Abstract
Aims
Atrial fibrillation (AF), the most common sustained cardiac arrhythmia, is an important cause of morbidity and mortality. A genetic mutation in the NPPA gene, which encodes the atrial natriuretic peptide, has been identified as the putative causative factor in a family with an autosomal dominant pattern of inheritance for AF. Two common single nucleotide polymorphisms (SNPs) in NPPA, rs5063 and rs5065, result in amino acid changes of the primary peptide and have been previously implicated in conditions associated with AF, including stroke and hypertension. Recently, the rs5063 SNP has been reported to confer an increased risk of AF development in a Chinese population. We sought to examine the associations of both rs5063 and rs5065 with AF in two separate North American cohorts of European ancestry.
Methods and results
Patients with early-onset AF, along with healthy controls, were recruited at the University of Ottawa Heart Institute (UOHI) and the Massachusetts General Hospital (MGH). Study participants were genotyped for rs5063 and rs5065 using a combination of restriction fragment length polymorphism analysis and DNA microarrays. The study genotyped a total of 620 AF cases and 2446 healthy controls. The UOHI arm of the study identified an odds ratio (OR) of 0.72 [95% confidence interval (CI): 0.42–1.24] for rs5063, whereas an OR of 1.33 (95% CI: 0.80–2.21) was observed in the MGH arm. The combined OR approximated unity (OR 0.99; 95% CI: 0.54–1.80). Analysis of rs5065 revealed an OR of 1.12 (95% CI: 0.84–1.48) in UOHI, 1.08 (95% CI 0.80–1.45) in MGH, and 1.10 (95% CI 0.90–1.35) when combined.
Conclusion
Common non-synonymous genetic variants within NPPA in these two large North American cohorts of European ancestry are not associated with the development of AF.
Keywords: Atrial fibrillation, Atrial natriuretic peptide, Single nucleotide polymorphism, Genetics
Introduction
Atrial fibrillation (AF) represents the most common sustained cardiac arrhythmia and is an important contributor to cardiovascular morbidity and mortality.1 Its ubiquitous presence within the general population is reflected by a 25% lifetime risk of developing the arrhythmia in those 40 years of age.2 The significance of this striking figure is amplified by a recent study suggesting that the number of Americans affected by AF may undergo a nearly seven-fold increase to as many as 16 million by the year 2050.3 AF is an independent risk factor for death4 and is commonly associated with stroke. The presence of AF confers an age-dependent increase in the attributable risk of stroke ranging from 1.5% in those aged 50–59 years to 23.5% in octogenarians.5 This is particularly relevant given the aging population and is further exacerbated by the modest therapeutic efficacy of current treatment regimens.
A detailed understanding of the pathophysiology underlying AF remains elusive; however, recent work suggests that genetics play an important role.6,7 This is particularly the case in lone AF, whereby the arrhythmia develops at a relatively early age in the absence of established risk factors.8,9 Over the past decade, genes encoding cardiac potassium channels, sodium channels, and connexins have been identified as predisposing factors for the arrhythmia in cases of lone AF.10–17 In the context of the more common forms of AF, recent genome-wide association studies have identified variants at three genetic loci that are associated with AF, 1q21, 4q25, and 16q22, although the culprit genes within these regions have yet to be identified.18–21
Recently, Hodgson-Zingman et al.22 reported that a mutation in the gene encoding atrial natriuretic peptide (ANP), NPPA, was responsible for an autosomal dominant form of familial lone AF (Figure 1). Unlike previous genetic associations with AF, this study implicated a circulating hormone as the causative factor. The reported frameshift mutation within NPPA resulted in the loss of a stop codon and extension of the reading frame leading to a mutant peptide that is 12 amino acids longer than the wild-type.22 This novel and presumably rare mutation implicates ANP in AF pathophysiology. The association of natriuretic peptides and the arrhythmia is further supported by recent work which demonstrated that increased B-type natriuretic peptide levels predicted the development of incident AF.23,24
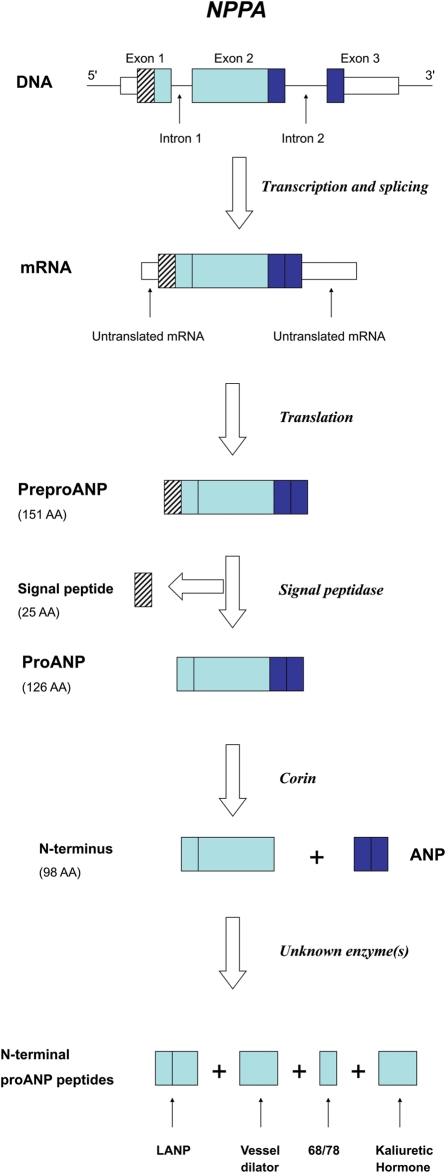
Figure 1.
