0 indicates perfect predictions [16]. The value of the Area Under the ROC Curve (AUC) for the HLA A*0201 analysis using Vaxitope is 0.929. Our analysis of 30 alleles indicates that Vaxitope is a very specific and sensitive method for MHC Class I and II binding epitope prediction (Table 2).
Table 2.
Epitope prediction performance by Vaxitope as measured by AUC values.
| # | MHC allele | Length | AUC | Sensitivity (P = .01) | Specificity (P = .01) | Sensitivity (P = .05) | Specificity (P = .05) | Sensitivity (P = .1) | Specificity (P = .1) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | HLA-A*0101 | 9 | 0.929 | .854 | .874 | .99 | .709 | 1 | .621 |
| 2 | HLA-A*0201 | 9 | 0.871 | .298 | .956 | .531 | .876 | .792 | .783 |
| 3 | HLA-A*0201 | 10 | 0.913 | .471 | .957 | .789 | .874 | .901 | .773 |
| 4 | HLA-A*0202 | 9 | 0.869 | .309 | .953 | .658 | .875 | .792 | .774 |
| 5 | HLA-A*0202 | 10 | 0.863 | .333 | .949 | .737 | .808 | .891 | .684 |
| 6 | HLA-A*0203 | 9 | 0.874 | .304 | .956 | .659 | .865 | .828 | .769 |
| 7 | HLA-A*0203 | 10 | 0.867 | .317 | .963 | .691 | .827 | .834 | .712 |
| 8 | HLA-A*0206 | 9 | 0.900 | .387 | .961 | .73 | .867 | .881 | .781 |
| 9 | HLA-A*0206 | 10 | 0.916 | .403 | .957 | .775 | .878 | .922 | .782 |
| 10 | HLA-A*0301 | 9 | 0.887 | .445 | .921 | .855 | .803 | .959 | .69 |
| 11 | HLA-A*0301 | 10 | 0.868 | .505 | .9 | .915 | .706 | .988 | .554 |
| 12 | HLA-A*1101 | 9 | 0.863 | .337 | .946 | .672 | .833 | .859 | .71 |
| 13 | HLA-A*1101 | 10 | 0.879 | .461 | .924 | .9 | .742 | .989 | .627 |
| 14 | HLA-A*2402 | 9 | 0.984 | .727 | .985 | .97 | .879 | 1 | .783 |
| 15 | HLA-A*3101 | 9 | 0.912 | .426 | .952 | .813 | .872 | .927 | .752 |
| 16 | HLA-A*3101 | 10 | 0.855 | .419 | .905 | .889 | .711 | .99 | .563 |
| 17 | HLA-A*3301 | 9 | 0.937 | .495 | .959 | .94 | .851 | .989 | .723 |
| 18 | HLA-A*3301 | 10 | 0.905 | .51 | .942 | .905 | .755 | .966 | .619 |
| 19 | HLA-A*6801 | 9 | 0.908 | .406 | .949 | .841 | .841 | .946 | .744 |
| 20 | HLA-A*6801 | 10 | 0.848 | .418 | .9 | .866 | .701 | .973 | .564 |
| 21 | HLA-A*6802 | 9 | 0.918 | .446 | .96 | .801 | .868 | .922 | .757 |
| 22 | HLA-A*6802 | 10 | 0.913 | .452 | .947 | .837 | .825 | .977 | .715 |
| 23 | HLA-A*6901 | 9 | 0.803 | .279 | .895 | .674 | .785 | .837 | .674 |
| 24 | HLA-B*0702 | 9 | 0.963 | .659 | .966 | .962 | .894 | .981 | .849 |
| 25 | HLA-B*1501 | 9 | 0.873 | .514 | .927 | .816 | .765 | .939 | .603 |
| 26 | HLA-B*3501 | 9 | 0.838 | .403 | .927 | .701 | .775 | .834 | .666 |
| 27 | HLA-B*5101 | 9 | 0.978 | .835 | .953 | 1 | .871 | 1 | .824 |
| 28 | HLA-B*5301 | 9 | 0.989 | .84 | .981 | .991 | .896 | 1 | .825 |
| 29 | HLA-B*5801 | 9 | 0.923 | .769 | .933 | .894 | .813 | .952 | .702 |
| 30 | H-2-Kb | 8 | 0.936 | .753 | .922 | .935 | .744 | .987 | .623 |
| 31 | H-2-IAd | —* | 0.928 | .582 | .992 | .705 | .959 | .82 | .926 |
| 32 | H-2-IEd | — | 0.977 | .903 | 1 | .935 | .935 | .935 | .887 |
| 33 | H-2-IEg7 | — | 0.998 | .993 | .989 | .993 | .945 | 1 | .893 |
| 34 | H-2-IEk | — | 0.940 | .775 | .95 | .875 | .9 | .9 | .813 |
| 35 | HLA-DPB1*0401 | — | 0.950 | .717 | .978 | .826 | .924 | .913 | .87 |
| 36 | HLA-DPB1*0901 | — | 0.978 | .739 | .989 | .891 | .913 | .913 | .859 |
| 37 | HLA-DR1 | — | 0.923 | .587 | .972 | .781 | .915 | .838 | .834 |
| 38 | HLA-DR7 | — | 0.990 | .976 | .988 | .976 | .905 | .976 | .845 |
| 39 | HLA-DRB1*1101 | — | 0.952 | .732 | .978 | .828 | .914 | .898 | .831 |
| 40 | HLA-DRB1*1501 | — | 0.951 | .846 | .981 | .897 | .942 | .91 | .872 |
Note: * means flexible length.
Figure 2.
ROC curve analysis of epitopes binding HLA A*0201.
