A method of identifying genes with significantly altered expression in persons with trichiasis is described.
Abstract
Purpose.
Several genes that are associated with protection from or susceptibility to trachomatous trichiasis (TT) have been identified through genetic association studies. Yet there have been few studies in which gene expression profiles were assessed in TT cases and disease-free controls. The purpose was to identify genes that are differentially expressed in the upper tarsal conjunctiva of subjects with TT.
Method.
Pathway-focused gene arrays were used to screen conjunctival RNA expression of 226 gene transcripts of interest. The screening was followed by validation of differentially expressed genes by qRT-PCR on an independent set of samples. Three different techniques were then used to test for quantitative differences in the recovered conjunctival protein fraction.
Results.
Focused arrays identified a set of 13 differentially expressed genes. Validation by qRT-PCR confirmed differential expression in four of these genes (COL1A1, COL7A1, MMP7, and TLR6). Increased expression of MMP7 was the only consistent differentially regulated gene in the conjunctival samples of trichiasis subjects. MMP7 was present in isolated conjunctival proteins and in the tissue culture supernatants of peripheral blood lymphocytes after stimulation.
Conclusions.
There is an imbalance in extracellular matrix turnover with minimal contribution of adaptive immune responses at this stage of trichiasis. There was little evidence of broad differential expression in genes characteristic of polar responses of adaptive T cells or macrophages. The control of the MMP7 response and its activity appears significant in the fibrotic changes observed in TT.
Trachoma, a disease caused by infection with Chlamydia trachomatis, is now predominantly found in the developing world. Recurrent infection with this bacterium leads to inflammation of the upper tarsal conjunctiva and eventually to scarring. Progressive scarring over time leads, in some individuals, to trachomatous trichiasis (TT), a morphologic change in the eyelid causing the lashes to rub against the globe. This process, which often occurs in the absence of detectable C. trachomatis infection, can result in corneal opacity and blindness.1 An estimated 40 million currently have active trachoma. Of those, 8.2 million have TT, and 1.3 million are irreversibly blind as a result.2
The SAFE strategy is recommended by the World Health Organization (WHO) for the control of blinding trachoma: surgery for trichiasis, antibiotics for infection, facial cleanliness, and environmental improvements to reduce transmission of infection. However, since fibrosis may continue to progress in the absence of current C. trachomatis infection, new cases of TT are likely to be seen in endemic communities after transmission of C. trachomatis has been controlled. Even after successful surgery, recurrence rates of up to 60% may be seen within 3 years.3 It is therefore important to understand the processes involved in the pathogenesis of TT by gaining a more complete understanding of the tissue-specific responses associated with the disease process.
It is well established that T helper type 1 (Th1) cells are associated with clearance of chlamydial infection.4,5 In particular, IFNγ plays an important role in the clearance of chlamydial infection in both mice and humans.6,7 However, the inflammatory response, to which IFNγ contributes, may also be the cause of disease if it is excessive or uncontrolled. Normally, the inflammatory response is counterbalanced by IL-10.8,9 More recently, regulatory T cells (Tregs) have been identified as important counterinflammatory mediators of disease in several chronic infections, especially at the site of infection.10,11 Tregs, which can be identified in the conjunctiva during ocular viral infection, may also play a part in the pathogenesis of trachoma.12,13 Additional counterbalancing mechanisms include the evolution of Th2 or type-2 responses. Type 2 responses are frequently associated with chronic inflammation and infection. Although there is some evidence that Th2 responses are contributory to chlamydia's effects,6,8,14 there is little convincing evidence that polar Th2 cell responses directly cause fibrosis.
On the other hand, the role of innate responses from epithelia and leukocytes are increasingly recognized as playing an important role in the pathologic inflammatory process. In particular, IL1β and -8 have both been identified in vitro (from C. trachomatis–infected cells15) and in vivo (from trichiasis patients with inflammation16,17), leading to the suggestion that these are critical factors in the development of inflammatory disease. There is now a much improved understanding of the interaction of innate responses, autoinflammatory cascades, and tissue homeostasis. In particular, the interaction of the matrix metalloprotease (MMP) family of proteins, proinflammatory cytokines, and innate antimicrobial peptides produced by T cells, NK cells, and macrophages.
Thus far, the investigation of the cellular responses and ECM composition of the conjunctiva or the tear film constituents of patients with trichiasis is limited to a few immunohistochemical and expression studies.17–21 Therefore, we investigated whether the expression of other genes involved in innate, adaptive, and extracellular matrix regulation were altered in conjunctival tissue from participants with TT. We show that at this stage of disease, in the almost complete absence of current chlamydial infection, there is little evidence of the involvement of factors produced by the specific adaptive immune response. The profile of responses is characterized by a dominance of gene expression of innate defense molecules and ECM components. In particular, the altered expression of MMP7 appears strongly associated with fibrotic disease.
Methods
Study Participants and Samples Collected
Informed consent was obtained from all study participants. The participants were recruited from rural and semi-urban areas within the Western and Lower River Regions of The Gambia. Trachoma was graded by a single experienced field supervisor according to the World Health Organization (WHO) simplified grading system. Subjects with TT (more than one eyelash touching the globe) were identified. For each TT case, an age-, sex-, and location-matched control subject without any signs of conjunctival scarring and who was not a member of the same family was also recruited. Participants were age matched within 5 years of one another. In a standardized manner, an ocular swab from the everted tarsal conjunctiva of each participant was collected into RNA stabilizer (RNAlater; Ambion Europe, Ltd., Huntingdon, UK) for the isolation of proteins and nucleic acids. Venous blood samples were obtained from a subgroup of these subjects. The study was conducted in accordance with the tenets of the Declaration of Helsinki. It was approved in The Gambia by the joint Ethics Committee of the Gambian government and the U.K. Medical Research Council and by the ethics committee of the London School of Hygiene and Tropical Medicine.
Isolation of DNA, Total RNA, and Total Protein from Ocular Swabs
Total RNA was isolated (RNeasy micro kits; Qiagen, Ltd., Crawley, UK) and used as a template to generate probes for miniarrays (SuperArray; Qiagen, Ltd.). Total RNA was eluted in 30 μL of RNase-free Tris. The total yield and purity of nucleic acid in Tris was estimated by spectrophotometer (NanoDrop ND-1000; Thermo Scientific, Wilmington, DE). RNA samples with low purity (260:280<1.8) or with evidence of impurities from the extraction procedure (230:260>1.8) were re-extracted by repetition of the extraction protocol with the addition of a twofold increase in wash buffers. These were then reassessed for purity and quantity. Only samples that had a spectrographic profile consistent with those outlined at the Genome Center Maastricht (RNA quality control: NanoDrop ND-1000; Thermo Scientific; http://biomedicalgenomics.org/RNA_quality_control.html) were used for the production of biotin UTP labeled-aRNA. Samples from 11 cases and controls with at least 100 ng total RNA were used as templates to generate aRNA. Assessment of the quality of RNA by microcapillary gels was not available. All subsequent ocular swab samples were extracted (All-Prep Kit; Qiagen, Ltd.), according to the manufacturer's instructions. To obtain maximum yields of DNA, RNA and protein from the ocular swabs, a modified initial step was used. Swabs were transferred to a 2-mL screw-cap tube containing sample lysis buffer. They were then vortexed vigorously and the swab discarded. The lysate was passed through a column that binds DNA. The column was washed, and the DNA was eluted in 100 μL of RNase-free water. Ethanol was added to the flow-through from the DNA column and added to a minispin column (RNeasy; Qiagen, Ltd.). Total RNA bound to the membrane was subjected to DNase I digestion. The total RNA was eluted in 60 μL of RNase-free water. Proteins were precipitated from the flow-through with buffer APP. The precipitated proteins were collected by centrifugation, and total protein was redissolved in 100 μL of 5% sodium dodecyl sulfate (SDS). DNA and protein samples were stored at −20°C and RNA at −70°C.
