Abstract
Most chronic infectious disease processes associated with bacteria are characterized by the formation of a biofilm which provides for bacterial attachment to the host tissue or implanted medical device. The biofilm protects the bacteria from the host’s adaptive immune response, as well as predation by phagocytic cells. However, the most insidious aspect of biofilm biology from the host’s point of view is that the biofilm provides an ideal setting for bacterial horizontal gene transfer (HGT). HGT provides for large-scale genome content changes in situ during the chronic infectious process. Obviously, for HGT processes to result in the reassortment of alleles and genes among bacterial strains the infection must be polyclonal (polymicrobial) in nature. In this review we marshal the evidence that all of the factors are present in biofilm infections to support HGT which results in the ongoing production of novel strains with unique combinations of genic characters and that the continual production of large numbers of novel, but related bacterial strains leads to persistence. This concept of an infecting population of bacteria undergoing mutagenesis to produce a ‘cloud’ of similar strains to confuse and overwhelm the host’s immune system parallels genetic diversity stratagies employed by viral and parasitic pathogens.
Keywords: chronic bacterial pathogenesis, bacteria, horizontal gene transfer, distributed genome hypothesis, supragenome, pangenome
Introduction
Biofilms serve as population-level virulence factors as they provide the resident bacteria with virulence attributes that single bacterium does not possess. Most of these biofilm-related population-level virulence traits are protective for the bacteria, permitting them to persist in the host in the face of both the innate and adaptive immune systems. Thus they are chiefly of a chronic nature as opposed to planktonic virulence factors, such as toxins, which make the host acutely ill. In addition to providing protection and enabling persistence, biofilms associated with the middle-ear mucosa also often induce the host to produce effusions, and/or to promote hyperplastic growth of the surrounding host tissue by down-regulating apoptosis [Post et al 2007, 2009]. Thus, there is inter-kingdom signaling that serves to provide a constant nutrient source for the biofilm bacteria which helps to maintain the infectious process.
Biofilms also provide an ideal setting for elevated levels of gene transfer among the resident bacteria, both among strains of a species and among related species (Wang et al 2002; Molin and Tolker-Nielsen 2003; Sorensen et al 2005). These gene transfers occur because nearly all of the chronic bacterial pathogens that form biofilms also contain inducible energy-requiring horizontal gene transfer (HGT) mechanisms that serve a nonnutritive purpose (as opposed to using the DNA simply as a food source). These microbial gene transfer capabilities have long been recognized by the infectious disease and clinical microbiological communities, but only in a very narrow sense. Capsular switching among the pneumococci was recognized as resulting from HGT among strains, as were differences in virulence associated with capsule type during epidemics (Ramirez and Tomasz 1999), but these observations were not generalized until 2001 when we advanced the distributed genome hypothesis (DGH) (Ehrlich 2001; Ehrlich et al 2005) which posits that bacterial biofilms associated with chronic infections are composed of multiple strains of a single species (as well as often being polymicrobial or polykingdom communities) and that real-time HGT among the component strains (and species) leads to the continuous generation of a cloud of new strains with novel combinations of genes, thereby providing the bacterial community with a means to thwart the adaptive immune response of the host.
Evolutionary Consequences of Horizontal Gene Transfer
Bacterial horizontal gene transfer (HGT) is defined as the movement of genes (almost always in a unidirectional manner) between two, often unrelated, bacterial cells. It is important to understand that the donor cell from which the horizontally transferred DNA arose does not have to be viable at the time of HGT, and in fact is definitely not the case in two of the three major HGT mechanisms employed by bacterial species. HGT mechanisms usually result in the transfer of one or more relatively small blocks of donor DNA into the recipient cell and thus provide for only the partial replacement of the receiving bacterium’s chromosome. The mean sizes of horizontally acquired gene blocks for those species such as Haemophilus influenzae, Streptococcus pneumoniae, and Staphylococcus aureus that have been studied extensively are usually only between 1 and 2 kilobases (Hogg et al 2007, Hiller et al 2007, Hall et al 2009, Boissy et al submitted), but larger horizontally acquired regions of 50 to 100 kilobases in size are not uncommon [Hiller et al submitted].
Detailed comparative whole chromosomal analyses among large numbers of strains of H. influenzae (Hogg et al 2007) and S. pneumoniae [Table 1] have revealed that on average each strain contains between 200 and 400 insertions/deletions (indels) throughout their chromosome relative to other strains of the species. Thus, each chromosome is highly mosaic with respect to the origin of its own component genes, and further, each strain’s chromosome is highly unique with respect to its gene possession complement. In fact, gene possession differences among the strains of a species account for the vast majority of the genetic heterogeneity within a species, and dwarf the amount of allelic differences observed within genes (Hall et al 2009). Exhaustive pair-wise comparisons among all of the genomically sequenced strains for each of the species Haemophilus influenzae, Streptococcus pneumoniae, Staphylococcus aureus, and Gardnerella vaginalis reveal that there are 385, 407, 246 and 608 gene possession differences, respectively, on average between every pair of strains that has been sequenced within these species (Hogg 2007; Hiller 2007). The 12 strain G. vaginalis supragenome (pangenome) contains 2248 genes of which only 719 are core with the remaining 1529 genes being distributed (non-core) among the 12 strains. Thus, more than two-thirds of the species’ genes are found in only a subset of strains. Since the average individual strain contains only 1257 genes, on average, each genome contains 538 (42%) distributed genes. These extraordinary gene possession differences can only arise via HGT mechanisms.
Table 1.