NPPA gene structure and expression. Transcription and translation of its three exons initially yields preproANP, a 151 AA protein. Post-translational modifications generate ANP along with the N-terminal proANP peptides. ANP, LANP, Vessel Dilator, and Kaliuretic Hormone are all felt to have biological activity. Adapted from Vesely.42
The possibility of a link between common genetic variants within NPPA and AF has been intriguing given the previous association of rs5063 (Val32Met) and rs5065 (X152Rext*3, reflecting a stop codon mutation and the addition of two additional amino acids to the peptide) with hypertension and stroke. Hypertension is a risk factor for AF, whereas stroke, as mentioned, is a common outcome of AF. Well-powered genetic association studies have suggested that the rs5063 minor allele may have a protective effect against the development and progression of hypertension.25,26 Given that hypertension is a risk factor for AF, it is conceivable that rs5063 may also provide a protective effect against the development of AF. In contrast, the minor alleles of both rs5063 and rs5065 have been suggested to increase the risk of stroke.27,28
A recent study in a Chinese population involving 384 AF cases and 844 controls initially found no relation between rs5063 and AF; however, when the analysis was restricted to the 160 cases of lone AF a significant association emerged.29 The presence of the rs5063 minor allele conferred an increased risk of developing lone AF with an odds ratio (OR) of 1.63 [95% confidence interval (CI): 1.09–2.43] which increased to 1.89 (95% CI: 1.26–2.81) following multivariable adjustment. This intriguing result suggesting a link between a common NPPA single nucleotide polymorphism (SNP) and AF would further support the role of natriuretic peptides in the pathophysiology of the arrhythmia and could have important clinical implications in the context of both primary prevention and medical treatment strategies. Accordingly, we sought to further investigate a potential relationship between common non-synonymous NPPA genetic variants, including rs5063, and early-onset AF in two large North American cohorts of European ancestry.
Methods
Clinical recruitment
University of Ottawa Heart Institute
Patients with lone AF or AF and hypertension were recruited from the referral base for AF management at the Arrhythmia Clinic at the University of Ottawa Heart Institute (UOHI). All AF cases had at least one episode of electrocardiographically documented AF characterized by erratic atrial activity without distinct P waves and irregularly irregular QRS intervals. Exclusion criteria consisted of risk factors for AF, aside from hypertension, including a history of coronary artery disease, left ventricular ejection fraction <50% or significant valvular disease on echocardiography. Patients with AF onset after age 60 years also were excluded to limit the influence of age-related factors and enhance the likelihood of genetic-based vulnerability to AF. Control subjects were drawn from the control arm of the Ottawa Heart Genomics Study (OHGS), a case–control genome-wide association study involving 1542 cases of coronary artery disease and 1455 healthy asymptomatic elderly persons without a history of cardiovascular disease.30 Controls from OHGS were excluded if they had a reported history of AF or were recruited prior to the age of 70 years.
All cases and controls were of European ancestry. Study participants provided written informed consent under a protocol approved by the Ethics Research Board at UOHI.