Quantification of Total Amounts of Nucleic Acid and Protein
Total RNA was quantified in a 2-μL sample by reading the absorbance at 260 nm (NanoDrop model ND-1000; Thermo Scientific). Total DNA was quantified by measuring the amount of mitochondrial DNA present in the sample using SYBR-green qPCR. Total protein was quantified in 10-μL samples with an enhanced bicinchoninic acid (BCA) assay (Pierce Chemical, Rockford, IL) on the spectrophotometer (NanoDrop ND-1000; Thermo Scientific).
Generation of Labeled aRNA Probes and Gene Array Analyses
Total RNA was amplified and labeled with biotin-UTP (Message Amp II kit; Ambion Europe Ltd.). Biotin-labeled aRNA was produced after two overnight rounds of in vitro transcription. Labeled aRNA was hybridized overnight to each membrane and two separate membranes (arrays) were used for each subject (with the exception of one sample from an affected case on one Th1/Th2/Th3 array). All arrays, hybridization solutions, wash, and probe detection kits were purchased (SuperArray Bioscience Corp., Frederick, MD). Each array consisted of 113 genes (two blanks, three artificial sequences, one bacterial plasmid sequence [PUC18], two biotin-coupled positive controls, and five reference genes covering seven positions) that were selected for involvement in pathways focused on human extracellular matrix and adhesion molecules (OHS-013) or human Th1/Th2/Th3 cells (OHS-034). Bound aRNA was detected by the addition of streptavidin-alkaline phosphatase (CDP-Star; Roche Diagnostics, Indianapolis, IN). Images of the membranes were captured with a chemiluminescence detection system (Chemi-Doc; Bio-Rad Laboratories, Ltd., Hemel Hempstead, UK) with a CCD camera after 100 seconds of exposure for ECM arrays and 200 seconds for Th1/Th2/Th3 arrays. Each 16-bit TIFF file was then downloaded into array-analysis software (GEArray Expression Analysis Suite 2.0; SuperArray) for image processing and array analysis. Local background correction for spot intensity was applied to each membrane and the raw spot intensities values were then exported to a spreadsheet along with all array meta-information (Excel; Microsoft, Redmond, WA). The raw spot intensities, images, and array information were then analyzed by using the image-processing and array tool features in commercial software (MatLab; The MathWorks, Natick, MA).
Array Normalization
We tested 11 different array normalization strategies, the most appropriate array normalization method was selected by testing each normalization with the technique described by Kroll and Wolfl.22 Normalization of the arrays with the global mean was selected as the best fitting normalization method. The filtered gene list of significantly differentially regulated gene transcripts was then produced based on rank differences in signal intensities. A distribution-free test of significance based on rank difference was used. Genes with rank differences which were significant at the 10% level, without adjustment of P-values for multiple testing, were considered differentially expressed. Only those in which at least five subjects had above-background expression values were selected for further analysis. Since multiple array normalization strategies were tested, only genes that appeared in global mean normalization and at least one of the other 10 normalization strategies tested were selected. Reproducibility and consistency between arrays was tested with a rank-based ANOVA. Last, a categorical nonquantitative analysis, based on presence or absence scoring, was also performed. These categorical data were analyzed by Fisher's exact test, and the results used to complement the quantitative analysis of differential gene expression. We calculated the percentage of false-positive, differentially expressed genes at the 10% and 5% level according to the method described by Stekel.23
Quantitative RT-qPCR
The PCR primer sequences used in this study are listed in Supplementary Table S1 (all Supplementary Tables are available at http://www.iovs.org/cgi/content/full/51/8/3893/DC1). qRT-PCR was performed using a two-step protocol previously described.24 Gene expression was quantified in duplicate by real-time qPCR (QuantiTect SYBR Green PCR kit; Qiagen, Ltd.) on a real-time thermal cycler (Rotor-Gene 6000; Corbett Research, Cambridge, UK). Standard curves were used to quantify the copy number in unknown or test samples, as described by Burton et al.20
Mitochondrial DNA qPCR
PCR was performed on genomic DNA with primers for human-specific hypervariable 1 (HV1) D-loop region mitochondrial DNA in conditions described by Harding-Esch et al.25 The amount of DNA in the sample was then expressed as follows: The sample with the highest cycle threshold (Ct1) was selected as the reference, and all other samples (n) were then estimated relative to this value by multiplying it by two to the power of the difference in the cycle threshold [Ct1 * 2(Ct1 − Ctn)].
Testing for C. trachomatis
A commercial assay (Amplicor CT/NG; Roche Diagnostics) was used for detection of C. trachomatis in cases and controls, as described elsewhere.26 The reaction buffer conditions required were attained by first diluting purified DNA (9 μL) in 94.5 μL of a 50:50 mixture of lysis and diluent buffers; 50 μL was then used in the standard assay. Positive and negative samples were assigned according to the manufacturer's instructions.
SDS-PAGE, Silver Staining, and Immunoblot Analysis
Total protein was pooled from each of the cases and controls, and 10 μg of protein was denatured by boiling in SDS-PAGE gel-loading buffer and loaded onto 5% to 15% polyacrylamide gradient gels (Bio-Rad Laboratories, Ltd.). Ten microliters of protein-marker IV (peqGOLD; Peqlab, Erlangen, Germany) was loaded as size markers. Total protein in SDS-PAGE gels was visualized by staining (ProteoSilver kit; Sigma-Aldrich, Poole, UK), per the manufacturers' protocol. Immunoblots were prepared as just described after the transfer of proteins from the SDS-PAGE gel to nitrocellulose membranes. The membranes were then preblocked in a TBS-T (Tris-buffered saline, 0.1% Tween) supplemented with 5% powdered milk solution overnight at 4°C before immunoblot analysis. The membranes were incubated with a primary anti-MMP7 antibody (MAB9071; 1:100; R&D Systems Europe Ltd, Abingdon, UK), followed by a secondary anti-mouse IgG horseradish peroxidase–conjugated antibody. Bound antibodies were detected with enhanced chemiluminescence (ECL Plus; GE Health Care, Amersham, UK), according to the manufacturer's instructions. Chemiluminescent output was then visualized with a 1-minute exposure using standard x-ray film. The films were photographed and scanned by a gel documentation system and analyzed (Gel-doc with Quantity-one software; Bio-Rad Laboratories, Ltd.).