Summary of Indels between the Reference strain R6 and 21Clinical Pneumococcal Isolates
| Strain | No. of Insertions | Median Insert Length (bp) | Mean Insert Length (bp) | Total Insert Length (Kb) | Max Insert Length (Kb) | No. of deletions | Median Deletion Length (bp) | Mean Deletion Length (bp) | Total Deletion Length (Kb) | Max Deletion Length (Kb) | Total No. of Indels |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BS70 | 85 | 295 | 1450 | 123 | 12 | 97 | 575 | 1641 | 159 | 19 | 182 |
| BS292 | 98 | 244 | 1508 | 148 | 16 | 102 | 515 | 2257 | 230 | 33 | 200 |
| BS293 | 106 | 277 | 1657 | 176 | 16 | 113 | 499 | 1965 | 222 | 26 | 219 |
| BS 397 | 106 | 277 | 1658 | 176 | 16 | 111 | 499 | 2040 | 227 | 33 | 217 |
| INV200 | 160 | 381 | 1480 | 237 | 16 | 121 | 289 | 1615 | 195 | 22 | 281 |
| BS74 | 68 | 232 | 1620 | 110 | 22 | 89 | 392 | 1685 | 150 | 37 | 157 |
| PAT 64 | 287 | 207 | 703 | 202 | 17 | 131 | 344 | 1204 | 158 | 28 | 418 |
| TIGR 4 | 63 | 243 | 1388 | 87 | 17 | 79 | 789 | 2363 | 187 | 37 | 142 |
| BS71 | 79 | 314 | 1717 | 136 | 15 | 71 | 343 | 1978 | 140 | 27 | 150 |
| OXC141 | 84 | 347 | 1667 | 140 | 15 | 114 | 343 | 1441 | 164 | 32 | 198 |
| BS75 | 85 | 225 | 1253 | 107 | 16 | 103 | 214 | 1434 | 148 | 13 | 188 |
| BS72 | 74 | 188 | 1334 | 99 | 13 | 84 | 223 | 1433 | 120 | 14 | 158 |
| BS68 | 91 | 304 | 1467 | 136 | 16 | 98 | 358 | 2190 | 215 | 36 | 189 |
| BS290 | 132 | 276 | 1189 | 157 | 16 | 88 | 240 | 2186 | 192 | 36 | 220 |
| BS73 | 81 | 209 | 951 | 77 | 10 | 87 | 306 | 2129 | 185 | 35 | 168 |
| SV35 | 85 | 299 | 1255 | 107 | 13 | 96 | 306 | 2376 | 228 | 33 | 181 |
| SV36 | 107 | 243 | 1132 | 121 | 13 | 91 | 348 | 2454 | 223 | 33 | 198 |
| 23F | 74 | 334 | 1440 | 107 | 13 | 84 | 424 | 3262 | 274 | 38 | 158 |
| INV104B | 91 | 294 | 1252 | 114 | 13 | 148 | 343 | 1384 | 205 | 36 | 239 |
| BS69 | 75 | 247 | 1446 | 108 | 15 | 100 | 528 | 1899 | 190 | 25 | 175 |
| TIGR 6706 | 73 | 247 | 1605 | 117 | 17 | 101 | 690 | 3060 | 309 | 48 | 174 |
| Averages | 100 | 271 | 1389 | 133 | 15 | 100 | 408 | 2000 | 196 | 31 | 201 |
Highlighted columns show the number of genomic insertions, deletions, and total indels/strain compared to the the S. pneumoniae reference strain, R6. The bottom row shows the 21-strain means for each measure.
No. = number; bp = base pairs; Kb = kilobase pairs; Indels = insertions and deletions.
HGT is defined in contrast to vertical gene transfer which is the standard mechanism by which a mother cell replicates her entire complement of DNA and then passes along identical (or nearly so) copies of each chromosome and plasmid to each of her daughter cells during cell division. Genes and chromosomes that are acquired solely though vertical transmission can be used to construct phylogenetic relationships among bacterial strains, species and higher taxa, however, genes that are acquired through HGT mechanisms produce mosaic chromosomes in which each part of the chromosome that was acquired horizontally has a different ancestry from every other part of the chromosome (unless there are two or more simultaneous transformative events arising from the uptake of DNA from a single donor/competence event – Hiller et al submitted) which therefore makes phylogenetics at the whole chromosome level very difficult. In other words for any set of strains containing mosaic chromosomes each individual gene that has been horizontally transferred and then used to build a phylogenetic tree will produce a different tree structure from the same set of strains (Figure 1) (Shen 2005; Hall 2009). Extensive HGT does not always completely obliterate the average chromosomal phylogenetic signal as has been recently demonstrated for S. pneumoniae [Donati et al submitted], however, because of extensive HGT, strains that are phylogenetically related may have profoundly different genic compositions and thus produce very different disease phenotypes (Buchinsky et al 2007).
Figure 1.
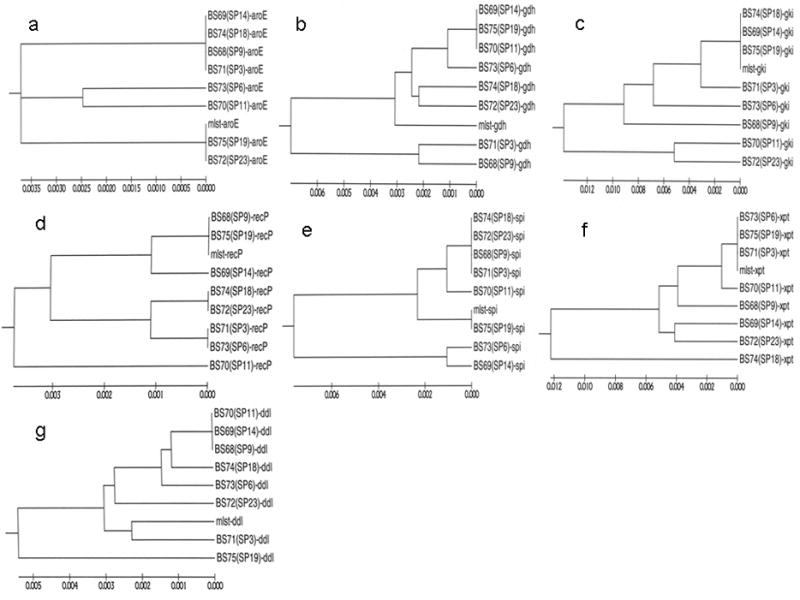
Phylogenetic trees were built for each of seven S. pneumoniae core genes using the same set of eight clinical strains. Each gene produces a different tree stucture (from the same set of strains) demonstrating the mosaic nature of the S. pneumoniae genome in general which results from extensive and continuous horizontal gene transfer. (a) – (g): Phylogenetic trees showing the relative distances of seven housekeeping loci among the eight clinical strains of S. pneumoniae including: a) the aroE gene encoding shikimate dehydrogenase; b) the gdh gene encoding glutamate dehydrogenase; c) the gki gene encoding glucose kinase; d) the recP gene encoding the RecP recombination protein; e) the spi gene encoding a signal peptidase; f) the xpt gene encoding xanthine phosphoribosyltransferase; and g) the ddl gene encoding D-alanyl-D-alanine ligase. Reference sequences were downloaded from http://www.mlst.net.mlst-aroE, mlst-gdh, mlst-recP, mlst-spi, mlst-xpt and mlst-ddl; h) Phylogenic tree reconstructed using concatenated sequences from all seven house-keeping genes (3,199 bp) of S. pneumoniae. Concatenated sequence mlst-7gene was made by using seven reference gene sequences from http://www.mlst.net.
