Abstract
Autophagy is a cellular process that has been defined and analyzed almost entirely by qualitative measures. In no small part, this is attributable to the absence of robust quantitative assays that can easily and reliably permit the progress of key steps in autophagy to be assessed. We have recently developed a cell-based assay that specifically measures proteolytic cleavage of a tripartite sensor protein by the autophagy protease ATG4B. Activation of ATG4B results in release of Gaussia luciferase from cells that can be non-invasively harvested from cellular supernatants. Here, we compare this technique to existing methods and propose that this type of assay will be suitable for genome-wide functional screens and in vivo analysis of autophagy.
Keywords: autophagy, assay, gaussia, luciferase, secretion, screen, in vivo
Background
Autophagy is a stress response that results in activation of a lysosomal degradation pathway. Though primarily viewed as a cellular starvation response, appropriate or inappropriate engagement of autophagy has been identified in diverse contexts important for human health,1,2 such as bacterial or viral infection,3,4 neurodegeneration,5-7 and cancer.8-12 A better understanding of autophagy may be important for the development of new therapeutic strategies to treat disease states arising in these settings.13 Methods to detect autophagy have been extensively reviewed14-16 and guidelines for their use and interpretation have been established.17 In general, autophagy assays are based either on changes in disposition of a specific protein component of the autophagy machinery or on morphological patterns associated with autophagosome formation. The elucidation of the autophagy machinery in yeast and more recently in mammalian cells has enabled the development of highly specific detection systems.18,19 In addition, directed ablation of genes contributing to autophagy in yeast and in mammals has contributed to a better understanding of this complex process. Recently, we have developed a luciferase-based assay that specifically measures the induction of autophagy by monitoring ATG4B proteolytic activity.20 In the following, we will compare this technique to common assays with respect to high-throughput screening and the in vivo analysis of autophagic pathways.
Quantitation of Autophagy
Table 1 summarizes and classifies common methods for detection of autophagy according to their suitability for large-scale screening, quantitation and in vivo applications. Historically, autophagy has been identified by the formation of a highly ordered structure in the cytoplasm, the autophagosome, which is definitively identified by electron microscopy.21,22 Autophagosome membranes are smooth double membranes that engulf cytoplasmic material including mitochondria and ribosomes.23 Quantitation based on electron microscopy is challenging but can be achieved by enumeration of autophagosome numbers or vacuole size.24 Although powerful and rigorous in its application, electron microscopy is widely thought to be most suitable for qualitative analyses.
Table 1.
Overview of cell-based autophagy assays
| Assay | Large scale |
Quantitation | In vivo assaya |
Specificity |
|---|---|---|---|---|
| Electron Microscopy | No | Semic | No | High |
| GFP-LC3 | Yes | Semic | Transgenic Mouse35 |
High |
| LC3 immunoblotting | No | No | No | High |
| Flow Cytometry | Yes | Yes | No | Indirectb |
| Monodansylcadaverine | Yes | Semic | Yes76 | Low |
| Amino acid and Sugar radiolabeling |
No | Yes | No | Indirectb |
| Lactate Dehydrogenase |
No | Yes | No | Indirectb |
| Luciferase Release | Yes | Yes | TBD | ATG4B cleavage |
in higher organisms;
requires the use of inhibitors to ensure specificity for autophagic processes;
quantitation of number or size of autophagosomes possible; (TBD, to be demonstrated).