Massachusetts General Hospital
The Massachusetts General Hospital (MGH) AF study8 is comprised of consecutive patients with early-onset AF referred to the arrhythmia service between 5 July 2001 and 19 February 2008. Patients with electrocardiographically documented AF and age <66 years at the time of AF onset were eligible for inclusion. Individuals with structural heart disease as assessed by echocardiography, hyperthyroidism, myocardial infarction, or heart failure were excluded. Each patient underwent a physical examination and standardized interview. All patients were evaluated by 12-lead electrocardiogram, echocardiogram, and laboratory studies. Referent subjects were comprised of unrelated subjects aged 18–74 years from the Framingham Heart Study (FHS),31–33 in whom follow-up data were available, with no history of AF at blood draw or in follow-up, and no history of myocardial infarction, heart failure, or valve disease at baseline.
Genetic analyses
Genomic DNA was extracted from blood samples using a commercially available kit (PUREGENE®, Gentra Systems Inc., Minneapolis, MN, USA). Genotyping of rs5063 and rs5065 for UOHI was performed via amplification of NPPA exons with the polymerase chain reaction using oligonucleotide primers based on previous work by Hodgson-Zingman et al.22 followed by restriction fragment length polymorphism (RFLP) analysis. The SNP identity was determined based on the RFLP profile after restriction enzyme digestion with BceAI (rs5063) and ScaI (rs5065) as previously described.34 Digestion products were resolved using 1.5% agarose gel electrophoresis and visualized with ethidium bromide. Genotyping for OHGS was performed using either the Affymetrix 5.0 or 6.0 arrays (Santa Clara, CA, USA). A subset of 91 control genotypes was additionally reacquired using RFLP to assess for concordance between genotyping methods. Whole genome SNP analysis was performed in MGH and FHS using the Affymetrix 6.0 and 5.0 arrays augmented with the 50 K Human Gene Focused Panel, respectively. Both rs5063 and rs5065 were directly genotyped on these platforms.
Statistical analysis
To ensure no genotyping errors, both SNPs were tested for deviation from Hardy–Weinberg equilibrium using an exact test. The association between each SNP and AF was determined by logistic regression with adjustment for sex and hypertension (UOHI/OHGS), or age, sex, and hypertension (MGH/FHS) assuming an additive genetic model. The associations between each SNP and AF were meta-analysed using the DerSimonian–Laird random-effects model using the inverse variance method to pool log OR.35 All statistical calculations were carried out using R.36
Power estimates were obtained for a χ2 additive test using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/).37 Estimates of SNP minor allele frequency were obtained from the HapMap CEU population.38 Power calculations were performed using relative risks assuming an additive effect between heterozygous and homozygous states. A power estimate of the meta-analysis was obtained by considering a sample size equivalent to pooling the UOHI/OHGS and MGH/FHS samples. Power was estimated based on a minor allele frequency of 6% for rs5063 and 14% for rs5065, assuming a prevalence for AF of 1% and an alpha of 0.025. For heterozygous relative risks of 1.25 and 1.5, estimates of the power for a pooled sample size were 0.328 and 0.8816 for rs5063 and 0.615 and 0.993 for rs5065, respectively.
Results
A total of 620 AF cases and 2446 healthy controls were genotyped in the study. The UOHI/OHGS arm of the study enrolled 245 patients with AF and 1338 controls, whereas 375 AF patients and 1108 referents were enrolled through MGH/FHS. The baseline clinical characteristics of these cohorts are summarized in Table 1.
Table 1.
Clinical characteristics of the AF and control cohorts from UOHI/OHGS and MGH/FHS
| Characteristics | UOHI/OHGS |
MGH/FHS |
||
|---|---|---|---|---|
| AF | Control | AF | Control | |
| Number | 245 | 1338 | 375 | 1108 |
| Agea (Mean ± SD) | 46.7 ± 10.9 | 75.5 ± 4.8 | 46.1 ± 11.7 | 59.4 ± 9.8 |
| Male (%) | 78.4 | 50.4 | 81 | 45 |
| Hypertension (%) | 23.3 | 36.1 | 22.6 | 39.6 |
aAge at onset of AF for cases or age at DNA collection for controls.