Bacterial Horizontal Gene Transfer (HGT) Mechanisms
HGT is accomplished largely through three fundamentally different mechanisms: competence and transformation, mating or conjugation, and viral transduction. Some species of bacteria use only one of these mechanisms whereas others utilize two or even all three. Transformation and mating are active processes and require significant energetic expenditures by the recipient and donor bacteria, respectively, as well as the maintenance of entire genetic regulons which encode the necessary machinery for the uptake and transfer of DNA, respectively. Thus, the bacteria that possess and maintain these systems must receive an evolutionary advantage in order for them to persist, particularly in the face of strong genomic deletatory mechanisms present in bacteria that are designed to minimize the genomic burden and eliminate unwanted foreign DNA – particularly that of bacteriophages (Brussow et al 2004). Viral transduction, on the other hand, is a passive process engendered by temperate phage. The widespread possession of HGT mechanisms among pathogenic bacterial species, regardless of phylogeny and gram-status, was one of the chief observational points on which the distributed genome hypothesis (DGH) was built (Ehrlich 2001; Shen et al 2003; 2005; 2006a; 2006b; Ehrlich et al 2005; 2008, Hu and Ehrlich 2008).
Historically, transformation was the first HGT mechanism identified. In 1928 Griffith reported ‘transformation’ of rough, avirulent live pneumococci into smooth, virulent pneumococci by the addition of factors from dead, smooth, virulent pneumococci (Griffith 1928). Thus, from its first recognition, transformation was demonstrated to be a population-level virulence factor (Hu and Ehrlich 2008), however, this very important clinical aspect of Griffith’s seminal work was over-shadowed for generations by the even larger basic science implications that derived from this same work. Griffith’s work also suggested the chemical nature of the gene and demonstrated conclusively that individual genes were not living entities in and of themselves. His observations also supported Mendel’s concept of there being discrete genes associated with specific phenotypes (Mendel 1866), but from a practical basis this work provided the means, through purification, to identify the hereditary molecule. In 1944, Avery, McLeod, and McCarty in a series of follow-up experiments to Griffith’s work demonstrated, to the surprise of the world at that time, that DNA, not protein, was the pneumococcal transforming substance (Avery et al 1944) and in so doing ushered in the era of mechanistic molecular biology.
Competence and transformation are actually two separate molecular processes. Competence is the metabolic state of being able to take up foreign DNA into the cell, and transformation results if and when foreign DNA is integrated into the host chromosome changing the genotype and ultimately the phenotype of the cell. In most bacterial species in which competence has been studied it has been determined to be an inducible phenomena associated with nutrient limitation or part of an SOS response (Herriott et al 1970; Prudhomme et al 2006; Håvarstein et al 2006; Kreth et al 2006 Claverys JP, Håvarstein LS 2007; Claverys et al 2007; Thomas et al 2009). Therefore, these processes, which greatly increase the probability of mutation, are triggered when the bacteria are under stress and indicate that bacteria can control their mutational rate based on environmental conditions. This is in stark contrast to the widely held view of evolution that mutational rates are invariant and are not able to be controlled by the organism. Viewed teleologically, the bacteria ‘realize’ that they must ‘change their spots’ to survive and thus activate an energetic system to increase the likelihood of genetic recombination and genic reassortment. In so doing they are utilizing a strategy very similar to that of sexual species in which they are recombining genes (all of which have proven their fitness under various conditions) in different combinations, to produce new organisms with the expectation that some of the recombinants will have a selective advantage under the prevailing conditions in the host, as well as ensuring the survival of the bacterial population and the bacterial gene pool as a whole.
Bacterial mating or ‘conjugation’ as it was dubbed by its discoverer, Joshua Lederberg, who was looking for a sexual phase in the life cycle of bacteria, can result in the transfer of either episomal (plasmid) elements and/or parts of the bacterial chromosome from a donor cell to a recipient cell (Lederberg and Tatum 1946) and unlike transformation requires cell:cell contact for transfer of the donated DNA (Davis 1950). Bacterial conjugation, like transformation is a bacterial equivalent of sex as both of these prokaryotic HGT mechanisms involve genetic exchange. However, neither of these processes include the entire genomes of the parental pair, but rather in both cases one bacterium serves as a donor which provides a section of DNA that, if chromosomal, replaces a section of the chromosomal DNA in the recipient strain, usually through homologous recombination. In the case of conjugation, as opposed to transformation where the donor cell must be dead, the conjugative donor must be viable as it contains either a conjugative plasmid, or mobilizable genetic element integrated into the chromosome, that encodes the molecular machinery to support the creation of a proteinaceous bridge, a pilus, through which the DNA is mobilized, as well as the enzymatic machinery to make a copy of the donor’s DNA for transport through the pilus into the recipient. For these reasons the bacteria initiating conjugation are referred to as male. This brings up a fundamental mechanistic dichotomy between these two energy-requiring bacterial HGT processes. In the case of transformation the recipient cell is the one expending energy and has evolved to either scavenge extracellular DNA (eDNA) or kill its neighbors to ensure an eDNA supply (vide infra), whereas with conjugation it is the donor cell that is expending most of the energy and thus its conjugative elements can be viewed as genetic parasites that evolved to spread themselves into new hosts. However, the conjugative elements often bring beneficial genes with them as well, including those encoding antibiotic and heavy metal resistances, the ability to utilize novel metabolites, or virulence determinants such as adhesins, iron acquisition systems, and serum tolerance.
Transduction, also first discovered in Lederberg’s lab (Zinder and Lederberg 1952), results when a temperate or lysogenic bacteriophage that has been integrated into the host chromosome excises itself and an adjacent section of the host chromosome as part of the lytic phase and then transfers the previous host’s chromosomal region to its next host upon chromosomal integration. Transduction, unlike competence/transformation and mating is a passive process on the part of both the donor and recipient bacteria as it does not require any energy expenditure or host mechanistic genes to accomplish.
Horizontal gene transfer in biofilms
There are four elements necessary for HGT to occur in situ within bacterial biofilms and result in the generation of diversity within the infecting bacterial population. First, it must be demonstrated that chronic infections, in general, are indeed associated with bacteria adopting a biofilm mode of growth. Second, it must be demonstrated that there is a supply or a means to generate a supply of DNA for HGT within the biofilm community. Third, there need to be mechanisms (vide supra) for the transfer of DNA into live organisms. Fourth, and perhaps most importantly, the infecting bacterial population must be polyclonal in nature, i.e. be made up of multiple independent strains of the same bacterial species that are present simultaneously. The necessity for polyclonality derives from the need to generate diversity. If the infection-colonization is monoclonal that means that each bacterium in the biofilm contains the same set of genes and the same set of allele forms of each gene, thus exchanging DNA between any two cells in such an environment would not produce a new strain with new combinations of genes and alleles. In such a case an extensive energy output would be rewarded with no possible gain in terms of creating a more competitive organism. Finally, it must be demonstrated that there is indeed gene exchange that occurs, in real time, among strains within a polyclonal biofilm population and that some of the recombinant strains persist and expand their presence over time (i.e. prove to have a reproductive advantage under the prevailing conditions in the host) and in turn serve as recipients or donors of DNA in further HGT processes.