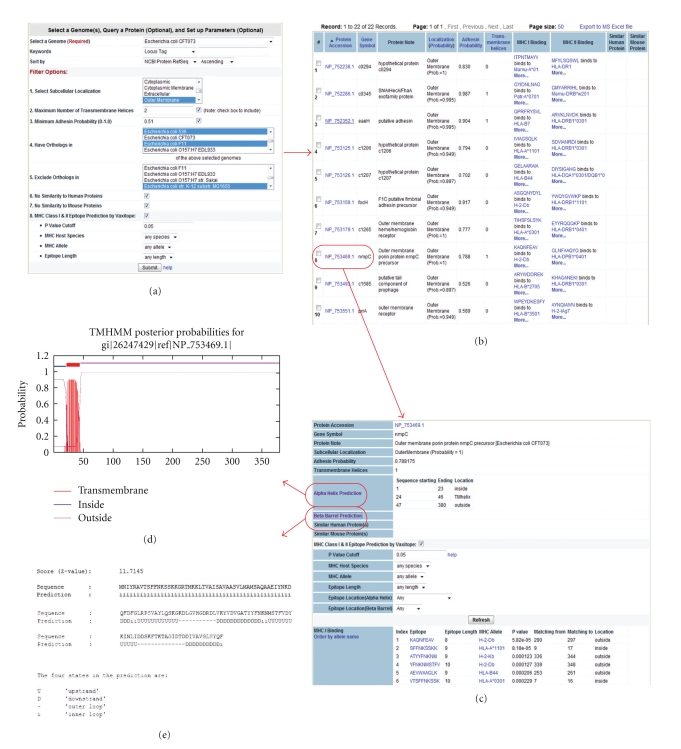
It is interesting to compare Vaxign and RANKPEP since both methods are based on PSSM. If only AUC values are taken into account, our prediction results are in general better than the results predicted by RANKPEP [15]. However, the results may not be comparable, since the data required to generate PSSMs might be different. Different from RANKPEP, which uses a percentage or top number as the cut off as shown in RANKPEP [15], Vaxitope defines statistical P-values based on a random sequence model that assumes each position in a random sequence is generated according to the average letter frequencies of all sequences in the NCBI peptide non-redundant database [39]. Our studies indicate that the P value of .05 provides a cutoff with high and balanced sensitivity and specificity (Table 2). Another unique feature in Vaxitope is that it integrates with other vaccine design components in Vaxign. For example, the input sequence of Vaxitope may come from those peptides that are part of an outer membrane protein and exposed outside the bacterial membrane (Figure 3). These protein peptides are predicted by Vaxign and easily available as input data for Vaxitope. Vaxitope also allows genome-wide query on different MHC host species.
Figure 3.
Vaxign prediction of UPEC vaccine targets. (a) Selection of E. coli CFT073 as a seed genome. Different filter options are applied: only outer membrane proteins, maximum of one transmembrane helix, minimum adhesin probability of.51, having orthologs in other three UPEC genomes but no orthologs in K-12 strain M1655, and no similarity to any human or mouse proteins. (b) In total 22 proteins were predicted to meet selected criteria. (c) Detailed results for a particular protein, for example, NmpC. MHC Class I and II epitope prediction results are also available and can be further filtered. (d) Prediction of α-helices and their possible cellular locations of NmpC. (e) Prediction of β-barrels and their possible cellular locations of NmpC.
Traditional reverse vaccinology does not consider prediction of epitopes. With the P value cut off of .05, 1436 epitopes from E. coli protein Hma for 39 MHC Class I alleles in 4 hosts and 515 epitopes for 23 MHC Class II alleles have been found in 4 hosts—human, mouse, macaque, and chimpanzee. It remains a challenge to rank and optimize the epitopes for vaccine development. Possible solutions to address this challenge are described in the Discussion.
3.3. User-Friendly Vaxign Web Interface
To make Vaxign easy to use, two methods of implementation have been developed. Users can either directly query precomputed prediction results from the Vaxign database, or request Vaxign to dynamically calculate results based on the users' input sequences. The prediction data from the precomputed Vaxign database can be easily queried using our Vaxign web query interface (Figure 3).
A simple web query interface is available for querying the precomputed Vaxign results from the protein level or genome level (Figure 3). Users are prompted to set up preferred query criteria; the output data are then provided. The query of precomputed Vaxign results is fast. A typical query involved in four genomes and all the steps as shown in our UPEC use case (Figure 3) takes approximately 2–5 seconds.
The other form is dynamic Vaxign analysis, which is similar to the precomputed Vaxign except that a user is prompted to provide information for up to 300 proteins at one time. The protein information may be protein sequences using FASTA format, NCBI protein GI, or RefSeq accession number. Vaxign predicts vaccine targets based on runtime execution. It typically takes 30–60 seconds to execute all the steps in run time for one single protein. Therefore, it would take 150–300 minutes to finish analysis of 300 proteins. Once all steps are finished, the web link of the predicted results will be sent to a registered user through email.
3.4. Vaxign Predicts 22 Outer Membrane Proteins as UPEC Vaccine Targets
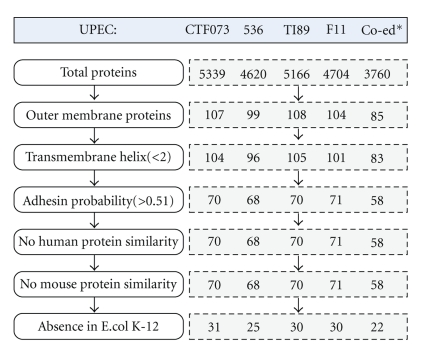
The genomes of all four UPEC strains (CFT073, 536, UT189, and F11) for which complete sequence data are available were analyzed by Vaxign (Figure 4). These four genomes contain 4704–5379 genes. Only outer membrane proteins (OMP) are predicted and analyzed. From the total 5379 proteins in UPEC strain CFT073, Vaxign detects 107 outer membrane proteins. Among the 107 proteins, three proteins contain more than one transmembrane helix. Vaxign further predicts 70 proteins from the 107 OMPs in strain CFT073 as possible adhesins or adhesin-like proteins [34]. These predicted adhesins are likely critical for colonization, a major challenge facing UPEC in the urinary tract. While some of these proteins, such as PapC [47], are adhesins, many of these 70 proteins (e.g., Hma, FepA) predicted to be adhesins are not typically considered as adhesins. The roles of these adhesin-like proteins in adhering to host cells require further investigation. None of these 70 proteins shows sequence similarity to any human or mouse proteins. Similar strategy was applied to obtain vaccine targets for the other three UPEC strains (Figure 4).
