This is a report of gene array analysis of glaucomatous retinal ganglion cells isolated by laser capture microdissection. Several novel and unexpected pathways were identified that are influenced by elevation of intraocular pressure.
Abstract
Purpose.
Intraocular pressure (IOP) is an important risk factor in glaucoma. Gene expression changes were studied in glaucomatous rat retinal ganglion cells (RGCs) to elucidate altered transcriptional pathways.
Methods.
RGCs were back-labeled with Fluorogold. Unilateral IOP elevation was produced by injection of hypertonic saline into the episcleral veins. Laser capture microdissection (LCM) was used to capture an equal number of RGCs from normal and glaucomatous retinal sections. RNA was extracted and amplified, labeled, and hybridized to rat genome microarrays, and data analysis was performed. After selected microarray data were confirmed by RT-qPCR and immunohistochemistry, biological pathway analyses were performed.
Results.
Significant changes were found in the expression of 905 genes, with 330 genes increasing and 575 genes decreasing in glaucomatous RGCs. Multiple cellular pathways were involved. Ingenuity pathway analysis demonstrated significant changes in cardiac β-adrenergic signaling, interferon signaling, glutamate receptor signaling, cAMP-mediated signaling, chemokine signaling, 14-3-3-mediated signaling, and G-protein-coupled receptor signaling. Gene set enrichment analysis showed that the genes involved in apoptotic pathways were enriched in glaucomatous RGCs. The prosurvival gene Stat3 was upregulated in response to elevated IOP, and immunohistochemistry confirmed that Stat3 and phosphorylated-Stat3 levels were increased in RGCs in experimental glaucoma. In addition, the expression of several prosurvival genes normally expressed in RGCs was decreased.
Conclusions.
There are extensive changes in gene expression in glaucomatous RGCs involving multiple molecular pathways, including prosurvival and prodeath genes. The alteration in the balance between prosurvival and prodeath may contribute to RGC death in glaucoma.
Glaucoma is the second leading cause of blindness worldwide.1 It is a neurodegenerative disease that is characterized by the slow and progressive degeneration of retinal ganglion cells (RGCs) and their axons, leading to atrophy of the optic nerve and loss of vision. Elevated intraocular pressure (IOP) is the leading risk factor for loss of RGCs and development of optic nerve atrophy. It is now clear that RGCs die by apoptosis in glaucoma.2–5 However, the trigger of the apoptosis is still unknown. To successfully target the mechanisms, we must to understand the molecular signaling networks in RGC death. We used laser capture microdissection (LCM) to specifically capture RGCs, and microarray technology, an accepted and powerful tool for large-scale gene expression profiling, to elucidate a global view of gene interaction networks in RGCs.
To date, several gene expression studies have been performed on the retina as a whole, to catalog changes in transcription that accompany glaucoma. The mRNA expression patterns in whole retina of several animal models of glaucoma, including DBA/2J mice,6 cynomolgus macaque monkeys,7 rat,8–11 and C57B16/J mice12 have been investigated. These studies revealed up- and downregulation of many genes in response to elevated IOP. However, the outcome and interpretation of these microarray studies can be strongly affected by inherent tissue heterogenicity, as the retina is a complex tissue composed of neuronal, glial, and vascular cell types. The RGCs comprise only 1% or less of the retinal cells. It is therefore difficult to causally link changes in gene expression patterns from whole retina to specific RGC gene expression, the associated pathways and subsequent physiological effects. In addition, site-specific, pathology-related gene expression changes can be diluted by the gene expression profile of other cell types in a whole retinal homogenate and thus not be readily detectable. Previous efforts to identify the complete picture of retinal gene expression may thus have missed important RGC genes, due to the low relative frequency of RGCs and, hence, their mRNA transcripts. LCM overcomes such obstacles.13 LCM allows morphologic preservation of tissue and the laser does not damage macromolecules such as DNA, RNA, or proteins.
In the present study, we sought to investigate the whole-genome regulation of RGCs under conditions of elevated IOP. We hypothesized that gene expression would change in glaucomatous RGCs compared with normal RGCs in a rat model of experimental glaucoma. To test our hypothesis, we applied LCM to capture an equal number of glaucomatous RGCs and normal RGCs and used microarray analysis to identify the genes and functional gene classes most altered in expression in the rat RGCs after exposure to experimentally elevated pressure. Then, we used real-time quantitative PCR (RT-qPCR) to validate selected changes in gene expression identified by microarray analysis. The JAK/STAT signaling pathway, especially the Stat3 protein, has been implicated in neurodegenerative and neuroprotective mechanisms.14 The phosphorylation of Stat3 is known to activate Stat3 and has been shown to be protective of RGCs after ischemic insult.15–17 The increased transcript expression of the prosurvival gene, signal transducer, and activator of transcription 3 (Stat3) was confirmed at the protein level in RGCs by immunohistochemistry (IHC). In addition, Stat3 phosphorylation was seen in RGCs after IOP elevation.
Methods
Study Design
The study was performed in five adult male Brown Norway rats. RGCs were back-labeled with Fluorogold. Seven days later, unilateral elevation of IOP was produced in one eye by injecting hypertonic saline into episcleral veins. The fellow eye served as the control. After 10 days of elevated IOP, the rats were killed, and the eyes were processed for LCM. First, to confirm the enrichment of RGCs, the gene expression of glial fibrillary acidic protein (Gfap, an astrocyte marker) and thymus cell antigen 1, theta (Thy1, an RGC marker) was compared between captured RGCs and captured ganglion cell layer (GCL) from normal eyes. Next, two thousand RGCs were captured from each normal eye and each eye with elevated IOP in three rats. Gene expression in the glaucomatous RGCs was compared with that in the fellow normal RGCs by gene microarray (Rat GeneChip 230.2; Affymetrix, Santa Clara, CA). Finally, an equal number of RGCs were captured from five rats, including the three rats from the microarray experiments. RT-qPCR was used to verify selected changes in gene expression initially identified by microarray analysis. Retinal tissue sections were also used to perform IHC localization of Stat3 and phosphorylated-Stat3.
Experimental Glaucoma
All animal-related procedures were performed in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Five adult male Brown Norway rats (300–450 g; Charles River, Wilmington, MA) were maintained in covered cages with a 12-hour light–dark cycle and fed with a standard rodent diet ad libitum.