Chronic Infectious Conditions Possess all the Elements Necessary for HGT
An examination of the conditions present during bacterial colonization of eukaryotic hosts, and during the subsequent chronic infectious disease processes, demonstrates that all of the criteria exist for fruitful genic reassortments.
Chronic Bacterial Infections are Characterized by Biofilms
Bacterial infections associated with chronic disease states are nearly universally found to have adopted a biofilm phenotype (Hu and Ehrlich 2008). The bacterially-elaborated extracellular matrix of the biofilm, associated with the final irreversible attachment of bacterial cells to a surface, is composed of multiple extracellular polymeric substances (EPS) including exopolysaccharides, eDNA, proteins and lipids, and provides a protective physical barrier for the bacteria within. The cooperative creation of the matrix on host tissues or implantable devices by a community of bacteria is a population-level virulence trait as it provides for a community of bacteria that are collectively more difficult for the host to eradicate than individual free-swimming or individual attached bacteria would be. Once initiated, a biofilm acts like a single dynamic living organism that can grow, change its physical properties in response to its environment, evolve through mutation to be better adapted to its environment (Boles et al 2004; Kraigsley and Finkel, 2009), and incorporate other pathogenic species into an integrated polymicrobial community. Importantly, not only can multiple species coexist with a biofilm, but within spatially structured environments they will mutate and evolve in such a way as to improve interaction with other resident species producing a more stable and productive community than their ancestral counterparts (Hansen et al 2007). The biofilm serves as a skeleton for large numbers of bacteria within a single structure and provides the population of interacting organisms with protection, one of the hallmarks of multicellular organisms. Extending the skeletal metaphor, the biofilm matrix also plays important roles in signaling control and nutrient availability. Rheological studies by Stoodley and co-workers have demonstrated that the hydrated EPS matrix is highly viscoelastic and can be rapidly remodeled in response to changes in shear and other environmental stressors (Klapper et al 2002; Dunsmore et al 2002; Stoodley et al 2002; Towler et al 2003; Shaw 2004). Thus, in this regard it displays similar qualities to endochondral bone in that the strength of the extracellular matrix is modifiable by the cellular component in accordance with the external load.
Detailed imaging and metabolic studies spurred by the development of the confocal microscope and PCR-based diagnostics have revealed that many disease conditions that were previously thought of as being chronic inflammatory in nature are actually indolent bacterial biofilm infections. These include: osteomyelitis associated with S. aureus and S. epidermidis (Marrie et al 1985; Costerton 2005); gall bladder disease (Sung et al 1991; Stewart et al 2007); various chronic middle-ear disease processes associated with H. influenzae, S. pneumoniae, Moraxella catarrhalis and P. aeruginosa including otitis media with effusion, recurrent otitis media, and otorrhea (Rayner et al 1998; Ehrlich et al 2002; Dohar et al 2005; Post et al 2004, 2007, 2009; Hall-Stoodley et al 2006; Bakaletz 2007; Kerschner et al 2007; Apicella 2009); chronic rhinosinusitis associated with H. influenzae, S. aureus and other bacteria (Sanderson et al 2006; Palmer et al 2006; Psaltis et al 2007, Prince et al 2008)[Ehrlich et al unpublished]; cholesteatoma associated with P. aeruginosa (Chole and Faddis 2002); tonsillitis (Chole and Faddis 2003); and adenoiditis associated with H. influenzae, S. pneumoniae, and Moraxella catarrhalis (Zuliani et al 2006; Nistico et al 2009). In addition there is substantial evidence to support a bacterial biofilm etiology for many chronic infections of the urogenital systems of both men and women including cystitis, prostatitis, vaginitis, and endometritis (Nickel et al 1994; Hua et al 2005; Swindinski et al 2008) [Ahmed et al submitted], and recently both S. aureus and S. epidermidis have been demonstrated to form biofilms at surgical site infections (Kathju et al 2009). Biofilms are also associated with dental infections including plaque, endodontitis (Carr et al 2009) and periodontitis (Marquis 1995; Paster et al 2006).
Moreover, biofilms represent the overwhelming bacterial phenotype associated with chronic non-healing wounds such as venous and diabetic ulcers, pressure sores, and burn wounds. These infections are often complex polymicrobial and polykingdom communities (Davis et al 2006; Wolcott and Ehrlich 2008). These chronic wound infections and foreign body infections associated with implantable medical devices and indwelling catheters (Ehrlich 2004, 2005, Stoodley 2005, 2008) are nearly impossible to eradicate without aggressive debridement and removal of the device, and have become the bane of many permanent and long term interventional strategies, including artificial joints, central vascular lines, urinary catheterizations, cardiac pace makers and defibrillators, ventricular-peritoneal shunts, and dialysis ports (reviewed in Ehrlich et al 2004).
Availability of bacterial DNA within biofilms
These observations of bacterial phenotype are important because both transformation and mating have been demonstrated to be up to 104-fold higher in biofilms than in planktonic forms (Molin and Tolker-Nielsen 2003; Sorenson et al 2005). High transformation rates in biofilms likely result from the fact that one of the major constituents of the biofilm matrix is eDNA (Figure 2), thus providing a ready source of genetic raw material. In the case of mating, the close spatial juxtaposition of bacterial cells in the biofilm and the physical stability imparted by the biofilm matrix likely support pilus attachment and reduce the likelihood that the conjugal bridges through which the donor DNA is exported will be broken due to hydrodynamic shear stresses. The Bakaletz lab has further demonstrated that the biofilm matrix of Haemophilus influenzae, in addition to containing DNA, also contains very high concentrations of type IV pili (Jurcisek and Bakaletz 2007). Subsequently, Juhas et al (2007a) demonstrated that some H. influenzae strains encode pilus genes which have been shown to support conjugal DNA transfer.
Figure 2.
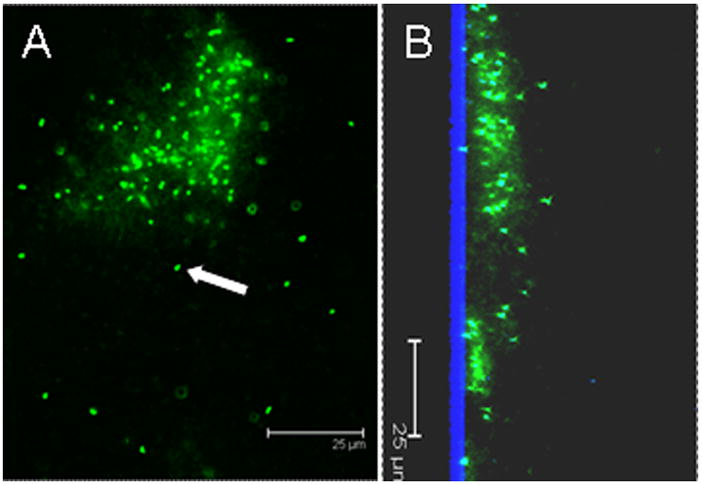
Confocal micrograph of a P. aeruginosa PAO1 biofilm grown on a glass substratum demonstrating extracellular DNA within the biofilm matrix surrounding the bacteria. A) plan view, and B) saggital section with the glass substratum is shown in blue by reflected light. The biofilm was stained with the DNA stain Syto9. The cells (an example of a non-matrix enclosed cell is shown with white arrow) were slightly overexposed to show the more diffuse signal from the extracellular DNA (eDNA) between the individual bacteria. This image demonstrates: 1) the close proximity of the cells within the biofilm; 2) that within biofilms, which are relatively quiescent (unlike laboratory planktonic cultures which are continually mixed), the cells have more time to interact (i.e. for conjugation), and 3) there is a pool of eDNA surrounding the cells which provides structural stability as well as serving as a source for transformation. Scale bars = 25 μm.