In Vitro Culture and Stimulation of PBMCs
PBMCs were isolated and cultured as described elsewhere.27 The cells were co-incubated with 5 μg/mL of pokeweed mitogen (PWM), serovar A C. trachomatis elementary bodies (EBs), or culture medium alone. The cells were cultured for 7 days. On day 6, 100 μL of culture supernatant was removed from each well, followed by the addition of 1 μCi/well [3H]thymidine for the last 18 hours of culture, to determine the proliferative index. The culture supernatant was frozen at −20°C until tested for cytokines by capture ELISA.
MMP7 Capture ELISA
MMP7 ELISA kits (Quantikine; R&D Systems) were used to measure amounts of pro- and active MMP7 according to the manufacturer's instructions for tissue culture supernatants. PBMC culture supernatants from 30 cases and controls stimulated for 6 days with pokeweed mitogen, C. trachomatis A EBs, or culture medium alone were tested. The quantity of MMP7 in nanograms per milliliter was then estimated from a standard curve performed at the time of the assay for each plate. The sensitivity of the assay was 0.094 ng/mL.
Statistical Analysis
For qRT-PCR and ELISA, a matched-pair analysis was performed using the Wilcoxon signed-rank test. For assays below sensitivity, the data were reclassified as positive or negative, and the pairing was maintained by using McNemar's χ2 test. We did not estimate the power of the study since, in the absence of any previous studies, it was not possible to estimate the size of the expected differences or the number of genes in a particular pathway that might be affected. Unadjusted P-values after multiple testing are presented with the significance level expected after Bonferroni correction.28
Results
Study Group and Sample Characteristics
Table 1 describes patient and sample characteristics of the individuals participating in the different parts of the study. Eleven case–control pairs were studied with gene microarrays (SuperArray), and a further 47 case–control pairs were studied with qRT-PCR. Cases and controls were matched on age, sex, location, and ethnicity, which act as a proxy control for infection exposure history but without the development of scarring disease. Only one case tested positive for C. trachomatis infection. Nucleic acid and proteins were isolated from the swabs of these subjects and the levels estimated by using the different assays. The levels of each of these fractions were not significantly different between the cases and controls and indicated no inherent fundamental differences or technical inconsistencies that might later affect measurements of differential gene expression. The total number of genes expressed on each array by each group of cases or controls was extracted from the arrays. No large-scale or significant differences were indicated in the total number of genes expressed between the cases and controls. This lack of significance suggested that the comparison of healthy to diseased tissue expression was appropriate and that altered expression levels of genes normally expressed in the conjunctiva, rather than aberrant gene expression, contribute to the pathologic changes observed.
Table 1.
Participant Characteristics for Each Part of the Study
| Cases | Controls | |
|---|---|---|
| Gene Array Samples | ||
| Age (range), y | 67 (49–80) | 65 (44–80) |
| Sex | ||
| Male | 3 | 3 |
| Female | 8 | 8 |
| Total RNA yield (ng/μL) | 18.72 (8.4–63) | 15.8 (10.2–64.4) |
| CT/NG Amplicor | NT | NT |
| Median number (range) of genes expressed per array type | ||
| ECM | 40 (29–64) | 49 (36–65) |
| Th1/Th2/Th3 | 33 (19–55) | 48 (21–63) |
| qRT-PCR Samples | ||
| Age (range), y | 54 (3–76) | 50 (3–75) |
| Sex | ||
| Male | 22 | 22 |
| Female | 25 | 25 |
| Total RNA yield (ng/μL) | 13.75 (6.4–29.2) | 13.64 (2.9–22.8) |
| Total DNA* (qPCR) | 3300 (67–13790) | 3536 (27–39840) |
| Total protein (ng/μL) | 309.8 (100–794) | 331.4 (119–791) |
| CT/NG Amplicor (n positive) | 1 | 0 |
n = 11 for both cases and controls for Gene Array; n = 47 for both groups for qRT-PCR. Data are expressed as median (range). NT, not tested.
Below measurable range (<2 ng/μL) of the spectrometer (OD 280 nm) and therefore estimated by relative quantification real-time PCR.
Genes Involved in ECM Breakdown, Remodeling, and Adhesion
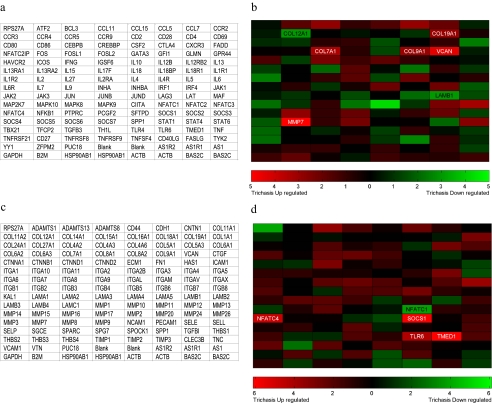
The conjunctival RNA samples of 22 subjects in total, representing 11 case–control, pair-matched samples, were tested on both the ECM and adhesion molecules array and on the Th1/Th2/Th3 array. There was a sufficient yield of aRNA from one sample after amplification to probe only the ECM array. The array platform layout and differential gene expression across the sum of these arrays is visually represented in Figure 1. Genes that were significantly differentially expressed from both array types are shown in Table 2, including accession numbers and brief descriptions of gene product function. In total, nine genes were considered significantly upregulated, and three were downregulated in the TT cases compared with the controls. One additional gene (COL1A1) was identified as significantly more frequently expressed in the TT cases than in the controls based on categorical scoring of spot presence or absence (Supplementary Table S2). The signal intensities of these spots are close to background. At P < 0.1 or < 0.05, the number of potential false-positive genes with differential gene expression was calculated as 24 and 0.6, respectively. The raw acquired images of the developed arrays are shown in Supplementary Figure S1 http://www.iovs.org/cgi/content/full/51/8/3893/DC1, and tables of the raw and background spot intensity values are given in Supplementary Table S3. There were no significant differences between the cases and controls in the total number of genes expressed on each type of array (reference genes and control sequences were excluded). A simple summary table of the genes expressed in the extracted conjunctival RNA is shown in Supplementary Table S4.
Figure 1.
Total RNA was isolated from conjunctival swabs of 11 trichiasis cases and 11 age-, sex-, and location-matched controls. mRNA was amplified, biotin labeled, and used as a probe on each array. The array layouts with gene symbols are shown in (a) for ECM and cell adhesion molecules and in (c) for Th1/Th2/Th3. (b, d) Genes in which the rank difference was significant at the 10% level are indicated by their designated gene symbol. (Red) Genes with increasing intensity of expression; (green) decreasing intensity of expression between cases and controls for each array type.
Table 2.