RGCs were bilaterally back-labeled by stereotaxic injection of 3% Fluorogold (Fluorochrome LLC, Denver, CO) into the superior colliculus of deeply anesthetized rats, as described previously.3 The animals were allowed to recover for a week to allow for adequate back-labeling of RGCs before experimental glaucoma was induced.
Anesthesia was induced with a mixture of acepromazine maleate (1.5 mg/kg), xylazine (7.5 mg/kg), and ketamine (75 mg/kg; all from Webster Veterinary Supply, Sterling, MA). To elevate IOP, hypertonic 1.9 M saline was injected into limbal aqueous humor–collecting veins of one eye, as originally described by Morrison et al.18 The fellow eye served as the control.
IOP was measured with a handheld tonometer (TonoPen XL; Medtronic Ophthalmics, Jacksonville, FL). Before the first hypertonic saline injection, baseline IOPs for both eyes were measured on three consecutive days for each rat. During the induction of experimental glaucoma, IOP in each eye was measured every other day while the rats were in the awake state. To minimize diurnal variability in IOP, measurements were taken between 10 AM and 2 PM. To assess total IOP exposure, we calculated integrated IOP as the area under the pressure–time curve (AUC = experimental eye − control eye), beginning with the day of the first saline injection. After IOP had been elevated for 10 days, the rats were killed by asphyxiation with carbon dioxide, the eyes were rapidly removed, and the eye cups were collected.
Tissue Preparation and Assessment
After the eyes were enucleated, the posterior segments were minimally fixed in 4% paraformaldehyde for an hour and cryoprotected with serial sucrose dilutions. Eye cups were frozen in optimal cutting temperature compound (OCT; Tissue-Tek; Miles Inc., Diagnostic Division, Elkhart, IN) and sectioned in their entirety at 16 μm. These sections were used for RGC counting, LCM of RGCs, and IHC.
Estimation of total RGC number in each retina was performed with statistically unbiased stereological protocols by using an optical dissector and a fractionator, as previously described.3,19 Fluorogold-positive cells were immunostained with an anti-Fluorogold antibody (1:200, Fluorochrome LLC) and counted (C.A.S.T. system; ver. 2.3.1.2; Olympus, Albertslund, Denmark).
Optic nerves were assessed in saline-injected eyes and fellow eyes. All corresponding optic nerve sections were taken from approximately 2 mm posterior to the globe, fixed in glutaraldehyde, embedded in plastic, stained with toluidine blue, and evaluated by light microscopy. Optic nerve damage was graded from 1 (no injury) to 5 (degeneration involving the total nerve area) by four masked observers who used a modification of the method described by Morrison.3,18
LCM of Fluorogold-Labeled RGCs
For LCM, a commercial system (PixCell IIe; Arcturus, Mountain View, CA) was used, as previously described.3,20 Briefly, the tissue sections were dehydrated by serial immersions in 75%, 95%, and 100% ethanol for 2 minutes, followed by a 2-minute immersion in xylene. Tissue sections were then air-dried for 2 to 5 minutes at room temperature. A cleanup pad (Cap-Sure; Arcturus) was used to remove any extra tissue on the slide. RGCs were identified by the presence of Fluorogold back-labeling (excitation filter, 330–380 nm; barrier filter, 400 nm). An equal number of RGCs from each eye were captured onto single caps (Cap-Sure MacroLCM Caps; Arcturus), by using the following parameters: spot size, 7.5 μm; power, 65 mW; pulse duration, 750 μs. For the microarray experiment, 2000 RGCs were captured from each eye. RGCs were captured from approximately 30 sections per eye. Because RGCs were captured from retinal cross sections, the sampled area included central and peripheral retina. The microdissected samples were collected in a noncontact and contamination-free manner, immediately lysed by 20 μL LCM extraction buffer (Arcturus). For RT-qPCR, 2600 RGCs were captured from each eye.
RGC Enrichment
To assess the enrichment of RGCs, LCM-captured RGCs from a normal eye were compared to LCM-captured GCL from the same normal eye. The gene expression of Gfap and Thy1 was compared between LCM-captured RGCs and LCM-captured GCL.
RNA Isolation and Microarray Experiments
For each eye of the rats (n = 5 eyes), the Fluorogold-labeled RGCs on each cap were lysed with 20 μL of extraction buffer for 30 minutes at 42°C, and centrifuged (12,500g) from the caps into microfuge tubes. Isolated RGCs were processed for RNA extraction (PicoPure RNA; Arcturus) according to the manufacturer's protocol. The total cellular RNA was finally resuspended in 14 μL elution buffer. One microliter of the isolate was used to determine the quality of the RNA by detecting 28S/18S ribosomal RNA peaks (RNA Pico 6000 LabChip with a model 2100 Expert Bioanalyzer; Agilent Technologies, Palo Alto, CA).
Isolated RNA was amplified by linear amplification (RiboAmp HS RNA amplification kit; Arcturus). One microliter of the resuspended isolate was run on the bioanalyzer, to estimate average fragment length of the aRNA molecules. The labeling and hybridization of the cRNA was performed at the NIH Neuroscience Microarray Consortium (Keck Microarray Resource, New Haven, Yale University School of Medicine, New Haven, CT). Briefly, 10 μg of total RNA was used for cRNA labeling, according to the recommended microarray protocol (Affymetrix) and hybridized to the rat gene chip (Rat 230.2 chip; Affymetrix) for 16 hours at 45°C in a rotisserie oven. After hybridization, the arrays were washed using the fluidics station and stained with streptavidin-phycoerythrin according to the technical manual (Affymetrix). Washed arrays were scanned (GeneChip Scanner 3000; Affymetrix). Scanned output files were visually inspected for hybridization artifacts and then analyzed (GeneChip Operation Software [GCOS]; Affymetrix). The software recorded intensities for perfect match (PM) and mismatch (MM) oligonucleotides, and determined whether genes were present (P), marginal (M), or absent (A).