The biofilm matrices of all bacterial species that have been characterized for molecular composition including P. aeruginosa, H. influenzae, S. pneumoniae, S. mutans, Staphylococcus aureus and Enterococcus faecalis contain large amounts of eDNA (Whitchurch et al 2002; Jurcisek and Bakaletz 2007; Hall-Stoodley et al 2008; Perry et al 2009; Thomas et al 2009). Even more interestingly, the laboratories of Shi, Clavery, Havarstein, Cvitkovitch and Hancock have convincingly demonstrated a temporal link between conspecific fratricide and the development of competence among the streptococci and the enterococci as a means to ensure a source of species-specific eDNA for those cells first becoming competent (able to take up foreign DNA). The streptococci just prior to their becoming competent produce and release bacteriocins which will kill their neighbors, thus ensuring a ready supply of DNA for transformation (Kreth et al 2006; Prudhomme et al 2006; Claverys and Håvarstein 2007; Perry et al 2009(, whereas the enterococci utilize a toxin-antitoxin system that kills quorum non-responders of their own species (Thomas 2009). Haemophilus influenzae and the other naturally competent Pasteurellaceae utilize a different mechanism to ensure they primarily take up DNA from their own and highly-related species. Within their genomes they have a highly repeated uptake signal sequence (USS) which is present at approximately one copy/gene and their competence apparatus has evolved to selectively take up only DNAs that contain their species-specific USS (Redfield 2005).
Third, and most importantly for HGT mechanisms, colonization is nearly always polyclonal, an observation which had long been missed due to the medical microbiology community’s adherence to Koch’s postulates which teach that a single clonal isolate must be obtained from an infected individual and subsequently demonstrated to cause the same disease in a second host to establish etiology. The mantra of always purifying a single clone put blinders on the medical microbiology community because any diversity which was present was never observed. Over the past decade and a half the laboratories of Smith-Vaughan, Murphy and Gilsdorf, have repeatedly demonstrated, by examining OM patients, COPD patients and the normal nasopharynx, respectively, that nearly all persons that are infected or colonized with Haemophilus influenzae are polyclonally infected – sometimes with more than twenty strains simultaneously (Smith-Vaughan et al 1995, 1996, 1997; Murphy et al 1999; Escevit 2004, 2005, Fargo et al 2004; Mukandun et al 2007, Lacross et al 2008) Similarly, the de Lencastre laboratory and independently Dowson’s group have observed polyclonal infection with pneumococcus (Sá-Leão et al 2002, 2006 & 2008; Jefferies et al 2004;), and Hoiby’s and Molin’s groups in Denmark have seen polyclonal Pseudomonas aeruginosa infections in the CF lung (Jelsbak et al 2007).
Polyclonality is critical to the DGH as it posits that at the species and local population levels there exists a supragenome (pangenome) which is much larger in terms of the total number of genes (not just alleles) than the genome of any single strain within that species, or population. Thus, under this rubric the majority of genes within a species are not possessed by all strains of that species, but rather each strain contains a unique distribution of noncore genes from the species-level supragenome, as well as the species core genome (those genes that are carried by all strains of a species). Thus, we predicted that the bacteria’s possession of HGT mechanisms and the polyclonality of chronic infections, would provide a setting in which new strains with unique combinations of distributed genes would be continually generated. Furthermore, some of these novel strains will have improved survival characteristics under the current prevailing conditions in the host.
Recently we have obtained direct evidence of massive and repeated HGT among pneumococcal strains during a polyclonal pediatric chronic infection which supports the above hypotheses. In this study we identified a dominant strain that over a period of seven months underwent more than a dozen transformation events leading to replacement of approximately 7% of its genome (Figure 3). The fact that we were able to recover multiple recombinant strains when isolating only one strain per time point suggests that these recombinant strains did indeed have a selective advantage in the host environment.
Figure 3.
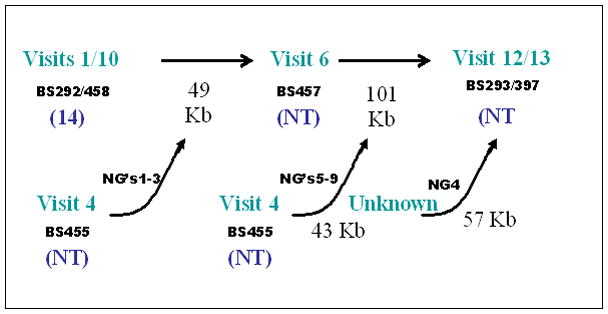
Schematic demonstrating the evolution of S. pneumoniae strains in situ during a naturally-occurring chronic polyclonal pediatric nasopharyngeal infection associated with multiple bouts of otitis media. Identical strains were isolated at visits 1 and 10, and at visits 12 and 13. The 1/10 isolates appear to be the ancestral strain which evolved to the visit 6 strain through the horizontal gene transfer (HGT) of three regions (NG’s) donated from the visit 4 strain. The visit 6 strain, in turn, evolved into the visit 12/13 strain by the HGT of five more NG’s from the visit 4 strain, as well as and an additional NG from an unrecovered strain.
Summary
Our laboratory, as well as those of our colleagues Tettelin et al (2005), Bentley (2010) Hall (2010), Donati [submitted] have used whole genome sequencing to characterize the sizes of the supragenomes, and determine the average number of gene possession differences of multiple independent clinical or environmental strains for over two dozen bacterial species including: Escherichia coli, H. influenzae, Pseudomonas fluorescens, S. pneumoniae, S. agalactiae, S. aureus, and G. vaginalis. These studies have validated the DGH for all species examined and demonstrated that the noncore genes in each strain comprise on average one fifth to one third of each strain’s genome, and that the species-level supragenomes are often 3–4 times the size of the core genomes (Tettelin et al 2005; Hogg et al 2007; Hiller et al 2007; Hall et al 2009) [Ahmed et al submitted; Donati et al submitted]. The predictions of the DGH and the observation that there are enormous gene possession differences among the strains of nearly all bacterial species combine to suggest that during chronic infections the bacteria, through HGT mechanisms, create a ‘cloud’ of related strains each with distinct antigenic and virulence profiles which serve to keep the bacterial population ‘one step ahead of the host’s immune system’. Such a strategy would be analogous to what has been demonstrated for other classes of chronic pathogens such as HIV and the trypanosomes which use error prone nucleic acid polymerases and programmed gene cassette swapping to generate a cloud of diverse strains to avoid immune clearance. Thus, it would appear that diversity generation, regardless of its precise mechanism, is key to the maintenance of a chronic infectious disease state.