Differentially Expressed Genes Identified from ECM and Th1/2/3 Arrays
| Gene Symbol | Accession No.* | Gene Name (Function) | Change Ratio† | Rank Difference | P |
|---|---|---|---|---|---|
| Extra Cellular Matrix and Adhesion Molecules | |||||
| Upregulated | |||||
| COL7A1 | NM_000094 | Collagen, type VII, alpha 1 (Adhesion molecule, basement membrane constituent, ECM structural constituent, ECM protease inhibitor) | 0.94 | +3.36 | 0.0981 |
| COL9A1 | NM_001895 | Collagen, type IX, alpha 1 (Adhesion molecule, ECM structural constituent) | 1.34 | +3.00 | 0.0695 |
| COL19A1 | NM_001858 | Collagen, type XIX, alpha 1 (Cell-cell adhesion, ECM structural constituent) | 0.99 | +3.00 | 0.0695 |
| MMP7 | NM_002423 | Matrix metallopeptidase 7 (ECM protease, epithelial cell inflammatory response to virulent bacteria, activation of α-defensins and other innate factors) | 4.30 | +5.00 | 0.0665 |
| VCAN | NM_004385 | Versican (Basement membrane connective tissue link protein, tumour metastasis) | 1.30 | +4.45 | 0.0404 |
| Downregulated | |||||
| COL12A1 | NM_004370 | Collagen, type XII, alpha 1 (Adhesion molecule, ECM structural constituent) | 0.53 | −3.00 | 0.0695 |
| LAMB1 | NM_002291 | Laminin, beta 1 (Structural basement membrane protein) | 0.79 | −3.00 | 0.0695 |
| Th1/Th2/Th3 | |||||
| Upregulated | |||||
| NFATC4 | NM_004554 | Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent-4 (Th2 and induction of IL4 and IL2) | 1.84 | +4.77 | 0.0783 |
| SOCS1 | NM_003745 | Suppressor of Cytokine signaling 1 (Th1 cytokine induced inhibition of IFNγ signaling) | 1.54 | +5.92 | 0.0262 |
| TLR6 | NM_006068 | Toll-like receptor 6 (innate inflammatory response to bacterial lipoproteins with TLR2) | 1.34 | +4.20 | 0.0763 |
| TMED1 | NM_006858 | Transmembrane emp24 protein transport domain containing 1 (Th2, NK2 and NKT2 surface-expressed marker) | 2.36 | +6.11 | 0.0242 |
| Downregulated | |||||
| NFATC1 | NM_172390 | Nuclear factor of activated T cells, cytoplasmic, calcineurin dependent-1 (Th2 cytokine gene expression) | 0.37 | −3.92 | 0.0399 |
http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD.
Change ratio of expression derived from raw non-normalized values from the array. Percentage of false positives at P < 0.05 = 15; P < 0.1 = 100.
Validation of Differentially Expressed Genes in Total RNA from Ocular Swabs from an Independent Set of TT Subjects and Controls
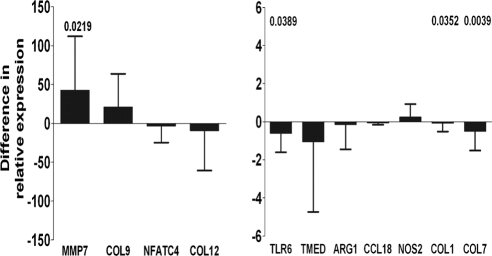
Initially, human ribosomal protein 1 (HuPO1), used in several other studies as a housekeeping gene,29,30 was selected as a reference gene for qRT-PCR normalization. However, the HuPO1 copy number per nanogram total RNA was not equal in cases and controls (data not shown) and so was disregarded. As an alternative normalization method, the amount of total RNA or DNA, which was not significantly different between cases and controls, was selected. Normalization by either of these latter measures was considered suitable, since these had minimal effects on the correlation structure of the data and have been used by others.31,32 Real-time quantitative PCR was then used in a larger set of independent samples to determine whether the 14 genes identified from the array screening would be differentially expressed when assayed with nonamplified total RNA as the starting template. Assays with sufficient specificity and sensitivity for COL19A1 were not developed. Three additional genes, ARG1, CCL18, and NOS2, characteristic of M1/M2 macrophage polarization, were screened at this stage (Fig. 2). Four genes were significantly differentially expressed (P < 0.05). MMP7 was upregulated and consistent with the array results, whereas COL1A1, COL7A1, and TLR6 were all found to be downregulated in cases by qRT-PCR and were inconsistent with array results. The assays of four genes (VCAN, LAMB1, SOCS1, and NFATC1) were not sensitive enough to provide sufficient quantitative data, and the results were therefore reclassified categorically as responder or nonresponder and analyzed by McNemar's χ2 test. None of the four genes was significantly differentially expressed between groups when analyzed in this way. Most of the genes identified from the array and tested by qRT-PCR (n = 7/12) were consistent in their expression. MMP7 was the only gene that was consistent in its significant differential expression by array and by qRT-PCR and was frequently identified as differentially expressed, irrespective of the array normalization method selected. We therefore focused on MMP7 expression in the recovered conjunctival protein fraction and on its production by in vitro cultured PBMCs stimulated with chlamydial antigens from the cases and controls.
Figure 2.
Median difference (with upper or lower quartile range) in relative expression from total RNA conjunctival samples of 47 cases and 47 controls measured by qRT-PCR with primers specific for each gene. Positive differences indicate an upregulation in disease and negative differences indicate a reduction in expression. Unadjusted significant P-values (P < 0.05), shown above each target gene, were calculated with Wilcoxon-signed rank test. Bars indicate significantly downregulated genes. Bonferroni correction for multiple testing estimates significant values needed to reach 0.05/16 (P < 0.003125).
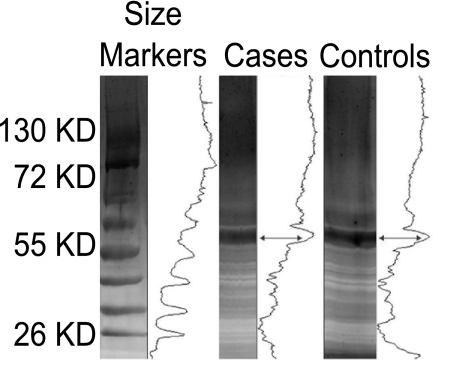
SDS-PAGE Resolution of Isolated Conjunctival Proteins
Silver staining was used to visualize the total protein content recovered from the conjunctival swabs of the cases and controls in SDS-PAGE gels (Fig. 3). The proteins were pooled from each of the 47 cases and controls, and equivalent amounts of protein were loaded on the gel. No gross differences in the number or the intensity of the protein bands were evident. Overall, the cases and controls appeared to have nine major bands. A dominant, intense band at ∼60 kDa was present in both groups. The gels were analyzed (Quantity-one software; Bio-Rad) and produced a density profile that suggested a small change between the cases and controls in abundance of proteins below the major band at ∼60 kDa.
Figure 3.
Conjunctival protein fraction from pooled cases (10 μg) and pooled controls (10 μg) run on 5% and 15% SDS-PAGE silver-stained gels. Density profiles are shown to the right of each lane, indicating a small change in profile shape and height below the major 60-kDa band (arrows).