Microarray Data Analysis
The output .CEL files obtained from the software server (GCOS; Affymetrix) were processed and normalized by dChip.21 The probe level analysis was performed by dChip, which normalizes arrays to the PM and MM probes before computing model-based expression levels. Briefly, invariant set normalization was performed, the PM-only model was used for generating gene signal intensities, and the significance of changes in gene expression due to elevated pressure was determined. The parameters were set with a false-discovery rate (FDR) of 5%, and a minimum change of twofold, to identify the genes with potential biological significance. Because of the small sample size, a threshold of the percentage of samples called “present” in the samples used in both groups was set as 30%, to filter the significantly changed genes. That means the significantly changed genes that meet dChip comparison criteria may be absent from some chips but not all chips. Biologically, it is reasonable that genes expressed in diseased cells may not be present in normal cells or vice versa. Therefore, only the genes absent in all samples were ignored. All microarray data have been deposited in the NIH/NLM Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/projects/geo/ provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD) and are accessible through GEO Series accession number GSE12596.
Then, the genes were subjected to bioinformatics analysis. Pathway analysis (Ingenuity Systems, Redwood City, CA) was performed to determine canonical pathways with expression that is differentially affected between glaucomatous RGCs and normal RGCs (www.ingenuity.com). Gene set enrichment analysis (GSEA) was used to assess whether a set of functionally related genes would show statistically significant differences between glaucomatous RGCs and normal RGCs.22,23
Real-Time Quantitative PCR
Total RNA was isolated from 2600 RGCs from five pairs of eyes, including the same set of eyes used for microarray analysis plus an additional two pairs (Table 1). The procedure is the same as the RNA extraction for microarrays. Reverse transcription (RT) was performed, according to the manufacturer's specifications (Invitrogen Corp., Carlsbad, CA), with 13 μL of extracted RNA from each LCM sample and 200 U of RT polymerase (Superscript; Invitrogen).
Table 1.
IOP Characteristics of the Rats
| Eye Pair | Total AUC | Average IOP* |
Peak IOP* |
||
|---|---|---|---|---|---|
| Control | Experimental | Control | Experimental | ||
| 1†‡ | 155 | 19.13 | 29.96 | 19.30 | 43.69 |
| 2†‡ | 195 | 17.78 | 34.73 | 18.4 | 44.87 |
| 3†‡ | 169 | 16.77 | 30.94 | 19.47 | 45.06 |
| 4‡ | 209 | 18.78 | 32.30 | 20.53 | 43.73 |
| 5‡ | 150 | 19.13 | 31.90 | 19.93 | 41.13 |
AUC, area under the time-pressure curve, used to assess exposure to IOP.
Both the average IOP and the peak IOP of experimental group are significantly higher than the control group (P < 0.0001).
Eye pairs 1–3 used for microarray experiment.
Eye pairs 1–5 used for RT-qPCR confirmation.
The amount of cDNA corresponding to 2600 RGCs was amplified with the primer pairs for Gapdh, Nrg1, Cpg15, Ywhab, Ywhaz, Grb2, Map2k1, C3, C1qa, C1qb, Stat3 and Stat5a (Table 2). mRNA extracted from Brown Norway rat brain was reverse transcribed and used as the reference sample. RT-qPCR was performed (iCycler iQ; Bio-Rad, Hercules, CA) with SYBR green PCR master mix (Bio-Rad). A computer program (SDS2.1 software; Bio-Rad) was used to visualize the data. The standard curve method was used to determine relative changes in gene expression levels with Gapdh serving as the reference, which did not change significantly in our samples. A Student's t-test was conducted to compare the normalized relative expression values between the control and high-IOP eyes, to determine statistical significance. To confirm the identity of real-time PCR reaction products, a DNA chip (Agilent Technologies) and a bioanalyzer (model 2100 Agilent Technologies, Palo Alto, CA) were used to verify the size of the products. The amplicons of selected genes, Nrg1 and Cpg15, were sequenced at the DNA Sequencing Center for Vision Research, Massachusetts Eye and Ear Infirmary, Boston, MA. Sequence data were aligned with sequence analysis software (Chromas, ver. 2.33; Tewantin, QLD, Australia) and compared with the published gene sequence by an NCBI BLAST search (www.ncbi.nlm.nih.gov/blast/ provided in the public domain by NCBI, Bethesda, MD).
Table 2.
Primers Used for RT-qPCR
| Gene Name | GenBank* | Primers Sequence | Amplicon Size (bp) |
|---|---|---|---|
| C1qa: complement component 1, q subcomponent, alpha polypeptide | NM_001008515 | F: TTG CAG GAA TTC CTG GCC GCC C | 207 |
| R: CTT TCA CAC CTT TCA ACC CTT GGG G | |||
| C1qb: complement component 1, q subcomponent, beta polypeptide | NM_019262 | F: CCT GGG TTT GCT CCA TGT CTC CT | 213 |
| R: AAA TTT GCC TGG GAT CCC AGG G | |||
| C3: complement component 3 | NM_016994 | F: CTC TGG GGA GAA AAG CCC AAT ACC | 97 |
| R: AAA CCA CCA TTG TTT CTG TGA ATG CCC | |||
| Cpg15: candidate plasticity gene 15 | NM_053346 | F: TAT CCA AGG CAG CTT ATT CG | 200 |
| R: ATG GCT CTT CCT CTC GAT TT | |||
| Gapdh: glyceraldehyde-3-phosphate dehydrogenase | NM_017008 | F: CCA ATG TAT CCG TTG TGG AT | 177 |
| R: CAT CAA AGG TGG AAG AAT GG | |||
| Gfap: glial fibrillary acidic protein | NM_017009 | F: TGG ACA CCA AAT CTG TGT CAG AAG G | 117 |
| R: ATC ACA TCC TTG TGC TCC TGC TTC G | |||
| Grb2: growth factor receptor bound protein 2 | NM_030846 | F: AAG GTG CTT CGT GAC GGA GCC | 180 |
| R: GGT CAA AGT CAA AGA GTG CCT GG | |||
| Map2k1: mitogen activated protein kinase kinase 1 | NM_031643 | F: TTG GAT CAA GTG CTG AAG AAA GCT GG | 135 |
| R: GTT GGA AGG CTT GAC ATC TCT GTG C | |||
| Nrg1: neuregulin 1 | NM_031588 | F: GGT GAA GGA CCT GTC AAA CCC G | 218 |
| R: GCT TCT GCC GCT GCT TCT TGG | |||
| Stat3: signal transducer and activator of transcription 3 | NM_012747.2 | F: TTA TCA GCT TAA AAT TAA AGT GTG C | 183 |
| R: ATT CCC ACA TCT CTG CTC CC | |||
| Stat5a: signal transducer and activator of transcription 5a | NM_017064.1 | F: CAG CAG ACC ATC ATC CTG G | 153 |
| R: ATC TGC TGC CGG TTC TGC | |||
| Thy1: thymus cell antigen 1, theta | NM_012673 | F: ACA TGT GTG AAC TTC GAG TCT CGG G | 99 |
| R: GCT TAT GCC ACC ACA CTT GAC CAG | |||
| Ywhab: tyrosin 3-monooxygenase/tyyptophan 5-monooxygenase activation protein, beta polypeptide | NM_019377 | F: TCC TGA AAA GGC CTG TAG CCT GG | 184 |
| R: TTC TCT CCC TCT CCA GCA TCT CC | |||
| Ywhaz: tyrosin 3-monooxygenase/tyyptophan 5-monooxygenase activation protein, zeta polypeptide | NM_013011 | F: ATC AGA CTG GGT CTG GCC CTC AA | 216 |
| R: TGC TTC GTC TCC TTG GGT ATC CG |
http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD.