These observations on diversity generation by bacteria during chronic infectious processes suggest that interventional therapeutic strategies could be developed to target this aspect of microbial pathogenesis. One such strategy would be STAMP (Specific Targeted Anti-Microbial Peptides) technology wherein a bifunctional peptide is constructed that contains a generic bacteriolytic segment and a species-specific ligand for targeting. By targeting the DNA uptake system of Streptococcus mutans the Shi laboratory has demonstrated a multi-log kill of S. mutans in the presence of other streptococcal species which were relatively unharmed (Eckert et al, 2006) providing a “magic bullet” for the pathogen of interest. This strategy is currently being adopted within our laboratory for S. pneumoniae and should be generally applicable to a broad array of pathogenic bacteria.
Acknowledgments
The authors thank Ms. Mary O’Toole for help in the preparation of this manuscript. This work was supported by Allegheny General Hospital, Allegheny Singer Research Institute, Grants from the Health Resources and Services Administration (HRSA); a system usage grant from the Pittsburgh Supercomputing Center (G.D.E.); and NIH grants DC04173 (G.D.E.), DC02148 (G.D.E.), DC02148-16S1 (G.D.E), and AI080935 (GDE).
References
- Aitken ML. Clinical trials of recombinant human DNase in cystic fibrosis patients. Monaldi Arch Chest Dis. 1993;48(6):653–656. [PubMed] [Google Scholar]
- Apicella MA. Bacterial Otitis Media, the Chinchilla Middle Ear, and Biofilms. J Infect Dis. 2009;199(6):786–794. doi: 10.1086/597043. [DOI] [PubMed] [Google Scholar]
- Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;89:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakaletz LO. Bacterial biofilms in otitis media: evidence and relevance. Pediatr Infect Dis J. 2007;26(10 Suppl):S17–9. doi: 10.1097/INF.0b013e318154b273. Review. [DOI] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci U S A. 2004 Nov 23;101(47):16630–5. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H, Canchaya C, Hardt W-C. Phages and the Evolution of Bacterial Pathogens: from Genomic Rearrangements to Lysogenic Conversion. Microbiology and Molecular Biology Reviews. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchinsky FJ, Forbes M, Hayes J, Hu FZ, Greenberg P, Post JC, Ehrlich GD. Phenotypic Plurality among Clincial Strains of nontypeable Haemophilus influenzae determined by symptom severity in the Chinchilla model of Otitis Media. BMC Microbiology. 2007;14(7):56. doi: 10.1186/1471-2180-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr GB, Schwartz RS, Schaudinn C, Gorur A, Costerton JW. Ultrastructural examination of failed molar retreatment with secondary apical periodontitis: an examination of endodontic biofilms in an endodontic retreatment failure. J Endod. 2009;35(9):1303–1309. doi: 10.1016/j.joen.2009.05.035. [DOI] [PubMed] [Google Scholar]
- Chole RA, Faddis BT. Evidence for microbial biofilms in cholesteatomas. Arch Otolaryngol Head Neck Surg. 2002;128(10):1129–1133. doi: 10.1001/archotol.128.10.1129. [DOI] [PubMed] [Google Scholar]
- Chole RA, Faddis BT. Anatomical evidence of microbial biofilms in tonsillar tissues: a possible mechanism to explain chronicity. Arch Otolaryngol Head Neck Surg. 2003;129(6):634–636. doi: 10.1001/archotol.129.6.634. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5(3):219–229. doi: 10.1038/nrmicro1613. Review. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Håvarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64(6):1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. Review. Erratum in: Mol Microbiol 65(1):230. [DOI] [PubMed] [Google Scholar]
- Davis BD. Non-filterability of the agents of genetic recombination in E. coli. J Bacteriol. 1950;60:507–508. doi: 10.1128/jb.60.4.507-508.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohar JE, Hebda PA, Veeh R, Awad M, Costerton JW, Hayes J, Ehrlich GD. Mucosal biofilm formation on middle-ear mucosa in a non-human primate model of chronic suppurative otitis media. Laryngoscope. 2005;115:1469–1472. doi: 10.1097/01.mlg.0000172036.82897.d4. [DOI] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Riley D, Covacci A, Bentley SD, Kilian M, Ehrlich GD, Hu FZ, Rappuoli R, Moxon ER, Masignani V. Structure and dynamics of the Streptococcus pneumoniae pan-genome: insights from comparative analysis with closely related species. (submitted) [Google Scholar]
- Ecevit IZ, McCrea KW, Pettigrew MM, Sen A, Marrs CF, Gilsdorf JR. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia Genes in Haemophilus influenzae Isolates. J Clin Microbiol. 2004;42(7):3065–3072. doi: 10.1128/JCM.42.7.3065-3072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecevit IZ, McCrea KW, Marrs CF, Gilsdorf JR. Identification of new hmwA alleles from nontypeable Haemophilus influenzae. Infect Immun. 2005;73(2):1221–1225. doi: 10.1128/IAI.73.2.1221-1225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother. 2006 Nov;50(11):3651–7. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich GD. The Biofilm and Distributed Genome Paradigms provide a new Theoretical Structure for Understanding Chronic Bacterial Infections. Interscience Conference on Antimicrobials Agents and Chemotherapy (ICAAC); Chicago, Illinois. December 18, 2001.2001. [Google Scholar]
- Ehrlich GD, Hu FZ, Post JC. Role for Biofilms in Infectious Disease. In: Ghannoum M, O’Toole GA, editors. Microbial Biofilms. Washington, DC: ASM Press; 2004. pp. 332–358. [Google Scholar]
- Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial Plurality as a General Mechanism Driving Persistence in Chronic Infections. Clin Orthop Relat Res. 2005;437:20–24. doi: 10.1097/00003086-200508000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich GD, Hiller NL, Hu FZ. What makes Pathogens Pathogenic. Genome Biol. 2008;9(6):225. doi: 10.1186/gb-2008-9-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo RS, Foxman B, Patel MJ, Zhang L, Pettigrew MM, McCoy SI, Marrs CF, Gilsdorf JR. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr Infect Dis J. 2004;23(1):41–46. doi: 10.1097/01.inf.0000106981.89572.d1. [DOI] [PubMed] [Google Scholar]