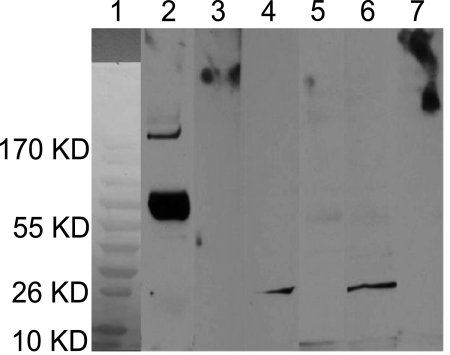
Detection of MMP7 in Recovered Conjunctival Proteins by Immunoblot Analysis
The molecular weight of pro-MMP7 is 28 kDa; however, the recombinant hMMP7 (MMP7 Quantikine kit; R&D Systems) is produced in carrier form with a larger molecular weight. Figure 4, lane 2, shows a large band at ∼70 kDa when probed with anti-MMP7 that served as a positive control reaction. When probed with secondary antibody alone (lane 3) there was no reactivity. A band of equal intensity was visible in lanes 4 (pooled cases) and 6 (pooled controls) at ∼26 kDa, when probed with monoclonal anti-MMP7 antibody. No bands were visible in lanes 5 (pooled cases) and 7 (pooled controls) when probed with secondary antibody alone.
Figure 4.
Immunoblot using conjunctival proteins recovered from cases and controls. Total pooled proteins (10 μg) were transferred to nitrocellulose membranes and probed with an anti-MMP7 primary (1°) antibody. Bound antibody was detected with a secondary (2°) antibody conjugated to HRP (anti-mouse IgG-HRP) and detected by ECL. Control lanes were probed with 2° antibody alone. Molecular weight markers were used in lane 1 to estimate the size of unknown bands from pooled samples. Lanes 2 and 3 were loaded with 0.1 ng of recombinant human MMP7 (rhMMP7) as a positive control (∼70 kDa when probed with anti-MMP7). Probing with 2° antibody alone (lane 3) elicited no reactivity. A band at ∼26 kDa was visible in lanes 4 (pooled cases) and 6 (pooled controls) when probed with monoclonal anti-MMP7 antibody. No bands were visible in lanes 5 (pooled cases) and 7 (pooled controls) when probed with 2° antibody alone.
Quantification of MMP7 by Quantikine Capture ELISA
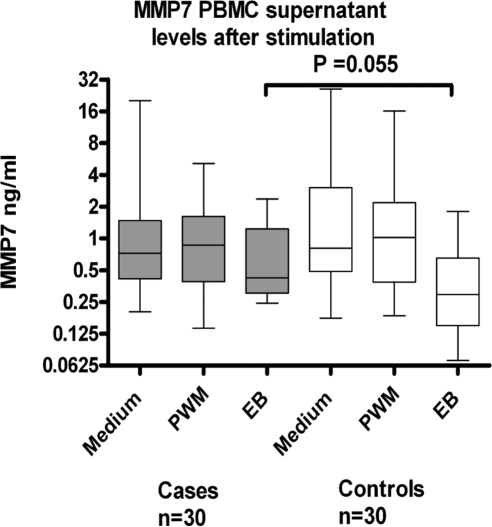
Since quantitative differences between the cases and controls could not be determined with immunoblot analysis, we attempted to measure MMP7 levels in the individual samples of recovered conjunctival proteins and in PBMC tissue culture supernatants. PBMCs were used as a source of cells that express abundant levels of MMP7, to complement the estimates of MMP7 levels directly recovered from the conjunctival proteins. However, the levels of MMP7 could not be measured in recovered total conjunctival protein collected from ocular swabs because of the inhibition of the assay by the samples. MMP7-spiked samples remained inhibited despite dilution of the samples and exchange of sample in appropriate buffers and diluents. MMP7 levels in culture supernatant of PBMCs appear to accumulate over time since at 2 days after culture, levels were not detectable (data not shown). By 6 days of culture spontaneous production of MMP7 in unstimulated cultures was highest in both the cases and controls, although highly variable (Fig. 5). Stimulation with either PWM or EBs inhibited or blocked the spontaneous production of MMP7 by PBMCs. This difference in levels reached borderline significance in response to stimulation with EBs, such that in the cases, stimulation with EBs had a smaller effect on the inhibition of MMP7 production than in the controls. The degree of inhibition or the amount of MMP7 production did not correlate (positively or negatively) with either the PBMC proliferative index or production of IFNγ (data not shown).
Figure 5.
Production of MMP7 (ng/mL) in tissue culture supernatants collected from cases and controls. Freshly isolated PBMCs from cases or controls were cultured with PWM, serovar A EBs, or culture medium alone for 6 days. Production of MMP7 by cells in culture medium alone was the highest in both cases and controls. Stimulation with either PWM or EB inhibited the spontaneous production of MMP7 by PBMCs. This difference in levels reached borderline significance in response to stimulation with EBs, such that in cases (median [IQR] = 0.48 [1.52–0.35] ng/mL) stimulation with EBs has a lesser effect on the inhibition of MMP7 production than in controls (median [IQR] = 0.35 [0.62–0.128] ng/mL).
Discussion
We investigated conjunctival gene expression in subjects with TT using low-density, focused gene expression arrays and qRT-PCR, targeting the genes involved in inflammation and ECM composition. Validation of focused array data by RT-PCR confirmed the differential expression of genes involved in ECM regulation (MMP7, COL1, and COL7) and innate immune responses (TLR6), but only MMP7 expression was consistent between results obtained with arrays and qRT-PCR. There was no evidence from qRT-PCR of differential regulation of Th2 transcription factors, NK2 cell markers, or suppression of IFNγ signaling by SOCS1. The addition of CCL18, NOS, and ARG to screen for macrophage polarization found no evidence of polarization.
MMP7 upregulation is widely found in other fibrotic diseases and tumor metastasis.33,34 MMP7 is naturally expressed in epithelia and injured tissue35 and is increased in alternatively activated (M2) macrophages.36,37 MMP7 has a wider role than ECM regulation, with an increasingly recognized position in innate defense by its action on latent TNF, α-defensins, and FAS ligand.38–40 Mice deficient in MMP7 (mmp7−/−) that lack functional intestinal α-defensins are highly susceptible to infection by enteric pathogens. They also have reduced levels of active FAS ligand and epithelial cell apoptosis. Furthermore, MMP7 is crucial in establishing chemokine gradients; mmp7−/− mice are unable to effectively recruit neutrophils to sites of pathogen entry or tissue damage,41 suggesting an important role in inflammation. It is also responsible for activation of pro-TNF on macrophages42 and has been identified as a biomarker of idiopathic pulmonary fibrosis.43
The role of MMPs in Chlamydia-induced diseases has been studied. MMP9 has received the most attention after reports that indicated increased conjunctival expression20,24 and enzymatic activity18 in trachoma patients. As yet, there are no small rodent models of ocular chlamydial infection that can reproduce the chronic changes observed in the human conjunctiva. Urogenital infection of the mouse offers the next best approximation in small rodents and does induce the expression of MMPs (MMP2, -9, and -1244). Administration of chemical inhibitors of MMPs protected mice from ascending infection and hydrosalpinx.45 Subsequent experiments in MMP9KO mice demonstrated that the ablation of MMP9 production was sufficient in this model to prevent the pathologic changes associated with ascending genital infection.46 Changes in MMP7 levels have not been directly reported in response to chlamydial disease, although the effect of infection in MMP7KO mice has been studied in relation to sequelae and the immune response. Deficiency in MMP7 had little effect on the magnitude or duration of infection, fertility rates, and hydrosalpinx formation,47 leading the authors to conclude that MMP7 was not a requirement in the progression or resolution of chlamydial infection in this model. However, the natural history of ocular infection in humans is markedly different from that in murine genital tract models of infection; thus, the contribution of MMP7 to fibrosis in trachoma is likely the result of repeated or prolonged inflammatory insults, which are often associated with colonization with multiple bacterial species.