Acquisition of 91 control genotypes for rs5063 and rs5065 in OHGS was performed using both RFLP and DNA microarrays. A concordance of 98 and 99% was observed between the two methods, respectively. Analysis for deviation from Hardy–Weinberg equilibrium in UOHI/OHGS was acceptable with a P-value of 0.074 for rs5063 and 0.078 for rs5065, whereas both SNPs from MGH/FHS met the pre-specified threshold suitable for study (P > 1 × 10−6).
The calculated OR for rs5063 in UOHI/OHGS was 0.72 (95% CI: 0.42–1.24), although the OR for MGH/FHS was 1.33 (95% CI: 0.8–2.21; Table 2). Combining the results from both centres yielded an overall OR of 0.99 (95% CI: 0.54–1.80) indicative of no association between rs5063 and AF (Table 2). Analysis of rs5065 in UOHI/OHGS produced an OR of 1.11 (95% CI: 0.84–1.47), whereas an OR of 1.08 (95% CI: 0.80–1.45) was observed in MGH/FHS (Table 2). When combined, the overall OR was 1.10 (95% CI: 0.90–1.35) indicating no association with rs5065 and AF (Table 2).
Table 2.
Genotyping results for rs5063 and rs5065 and their association with AF
| Cases/Controls | rs5063 |
rs5065 |
||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | |
| UOHI/OHGS | 0.72 | 0.42–1.24 | 0.24 | 1.12 | 0.84–1.48 | 0.44 |
| MGH/FHS | 1.33 | 0.80–2.21 | 0.27 | 1.08 | 0.80–1.45 | 0.63 |
| Combined | 0.99 | 0.54–1.80 | 0.96 | 1.10 | 0.90–1.35 | 0.36 |
ORs, CIs, and P-values are derived from logistic regression adjusted for gender and hypertension (UOHI/OHGS), and gender, hypertension and age (MGH/FHS). Meta-analysis (UOHI/OHGS+MGH/FHS) was calculated using the DerSimonian–Laird random-effects model.
Discussion
We performed a case–control study of non-synonymous or amino acid altering SNPs within the NPPA gene in 620 cases of lone AF or AF with hypertension and 2446 healthy controls recruited from two North American centres. These investigations were initiated following work which identified a frameshift mutation within NPPA as the cause of an autosomal dominant form of lone AF. Although this mutation is likely rare and not a common contributor to AF, the recognition that ANP may influence AF pathophysiology raised the possibility that common variants within NPPA may predispose to the arrhythmia. This notion was further fuelled by the findings of a recent study suggesting that rs5063, a common NPPA SNP, was associated with a significantly increased risk of developing the arrhythmia in Chinese patients with lone AF. Nonetheless, in this study involving two large North American cohorts with early-onset AF we found no association of the arrhythmia with either rs5063 or rs5065.
There are a variety of potential explanations that may account for the discordant results between our work and the Chinese group. Firstly, their relatively small sample size and lack of a replication sample may have led to a spurious finding, especially given the low minor allele frequency of rs5063 in the range of 5–10%.29,39 Our inability to replicate their finding with a much larger number of AF cases and controls increases the likelihood that their apparent association may have been a false positive result. Alternatively, differences in linkage disequilibrium patterns between Chinese and European ancestral populations may result in the tested SNPs being differentially linked to a common causal element, such that an association is only observed among those of Chinese ancestry. Lastly, the susceptibility loci for AF may differ to some degree in these two ancestral populations. The differences in genetic background between subjects of European and Chinese descent could alter the biologic importance of a SNP with AF pathophysiology and may contribute to the conflicting results.
It should also be noted that there were minor differences in the baseline clinical characteristics of the patients recruited into each study. The average age of AF onset in our study was 46 years compared with 55 years in the Chinese study. In addition, ∼23% of our patients had hypertension whereas only lone AF cases were analysed in the Chinese group. These discrepancies, although unlikely, could conceivably have resulted in cohorts with different forms or sub-phenotypes of AF driven by distinct processes.40 In this context, it is conceivable that a particular SNP could increase the risk of one form of AF, but have no effect on another AF sub-phenotype. This is of particular interest given the reported association between rs5063 and hypertension and the possibility that the SNP may influence AF pathophysiology through a hypertension-induced tissue alterations that may promote AF. Given that only lone AF cases without hypertension were examined in the Chinese group, this should not account for their reported association. A potential hypertensive mediated relation between rs5063 and AF in the AF cases with hypertension in our study could have been obscured by adjusting for hypertension in our analysis; however, this was excluded when an unadjusted model also failed to identify an association.