- Griffith F. The significance of pneumococcal types. J Hyg. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG, Ehrlich GD, Hu FZ. Pan-genome analysis provides much higher strain typing resolution than does MLST. Microbiology. 2009 Dec 17; doi: 10.1099/mic.0.035188-0. [Epub ahead of print] PubMed PMID: 20019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Nistico L, Dice B, Nguyen D, Mershon WJ, Johnson C, Hayes J, Hu FZ, Post JC, Stoodley P, Ehrlich GD. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiology. 2008;8(1):173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007 Feb 1;445(7127):533–6. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010 Jan 22;327(5964):469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. New insights into the peumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol. 2006;59(4):1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- Herriott RM, Meyer EM, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. Comparative Genomic Analyses of Seventeen Streptococcus pneumoniae Strains: Insights into the Pneumococcal Supragenome. J Bacteriol. 2007;189(22):8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Ahmed A, Powell E, Eutsey RE, Earl J, Martin D, Janto B, Hogg JS, Boissy R, Barbadora K, Post JC, Hu FZ, Ehrlich GD. Generation of genic diversity of Streptococcus pneumoniae via horizontal gene transfer during a chronic polyclonal pediatric infection. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JS, Hu FZ, Janto B, Boissy R, Gladitz J, Swierczek N, Hayes J, Keefe R, Yu S, Post JC, Hu FZ, Ehrlich GD. Characterization and Modeling of the Haemophilus influenzae Core and Supra-Genome based on the Complete Genomic Sequences of Rd and 12 clinical Nontypeable16 Strains. Genome Biology. 2007;8(6):R103. doi: 10.1186/gb-2007-8-6-r103. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FZ, Ehrlich GD. Population-level virulence factors amongst pathogenic bacteria: relation to infectious outcome. Future Medicine. 2008;3:31–42. doi: 10.2217/17460913.3.1.31. [DOI] [PubMed] [Google Scholar]
- Hua VN, Williams DH, Schaeffer AJ. Role of bacteria in chronic prostatitis/chronic pelvic pain syndrome. Curr Urol Rep. 2005;6(4):300–306. doi: 10.1007/s11934-005-0028-z. Review. [DOI] [PubMed] [Google Scholar]
- Jefferies JM, Smith A, Clarke SC, Dowson C, Mitchell TJ. Genetic analysis of diverse disease-causing pneumococci indicates high levels of diversity within serotypes and capsule switching. J Clin Microbiol. 2004;42(12):5681–5688. doi: 10.1128/JCM.42.12.5681-5688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsbak L, Johansen HK, Frost AL, Thogersen R, Thomsen LE, Ciofu O, Yang L, Haagensen JA, Hoiby N, Molin S. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect Immun. 2007;75(5):2214–2224. doi: 10.1128/IAI.01282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Power PM, Harding RM, Ferguson DJ, Dimopoulou ID, Elamin AR, Mohd-Zain Z, Hood DW, Adegbola R, Erwin A, Smith A, Munson RS, Harrison A, Mansfield L, Bentley S, Crook DW. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007;8(11):R237. doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJ, Hood DW. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol. 2007;189(3):761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189(10):3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathju S, Nistico L, Hall-Stoodley L, Post JC, Ehrlich GD, Stoodley P. Chronic surgical site infection due to suture-associated polymicrobial biofilm. Surg Infect. 2009;10(5):457–461. doi: 10.1089/sur.2008.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschner JE, Ehrlich GD, Post JC. The role of biofilms in otitis media. ENT News. 2007;16(3):74–76. [Google Scholar]
- Kraigsley AM, Finkel SE. Adaptive evolution in single species bacterial biofilms. FEMS Microbiol Lett. 2009 Apr;293(1):135–40. doi: 10.1111/j.1574-6968.2009.01526.x. [DOI] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Zhu L, Shi W, Qi F. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett. 2006;265(1):11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Lacross NC, Marrs CF, Patel M, Sandstedt SA, Gilsdorf JR. High genetic diversity of nontypeable Haemophilus influenzae isolates from two children attending a day care center. J Clin Microbiol. 2008;46(11):3817–3821. doi: 10.1128/JCM.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Hendrickson H. Genome evolution in bacteria: order beneath chaos. Curr Opin Microbiol. 2005;8(5):572–578. doi: 10.1016/j.mib.2005.08.005. Review. [DOI] [PubMed] [Google Scholar]
- Lederberg J, Tatum EL. Gene recombination in E. coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- Lee HY, Perelson AS, Park S-C, Leitner T. Dynamic Correlation between Intrahost HIV-1 Quasispecies Evolution and Disease Progression. PloS Computational Biol. 2008;4(12):e1000240. doi: 10.1371/journal.pcbi.1000240. doi: 1371/journla.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;9;4(6):e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan H, Redfield RJ. Tracing the evolution of competence in H. influenzae. PLoS One. 2009;10;4(6):e5854. doi: 10.1371/journal.pone.0005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel JG. Versuche über Plflanzenhybriden Verhandlungen des naturforschenden Vereines in Brünn, Bd. IV für das Jahr, 1865 Abhandlungen. 1866:3–47. [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14(3):255–261. doi: 10.1016/s0958-1669(03)00036-3. Review. [DOI] [PubMed] [Google Scholar]
- Mukundan D, Ecevit Z, Patel M, Marrs CF, Gilsdorf JR. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J Clin Microbiol. 2007;10(5):3207–3217. doi: 10.1128/JCM.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Graf CD, Whatmore AM, King SJ, Trzcinski K, Pickerill AP, Doherty N, Paul J, Griffiths D, Crook D, Dowson CG. Population biology of Streptococcus pneumoniae isolated from oropharyngeal carriage and invasive disease. Microbiology. 1999;(Pt 11):3283–3293. doi: 10.1099/00221287-145-11-3283. [DOI] [PubMed] [Google Scholar]
- Murphy TF, Sethi S, Klingman KL, Brueggemann AB, Doern GV. Simultaneous respiratory tract colonization by multiple strains of nontypeable haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J Infect Dis. 1999;180(2):404–409. doi: 10.1086/314870. [DOI] [PubMed] [Google Scholar]
- Nickel JC, Costerton JW, McLean RJ, Olson M. Bacterial biofilms: influence on the pathogenesis, diagnosis and treatment of urinary tract infections. J Antimicrob Chemother. 1994;33(Suppl A):31–41. doi: 10.1093/jac/33.suppl_a.31. Review. [DOI] [PubMed] [Google Scholar]