Staining methods, e.g., the use of monodansylcadaverine (MDC)25,26 or mitotracker red, provide an alternative that can be easily integrated with high-throughput screening approaches. These reagents stain acidified or low electrochemical potential compartments and when combined with selective inhibitor treatments can overcome problems with non-specific background staining.24 An alternative is the visualization of green fluorescent protein (GFP)-tagged components of the autophagy machinery. Microtubule associated protein 1 light chain 3 (LC3) is a marker protein of the autophagosomal membrane that is modified during the induction of autophagy by several processing steps including cleavage by the protease ATG4B to generate LC3-I, ATG3/7-mediated ligation of phosphatidyl-serine or phosphatidyl-ethanolamine to generate LC3-II27 and translocation to the autophagosomal membrane.28 A second reaction catalyzed by ATG4B removes LC3-II from the autophagosomal membrane by delipidation.28 Translocation of GFP-LC3 can be identified by puncta that appear following engagement of the autophagic response and are readily visible by light microscopy.28 Other GFP fusions to candidate marker proteins such as ATG18 and RAB24 have been examined for their suitability as autophagy reporters,29,30 but to date GFP-LC3 is the best characterized. It has been noted that GFP-LC3 can be observed to undergo autophagy-independent or saponin-induced aggregation.31,32 Further refinements such as a tandem fluorescent-tagged LC3 protein to distinguish between autophagy induction and lysosomal fusion33 and deletion mutants of LC3 that are not subject to lipidation have been proposed to reduce background aggregation and enhance specificity.34 The development of GFP-LC3 transgenic mice has been a major breakthrough for our understanding of the in vivo function of autophagy.35
GFP-LC3 fluorescence localization studies can be complemented by immunoblotting of LC3.36 Lipidated LC3 (LC3-II) migrates at a different size by polyacrylamide gel electrophoresis and can be distinguished from unprocessed LC3 (LC3-I).28 Immunoblotting of LC3 allows further discrimination of specific aspects in autophagy such as the determination of autophagic flux.37 An alternative for detection of the autophagic flux is measuring p62 (SQSTM1/sequestosome 1) protein levels.38 Inhibition of autophagy correlates with increased p62 expression,39 but transcriptional upregulation of this gene might also be autophagy-independent.
For high-throughput analysis of autophagy, GFP-LC3 localization is an attractive option. Recently, a siRNA-based screen has been performed using 293 cells stably expressing GFP-LC3 and kinases required for starvation-induced autophagy were identified.40 Another image-based screen identified small molecule compounds that induce autophagy in the glioblastoma H4 cell line.41 One consideration is that morphological assays such as fluorescence puncta formation have a limited potential for quantitation. The number or size of autophagosomes can be determined and quantified using appropriate software,40,42 but these assays do not have a broad dynamic range and are to some degree subjective. A flow cytometry assay has been developed that is based on loss of GFP-LC3 fluorescence during autophagy43 and meets many of the requirements for a practical quantitative assay, but as formulated, the assay requires the use of lysosomal and autophagic inhibitors for correct interpretation of results. In addition cytometric methods typically have relatively limited dynamic range compared to luciferase assays.
Monitoring enzymatic activities associated with autophagy allows better quantitation. For instance, lactate dehydrogenase is a specific indicator of autophasosomal-lysosomal transfer.44 Other assays based on administration of radio-labeled metabolites such as amino acids provide an alternative but are less attractive for large-scale screening for practical reasons as they require injection of radioactive molecules.45,46
The Luciferase Release Assay
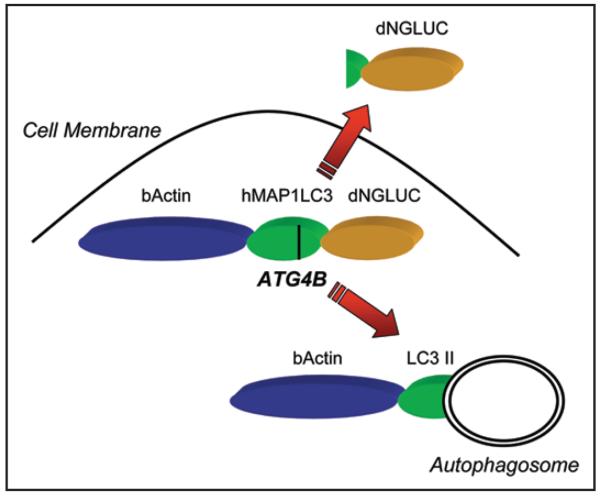
Recently, a simple cell-based autophagy sensor has been described that is highly specific for LC3 cleavage and amenable to large-scale screening approaches.20 This sensor is based on ATG4B-dependent release of Gaussia luciferase (GLUC) from cells. GLUC is a reporter enzyme from the marine copepod Gaussia princeps that is secreted from cells by signal-peptide mediated secretion.47,48 Surprisingly, GLUC lacking the signal peptide rapidly exits the cell by a non-conventional pathway.20 This pathway can be frustrated by anchoring an N-terminally deleted form of GLUC (dNGLUC) to the actin cytoskeleton. If a protease-specific linker is introduced between the actin anchor and dNGLUC, secretion can be made dependent on cleavage of the linker.20 By insertion of the full-length open reading frame of LC3 between β-actin and dNGLUC, LC3 cleavage can be monitored by harvesting the supernatant of cells (Fig. 1). Both ATG4B expression and shRNA mediated knockdown modulate the luciferase levels released from cells and allow the detection of events that control ATG4B activity. Inhibition of mTOR either by treatment with rapamycin or by inhibition of AKT1 results in activation of autophagy49 and can be detected using this reporter.20 This assay system is a potentially useful new tool to elucidate signaling pathways that lead to autophagy.