Immunohistochemistry
The avidin–biotin–peroxidase complex and 3,3-diaminobenzidine tetrahydrochloride (ABC-DAB) procedure to determine Stat3 expression was performed on retinal sections from normal and elevated-IOP eyes. The 16-μm-thick cryostat sections were thawed and air dried for 10 minutes at room temperature. The sections were incubated in 0.3% H2O2 for 10 minutes, washed in 0.1 M phosphate-buffered saline (PBS), blocked, and incubated with Stat3 (1:100; Cell signaling) for 48 hours. Biotinylated anti-rabbit IgG was used as the secondary antibody. After they were washed with PBS, the sections were treated in ABC (Vectastain Elite ABC kit, Vector Laboratories, Burlingame, CA) solution according to the instructions of the supplier. After ABC treatment and washing in PBS, the slides were treated with a DAB Kit (Vector Laboratories) for 2 to 3 minutes. The reaction was terminated by rinsing in water. After several washes in distilled water, the slides were dehydrated in ethanol, cleared in xylene, and coverslipped (Permount; Fisher Scientific, Pittsburgh, PA).
Immunofluorescence staining for Stat3 and phosphorylated-Stat3 was also performed on retinal sections from normal and elevated-IOP eyes. Sections were incubated with a blocking solution for 1 hour at room temperature and incubated with primary antibodies specific for Stat3 (1:400, Cell Signaling, Beverly, MA) or phosphorylated-Stat3 (1:250, cell signaling) at 4°C overnight. Anti-goat antibodies (Alexa 594; Molecular Probes, Eugene, OR) were used as secondary antibodies with 1-hour incubation at room temperature. Fluorescence staining was detected with a fluorescence microscope (Olympus, Tokyo, Japan).
Results
Evaluation of the High-IOP–Induced Glaucoma Model
To quantify glaucoma progression, we calculated the integrated IOP (AUC), assessed RGC loss, and graded optic nerve damage. Table 1 shows the data for all samples processed for microarray and RT-qPCR. Animals with similar IOP histories were chosen. The average RGC loss was 33%. Two available optic nerves were graded at 3.49 and 3.50, suggesting moderate retina damage (scale: 1, no damage; 5, severe damage).
RGC Enrichment
RGCs were identified by the presence of Fluorogold back-labeling and captured for microarray analysis. Because astrocytes and amacrine cells occupy the ganglion cell layer along with RGCs, we sought to assess the efficiency of LCM in enriching each sample for RGCs. We determined the specific mRNA expression of Gfap (astrocyte marker) and Thy1 (RGCs marker) between LCM-captured RGCs and LCM-captured GCL (Table 3). Compared with LCM-captured GCL, Gfap was 65.04-fold less abundant in LCM-captured RGCs (P = 0.006). No significant differences in Thy-1 levels were found between LCM captured-RGCs and LCM-captured GCL (P = 0.07). In addition, our microarray findings showed that the transcriptional expression of macrophage markers (CD14, CD68, and epidermal growth factor module-containing mucin-like receptor 1 [EMR1]) and an amacrine cell marker (HPC-1/syntaxin 1A) were not significantly different between normal and glaucomatous RGCs. The change ratios of CD14, CD68, EMR1, and syntaxin 1A were 1.23, 1.04, −1.12, and 1.14, respectively. These results indicate that although some astrocytes were captured along with the RGCs, Fluorogold labeling effectively guided LCM harvesting of the RGCs. LCM capture successfully captured RGCs and effectively excluded most glial cells, macrophages, and amacrine cells.
Table 3.
The mRNA Expression of Gfap and Thy1 among LCM-Captured RGC and GCL*
| Marker | LCM RGC (n = 3) | LCM GCL (n = 3) | P | GCL:RGC Ratio |
|---|---|---|---|---|
| Gfap | 0.49 ± 0.05 | 31.87 ± 4.79 | 0.006 | 65.04 |
| Thy1 | 47.00 ± 1.97 | 29.13 ± 4.86 | NS | 0.62 |
The relative expression level (mean ± SD) of each gene was calculated based on the standard curve, with Gapdh used as a reference. Gapdh did not change significantly in the samples.
Identification of Differentially Expressed Genes
Microarrays were used to survey the change in gene expression of RGCs from normal eyes and high-IOP eyes. A total of 1532 genes were significantly changed (Supplementary Table S1, http://www.iovs.org/cgi/content/full/51/8/4084/DC1). After FDR correction for multiple testing, 905 genes remained significant with a change of twofold or more in expression, with 330 genes increasing and 575 genes decreasing in glaucomatous RGCs (Supplementary Table S2, http://www.iovs.org/cgi/content/full/51/8/4084/DC1). Analysis of the main gene ontology categories revealed that the genes involved in apoptosis and response to oxidative stress were among the most upregulated (Table 4), including Angptl4, Ednrb, Pnkp, Gpx1, Ca3, RT1-Aw2, and Nek6. On the other hand, the genes vital for neuron, axon, and dendrite development were among the most downregulated (Table 5), which include Gja1, Snap25, Cit, Nrep, Gabrg2, and Vdac3.