Figure 4.
Prediction of UPEC vaccine targets conserved in four sequenced UPEC genomes using Vaxign. Note: * Co-ed represents the conserved proteins.
Ortholog analysis was then applied to obtain conserved vaccine targets from four UPEC strains. In total, 85 OMPs were found to be conserved across all four pathogenic UPEC strains (Figure 4). Among these 85 OMPs, two proteins (NP_755264.1, NP_756232.1) are predicted to contain three transmembrane helixes. Multiple transmembrane helixes make it difficult to purify recombinant proteins [48]. Therefore, these two proteins may not be good vaccine targets as whole protein antigens. When adhesin probability is taken into account, 58 out of the 83 proteins have an adhesin probability of ≥.051.
Functional gene enrichment analysis was performed to classify the roles of these 58 OMPs using the software DAVID (Table 3). Only 48 genes have annotation in DAVID and thus included in the DAVID analysis. Among these 48 genes, significantly enriched function annotations are in the areas of transport activities, TonB-dependent receptor (beta-barrel), Gram-negative porin, iron ion transmembrane transporter activity, and fimbrial biogenesis in outer membrane (Table 3).
Table 3.
Selected function annotations significantly enriched for UPEC vaccine candidates based on DAVID analysis.
| Category | Term | # of Genes | % | P-value | Benjamini P-value |
|---|---|---|---|---|---|
| GO MF* | Transport activity | 22 | 45.8 | 1.8E−14 | 2.7 E−11 |
| Interpro | TonB-dependent receptor, beta-barrel | 10 | 20.8 | 1.0 E−13 | 4.3 E−10 |
| Interpro | Porin, Gram-negative type | 8 | 16.7 | 1.3 E−12 | 1.8 E−9 |
| GO MF | Iron ion transmembrane transporter activity | 5 | 10.4 | 6.9 E−6 | 1.4 E−3 |
| Interpro | Fimbrial biogenesis outer membrane | 5 | 10.4 | 9.3 E−5 | 4.1 E−2 |
*: GO MF, the Molecular Function (MF) branch of the Gene Ontology (GO).
Of these 58 outer membrane proteins identified by Vaxign, 36 were further found to be present in the non-pathogenic E. coli K-12 strain MG1655 [43]. K-12 is used to remove those proteins that have been exposed to the host environment (e.g., gut) and may be tolerant by the host [49]. Only 22 proteins have been identified to be unique to the pathogenic UPEC strains (Figure 4).
A table of genes in different categories were further generated based on the Figure 4 and Table 3 and manual curation of literature data (Table 4). Eight E. coli proteins are predicted to contain iron-binding and iron siderophore transporter activity. Ten proteins are associated with a TonB box [50], and thus may play a role in iron acquisition by the bacterium. Another eight proteins are fimbrial biogenesis outer membrane usher proteins. Nine proteins are related to porin and ion transport. Indeed, many proteins in the list participate in transporter activity. Many lipoproteins have also been found. All of these targets would be logical selections. Many hypothetic proteins have been found with no defined functions or annotations.
Table 4.
Conserved UPEC outer membrane proteins predicted by Vaxign.
| # | Protein RefSeq | Symbol | Adhesin | TMH | Not in K-12 | Micro-array | Prote-omics | Protein Name |
|---|---|---|---|---|---|---|---|---|
| Iron ion binding and iron/siderophore transporter activity | ||||||||
| 1 | NP_752820.1 | ybiL | 0.857 | 1 | − | − | Catecholate siderophore receptor fiu precursor (TonB-dependent receptor fiu) (Ferric iron uptake protein) | |
| 2 | NP_754328.1 | c2436 | 0.473 | 0 | − | − | putative pesticin receptor precursor (tonB-dependent receptor) | |
| 3 | NP_754406.1 | c2518 | 0.83 | 0 | X | − | − | TonB dependent receptor |
| 4 | NP_752238 | c0294 | 0.83 | 0 | X | − | − | TonB dependent receptor |
| 5 | NP_753164.1 | iroN | 0.672 | 0 | + | + | Siderophore receptor iron | |
| 6 | NP_752600.1 | fepA | 0.792 | 0 | + | − | ferrienterobactin receptor (TonB-dependent receptor) | |
| 7 | NP_752135.1 | fhuA | 0.746 | 0 | + | + | ferrichrome outer membrane transporter | |
| 8 | NP_754374.1 | Hma (c2482) | 0.772 | 0 | X | + | + | outer membrane heme acquisition protein |
| 9 | NP_756170 | chuA | 0.846 | 0 | X | + | + | Outer membrane heme/hemoglobin receptor |
| 10 | NP_753551.1 | prrA | 0.589 | 0 | X | − | − | Putative TonB-dependent outer membrane receptor |
| 11 | NP_753179 | c1265 | 0.777 | 0 | X | − | − | Outer membrane heme/ hemoglobin receptor |
| 12 | NP_753125 | c1206 | 0.794 | 0 | X | − | − | Outer membrane heme/haemoglobin receptor |