- Nistico L, Gieseke A, Stoodley P, Hall-Stoodley L, Kerschner JE, Ehrlich GD. Fluorescence in situ hybridization for the detection of biofilm in the middle ear and upper respiratory tract mucosa. Methods Mol Biol. 2009;493:191–215. doi: 10.1007/978-1-59745-523-7_12. [DOI] [PubMed] [Google Scholar]
- Palmer J. Bacterial biofilms in chronic rhinosinusitis. Ann Otol Rhinol Laryngol Suppl. 2006;196:35–39. doi: 10.1177/00034894061150s906. Review. [DOI] [PubMed] [Google Scholar]
- Perry JA, Cvitkovitch DG, Lévesque CM. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett. 2009;299(2):261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post JC, Ehrlich GD. Biofilms in Otolaryngologic Infections. In: Snow J, Ballanger JJ, editors. Ballenger’s Otorhinolaryngology: Head and Neck Surgery. 16 B.C. Decker; 2007. [Google Scholar]
- Post JC, Ehrlich GD. Biofilms and their role in ear and respiratory infections. In: Snow JB, Wackym PA, editors. Otorhinolaryngology Head and Neck Surgery. 17. Hamilton, Ontario: BC Decker Publisher; 2009. pp. 839–845. [Google Scholar]
- Prince AA, Steiger JD, Khalid AN, Dogrhamji L, Reger C, Eau Claire S, Chiu AG, Kennedy DW, Palmer JN, Cohen NA. Prevalence of biofilm-forming bacteria in chronic rhinosinusitis. Am J Rhinol. 2008;22(3):239–245. doi: 10.2500/ajr.2008.22.3180. [DOI] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313(5783):89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- Psaltis AJ, Ha KR, Beule AG, Tan LW, Wormald PJ. Confocal scanning laser microscopy evidence of biofilms in patients with chronic rhinosinusitis. Laryngoscope. 2007;117(7):1302–1306. doi: 10.1097/MLG.0b013e31806009b0. [DOI] [PubMed] [Google Scholar]
- Ramirez M, Tomasz A. Acquisition of new capsular genes among clinical isolates of antibiotic-resistant Streptococcus pneumoniae. Microb Drug Resist. 1999;5(4):241–246. doi: 10.1089/mdr.1999.5.241. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Leão R, Tomasz A, Santos Sanches I, de Lencastre H. Pilot study of the genetic diversity of the pneumococcal nasopharyngeal flora among children attending day care centers. J Clin Microbiol. 2002 Oct;40(10):3577–3585. doi: 10.1128/JCM.40.10.3577-3585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Leão R, Nunes S, Brito-Avô A, Alves CR, Carriço JA, Saldanha J, Almeida JS, Santos-Sanches I, de Lencastre H. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J Clin Microbiol. 2008;46(1):225–234. doi: 10.1128/JCM.01551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Leão R, Simões AS, Nunes S, Sousa NG, Frazão N, de Lencastre H. Identification, prevalence and population structure of non-typable Streptococcus pneumoniae in carriage samples isolated from preschoolers attending day-care centres. Microbiology. 2006;152(Pt 2):367–376. doi: 10.1099/mic.0.28596-0. [DOI] [PubMed] [Google Scholar]
- Shen K, Wang X, Post JC, Ehrlich GD. Molecular and Translational Research Approaches for the study of Bacterial Pathogenesis in Otitis Media. In: Rosenfeld R, Bluestone CD, editors. Evidence-based Otitis Media. 2. Hamilton: B.C. Decker Inc; 2003. pp. 91–119. [Google Scholar]
- Shen K, Antalis P, Gladitz J, Sayeed S, Ahmed A, Yu S, Hayes J, Johnson S, Dice B, Dopico R, Keefe R, Janto B, Chong W, Goodwin J, Wadowsky RW, Erdos G, Post JC, Ehrlich GD, Hu FZ. Identification, Distribution, and Expression of Novel (nonRd) Genes in Ten Clinical Isolates of Nontypeable Haemophilus influenzae. Infect Immun. 2005;73:3479–3491. doi: 10.1128/IAI.73.6.3479-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Sayeed S, Antalis P, Gladitz J, Ahmed A, Dice B, Janto B, Dopico R, Keefe R, Hayes J, Johnson S, Yu S, Ehrlich N, Jocz J, Kropp L, Tadique E, Wong R, Wadowsky RM, Slifkind M, Preston RA, Erdos G, Post JC, Ehrlich GD, Hu FZ. Extensive genomic plasticity in Pseudomonas aeruginosa revealed by identification and distribution studies of novel (nonPAO1) genes among clinical isolates. Infect Immun. 2006;74(9):5272–5283. doi: 10.1128/IAI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Vaughan HC, Sriprakash KS, Mathews JD, Kemp DJ. Long PCR-ribotyping of nontypeable Haemophilus influenzae. J Clin Microbiol. 1995;33(5):1192–1195. doi: 10.1128/jcm.33.5.1192-1195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Vaughan HC, Leach AJ, Shelby-James TM, Kemp K, Kemp DJ, Mathews JD. Carriage of multiple ribotypes of non-encapsulated Haemophilus influenzae in aboriginal infants with otitis media. Epidemiol Infect. 1996;116(2):177–183. doi: 10.1017/s0950268800052419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Vaughan HC, Sriprakash KS, Mathews JD, Kemp DJ. Nonencapsulated Haemophilus influenzae in Aboriginal infants with otitis media: prolonged carriage of P2 porin variants and evidence for horizontal P2 gene transfer. Infect Immun. 1997;65(4):1468–1474. doi: 10.1128/iai.65.4.1468-1474.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in in situ a critical review. Nature Reviews Microbiology. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Kathju S, Hu FZ, Erdos G, Levenson JE, Mehta N, Dice B, Johnson S, Hall-Stoodley L, Nistico L, Sotereanos N, Sewecke J, Post JC, Ehrlich GD. Molecular and Imaging Techniques for Bacterial Biofilms in Joint Arthroplasty Infections. Clin Orthop Relat Res. 2005;437:31–40. doi: 10.1097/01.blo.0000175129.83084.d5. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Nistico L, Johnson S, Lasko L-A, Baratz M, Ehrlich GD, Kathju S. Direct demonstration of viable S. aureus biofilms in an infected total elbow arthroplasty. J Bone Joint Surg Am. 2008;90(8):1751–1758. doi: 10.2106/JBJS.G.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Mendling W, Loening-Baucke V, Swidsinski S, Dörffel Y, Scholze J, Lochs H, Verstraelen H. An adherent Gardnerella vaginalis biofilm persists on the vaginal epithelium after standard therapy with oral metronidazole. Am J Obstet Gynecol. 2008;198(1):97.e1–6. doi: 10.1016/j.ajog.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72(4):1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Chi B, Kuramitsu HK. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol Immunol. 2002 Apr;17(2):108–12. doi: 10.1046/j.0902-0055.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Wolcott RD, Ehrlich GD. Biofilms and Chronic Infections. 2008;299(22):2682–2684. doi: 10.1001/jama.299.22.2682. [DOI] [PubMed] [Google Scholar]
- Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]