Figure 1.
The Luciferase Release Assay. Secreted Gaussia Luciferase (GLUC) can be anchored inside cells by deletion of the signal peptide and fusion to β-actin. We have inserted full-length hMAP1LC3 as linker between β-actin and GLUC. Upon proteolytic cleavage of LC3 by ATG4B, the dNGLUC fragment is released and exported across the cellular membrane. GLUC activity can be non-invasively harvested from cell culture supernatants and directly correlates to cleavage of hMAP1LC3.
The luciferase release assay non-invasively measures protein cleavage over time in the context of the complex physiology of intact living cells and is compatible with high-throughput screening methodologies. Several considerations require additional controls when using this assay. To ensure that cleavage is specific, mutations of the cleavage site can be engineered and tested with the same system. Since β-actin itself is subject to proteolytic processing under certain conditions,50 it is advisable to measure release of luciferase activity from Actin-dNGLUC in parallel. It should be noted that the luciferase release system can detect cleavage of short peptides as well as cleavage of full-length proteins. Therefore, the luciferase release assay can be further modulated by replacing the LC3 gene with a short peptide motif or a different protease target gene such as GABARAP. For instance, calpain-mediated cleavage of ATG5 results in a switch from autophagy to apoptosis51 and thus a calpain-sensitive linker introduced between β-actin and dNGLUC may be useful to study this specific aspect.
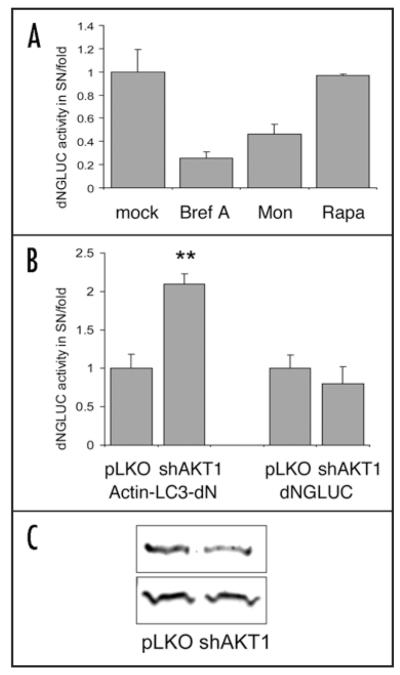
The mechanism of dNGLUC secretion is presently poorly characterized at a molecular level. One hypothetical form of interference with the luciferase assay could take the form of an interaction between elements of the autophagosome and uncharacterized components of the luciferase secretion apparatus. To explore this possibility, cells transfected with dNGLUC-expressing constructs were treated with Rapamycin or transfected with shRNA-expressing constructs mediating knockdown of AKT1, two treatments that should have the effect of potentiating autophagic responses. Such treatments had no effect on the constitutive secretion of dNGLUC, yet resulted in robust activation of the Actin-LC3-dNGLUC reporter (Fig. 2). Secretion of GLUC is blocked by treatment with Brefeldin A,52 even in the absence of a signal peptide (Fig. 2A). Hence, the luciferase release assay may not be suited for the study of reticulophagy (autophagy of the endoplasmic reticulum) that can be induced by inhibition of p5353 or chemical treatments resulting in ER stress.14 A different GLUC-based approach has recently been designed that is better suited for the study of ER stress.52
Figure 2.