| 13 | NP_755646 | c3775 | 0.79 | 0 | X | − | − | putative iron compound receptor |
| 14 | NP_753820.1 | yddB | 0.765 | 0 | − | − | hypothetical protein c1924 (tonB-dependent receptor family) | |
| Fimbrial biogenesis outer membrane usher protein | ||||||||
| 15 | NP_757244.1 | fimD | 0.744 | 1 | + | − | Outer membrane usher protein fimD precursor | |
| 16 | NP_757034 | papC_2 | 0.674 | 1 | X | − | − | PapC protein |
| 17 | NP_755465 | papC | 0.666 | 0 | X | − | − | PapC protein |
| 18 | NP_754524.1 | yehB | 0 | 0 | − | − | Outer membrane usher protein yehB precursor | |
| 19 | NP_752120.1 | htrE | 0.643 | 0 | − | − | Putative outer membrane usher protein | |
| 20 | NP_753830.1 | c1934 | 0.856 | 1 | − | − | Outer membrane usher protein fimD precursor | |
| 21 | NP_754765.1 | yfcU | 0.563 | 0 | X | − | − | Fimbrial export usher family protein |
| 22 | NP_753156.1 | focD | 0.854 | 0 | − | − | F1C fimbrial usher | |
| 23 | NP_756076.1 | ycbS | 0.559 | 1 | − | − | Outer membrane usher protein ycbS precursor | |
| 24 | NP_753159 | focH | 0.917 | 0 | X | − | − | F1C putative fimbrial adhesin precursor |
| Porin and ion transport | ||||||||
| 25 | NP_753469.1 | nmpC | 0.788 | 1 | X | − | + | Outer membrane porin protein nmpC precursor |
| 26 | NP_754644.1 | ompC | 0.688 | 0 | − | + | Outer membrane porin protein C | |
| 27 | NP_756858.1 | lamB | 0.806 | 0 | − | + | maltoporin | |
| 28 | NP_752996.1 | ompF | 0.614 | 0 | − | + | Outer membrane protein F (Porin family) | |
| 29 | NP_754240.1 | c2348 | 0.759 | 0 | X | − | − | Outer membrane porin protein nmpC precursor |
| 30 | NP_752325.1 | phoE | 0.729 | 0 | − | − | Outer membrane phosphoporin protein E | |
| 31 | NP_753724.1 | ompN | 0.751 | 0 | − | − | Outer membrane protein N precursor | |
| 32 | NP_754275.1 | c2383 | 0.597 | 0 | X | − | − | Outer membrane protein N precursor |
| 33 | NP_754771.1 | fadL | 0.871 | 0 | − | + | Long-chain fatty acid outer membrane transporter | |
| 34 | NP_756025.1 | hofQ | 0.186 | 0 | + | − | Outer membrane porin HofQ | |
| Other transport proteins | ||||||||
| 35 | NP_756748.1 | c4894 | 0.81 | 0 | X | − | − | Nucleoside-specific channel-forming protein tsx precursor |
| 36 | NP_756500.1 | c4642 | 0.694 | 0 | − | − | Putative outer membrane Protein yieC | |
| 37 | NP_753669.1 | c1765 | 0.437 | 0 | − | − | Partial putative outer membrane channel protein | |
| 38 | NP_752455.1 | tsx | 0.833 | 0 | − | + | Nucleoside-specific channel-forming protein tsx precursor | |
| Lipoproteins | ||||||||
| 39 | NP_756849.1 | yjbH | 0.651 | 0 | − | − | Lipoprotein yjbH precursor | |
| 40 | NP_755008.1 | yfiB | 0.564 | 0 | − | − | Putative outer membrane lipoprotein | |
| 41 | NP_756936.1 | yjcP | 0.185 | 0 | − | − | Putative outer membrane efflux protein MdtP | |
| 42 | NP_752589.1 | cusC | 0.516 | 0 | − | − | Copper/silver efflux system outer membrane protein CusC | |
| 43 | NP_754925.1 | yfhM | 0.224 | 0 | − | − | Lipoprotein yfhM precursor | |
| 44 | NP_756232.1 | yiaD | 0.526 | 3 | + | − | Putative outer membrane lipoprotein | |
| Other outer membrane proteins | ||||||||
| 45 | NP_752286.1 | c0345 | 0.987 | 1 | X | − | − | ShlA/HecA/FhaA exofamily protein |
| 46 | NP_752352 | eaeH | 0.904 | 1 | X | − | − | Putative adhesin |
| 47 | NP_753126 | c1207 | 0.702 | 0 | X | − | − | Hypothetical protein |
| 48 | NP_753493 | c1585 | 0.526 | 0 | X | − | − | Putative tail component of prophage |
| 49 | NP_754912 | c3030 | 0.71 | 1 | X | − | − | SinI-like protein |
| 50 | NP_756286 | c4424 | 0.99 | 0 | X | − | − | Putative adhesin |
| 51 | NP_752116.1 | yadC | 0.81 | 1 | − | − | Putative fimbrial-like adhesin protein | |
| 52 | NP_752162.1 | yaeT | 0.637 | 0 | + | + | Outer membrane protein assembly factor YaeT | |
| 53 | NP_752163.1 | hlpA | 0.587 | 0 | − | − | Periplasmic chaperone | |
| 54 | NP_752339.1 | yagX | 0.571 | 0 | − | − | Hypothetical protein c0402 | |
| 55 | NP_752642.1 | crcA | 0.739 | 1 | − | − | Palmitoyl transferase for Lipid A | |
| 56 | NP_752830.1 | ompX | 0.818 | 1 | + | + | Outer membrane protein X | |
| 57 | NP_753262.1 | flgK | 0.84 | 0 | − | − | Flagellar hook-associated protein FlgK | |
| 58 | NP_753627.1 | ompW | 0.848 | 0 | − | + | Outer membrane protein W | |
| 59 | NP_753695.1 | ompG | 0.618 | 0 | − | − | Outer membrane protein G precursor | |
| 60 | NP_754014.1 | ydiY | 0.708 | 0 | − | − | Hypothetical protein c2120 | |
| 61 | NP_754081.1 | yeaF | 0.852 | 0 | + | + | MltA-interacting protein precursor | |