Table 4.
The Thirty Most Upregulated Genes in RGCs with Elevated IOP
| GenBank* | Gene Name | Change Ratio | GO Biological Process |
|---|---|---|---|
| NM_017009 | Gfap: glial fibrillary acidic protein | 19.9 | Intermediate filament-based process |
| AA945643 | Chi3l1: chitinase 3-like 1 | 10.4 | Carbohydrate metabolic process |
| AI176253 | Nyw1: Ischemia related factor NYW-1 | 8.17 | Unknown |
| NM_021584 | Ania4: activity and neurotransmitter-induced early gene protein 4 (ania-4) | 6.38 | Unknown |
| NM_022512 | Acads: acetyl-Coenzyme A dehydrogenase, short chain | 5.51 | Metabolic process, response to glucocorticoid stimulus, oxidation reduction |
| NM_013154 | Cebpd: CCAAT/enhancer binding protein (C/EBP), delta | 4.67 | Transcription, regulation of transcription, DNA-dependent |
| AI170182 | Man1a2_predicted: mannosidase, alpha, class 1A, member 2 (predicted) | 4.07 | Unknown |
| NM_031140 | Vim: vimentin | 3.86 | Intermediate filament-based process |
| BF415939 | Fos: FBJ murine osteosarcoma viral oncogene homolog | 3.83 | Regulation of transcription, DNA-dependent, nervous system development |
| BG380285 | Ifitm1_predicted: interferon induced transmembrane protein 1 (predicted) | 3.81 | Response to biotic stimulus |
| AA818262 | Angptl4: angiopoietin-like 4 | 3.75 | Angiogenesis, response to hypoxia, signal transduction, negative regulation of apoptosis |
| X57764 | Ednrb: endothelin receptor type B | 3.71 | Neural crest cell migration, protein amino acid phosphorylation, signal transduction, apoptosis, pigmentation |
| NM_016994 | C3: complement component 3 | 3.66 | GPCR protein signaling pathway, complement activation, inflammatory response |
| AW520745 | Pnkp: polynucleotide kinase 3′-phosphatase | 3.69 | DNA repair, response to oxidative stress, dephosphorylation, nucleotide phosphorylation |
| AI317842 | Kif22: kinesin family member 22 | 3.60 | Microtubule-based movement |
| S41066 | Gpx1: glutathione peroxidase 1 | 3.54 | Release of cytochrome c from mitochondria, regulation of inflammatory response to antigenic stimulus, metabolic process, regulation of caspase activity, regulation of neuronal apoptosis |
| AB030829 | Ca3: carbonic anhydrase 3 | 3.48 | Metabolic process, response to oxidative stress |
| AI070137 | Aoc3: amine oxidase, copper containing 3 | 3.43 | Cell adhesion, amine metabolic process |
| BE102350 | RT1-Aw2: RT1 class Ib, locus Aw2 | 3.31 | Apoptosis, inflammatory response, calcium-mediated signaling, regulation of synapse structure and activity, regulation of axonogenesis |
| NM_019144 | Acp5: acid phosphatase 5, tartrate resistant | 3.30 | Bone resorption |
| BI300770 | Igtp: interferon gamma induced GTPase | 3.30 | Unknown |
| NM_019358 | Pdpn: podoplanin | 3.30 | Cell morphogenesis, prostaglandin metabolic process, transport, signal transduction, cell proliferation |
| NM_031832 | Lgals3: lectin, galactose binding, soluble 3 | 3.29 | Extracellular matrix organization and biogenesis |
| BF411958 | Rufy1: RUN and FYVE domain containing 1 | 3.20 | Protein transport, regulation of endocytosis |
| AI407092 | Rdx: Radixin | 3.18 | Microvillus biogenesis, apical protein localization |
| BF281577 | Rcn1_predicted: Reticulocalbin 1 (predicted) | 3.17 | Ciliary or flagellar motility |
| AI072892 | Frzb: frizzled-related protein | 3.10 | Regulation of Wnt receptor signaling pathway |
| NM_133609 | Eif2b3: eukaryotic translation initiation factor 2B, subunit 3 gamma | 3.07 | Regulation of Rho protein signal transductioniosynthetic process, oligodendrocyte, Regulation of translational initiation in response to stress |
| BE113920 | Stat3: signal transducer and activator of transcription 3 | 3.06 | Eye photoreceptor cell differentiation, transcription, acute-phase response, signal transduction, JAK-STAT cascade |
| BF282365 | Nek6: NIMA (never in mitosis gene a)-related expressed kinase 6 | 2.90 | Protein amino acid phosphorylation, apoptosis, regulation of I-κB kinase/NF-κB cascade |
http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD.
Table 5.