MMP7 is one of several genes whose expression is controlled via Wnt (wingless, int) signaling. At least seven recognized terminal target genes of canonical Wnt signaling have been identified in fibrosis (BMP, MYC, CD44, NOS2, MMP7, PPAR, and FN1).48 Four of these genes (CD44, NOS2, MMP7, and FN1) were upregulated in TT cases on the array. A limited number of genes involved in the Wnt signaling pathway were covered on the array, some of which are involved in E-cadherin signaling and the maintenance of the epithelial cell barrier.
Immune-mediated fibrotic diseases, such as schistosomula-induced hepatic fibrosis, are dominated by Th2-cytokine responses where IL13 and the decoy receptor IL13Rα2 are key.49 IL-11 and its receptors are also clearly identified as playing a significant role in some immune-mediated fibrotic diseases.50 More recently polarized macrophages that favor deposition of collagens and fibrosis have been demonstrated to be important in the development of disease (e.g., pulmonary fibrosis51 and responses to chronic helminth infection52). There was no evidence of Th1/Th2/Th3 polarization, although altered expression of NFATC1/4, SOCS1, and TMED1 may suggest some evidence of inhibition of IFNγ signaling and stimulation of Th2, NK2 cells. We did not identify any of the factors found in previous studies on trichiasis patients as having significantly altered expression (IL1B, TNF, MMP1:TIMP1). Nor were genes that have been identified in other infection or immune-driven fibrotic diseases differentially expressed. This does not rule out the involvement of these genes in the disease process. The small number of subjects studied and the sensitivity of the array could account for the lack of differences. In addition, the simplified WHO grading system used to identify cases and controls, although satisfactory for rapid population screening by national control programs, does not record additional important information for detailed research that may be required. Taken together, these factors may explain why these genes were not identified as differentially expressed. Within these limitations, the results identify the major changes in gene expression in this group of subjects and point to changes in MMP7 as the major gene with altered expression.
Since MMP7 gene expression was upregulated in the TT cases, we investigated whether the change would be reflected in the proteins recovered from the same conjunctival samples. The total protein showed some evidence of changes in the abundance of proteins below 60 kDa. This result warrants further study by two-dimensional SDS-PAGE. Using immunoblot analysis, we demonstrated the presence of MMP7, but attempts to quantify the amount of protein by capture ELISA were inhibited by the sample. The assay used to quantify MMP7 was developed to be performed in the presence of TIMP, the natural inhibitor of MMP7; thus, inhibition was due to an unknown factor. The lack of differences in MMP7 in pooled samples by immunoblot analysis may be masked the natural biological variation of the samples and the semiquantitative nature of immunoblot analysis techniques. Stimulation of PBMCs from cases and controls with C. trachomatis EBs resulted in a reduction in the inhibition of spontaneous production of MMP7 in the cases compared with that in the controls (i.e., higher levels of MMP7 remained in the supernatants of cases after stimulation). This observation is difficult to interpret and relate to the increased MMP7 gene expression observed in the conjunctiva of the TT cases. However, PBMCs are a recognized source of MMP7 in several studies, including the observation of differing levels of spontaneous production between clinical groups.53,54
RT-PCR in 47 case–control pairs showed that TLR6 expression was reduced in TT cases, suggesting that the host's recognition of bacterial ligands is affected in TT cases. Alternatively, reduced TLR6 expression could be due to a reduced number of TLR6-expressing cells or changes caused by the disease process that result in the well-described increased susceptibility to other bacterial infections.55 In the mouse, reduced Tlr6 expression has been shown in macrophages on stimulation with TNFα or IFNγ,56 and a proinflammatory environment has also been shown to reduce TLR6 expression in epithelial cells.57
Reduced transcripts of two collagen proteins in subjects with scarring trachoma suggest that the composition of the extracellular matrix is altered. Collagen type I (COL1A1) and VII (COL7A1) transcripts were downregulated in individuals with TT. Many factors including cytokines affect expression of COL1A1. TGFβ, IL-1β, TNFα, and IFNγ have all been shown to affect expression of COL1A1.58 COL7A1 gene expression is also regulated by proinflammatory cytokines (TNFα, IL-1β, and TGFβ), expression is up- or downregulated depending on the cell-type and the cytokines present,59,60 indicating a complex biology. Transcription of COL7A1 is important in the formation of anchoring fibrils, and this may play a role in the morphologic changes that take place in the conjunctiva of subjects with trichiasis.
C. trachomatis DNA was detected in only one individual, consistent with the low prevalence of C. trachomatis infection normally seen in subjects with TT in The Gambia and elsewhere.61 We did not collect additional swabs for standard microbial cultures. Publications in the literature suggest that ∼40% of subjects with TT are colonized with at least one nonchlamydial bacterial species.62 The contribution of additional species and changes in the ocular microbiome are therefore important, since they directly influence the host's expression profile and response to further challenge by infection.63 The altered PBMC response levels of MMP7 in trichiasis may be a result of inherited genetic polymorphisms that predisposes the host to inflammation-driven fibrosis. Investigation of the signaling pathways and downstream targets of MMP7, along with MMP7 polymorphisms (some of which have already been identified as risk factors for fibrosis64,65), may yield important information in the etiology of the scarring process.
We ultimately found four genes with altered expression: MMP7, COL1A1, COL7A1, and TLR6. We accepted a high rate of potential false-positive association when screening genes by the array and subsequently during validation by qRT-PCR, since the normal methods of correcting P-values and the general applicability of adjusting for multiplicity of tests remain matters of contention among biostatisticians.23,28 We therefore leave our colleagues to balance the significance of the findings against the interpretation of the study results. An independent replication study with a larger number of subjects studied by each method is the most appropriate manner of testing these associations. We suggest that in geographic regions where active trachoma and infection are hypoendemic, the C. trachomatis–specific adaptive and regulatory immune responses exert their major effect, if any, during the active stages of the disease when conjunctival scarring is in its early stages. The later stages of disease, such as those seen in adult TT cases, which are characterized by innate proinflammatory and fibrotic responses, continue to progress once the inflammatory environment is established and maintained by other nonchlamydial inflammatory insults. In populations, where rapidly advancing TS and TT can be observed in children and young adults, the role of C. trachomatis–infection and the inflammatory immune response in the acceleration of the disease process requires closer inspection with longitudinal follow-up of participants and repeated measures of gene expression.