| 62 | NP_754523.1 | yehA | 0.847 | 0 | − | − | Hypothetical protein c2635 | |
| 63 | NP_754661.1 | yfaL | 0.949 | 0 | − | − | Adhesin | |
| 64 | NP_755530.1 | c3655 | 0.962 | 0 | − | + | Antigen 43 precursor | |
| 65 | NP_756601.1 | pldA | 0.756 | 0 | − | − | Phospholipase A | |
| 66 | NP_754578.1 | cirA | 0.51 | 0 | − | − | Colicin I receptor | |
| 67 | NP_755652.2 | tolC | 0 | 0 | − | + | Outer membrane channel protein | |
| 68 | NP_752583.1 | ompT | 0 | 0 | + | + | Outer membrane protease | |
| 69 | NP_756783.1 | btuB | 0 | 0 | − | + | Vitamin B12/cobalamin outer membrane transporter | |
| 70 | NP_754246.1 | fliF | 0.213 | 2 | − | − | Flagellar MS-ring protein | |
| 71 | NP_755960.1 | yheF | 0.239 | 0 | − | − | General secretion pathway protein D precursor | |
| 72 | NP_752585.1 | nfrA | 0.298 | 0 | − | − | Bacteriophage N4 receptor, outer membrane subunit | |
| 73 | NP_753902.1 | uidC | 0.304 | 1 | − | − | Putative outer membrane porin protein | |
| 74 | NP_754656.1 | c2770 | 0.348 | 0 | − | − | Hypothetical protein c2770 | |
| 75 | NP_757165.1 | ytfM | 0.4 | 0 | − | − | Hypothetical protein c5318 | |
| 76 | NP_753730.1 | ydbH | 0.401 | 1 | − | − | Hypothetical protein c1828 | |
| 77 | NP_756204.1 | yhjL | 0.408 | 0 | − | − | Cellulose synthase subunit BcsC | |
| 78 | NP_751977.1 | c0021 | 0.418 | 1 | − | − | Hypothetical protein c0021 | |
| 79 | NP_757166.1 | ytfN | 0.426 | 1 | − | − | Hypothetical protein c5319 | |
| 80 | NP_754559.2 | yohG | 0.427 | 0 | + | − | Multidrug resistance outer membrane protein MdtQ | |
| 81 | NP_756598.1 | c4739 | 0.43 | 0 | − | − | Hypothetical protein c4739 | |
| 82 | NP_753585.1 | ychP | 0.435 | 0 | − | − | Hypothetical protein c1680 | |
| 83 | NP_754913.1 | c3031 | 0.448 | 0 | − | − | SinH-like protein | |
| 84 | NP_752491.1 | ybaU | 0.464 | 1 | − | − | Peptidyl-prolyl cis-trans isomerase (rotamase D) | |
| 85 | NP_755264.1 | c3389 | 0.282 | 3 | X | − | − | Hypothetical protein |
Note: The protein RefSeq numbers are from UPEC strain CFT073. All 85 proteins are conserved across total four UPEC genomes. This table also shows the adhesin probability, number of transmembrane helixes, and absence in nonpathogenic K-12 MG1655. TMH, transmembrane alpha helix prediction. Microarray, transcriptomic mRNA results (+ for up-regulation in vivo). Proteomics, protein expression results (+ for up-regulation in urine or in vivo).
3.5. Comparison of Vaxign Prediction Results and other Methods
The predicted results based on DNA sequence analysis are compared with data from transcriptomic microarray data [34], mass spectrometry proteomic studies [35, 51], and antigenicity analysis [36]. Out of 85 predicted outer membrane proteins that are conserved among four UPEC strains, 23 proteins have been found upregulated in vivo or in urine at the mRNA and/or protein levels (Table 4). It was found that many proteins with upregulated gene expression belong to iron ion binding proteins and porin family. However, only one protein (FimD) from fimbrial biogenesis outer membrane protein family was shown to be upregulated in DNA microarray analysis (Table 4) [34].
Five out of 14 iron binding proteins (IroN, FepA, FhuA, Hma, and ChuA) discovered by Vaxign have been found to be upregulated in vivo or in urine (Table 4) [34–36, 51]. Since iron metabolism is critical for UPEC pathogenesis, these proteins are important vaccine targets. Five proteins from porin family have also been found upregulated in vivo or in urine, including NmpC, OmpC, LamB, OmpF, and FadL (Table 4). Limited study has been performed to investigate the roles of these porin proteins in induction of protective immunity against UPEC infection.
3.6. Verification of Vaxign Predicted Results
Iron binding proteins were chosen for development of UPEC subunit vaccines. These proteins are typically outer membrane β-barrel proteins that function as receptors for iron-containing compounds. This group of proteins were predicted by Vaxign (Table 4) and significantly enriched based on gene enrichment analysis (Table 3). The antigen c2482 (renamed Hma for heme acquisition), a heme-binding protein, was first cloned and purified, and used for in vivo mouse testing. It was found that intranasal immunization with Hma generated an antigen-specific humoral response, antigen-specific production of IL-17 and IFN-γ, and provided significant protection against experimental infection with UPEC strain CFT073 [33].