Rapamycin or shRNA-mediated knockdown of AKT does not affect secretion of dNGLUC. (A) 293ET cells transfected with dNGLUC were treated for 6 h with 10 μg/ml Brefeldin A (Bref), 7 μM Monensin (Mon) and 200 nM Rapamycin (Rapa) or left untreated and assayed for Gaussia Luciferase activity in SN and cell lysates. The release of dNGLUC activity in SN was normalized to total activity in SN and cell lysates. Shown is the percentage inhibition of secreted dNGLUC activity from three independent transfections. Inhibition of ER/Golgi trafficking by Brefeldin and Monensin reduced dNGLUC secretion, while treatment with Rapamycin had no effect. (B) 293ET cells transfected with shRNA targeting AKT1,20 or the parental pLKO vector plus Actin-LC3-dNGLUC or dNGLUC were cultured for 72 h prior to harvesting of SN and cell lysis. Shown is the fold release of GLUC activity of secreted activity as ratio of total activity. Significances were calculated with a two-sided paired T-Test from three independent transfections (**p < 0.01). shAKT1 resulted in a 2-fold increase in luciferase release compared to vector control while dNGLUC secretion was not affected by shAKT1 knockdown. C, shRNA-mediated knockdown reduces expression of AKT1 but not β-actin. Whole cell lysates were resolved by 10% SDS-PAGE, immunoblotted with antibodies against AKT (Cell Signaling) and β-Actin (Abcam) and detected using the Odyssey Infrared Imaging System (LiCor). For technical details see Reference Ketteler et al.20
Regulation of ATG4B activity
The release of luciferase is highly sensitive over a broad range, robust and directly dependent on the action of ATG4B protease on a peptide linker drawn from the natural substrate, LC3. The constitutive proteolytic processing of intact LC3 is thought to occur shortly after translation, based on evidence that the uncleaved form of LC3 could not be detected in cells.28 ATG4B can also cleave the lipid anchor (phosphatidylserine or phosphatidylethanolamine) from lipidated LC3 (LC3-II) and forced expression of ATG4B has been shown to reduce the amount of this form of the protein, whereas coexpression of ATG4B with LC3 under nutrient depletion conditions reduces the punctate localization pattern of LC3.54 Similarly, knockdown of ATG4B increases the level of lipidated LC3-II in 293 cells.54 In embryonic stem cell lysates, ATG4B expression results in an increase of cleaved LC3-I which was attributed to LC3-II deconjugation.55 In the luciferase release reporter, the fusion of LC3 to β-actin and Gaussia luciferase stabilizes LC3 in a form that can be detected as an uncleaved pro-peptide of 83 kDa.20 When expressed in cells, cleavage by ATG4B results in luciferase release that corresponds to increases or decreases in cellular ATG4B activity. The relative resistance of the chimeric reporter substrate to the action of ATG4B under basal conditions suggests the susceptibility of substrate to ATG4B may more closely resemble that of GABARAP, another ATG4B target, that is readily detectable in the unprocessed form by immunoblotting of cell lysates.55
Recent studies have highlighted a potentially complex control of ATG4B activity by post-translational modifications as well as at the level of transcription. In both humans and rats, ATG4B expression varies widely between different tissues.56,57 ATG4B expression is very high in human heart and skeletal muscle but low in other tissues such as lung and fetal kidney.56 It has been shown that activation of the transcription factor FOXO3A leads to induction of autophagy genes including ATG4B in skeletal muscle.58,59 FOXO3A is inhibited by the serine/threonine kinase AKT.60 Thus, inhibition or ablation of AKT may lead to activation of FOXO3A and upregulation of autophagy genes. In addition, ATG4B levels rapidly increase under conditions of starvation,61-63 but the molecular mechanism for this regulation is not well understood. The rapidity of the increase in ATG4B levels supports the view that ATG4B activity may be a sensitive indicator for the induction of autophagy.