Supplementary Material
Acknowledgments
The authors thank the ophthalmic nurses of the Gambian National Eye Care Programme and the field staff from the Medical Research Council Laboratories for their hard work and dedication.
Footnotes
Supported by a grant from the Wellcome Trust (079246/Z/06/Z) with additional support from the Medical Research Council, UK, in The Gambia. The funders had no part in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Disclosure: M.J. Holland, None; D. Jeffries, None; M. Pattison, None; G. Korr, None; A. Gall, None; H. Joof, None; A. Manjang, None; M.J. Burton, None; D.C.W. Mabey, None; R.L. Bailey, None
References
- 1.Mabey DC, Solomon AW, Foster A. Trachoma. Lancet 2003;362:223–229 [DOI] [PubMed] [Google Scholar]
- 2.Mariotti SP, Pascolini D, Rose-Nussbaumer J. Trachoma: global magnitude of a preventable cause of blindness. Br J Ophthalmol 2009;93:563–568 [DOI] [PubMed] [Google Scholar]
- 3.Khandekar R, Mohammed AJ, Courtright P. Recurrence of trichiasis: a long-term follow-up study in the Sultanate of Oman. Ophthalmic Epidemiol 2001;8:155–161 [DOI] [PubMed] [Google Scholar]
- 4.Igietseme JU, Ramsey KH, Magee DM, Williams DM, Kincy TJ, Rank RG. Resolution of murine chlamydial genital infection by the adoptive transfer of a biovar-specific, Th1 lymphocyte clone. Reg Immunol 1993;5:317–324 [PubMed] [Google Scholar]
- 5.Perry LL, Feilzer K, Caldwell HD. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol 1997;158:3344–3352 [PubMed] [Google Scholar]
- 6.Wang S, Fan Y, Brunham RC, Yang X. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur J Immunol 1999;29:3782–3792 [DOI] [PubMed] [Google Scholar]
- 7.Cohen CR, Koochesfahani KM, Meier AS, et al. Immunoepidemiologic profile of Chlamydia trachomatis infection: importance of heat-shock protein 60 and interferon-gamma. J Infect Dis 2005;192:591–599 [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Gartner J, Zhu L, Wang S, Brunham RC. IL-10 gene knockout mice show enhanced Th1-like protective immunity and absent granuloma formation following Chlamydia trachomatis lung infection. J Immunol 1999;162:1010–1017 [PubMed] [Google Scholar]
- 9.Wang C, Tang J, Geisler WM, Crowley-Nowick PA, Wilson CM, Kaslow RA. Human leukocyte antigen and cytokine gene variants as predictors of recurrent chlamydia Trachomatis infection in high-risk adolescents. J Infect Dis 2005;191:1084–1092 [DOI] [PubMed] [Google Scholar]
- 10.Suffia IJ, Reckling SK, Piccirillo CA, Goldszmid RS, Belkaid Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J Exp Med 2006;203:777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott-Browne JP, Shafiani S, Tucker-Heard G, et al. Expansion and function of Foxp3-expressing T regulatory cells during tuberculosis. J Exp Med 2007;204:2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesburn AB, Bettahi I, Dasgupta G, et al. Functional Foxp3+ CD4+ CD25(Bright+) “natural” regulatory T cells are abundant in rabbit conjunctiva and suppress virus-specific CD4+ and CD8+ effector T cells during ocular herpes infection. J Virol 2007;81:7647–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faal N, Bailey RL, Jeffries D, et al. Conjunctival FOXP3 expression in trachoma: do regulatory T cells have a role in human ocular Chlamydia trachomatis infection? PLoS Med 2006;3:e266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gondek DC, Roan NR, Starnbach MN. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. J Immunol 2009;183:1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen SJ, Eckmann L, Quayle AJ, et al. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest 1997;99:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bobo L, Novak N, Mkocha H, Vitale S, West S, Quinn TC. Evidence for a predominant proinflammatory conjunctival cytokine response in individuals with trachoma. Infect Immun 1996;64:3273–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skwor TA, Atik B, Kandel RP, Adhikari HK, Sharma B, Dean D. Role of secreted conjunctival mucosal cytokine and chemokine proteins in different stages of trachomatous disease. PLoS Negl Trop Dis 2008;2:e264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Asrar AM, Geboes K, Al-Kharashi SA, et al. Expression of gelatinase B in trachomatous conjunctivitis. Br J Ophthalmol 2000;84:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Asrar AM, Van den Oord JJ, Geboes K, Missotten L, Emarah MH, Desmet V. Immunopathology of trachomatous conjunctivitis. Br J Ophthalmol 1989;73:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burton MJ, Bailey RL, Jeffries D, et al. Conjunctival expression of matrix metalloproteinase and proinflammatory cytokine genes after trichiasis surgery. Invest Ophthalmol Vis Sci 2010;51:3583–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satici A, Guzey M, Dogan Z, Kilic A. Relationship between tear TNF-alpha, TGF-beta1, and EGF levels and severity of conjunctival cicatrization in patients with inactive trachoma. Ophthalmic Res 2003;35:301–305 [DOI] [PubMed] [Google Scholar]
- 22.Kroll TC, Wolfl S. Ranking: a closer look on globalisation methods for normalisation of gene expression arrays. Nucleic Acids Res 2002;30:e50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stekel D. Microarray Bioinformatics New York: Cambridge University Press; 2003:263 [Google Scholar]
- 24.Burton MJ, Bailey RL, Jeffries D, Mabey DC, Holland MJ. Cytokine and fibrogenic gene expression in the conjunctivas of subjects from a Gambian community where trachoma is endemic. Infect Immun 2004;72:7352–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding-Esch EM, Edwards T, Sillah A, et al. Active trachoma and ocular chlamydia trachomatis infection in The Gambia: on course for elimination by 2020? PLoS Negl Trop Dis 2009;22;3(12):e573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton MJ, Holland MJ, Faal N, et al. Which members of a community need antibiotics to control trachoma?—conjunctival Chlamydia trachomatis infection load in Gambian villages. Invest Ophthalmol Vis Sci 2003;44:4215–4222 [DOI] [PubMed] [Google Scholar]
- 27.Holland MJ, Bailey RL, Conway DJ, et al. T helper type-1 (Th1)/Th2 profiles of peripheral blood mononuclear cells (PBMC); responses to antigens of Chlamydia trachomatis in subjects with severe trachomatous scarring. Clin Exp Immunol 1996;105:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feise RJ. Do multiple out come measures require p-value adjustment? BMC Med Res Methodol 2002;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burl S, Hill PC, Jeffries DJ, et al. FOXP3 gene expression in a tuberculosis case contact study. Clin Exp Immunol 2007;149:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dheda K, Chang JS, Breen RA, et al. Expression of a novel cytokine, IL-4delta2, in HIV and HIV-tuberculosis co-infection. AIDS 2005;19:1601–1606 [DOI] [PubMed] [Google Scholar]
- 31.Dheda K, Huggett JF, Chang JS, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 2005;344:141–143 [DOI] [PubMed] [Google Scholar]