ChuA was another heme/hemoglobin receptor that was also present in microarray & proteomics studies (Table 4) [34, 35]. Our experimental studies found that recombinant ChuA induced severe sickness in mice. Mice that recovered from the ChuA vaccination were challenged with strain CFT073, but were not protected (data not shown).
IroN has been found to be a protective antigen [49]. However, our study did not find significant protection stimulated by IroN [33]. This protein also exists in E. coli K-12, which may bring a discussion about whether it is needed to use this cutoff.
Two other proteins, IreA (NP_757022.1, c5174) and IutA (NP_755498.1), were also tested based on six independent screens [33]. Both are putative iron-regulated outer membrane virulence proteins. Our studies found that IreA and IutA were able to independently stimulate protective immunity in mouse bladder against challenge with UPEC strain CFT073 [33]. These two proteins were not shown in our final list of vaccine candidates predicted by our Vaxign analysis pipeline because they were filtered out due to their absence in the other three UPEC genomes.
4. Discussion
Vaxign is the first web-based vaccine design software program freely available for the purpose of facilitating reverse vaccinology. Vaxign optimizes the conditions and performance of many public tools and provides new programs in a way optimal for analyzing high throughput data. The seamless integration makes Vaxign a user-friendly environment specific for reverse vaccinology. Our analysis indicates that Vaxign specifically and sensitively predicts known vaccine targets and also provides new vaccine target candidates deserving further wet lab confirmation. Vaxign is expected to become a publically available web-based program for vaccine researchers to efficiently design vaccine targets and develop vaccines using a rationale reverse vaccinology strategy.
To test whether Vaxign is capable of predicting those protective antigens that have been validated based on wet laboratory experiments, we have curated the literature and obtain a list of proteins and used Vaxign to analyze those protective antigens. Vaxign has also been used to predict vaccine targets using other bacteria such as Brucella spp., Neisseria meningitides, and Mycobacterium tuberculosis. Our studies indicated that Vaxign predicted results are consistent with existing reports [37].
We showed in this report that Vaxign can be successfully used for prediction of UPEC vaccine candidates. While UPEC FimH was reported to be a protective antigen [52], it was not included in our list of predicted genes (Table 4). FimH is predicted by Vaxign as an adhesin with an adhesin probability of .96. This prediction is consistent with current knowledge about this protein [52]. Based on an X-ray structure analysis, FimH is folded into two domains of the all-beta class connected by a short extended linker [53]. FimH was not shown in our final predicted list since its subcellular localization was predicted unknown (Probability = .2). If only a high adhesin probability is considered, FimH would be included in our prediction list. This also indicates different Vaxign options selected by a user would change the results. However, we identified another protein in that complex (FimD) (Table 4). Vaxign identified IroN, Hma, and ChuA (Table 4) which were selected as possible protective antigens after lengthy experimental assessment [33]. Our study found that Hma induced protection in mice from transurethral challenge with UPEC. Another independent study indicated that subcutaneous immunization with denatured IroN conferred significant protection against renal, but not bladder, urinary tract infection in a mouse model [54].
While recombinant ChuA induced severe sickness in mice, the immunized mice did not protect against virulent UPEC infection. This sickness was probably due to its Heme-binding activity. The possible release of high levels of inflammatory cytokines and innate immune response might lead to mouse death. It is likely that ChuA contains some immunodominant T cell epitope(s) that activates effector (inflammatory) T cell immunity [46]. In many cases, subdominant epitopes that induce subdominant responses may be important components of an effective immune defence [19]. Immunization with subdominant but optimal epitopes can often induce T cell responses that are more effective than immunodominant epitopes. A more advanced in silico prediction would be able to predict and optimize epitopes for vaccine development.
Our study also indicated that microarray and proteomics gene expression data were complementary to DNA sequence-based analysis in predicting vaccine targets (Table 4). Future directions of further Vaxign development may include addition of other components such as analysis of high throughput transcriptomic (e.g., DNA microarray and superarray) and proteomic data for vaccine target prediction. Predicted vaccine targets can also be analyzed based on gene annotation enrichment to further refine vaccine targets using tools such as DAVID. The gene enrichment results combined with predictions based on DNA sequence analysis as well as mRNA and protein gene expression allowed us to focus on the group of iron binding proteins for experimental testing.
More than 700 microbial genomes have been sequenced and analyzed, which provide a foundation for scientists to develop vaccines using the reverse vaccinology. Reverse vaccinology shortens the period of vaccine target discovery and evaluation to 1-2 years [1]. This new strategy also revolutionizes new vaccine development against pathogens for which the applications of Pasteur's principles have failed.
The use of proteins is a common approach for genetically engineered vaccine development. However, generating epitope vaccines has many advantages and is currently an active research area. To give the most simplified example, if only one epitope of a large protein is protective, using the peptide epitope would allow the delivery of much higher dose of the key epitope during vaccination. Therefore, prediction of a successful epitope would increase efficacy for the vaccine.
Our studies found that Vaxitope is a sensitive and specific program for predicting immune epitopes that provide good candidates for epitope vaccine development. We are in the processing of designing and evaluating epitope-based UPEC vaccines using Vaxign. We will first target to predict epitopes from antigens (e.g., Hma) that have proven able to induce protective immunity.
It often occurs that many epitopes can be predicted from one specific protein. It is often challenging to rank predicted epitopes for vaccine testing. The epitope ranking can also be used to rank proteins. Many programs, such as EpiAssembler by EpiVax [55], allow epitope content ranking. It is known that the best T cell epitopes tend to contain “clusters” of MHC binding motifs, and the clustering is highly correlated with the immunogenicity [46]. Therefore, it is more effective to design a peptide(s) containing clustered epitopes for induction of better immunogenicity in rational vaccine development. Promiscuous epitopes are those MHC ligands or T-cell epitopes that are recognized in the context of more than one MHC molecule and recognized by more than one T-cell clone. Many software programs, such as TEPITOPE [56], enable the computational identification of promiscuous MHC ligands. The prediction of promiscuous epitopes is also an important feature for epitope-based vaccine design.