The Thirty Most Downregulated Genes in RGCs with Elevated IOP
| GenBank* | Gene Name | Change Ratio | GO Biological Process |
|---|---|---|---|
| AI411352 | Gja1: gap junction membrane channel protein alpha 1 | −10.46 | Embryonic development, neuronal migration, cell communication, cell-cell signaling, positive regulation of I-κB kinase/NF-κB cascade, neurite morphogenesis, protein oligomerization, regulation of calcium ion transport |
| D13921 | Acat1: acetyl-coenzyme A acetyltransferase 1 | −10.26 | Metabolic process |
| NM_030991 | Snap25: synaptosomal-associated protein 25 | −9.16 | Exocytosis, neurotransmitter secretion, axonogenesis, regulation of synaptogenesis |
| AI411995 | Ube2d3: Ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | −9.00 | Ubiquitin-dependent protein catabolic process |
| AI717472 | Tyrp1: tyrosinase-related protein 1 | −8.07 | Melanin metabolic process, pigmentation |
| AA957183 | Cit: Citron | −7.89 | Mitotic sister chromatid segregation, dendrite development, generation of neurons |
| BE105102 | Glrx2: Glutaredoxin 2 (thioltransferase) | −7.74 | Transport, response to organic substance, oxidation reduction |
| AW529483 | Slc4a5: Solute carrier family 4, sodium bicarbonate cotransporter, member 5 | −7.67 | Unknown |
| NM_057205 | Ube1c: ubiquitin-activating enzyme E1C | −6.73 | Mitotic cell cycle, protein modification process, metabolic process, regulation of cell cycle |
| AA851874 | Spg3a: spastic paraplegia 3A homolog (human) | −6.64 | Unknown |
| BG380454 | Basp1: brain abundant, membrane attached signal protein 1 | −6.45 | Unknown |
| BE104618 | Dhrs1: Dehydrogenase/reductase (SDR family) member 1 | −6.34 | Metabolic process |
| BE107450 | Nrep: Neuronal regeneration related protein | −5.95 | Regulation of TGFβ receptor signaling pathway, axon regeneration, regulation of neuronal differentiation |
| BG372673 | Trim32: tripartite motif protein 32 | −5.92 | Unknown |
| AI408819 | H3f3b: H3 histone, family 3B | −5.91 | Nucleosome assembly |
| BE120391 | Gabrg2: gamma-aminobutyric acid A receptor, gamma 2 | −5.79 | Transport, synaptic transmission, post-embryonic development |
| AI169170 | Eif4a2: eukaryotic translation initiation factor 4A2 | −5.68 | Translation |
| NM_013128 | Cpe: carboxypeptidase E | −5.61 | Proteolysis, insulin processing |
| BF284830 | Yipf3: Yip1 domain family, member 3 | −5.53 | Cell differentiation |
| NM_013219 | Cadps: Ca2+-dependent secretion activator | −5.24 | Transport, vesicle fusion, positive regulation of calcium ion-dependent exocytosis |
| AI029445 | Ldb2_predicted: LIM domain binding 2 (predicted) | −5.17 | Multicellular organismal development |
| AI172276 | Ppp1r2: protein phosphatase 1, regulatory (inhibitor) subunit 2 | −5.13 | Metabolic process, nervous system development, regulation of signal transduction, regulation of phosphoprotein phosphatase activity |
| BM388861 | Col9a1: procollagen, type IX, alpha 1 | −5.13 | Chondrocyte differentiation, phosphate transport |
| NM_053562 | Rpe65: retinal pigment epithelium 65 | −5.07 | Eye development, regulation of rhodopsin gene expression, retinal metabolic process |
| NM_057104 | Enpp2: ectonucleotide pyrophosphatase/phosphodiesterase 2 | −5.05 | Chemotaxis, metabolic process, lipid catabolic process |
| NM_013011 | Ywhaz: tyrosin 3-monooxygenase/tyyptophan 5-monooxygenase activation protein, zeta polypeptide | −5.03 | Anti-apoptosis, protein targeting, signal transduction |
| AI145413 | Gabra1: gamma-aminobutyric acid A receptor, alpha 1 | −4.94 | Transport, gamma-aminobutyric acid signaling pathway development |
| NM_031825 | Fbn1: fibrillin 1 | −4.91 | |
| AF439397 | Zwint: ZW10 interactor | −4.90 | Exocytosis, cell cycle, mitosis, cell division |
http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD.
The expression of several prosurvival genes was altered in glaucomatous RGCs. Several prosurvival genes that are present in normal RGCs were significantly downregulated in RGCs from eyes with high IOP (Table 6), including Cpg15, Nrg1, Map2k1, Ywhab, Ywhaz, Vsnl1, Nfl, Stmn2, Nell2, and Grb2 (P < 0.05). On the other hand, the message levels of two prosurvival genes, Stat3 (3.06 ± 0.53, P < 0.05) and Stat5a (2.26 ± 0.55, P < 0.05), were increased in RGCs in response to elevated IOP.
Table 6.
Prosurvival and Neurogenesis Genes with Decreased Expression in Glaucomatous RGCs
| Gene | Function | Glaucomatous RGCs/Normal RGCs (Change Ratio) |
|---|---|---|
| Stmn2 | Microtubule depolymerization and synaptic plasticity | −6.26* |
| Ywhaz | Mitochondrial import stimulation factor | −5.03* |
| Vsnl1 | Calcium dependent signaling | −3.45* |
| Ywhab | Signal transduction | −3.33* |
| Ret | Neurogenesis | −2.72 |
| Cpg15 | Promotes neurite outgrowth | −2.61* |
| Nfl | Cytoskeleton organization | −2.45* |
| Grb2 | Signal transduction | −2.34* |
| Nell2 | Growth and differentiation of neural cells | −2.25* |
| Calb2 | Calcium ion binding | −2.08 |
| NCAML1 | Neural and glial cell adhesion | −2.08 |
| Map2k1 | Signal transduction | −2.06* |
| Nrg1 | Neural and organ development | −2.06* |
| Nrp | Neuronal development | −2.01 |
| Rgs4 | G-protein coupled receptor signaling | −2.01 |
| Ywhag | Signal transduction | −1.74 |
| Coch | Perception of sound | −1.68 |
| Pik3r1 | Negative regulation of apoptosis | −1.56 |
| Plcb1 | Production of the second messenger molecules | −1.56 |
| Gap43 | Neurite outgrowth and nerve regeneration | −1.47 |
| Rgs3 | G-protein coupled receptor signaling | −1.47 |
| Nef3 | Cytoskeleton organization | −1.4 |
| Rnd1 | Neuritic process formation | −1.4 |
| Inpp4a | Phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity | −1.38 |
| Ywhah | Negative regulation of apoptosis | −1.26 |
| Sema6b | Neurogenesis and axon guidance | −1.22 |
| Prph1 | Cytoskeleton organization | −1.17 |
| Fbxo2 | Maintaining neurons in a postmitotic state | −1.15 |
| Ptpn5 | Regulation the duration of ERK activation | 1.12 |
P < 0.05.
Two methods of biological pathway analysis were used to further analyze our data. First, pathway analysis (IPA; Ingenuity) yielded distinct canonical pathways related to the RGC response to elevated IOP, including cardiac β-adrenergic signaling, interferon signaling (the JAK/STAT pathway exists within the interferon signaling pathway), antigen presentation pathway, lysine biosynthesis, glutamate receptor signaling, cAMP-mediated signaling, chemokine signaling, leukocyte extravasation signaling, 14-3-3-mediated signaling, synaptic long-term potentiation, and G-protein-coupled receptor signaling (Table 7).