- 32.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalization: strategies and considerations. Genes Immun 2005;6:279–284 [DOI] [PubMed] [Google Scholar]
- 33.McGuire JK, Li Q, Parks WC. Matrilysin (matrix metalloproteinase-7) mediates E-cadherin ectodomain shedding in injured lung epithelium. Am J Pathol 2003;162:1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura T, Biyajima K, Aoki M, Oshima M, Taketo MM. Matrix metalloproteinase 7 is required for tumor formation, but dispensable for invasion and fibrosis in SMAD4-deficient intestinal adenocarcinomas. Lab Invest 2009;89:98–105 [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Boado YS, Wilson CL, Hooper LV, Gordon JI, Hultgren SJ, Parks WC. Bacterial exposure induces and activates matrilysin in mucosal epithelial cells. J Cell Biol 2000;148:1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginderachter J, Liu Y, Devoogdt N, et al. Classical and alternative activation of macrophages: different pathways of macrophage-mediated tumor promotion In: Kaiser HE, Nasir A. eds. Selected Aspects of Cancer Progression: Metastasis Apoptosis Immune Response 2008:139–156 [Google Scholar]
- 37.Shaykhiev R, Krause A, Salit J, et al. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol 2009;183:2867–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell WC, Fingleton B, Wilson CL, Boothby M, Matrisian LM. The metalloproteinase matrilysin proteolytically generates active soluble Fas ligand and potentiates epithelial cell apoptosis. Curr Biol 1999;9:1441–1447 [DOI] [PubMed] [Google Scholar]
- 39.Wilson CL, Ouellette AJ, Satchell DP, et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999;286:113–117 [DOI] [PubMed] [Google Scholar]
- 40.Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest 2000;105:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 2002;111:635–646 [DOI] [PubMed] [Google Scholar]
- 42.Churg A, Wang RD, Tai H, et al. Macrophage metalloelastase mediates acute cigarette smoke-induced inflammation via tumor necrosis factor-alpha release. Am J Respir Crit Care Med 2003;167:1083–1089 [DOI] [PubMed] [Google Scholar]
- 43.Rosas IO, Richards TJ, Konishi K, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med 2008;5:e93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey KH, Sigar IM, Schripsema JH, Shaba N, Cohoon KP. Expression of matrix metalloproteinases subsequent to urogenital Chlamydia muridarum infection of mice. Infect Immun 2005;73:6962–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imtiaz MT, Schripsema JH, Sigar IM, Kasimos JN, Ramsey KH. Inhibition of matrix metalloproteinases protects mice from ascending infection and chronic disease manifestations resulting from urogenital Chlamydia muridarum infection. Infect Immun 2006;74:5513–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imtiaz MT, Distelhorst JT, Schripsema JH, et al. A role for matrix metalloproteinase-9 in pathogenesis of urogenital Chlamydia muridarum infection in mice. Microbes Infect 2007;9:1561–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal S, Schmidt AP, Peterson EM, Wilson CL, de la Maza LM. Role of matrix metalloproteinase-7 in the modulation of a Chlamydia trachomatis infection. Immunology 2006;117:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Toerne C, Schmidt C, Adams J, et al. Wnt pathway regulation in chronic renal allograft damage. Am J Transplant 2009;9:2223–2239 [DOI] [PubMed] [Google Scholar]
- 49.Chiaramonte MG, Schopf LR, Neben TY, Cheever AW, Donaldson DD, Wynn TA. IL-13 is a key regulatory cytokine for Th2 cell-mediated pulmonary granuloma formation and IgE responses induced by Schistosoma mansoni eggs. J Immunol 1999;162:920–930 [PubMed] [Google Scholar]
- 50.Chen Q, Rabach L, Noble P, et al. IL-11 receptor alpha in the pathogenesis of IL-13-induced inflammation and remodeling. J Immunol 2005;174:2305–2313 [DOI] [PubMed] [Google Scholar]
- 51.Prasse A, Pechkovsky DV, Toews GB, et al. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am J Respir Crit Care Med 2006;173:781–792 [DOI] [PubMed] [Google Scholar]
- 52.Babu S, Kumaraswami V, Nutman TB. Alternatively activated and immunoregulatory monocytes in human filarial infections. J Infect Dis 2009;199:1827–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anand SP, Selvaraj P. Effect of 1, 25 dihydroxyvitamin D(3) on matrix metalloproteinases MMP-7, MMP-9 and the inhibitor TIMP-1 in pulmonary tuberculosis. Clin Immunol 2009;133:126–131 [DOI] [PubMed] [Google Scholar]
- 54.Kouwenhoven M, Ozenci V, Gomes A, et al. Multiple sclerosis: elevated expression of matrix metalloproteinases in blood monocytes. J Autoimmun 2001;16:463–470 [DOI] [PubMed] [Google Scholar]
- 55.Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A 2000;97:13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patole PS, Pawar RD, Lech M, et al. Expression and regulation of Toll-like receptors in lupus-like immune complex glomerulonephritis of MRL-Fas(lpr) mice. Nephrol Dial Transplant 2006;21:3062–3073 [DOI] [PubMed] [Google Scholar]
- 57.Ritter M, Mennerich D, Weith A, Seither P. Characterization of Toll-like receptors in primary lung epithelial cells: strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins and inflammatory response. J Inflamm 2005;2:16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh AK. Factors involved in the regulation of type I collagen gene expression: implication in fibrosis. Exp Biol Med (Maywood) 2002;227:301–314 [DOI] [PubMed] [Google Scholar]
- 59.Takeda H, Kon A, Ito N, et al. Keratinocyte-specific modulation of type VII collagen gene expression by pro-inflammatory cytokines (tumor necrosis factor-alpha and interleukin-1beta). Exp Dermatol 2005;14:289–294 [DOI] [PubMed] [Google Scholar]
- 60.Calonge MJ, Seoane J, Massague J. Opposite Smad and chicken ovalbumin upstream promoter transcription factor inputs in the regulation of the collagen VII gene promoter by transforming growth factor-beta. J Biol Chem 2004;279:23759–23765 [DOI] [PubMed] [Google Scholar]
- 61.Burton MJ, Kinteh F, Jallow O, et al. A randomised controlled trial of azithromycin following surgery for trachomatous trichiasis in the Gambia. Br J Ophthalmol 2005;89:1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burton MJ, Adegbola RA, Kinteh F, et al. Bacterial infection and trachoma in The Gambia: a case control study. Invest Ophthalmol Vis Sci 2007;48:4440–4444 [DOI] [PubMed] [Google Scholar]
- 63.Mans JJ, von Lackum K, Dorsey C, et al. The degree of microbiome complexity influences the epithelial response to infection. BMC Genomics 2009;10:380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tzu-Min H, Shin CC, Wei-Hsuan Y, et al. A novel nonsynonymous variant of matrix metalloproteinase-7 confers risk of liver cirrhosis. Hepatology 2009;9999:NA [DOI] [PubMed] [Google Scholar]
- 65.Beeghly-Fadiel A, Shu XO, Long J, et al. Genetic polymorphisms in the MMP-7 gene and breast cancer survival. Int J Cancer 2009;124:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.