The benefits of unravelling the genetics underlying AF will hopefully include the ability to identify individuals at high risk of developing the arrhythmia. In combination with current prediction models guided by clinical criteria, identification of at-risk individuals may facilitate implementation of primary preventive measures that may curb the rising incidence of AF.41 A second beneficial outcome to clinical practice that may be derived from an improved genetic understanding of AF is the ability to identify the underlying pathophysiologic factors driving the arrhythmia in a particular patient. Given the heterogeneous nature of AF, a targeted form of therapy that specifically addresses the driving factors within a given patient should maximize treatment efficacy and reduce adverse effects. This pharmacogenomic paradigm should serve to improve the modest efficacy of contemporary treatment strategies. However, prior to the development of a genetic risk score or implementation of a pharmacogenomic strategy for AF in routine clinical practice, it remains necessary to identify relatively common genetic variants that predispose to the arrhythmia. Although NPPA has been implicated in the development of AF, our study does not suggest a strong influence of the common non-synonymous genetic variants within NPPA on the risk of AF. Therefore, in the absence of an autosomal dominant form of the arrhythmia, genetic variation within NPPA may not be useful in the development of clinical tools or treatment strategies for AF.
This case–control study possesses important limitations including a modest power given the low minor allele frequencies for both SNPs. Although our study was well-powered to detect a moderate-sized association with AF, we cannot rule out a potential weak relationship. In addition, subjects were limited to subjects of European ancestry and the results may not be reflective of other ethnicities. Finally, this study examined lone and early-onset AF and may not be generalizable to other subtypes of AF such as AF associated with structural heart disease.
Conclusions
Our data extend prior knowledge by examining the association of common, non-synonymous genetic variants in NPPA in patients with early-onset AF. Despite prior reports, we have found no relation for the amino acid altering rs5063 and rs5065 SNPs in NPPA and the risk of AF development.
Conflict of interest: none declared.
Funding
This work was supported by grants from the Heart and Stroke Foundation of Ontario and Canadian Institutes of Health Research to M.H.G. and grants from the NIH: HL092577 to P.T.E. and E.J.B.; RC1-HL01056 to E.J.B.; and DA027021 to P.T.E.; S.A.L. was supported by a training grant from the NIH (T32HL00757).
References
- 1.Chugh SS, Blackshear JL, Shen WK, Hammill SC, Gersh BJ. Epidemiology and natural history of atrial fibrillation: clinical implications. J Am Coll Cardiol. 2001;37:371–8. doi: 10.1016/s0735-1097(00)01107-4. doi:10.1016/S0735-1097(00)01107-4. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Wang TJ, Leip EP, Larson MG, Levy D, Vasan RS, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–6. doi: 10.1161/01.CIR.0000140263.20897.42. doi:10.1161/01.CIR.0000140263.20897.42. [DOI] [PubMed] [Google Scholar]
- 3.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna WP, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980–2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–25. doi: 10.1161/CIRCULATIONAHA.105.595140. doi:10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin EJ, Wolf PA, D'Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–52. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 5.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke. 1991;22:983–8. doi: 10.1161/01.str.22.8.983. [DOI] [PubMed] [Google Scholar]
- 6.Fox CS, Parise H, D'Agostino RB, Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. J Am Med Assoc. 2004;291:2851–5. doi: 10.1001/jama.291.23.2851. doi:10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 7.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–12. doi: 10.1093/eurheartj/ehi727. doi:10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 8.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;188:179–84. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 9.Christophersen IE, Ravn LS, Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH, et al. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol. 2009;2:378–83. doi: 10.1161/CIRCEP.108.786665. doi:10.1161/CIRCEP.108.786665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YH, Xu WJ, Bendahhou S, Wang XL, Wang Y, Xu WY, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–4. doi: 10.1126/science.1077771. doi:10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. doi:10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia M, Jin Q, Bendahhaou S, He Y, Larroque MM, Chen Y, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–9. doi: 10.1016/j.bbrc.2005.05.054. doi:10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 13.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. doi:10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35. doi: 10.1161/CIRCULATIONAHA.107.757955. doi:10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, et al. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–34. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Huang H, Liu G, Lam K, Rutberg J, Green MS, et al. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem Biophys Res Commun. 2009;380:132–7. doi: 10.1016/j.bbrc.2009.01.052. doi:10.1016/j.bbrc.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–88. doi: 10.1056/NEJMoa052800. doi:10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 18.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4. doi: 10.1038/ng.537. doi:10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. doi:10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81. doi: 10.1038/ng.416. doi:10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. doi:10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–65. doi: 10.1056/NEJMoa0706300. doi:10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patton KK, Ellinor PT, Heckbert SR, Christenson RH, DeFilippi C, Gottdiener JS, et al. N-terminal pro-B-type natriuretic peptide is a major predictor of the development of atrial fibrillation. The Cardiovascular Health Study. Circulation. 2009;120:1768–74. doi: 10.1161/CIRCULATIONAHA.109.873265. doi:10.1161/CIRCULATIONAHA.109.873265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnabel RB, Larson MG, Yamamoto JF, Sullivan LM, Pencina MJ, Meigs JB, et al. Relations of biomarkers of distinct pathophysiological pathways and atrial fibrillation incidence in the community. Circulation. 2010;121:200–7. doi: 10.1161/CIRCULATIONAHA.109.882241. doi:10.1161/CIRCULATIONAHA.109.882241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conen D, Glynn RJ, Buring JE, Ridker PM, Zee RY. Natriuretic Peptide Precursor A gene polymorphisms and risk of blood pressure progression and incident hypertension. Hypertension. 2007;50:1114–9. doi: 10.1161/HYPERTENSIONAHA.107.097634. doi:10.1161/HYPERTENSIONAHA.107.097634. [DOI] [PubMed] [Google Scholar]
- 26.Conen D, Cheng S, Steiner LL, Buring JE, Ridker PM, Zee RY. Association of 77 polymorphisms in 52 candidate genes with blood pressure progression and incident hypertension: the Women's Genome Health Study. J Hypertens. 2009;27:476–83. doi: 10.1097/hjh.0b013e32832104c8. doi:10.1097/HJH.0b013e32832104c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubattu S, Ridker P, Stampfer MJ, Volpe M, Hennekens CH, Lindpaintner K. The gene encoding atrial natriuretic peptide and the risk of human stroke. Circulation. 1999;100:1722–6. doi: 10.1161/01.cir.100.16.1722. [DOI] [PubMed] [Google Scholar]
- 28.Rubattu S, Stanzione R, Di Angelantonio E, Zanda B, Evangelista A, Tarasi D. Atrial natriuretic peptide gene polymorphisms and risk of ischemic stroke in humans. Stroke. 2004;35:814–8. doi: 10.1161/01.STR.0000119381.52589.AB. doi:10.1161/01.STR.0000119381.52589.AB. [DOI] [PubMed] [Google Scholar]
- 29.Ren X, Xu C, Zhan C, Yang Y, Shi L, Wang F, et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin Chim Acta. 2010;411:481–5. doi: 10.1016/j.cca.2009.12.019. doi:10.1016/j.cca.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Stewart AF, Dandona S, Chel L, Assogba O, Belanger M, Ewart G, et al. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the Ottawa Heart Genomics Study. J Am Coll Cardiol. 2009;53:1471–2. doi: 10.1016/j.jacc.2008.12.051. doi:10.1016/j.jacc.2008.12.051. [DOI] [PubMed] [Google Scholar]
- 31.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–81. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–90. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 33.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, et al. The third generation cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–35. doi: 10.1093/aje/kwm021. doi:10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 34.Masharain U, Nakashima PF, Lim DW, Frossard PM. Nsi I and Sca I restriction fragment length polymorphisms at the atrial natriuretic peptide (ANP) gene locus. Hum Genet. 1988;80:307. doi: 10.1007/BF01790105. [DOI] [PubMed] [Google Scholar]
- 35.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. doi:10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 36.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. ISBN 3-900051-07-0, URL http://www.R-project.org . [Google Scholar]
- 37.Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50. doi: 10.1093/bioinformatics/19.1.149. doi:10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 38.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. doi:10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Roberts JD, Gollob MH. Impact of genetic discoveries on the classification of lone atrial fibrillation. J Am Coll Cardiol. 2010;55:705–12. doi: 10.1016/j.jacc.2009.12.005. doi:10.1016/j.jacc.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D'Agostino RB, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–45. doi: 10.1016/S0140-6736(09)60443-8. doi:10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vesely DL. Atrial Natriuretic Hormones. Englewood Cliffs, NJ: Prentice Hall; 1992. [Google Scholar]