Table 7.
Top Canonical Pathways that Changed Significantly in RGC from Elevated IOP Eyes
| Pathway | P* | Ratio (%)† |
|---|---|---|
| Cardiac β-adrenergic Signaling | <0.0001 | 33.60 |
| Interferon Signaling (includes JAK/STAT) | 0.001 | 43.30 |
| Antigen presentation pathway | 0.001 | 30.80 |
| Lysine biosynthesis | 0.011 | 7.81 |
| Glutamate receptor signaling | 0.018 | 29.90 |
| cAMP-mediated signaling | 0.025 | 29.20 |
| Chemokine signaling | 0.026 | 31.60 |
| Leukocyte extravasation signaling | 0.030 | 24.5 |
| 14-3-3-mediated signaling | 0.035 | 28.20 |
| Synaptic long term potentiation | 0.035 | 29.5 |
| G-Protein coupled receptor signaling | 0.039 | 27.10 |
The P-value suggests whether there is an association between a specific pathway and uploaded dataset and whether it is due to chance.
The ratio is the number of molecules in a given pathway that meet cutoff criteria (twofold change), divided by total number of molecules that make up that pathway. The ratio is good for looking at which pathway has been affected the most based on the percentage of genes uploaded into IPA.
Second, GSEA demonstrated significant enrichment in complement coagulation cascades, Fas pathway, prostaglandin, and leukotriene metabolism, JAK/STAT signaling pathway, G-protein-coupled receptor database class (GPCRDB) A rhodopsin-like, prostaglandin synthesis regulation, and caspase pathway expression, whereas significant depletion in the citrate cycle, cholesterol biosynthesis, and G-protein-coupled receptor (GPCR) pathways was found in RGCs from high-IOP eyes (Table 8).
Table 8.
Gene Sets Enriched/Depleted in Glaucomatous RGC*
| Gene Set | Enrichment Score† | False-Discovery Rate q‡ |
|---|---|---|
| Enriched in glaucomatous RGCs | ||
| Complement coagulation cascades | 0.45 | <0.0001 |
| Fas pathway | 0.50 | <0.0001 |
| Prostaglandin and leukotriene metabolism | 0.44 | <0.0001 |
| JAK/STAT signaling | 0.42 | <0.0001 |
| G protein coupled receptor database class A rhodopsin-like | 0.35 | <0.0001 |
| Prostaglandin synthesis regulation | 0.47 | <0.0001 |
| Caspase pathway | 0.47 | <0.0001 |
| Depleted in Glaucomatous RGC | ||
| Citrate cycle | 0.55 | <0.0001 |
| Cholesterol biosynthesis | 0.81 | <0.0001 |
| G-protein-coupled receptor pathway | 0.57 | <0.0001 |
A total of 639 canonical pathways gene sets (http://www.broad.mit.edu/gsea/msigdb/genesets.jsp?collection=CP#; provided in the public domain by the Broad Institute, Massachusetts of Technology, Cambridge, MA) were calculated by gene set enrichment analysis.
Enrichment score (ES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes.
False-discovery rate is the estimated probability that a gene set with a given normalized ES represents a false-positive finding.
Validation of Selected Differentially Expressed Genes by RT-qPCR
To confirm selected microarray results, we examined expression levels by RT-qPCR after LCM in five pairs of eyes. Based on a combination of statistical analysis of microarray data and potential biological importance of the genes in question, 11 genes were chosen for RT-qPCR confirmation. These 11 genes represent the survival genes (Ywhaz, Ywhab, Cpg15, Grb2, Nrg1, Map2k1, Stat3, and Stat5a) and complement component genes (C3, C1qa, and C1qb). The change obtained by RT-qPCR correlated well with the direction of change obtained from the microarray data, confirming the validity of the microarray gene expression patterns (Table 9). Because RT-qPCR is more sensitive than microarray, RT-qPCR identified greater changes than microarray for these genes. In addition, several genes such as Stat5a and Ywhab were absent in some chips. RT-PCR confirmed the presence of these genes in both normal RGCs and glaucomatous RGCs. Direct sequencing confirmed the identity of all the genes selected to validate microarray results.
Table 9.
Validation of Microarray Gene Expression by Real-Time Quantitative PCR
| Gene | Ratio of Change (Mean ± SD) |
|
|---|---|---|
| Microarray (n = 3) | RT-qPCR (n = 5) | |
| Ywhaz: 14–3-3 family, zeta polypeptide | −5.03 ± 0.89* | −25.77 ± 5.42* |
| Ywhab: 14–3-3 family, beta polypeptide | −3.33 ± 0.55* | −23.14 ± 6.26* |
| Cpg15: candidate plasticity gene 15 | −2.61 ± 0.30* | −20.93 ± 9.79* |
| Grb2: growth factor receptor bound protein 2 | −2.34 ± 0.32* | −24.59 ± 10.43* |
| Nrg1: neuregulin 1 | −2.06 ± 0.30* | −6.56 ± 0.47* |
| Map2k1: mitogen activated protein kinase kinase 1 | −2.06 ± 0.23* | −9.73 ± 3.43* |
| C3: complement component 3 | 3.66 ± 1.01* | 5.07 ± 1.55* |
| C1qa: complement component 1, q subcomponent, alpha polypeptide | 1.39 ± 0.32 | 1.89 ± 0.45 |
| Stat3: signal transducer and activator of transcription 3 | 3.06 ± 0.53* | 7.28 ± 1.27* |
| Stat5a: signal transducer and activator of transcription 5a | 2.26 ± 0.55* | 2.74 ± 0.90* |
| C1qb: complement component 1, q subcomponent, beta polypeptide | 1.28 ± 0.11 | 2.13 ± 0.56* |
P < 0.05.