It is often that a vaccine candidate that is effective in a mouse model is not effective in human. If the epitopes are designed for human use, the mice used for testing the epitope vaccine usually need to be transgenic. Generating HLA transgenic mice is costly and time consuming. It is possible, however, to design epitopes that are effective for both mouse and human. For example, it was reported that an epitope in human immunodeficiency virus 1 reverse transcriptase was recognized by both mouse and human cytotoxic T lymphocytes [57]. Prediction and screening of such epitopes would simplify our testing of human vaccine candidates in the mouse model.
The molecular mimicry or the cross-reactivity between self epitopes and pathogen epitopes has been found a common reason for many pathogen-induced autoimmune diseases [46]. Many pathogens, such as Klebsiella pneumoniae, Proteus mirabilis, human coronavirus, and Lyme disease spirochete Borrelia burgdorferi carry antigens which cross-react with human antigens [44, 46]. For example, the oligopeptide QTDRED is common to both K. pneumoniae and HLA-B27 nitrogenase reductase enzyme. This sequence similarity appears to cause ankylosing spondyltis. Proteus mirabilis hemolysin contains a molecular mimicry sequence ESRRAL that has the same shape and charge distribution as the rheumatoid arthritis susceptibility sequence EQRRAA. Antibody levels against P. mirabilis hemolysin and a synthetic peptide ESRRAL were significantly higher in rheumatoid arthritis patients [44]. To avoid the autoimmunity, it is important to eliminate the epitopes that are conserved. Currently Vaxign provides a genome-wide sequence similarity analysis at protein levels. Many programs, such as Conservatrix [19] and IEDB Sequence Mapping tool (http://tools.immuneepitope.org/esm/esmhelp.jsp?tab=help), have been developed to map epitope sequences. We plan to develop such epitope sequence mapping tool in Vaxign in the future.
Vaxign is part of VIOLIN, a web-based vaccine database and analysis resource [58]. The predicted vaccine targets from Vaxign will also integrate with those manually annotated vaccine data available in VIOLIN. An literature mining program based on the Vaccine Ontology (http://www.violinet.org/vaccineontology) is also being developed to facilitate automated literature data processing and inference for the purpose of retrieving valuable data for rational vaccine design.
Acknowledgments
This paper was supported by a pilot research Grant to YH and HM at the Center for Computational Medicine and Bioinformatics (CCMB) at the University of Michigan Medical School, Michigan, USA, and Public Health Service grants AI43363 and AI081062 from the National Institutes of Health.
References
- 1.Rappuoli R. Reverse vaccinology. Current Opinion in Microbiology. 2000;3(5):445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 2.Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 3.Ariel N, Zvi A, Grosfeld H, et al. Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infection and Immunity. 2002;70(12):6817–6827. doi: 10.1128/IAI.70.12.6817-6827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross BC, Czajkowski L, Hocking D, et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19(30):4135–4142. doi: 10.1016/s0264-410x(01)00173-6. [DOI] [PubMed] [Google Scholar]
- 5.Montigiani S, Falugi F, Scarselli M, et al. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infection and Immunity. 2002;70(1):368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wizemann TM, Heinrichs JH, Adamou JE, et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infection and Immunity. 2001;69(3):1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti DN, Fiske MJ, Fletcher LD, Zagursky RJ. Application of genomics and proteomics for identification of bacterial gene products as potential vaccine candidates. Vaccine. 2000;19(6):601–612. doi: 10.1016/s0264-410x(00)00256-5. [DOI] [PubMed] [Google Scholar]
- 8.Betts JC. Transcriptomics and proteomics: tools for the identification of novel drug targets and vaccine candidates for tuberculosis. IUBMB Life. 2002;53(4-5):239–242. doi: 10.1080/15216540212651. [DOI] [PubMed] [Google Scholar]
- 9.De Groot AS. Immunomics: discovering new targets for vaccines and therapeutics. Drug Discovery Today. 2006;11(5-6):203–209. doi: 10.1016/S1359-6446(05)03720-7. [DOI] [PubMed] [Google Scholar]
- 10.Gardy JL, Laird MR, Chen F, et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21(5):617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 11.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Research. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachdeva G, Kumar K, Jain P, Ramachandran S. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005;21(4):483–491. doi: 10.1093/bioinformatics/bti028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henikoff S, Henikoff JG, Pietrokovski S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999;15(6):471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- 15.Reche PA, Glutting J-P, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56(6):405–419. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 16.Peters B, Bui HH, Frankild S, et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Computational Biology. 2006;2(6 , article e65) doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rammensee H-G, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 18.Vivona S, Bernante F, Filippini F. NERVE: new enhanced reverse vaccinology environment. BMC Biotechnology. 2006;6, article 35 doi: 10.1186/1472-6750-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Groot AS, Sbai H, Aubin CS, McMurry J, Martin W. Immuno-informatics: mining genomes for vaccine components. Immunology and Cell Biology. 2002;80(3):255–269. doi: 10.1046/j.1440-1711.2002.01092.x. [DOI] [PubMed] [Google Scholar]
- 20.DeLisi C, Berzofsky JA. T-cell antigenic sites tend to be amphipathic structures. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(20):7048–7052. doi: 10.1073/pnas.82.20.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sette A, Buus S, Appella E, et al. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(9):3296–3300. doi: 10.1073/pnas.86.9.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee H-G. Exact prediction of a natural T cell epitope. European Journal of Immunology. 1991;21(11):2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- 23.Sette A, Sidney J, Oseroff C, et al. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. Journal of Immunology. 1993;151(6):3163–3170. [PubMed] [Google Scholar]