Post-translational modifications that result in changes in ATG4B activity can be assessed by this method. For example, the oxidation of cysteine residues of ATG4B results in reduced deconjugating activity,64 and this mechanism has been proposed as a signaling switch for ATG4B activity.65 In addition, ATG4B is phosphorylated at Ser-34,66 Ser-383 and Ser-39267-69 in vivo suggesting that ATG4B may be a target of kinases and phosphatases regulating its activity. To date, the kinases that use ATG4B as a substrate have not been identified.
Based on enhanced luciferase release upon inhibition of mTOR20 or shRNA-mediated ablation of AKT1 (Fig. 2), we suggest that ATG4B activity is controlled by signal transduction cascades similar to those that induce autophagosome formation. This hypothesis is supported by studies indicating that insulin- and Akt-dependent signaling inhibits autophagy in human myeloblastic cell lines.70 Interestingly, in PTEN deficient cells that have elevated Akt signaling, autophagosome formation has been reported to be strongly inhibited while lipidation of LC3 was not affected,71 indicating that early steps in autophagosome formation can be inhibited by Akt signaling. In contrast, a recent study has shown enhanced autophagic cell death in the setting of increased AKT signaling following siRNA-mediated EGFR knockdown72 that is thought to decrease glucose flux into the cell. Lysosomal clearance of huntingtin aggregates by autophagy has been shown in response to insulin stimulation and activation of AKT73 indicating that AKT has opposing effects on early and late stages of autophagy.
The luciferase release assay may be well-suited for the study of signaling cascades that activate autophagy as the proteolytic activation of LC3 is thought to measure an early event of autophagy (Fig. 3). The crystal structure of ATG4B shows that pro-LC3 and LC3-II are processed by the same molecular mechanism and in both cases the same LC3 core is recognized as substrate.74,75 Thus, the luciferase release assay likely measures the proteolytic and deconjugating activity of ATG4B (see Fig. 3). Other events such as analysis of autophagosome formation or lysosomal degradation may be decoupled from the early phases of the response and if so, their study will likely require the application of different methods. The appearance of LC3-II can be studied by immunoblotting and punctate localization of GFP-LC3, whereas the degradation of LC3 after autophagosomal-lysosomal fusion can be studied by flow cytometry in combination with lysosomal inhibitor treatments (Fig. 3). The luciferase release assay measures the steady-state level of LC3 cleavage. The release of luciferase activity from cells allows non-invasive harvesting of supernatant at several time points, thus permitting time kinetic measurements. The time-resolved activation of LC3 cleavage in combination with live-cell imaging of GFP-LC3 will greatly improve the determination of autophagic flux. In addition, the assay should facilitate the search for states akin to autophagy that comprise activation of ATG4B or cleavage of LC3 without committing the cell to the canonical pathway of cellular autocatabolism.
Figure 3.
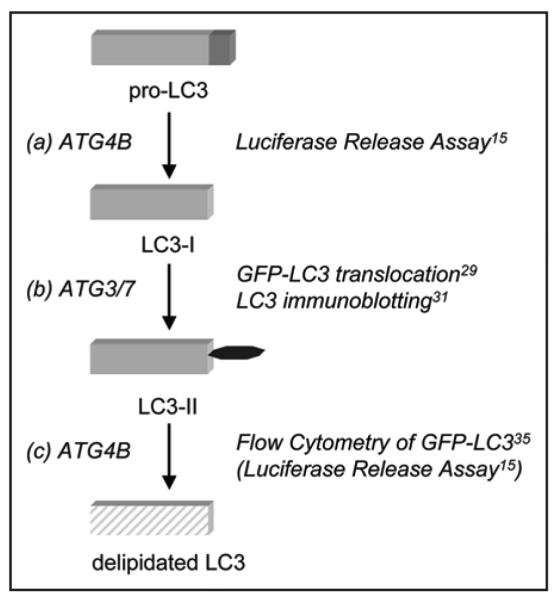
Sequential analysis of LC3 processing during autophagy. LC3 undergoes three distinct processing steps: ATG4B-mediated cleavage of the C-terminus (A), conjugation of phosphatidylethanolamine by ATG3/7 (B) and delipidation by ATG4B (C) resulting in lysosomal degradation (shaded box). Specific assays to measure these events are indicated. The luciferase release assay measures proteolytic cleavage of LC3, but may also be subject to inputs that regulate delipidation of LC3.
Conclusion and Outlook: In vivo Analysis of Autophagy
Inferences based on the analysis of mice bearing targeted disruptions of candidate genes have helped to identify fundamental aspects of autophagy in vivo. To analyze autophagy in whole animals, genetically encoded reporter assays are highly attractive. Such assays would have advantages in simplicity and ease of quantitation compared to measures that rely on the morphological analysis of tissues or cells extracted from study animals. Monodansylcadaverine can be administered to whole organisms such as plants26 and mice,76 and accumulation of the dansyl fluorophore in certain tissues is indicative of increased autophagy. However, genetically encoded reporter systems such as the GFP-LC3 or mCherry-LC3 transgenic mouse35,76 offer many advantages in terms of handling and analysis.
In the future, transgenic mice expressing the luciferase release reporter system discussed here may provide convenient, tractably quantitative, mouse models of autophagy. The detection of Gaussia luciferase in whole animals can be easily accomplished using CCD-cameras.48,77 In addition, it has been demonstrated that Gaussia luciferase secreted from tumor cells can be harvested from the blood and urine of mice78 suggesting that the feasibility of a system that allows the detection and monitoring of autophagic pathways in whole animals by harvesting body fluids. Such a system could also be applied in animals bearing experimental tumors that have been engineered to express the sensor. This would eliminate the need to sacrifice animals for analysis, allowing considerable savings in time, money, and biological material. A careful evaluation of necrotic cell death as a confounding factor for luciferase release is required, though we have observed that extensive cell death results in diminished expression and decreased luciferase release from cultured cells.
The potential therapeutic benefits that may proceed from a deeper understanding of the autophagy pathway appear promising. With novel methods that have recently been described and in vivo animal models, further studies may contribute to the understanding of autophagy and help identify novel targets for therapeutic approaches.
Acknowledgements
R.K. was supported by the Deutsche Forschungsgemeinschaft.
References
- 1.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colombo MI. Pathogens and autophagy: subverting to survive. Cell Death Differ. 2005;12:1481–3. doi: 10.1038/sj.cdd.4401767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 7.Klionsky DJ. Neurodegeneration: good riddance to bad rubbish. Nature. 2006;441:819–20. doi: 10.1038/441819a. [DOI] [PubMed] [Google Scholar]
- 8.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4:422–4. doi: 10.1016/s1535-6108(03)00306-4. [DOI] [PubMed] [Google Scholar]
- 12.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 13.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271–9. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 14.Tasdemir E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76. doi: 10.1007/978-1-59745-157-4_3. [DOI] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 20.Ketteler R, Sun Z, Kovacs KF, He WW, Seed B. A pathway sensor for genome-wide screens of intracellular proteolytic cleavage. Genome Biol. 2008;9:64. doi: 10.1186/gb-2008-9-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733. [PMC free article] [PubMed] [Google Scholar]
- 22.Arstila AU, Trump BF. Autophagocytosis: origin of membrane and hydrolytic enzymes. Virchows Arch B Cell Pathol. 1969;2:85–90. doi: 10.1007/BF02889572. [DOI] [PubMed] [Google Scholar]
- 23.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 24.Munafo DB, Colombo MI. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–29. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 25.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 26.Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
- 27.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP and GATE-16. J Biol Chem. 2006;281:3017–24. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 28.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–82. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 30.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 31.Ciechomska IA, Tolkovsky AM. Non-autophagic GFP-LC3 puncta induced by saponin and other detergents. Autophagy. 2007;3:586–90. doi: 10.4161/auto.4843. [DOI] [PubMed] [Google Scholar]