- 24.Davenport MP, Shon IAPH, Hill AVS. An empirical method for the prediction of T-cell epitopes. Immunogenetics. 1995;42(5):392–397. doi: 10.1007/BF00179401. [DOI] [PubMed] [Google Scholar]
- 25.De Groot AS, Jesdale BM, Szu E, Schafer JR, Chicz RM, Deocampo G. An interactive web site providing major histocompatibility ligand predictions: application to HIV research. AIDS Research and Human Retroviruses. 1997;13(7):529–531. doi: 10.1089/aid.1997.13.529. [DOI] [PubMed] [Google Scholar]
- 26.Brusic V, Rudy G, Harrison LC. MHCPEP: a database of MHC-binding peptides. Nucleic Acids Research. 1994;22(17):3663–3665. doi: 10.1093/nar/22.17.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters B, Sidney J, Bourne P, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biology. 2005;3(3, article e91):379–381. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litwin MS, Saigal CS, Yano EM, et al. Urologic diseases in America project: analytical methods and principal findings. Journal of Urology. 2005;173(3):933–937. doi: 10.1097/01.ju.0000152365.43125.3b. [DOI] [PubMed] [Google Scholar]
- 29.Connell I, Agace W, Klemm P, Schembri M, Mårild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langermann S, Möllby R, Burlein JE, et al. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. Journal of Infectious Diseases. 2000;181(2):774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]
- 31.Uehling DT, Hopkins WJ, Elkahwaji JE, Schmidt DM, Leverson GE. Phase 2 clinical trial of a vaginal mucosal vaccine for urinary tract infections. Journal of Urology. 2003;170(3):867–869. doi: 10.1097/01.ju.0000075094.54767.6e. [DOI] [PubMed] [Google Scholar]
- 32.Kumar V, Ganguly NK, Joshi K, et al. Protective efficacy and immunogenicity of Escherichia coli K13 diphtheria toxoid conjugate against experimental ascending pyelonephritis. Medical Microbiology and Immunology. 2005;194(4):211–217. doi: 10.1007/s00430-005-0241-x. [DOI] [PubMed] [Google Scholar]
- 33.Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HLT. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathogens. 2009;5(9) doi: 10.1371/journal.ppat.1000586. Article ID e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder JA, Haugen BJ, Buckles EL, et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infection and Immunity. 2004;72(11):6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alteri CJ, Mobley HLT. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infection and Immunity. 2007;75(6):2679–2688. doi: 10.1128/IAI.00076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan EC, Mobley HLT. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infection and Immunity. 2007;75(8):3941–3949. doi: 10.1128/IAI.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang Z, He Y. Vaxign: a web-based vaccine target design program for reverse vaccinology. Procedia in Vaccinology. 2009;1(1):23–29. [Google Scholar]
- 38.Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Research. 2004;32(8):2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14(1):48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 40.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 41.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blattner FR, Plunkett G, III, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 44.Wilson C, Tiwana H, Ebringer A. Molecular mimicry between HLA-DR alleles associated with rheumatoid arthritis and Proteus mirabilis as the aetiological basis for autoimmunity. Microbes and Infection. 2000;2(12):1489–1496. doi: 10.1016/s1286-4579(00)01303-4. [DOI] [PubMed] [Google Scholar]
- 45.Nachamkin I, Allos BM, Ho T. Campylobacter species and Guillain-Barre syndrome. Clinical Microbiology Reviews. 1998;11(3):555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber CA, Mehta PJ, Ardito M, Moise L, Martin B, De Groot AS. T cell epitope: friend or foe? Immunogenicity of biologics in context. Advanced Drug Delivery Reviews. 2009;61(11):965–976. doi: 10.1016/j.addr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naves P, del Prado G, Huelves L, et al. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microbial Pathogenesis. 2008;45(2):86–91. doi: 10.1016/j.micpath.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Serruto D, Rappuoli R, Pizza M. Meningococcus B: from genome to vaccine. In: Grandi G, editor. Genomics, Proteomics and Vaccines. John Wiley & Sons; 2004. pp. 185–201. [Google Scholar]
- 49.Durant L, Metais A, Soulama-Mouze C, Genevard J-M, Nassif X, Escaich S. Identification of candidates for a subunit vaccine against extraintestinal pathogenic Escherichia coli. Infection and Immunity. 2007;75(4):1916–1925. doi: 10.1128/IAI.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infection and Immunity. 2001;69(10):6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters MS, Mobley HLT. Identification of uropathogenic Escherichia coli surface proteins by shotgun proteomics. Journal of Microbiological Methods. 2009;78(2):131–135. doi: 10.1016/j.mimet.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langermann S, Palaszynski S, Barnhart M, et al. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276(5312):607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 53.Choudhury D, Thompson A, Stojanoff V, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285(5430):1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 54.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Olson R, Wilding GE. The Siderophore receptor IroN of extraintestinal pathogenic Escherichia coli is a potential vaccine candidate. Infection and Immunity. 2003;71(12):7164–7169. doi: 10.1128/IAI.71.12.7164-7169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Groot AS, Marcon L, Bishop EA, et al. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005;23(17-18):2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 56.Sturniolo T, Bono E, Ding J, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nature Biotechnology. 1999;17(6):555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 57.Hosmalin A, Clerici M, Houghten R, et al. An epitope in human immunodeficiency virus 1 reverse transcriptase recognized by both mouse and human cytotoxic T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(6):2344–2348. doi: 10.1073/pnas.87.6.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Z, Todd T, Ku KP, et al. VIOLIN: vaccine investigation and online information network. Nucleic Acids Research. 2008;36(1):D923–D928. doi: 10.1093/nar/gkm1039. [DOI] [PMC free article] [PubMed] [Google Scholar]