- 32.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–8. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 33.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 34.Tanida I, et al. Consideration about negative controls for LC3 and expression vectors for four colored fluorescent protein-LC3 negative controls. Autophagy. 2008;4:131–4. doi: 10.4161/auto.5233. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karim MR, et al. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy. 2007;3:553–60. doi: 10.4161/auto.4615. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 38.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 39.Wang QJ, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–68. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–74. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104:19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4 doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 44.Hoyvik H, Gordon PB, Seglen PO. Use of a hydrolysable probe, [14C]lactose, to distinguish between pre-lysosomal and lysosomal steps in the autophagic pathway. Exp Cell Res. 1986;166:1–14. doi: 10.1016/0014-4827(86)90503-3. [DOI] [PubMed] [Google Scholar]
- 45.Mortimore GE, Mondon CE. Inhibition by insulin of valine turnover in liver. Evidence for a general control of proteolysis. J Biol Chem. 1970;245:2375–83. [PubMed] [Google Scholar]
- 46.Seglen PO, Grinde B, Solheim AE. Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur J Biochem. 1979;95:215–25. doi: 10.1111/j.1432-1033.1979.tb12956.x. [DOI] [PubMed] [Google Scholar]
- 47.Knappskog S, et al. The level of synthesis and secretion of Gaussia princeps luciferase in transfected CHO cells is heavily dependent on the choice of signal peptide. J Biotechnol. 2007;128:705–15. doi: 10.1016/j.jbiotec.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 48.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–43. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cog-wheels of cancer. Autophagy. 2006;2:67–73. doi: 10.4161/auto.2.2.2458. [DOI] [PubMed] [Google Scholar]
- 50.Vande Walle L, et al. Proteome-wide Identification of HtrA2/Omi Substrates. J Proteome Res. 2007;6:1006–15. doi: 10.1021/pr060510d. [DOI] [PubMed] [Google Scholar]
- 51.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–32. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 52.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2:571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008 doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–10. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 55.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 56.Marino G, et al. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 57.Yoshimura K, et al. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy. 2006;2:200–8. doi: 10.4161/auto.2744. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 61.Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell. 2006;98:53–67. doi: 10.1042/BC20040516. [DOI] [PubMed] [Google Scholar]
- 62.Yoshimoto K, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma J, et al. An interrelationship between autophagy and filamentous growth in budding yeast. Genetics. 2007;177:205–14. doi: 10.1534/genetics.107.076596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371–3. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 66.Dai J, et al. Protein phosphorylation and expression profiling by Yin-yang multidimensional liquid chromatography (Yin-yang MDLC) mass spectrometry. J Proteome Res. 2007;6:250–62. doi: 10.1021/pr0604155. [DOI] [PubMed] [Google Scholar]
- 67.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 68.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA. 2007;104:1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gnad F, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saeki K, et al. Insulin-dependent signaling regulates azurophil granule-selective macroautophagy in human myeloblastic cells. J Leukoc Biol. 2003;74:1108–16. doi: 10.1189/jlb.0503211. [DOI] [PubMed] [Google Scholar]
- 71.Ueno T, et al. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy. 2008;4 doi: 10.4161/auto.6085. [DOI] [PubMed] [Google Scholar]
- 72.Weihua Z, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–93. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–31. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugawara K, et al. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem. 2005;280:40058–65. doi: 10.1074/jbc.M509158200. [DOI] [PubMed] [Google Scholar]
- 75.Kumanomidou T, et al. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J Mol Biol. 2006;355:612–8. doi: 10.1016/j.jmb.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 76.Iwai-Kanai E, et al. A Method to Measure Cardiac Autophagic Flux in vivo. Autophagy. 2008;4 doi: 10.4161/auto.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venisnik KM, Olafsen T, Gambhir SS, Wu AM. Fusion of Gaussia luciferase to an engineered anti-carcinoembryonic antigen (CEA) antibody for in vivo optical imaging. Mol Imaging Biol. 2007;9:267–77. doi: 10.1007/s11307-007-0101-8. [DOI] [PubMed] [Google Scholar]
- 78.Wurdinger T, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5:171–3. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]