Upregulated Stat3 and Phosphorylated-Stat3 Protein in Glaucomatous RGCs
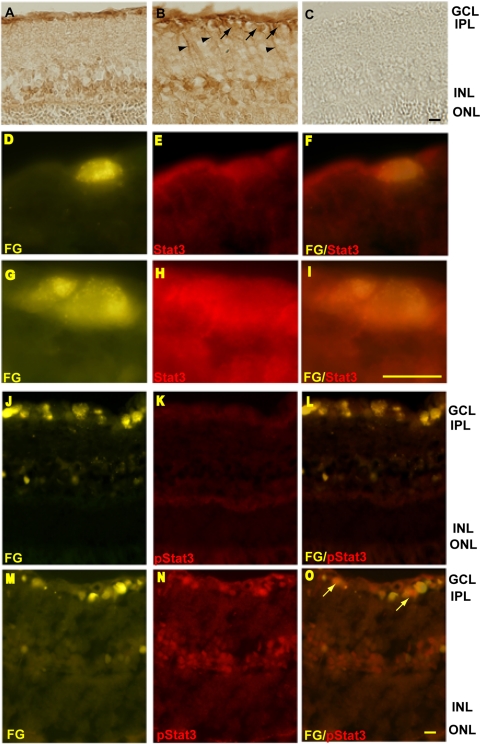
Stat3 was detected in the ganglion cells layers and inner nuclear layer (Fig. 1A) as previously described.24 Under conditions of high IOP, increased Stat3 immunoreactivity was seen in the GCL, inner nuclear layer, and inner plexiform layer, as well as in the cell processes (Fig. 1B). To further localize the increases in Stat3 and test whether Stat3 is activated in glaucomatous RGCs, the RGCs were back-labeled with Fluorogold, and the sections were fluorescently immunostained with Stat3 and phosphorylated-Stat3 antibodies. Figure 1F shows colocalization of Fluorogold and Stat3 in normal retina and Figure 1I shows increased Stat3 in an RGC under conditions of high IOP. Phosphorylated-Stat3 is expressed at low levels in normal RGCs (Fig. 1K), whereas it is increased in glaucomatous RGCs (Figs. 1N, 1O).
Figure 1.
Stat3 protein increased in the glaucomatous retina and in glaucomatous RGCs, and phospho-Stat3 was also increased in glaucomatous RGCs. (A) In the normal retina, positive staining for Stat3 was seen in the inner nuclear layer (INL) and GCL. (B) The expression of Stat3 was higher in the GCL in the high-IOP retina. (C) Negative control. (E) Low levels of Stat3 immunoreactivity were seen in Fluorogold-labeled RGCs in a retina with normal IOP. (G–I) In retinas with high IOP, increased Stat3 expression was seen in Fluorogold-labeled RGCs. (K–N) Phosphorylated-Stat3 was clearly increased in the GCL, including in RGCs (O compared with L). FG, Fluorogold. Arrow: retinal ganglion cell; Arrowhead: cell processes. Scale bars: 20 μm.
Discussion
Apoptosis is thought to be an acute or subacute event, yet in glaucoma, it takes weeks to years of elevated IOP to trigger RGC death. In an attempt to better understand upstream IOP-induced changes that may contribute to RGC dysfunction or apoptotic degeneration, we studied RGCs in animals exposed to IOP for 10 days, at a time point at which some, but not all, RGCs were likely to have died. Since RGCs account for only a small percentage of the cells in the retina and since it is these cells that are uniquely vulnerable to the consequences of elevated IOP in glaucoma, we developed a technique using LCM to isolate RGCs for microarray analysis.
Our work had several novel outcomes. In studies of changes in transcript profiles in experimental glaucoma, whole retina was used and a relatively modest number and diversity of gene expression changes were recorded. In comparison with these studies, we found detectable changes in nearly 1000 transcripts: 330 upregulated and 575 downregulated. The pattern of these changes highlights and reinforces several broad themes, including downregulation of many neuronal prosurvival genes and upregulation of apoptosis-related pathways and markers of inflammation, including complement pathways. We describe, for example, gene expression changes in Stat3 and extend these results to include evidence of increased protein levels of Stat3. In addition, we demonstrated an increase in Stat3 phosphorylation, which leads to the activation of Stat3 and may be an important prosurvival mediator. In addition, several unexpected but robust changes in patterns of gene expression emerged, including alterations in the GPCRs. These changes suggest new therapeutic targets for neuroprotection in glaucoma distinct from direct pressure-lowering approaches. The large number of individual transcripts and pathways altered in RGCs by exposure to elevated IOP suggest that the pathophysiological processes that ultimately are manifest by RGC degeneration are proceeded by a complex, likely integrated, set of changes in gene expression. The evidence implies that elevated IOP initiates transcriptional cascades that most likely contribute to neurodegeneration.
RGCs exist in a complex milieu and receive input from multiple different cell types, any of which may be influenced by elevated IOP. It is therefore important to study RGCs in situ to truly understand gene expression changes as a result of increased IOP. Whole retinal PCR is frequently used to study retinal gene responses to increased IOP. However, this technique does not differentiate responses of RGCs from other cell types. LCM, coupled with microarray analysis, is ideally suited to this challenge. Fluorogold back-labeling can effectively guide microscopic laser capture of neurons, as has been reported.25 In the present study, RGCs were identified by the presence of Fluorogold back-labeling and captured by LCM. Our results indicate that some astrocytes were probably captured along with the RGCs. Similar findings have been reported for LCM of substantia nigra neurons in which oligodendrocyte genes were found in the microarray results in histologic sections of a thickness similar to that of those used in our studies.25 Yao et al.25 suggest that the co-capture of neurons along with the surrounding glial cells may underscore the close association of these two cell types. Although we cannot exclude the possibility that some of the pathways identified as changing are changing in glial cells (astrocytes and/or Müller cell processes), in our control experiments we found that expression of the glial cell gene Gfap was 65.04-fold less abundant in LCM-captured RGCs than in LCM of the entire GCL. These results indicate that LCM results in a highly enriched sample of RGCs.
In this article, we report data from three pairs of eyes. Because of the small sample, we may have underestimated the actual changes, although publications of studies in which a similar number of paired samples was used have reported important data in other systems.26 qRT-PCR of independently laser-captured RGCs from five pairs of eyes confirmed the direction of change found in the microarray data in all cases.
Microarray analysis of isolated RGCs refines detection of gene responses to increased IOP. The current microarray study of RGC RNA yielded 905 genes with altered expression, many more than the number of significantly changed genes found in whole retina analyses from different glaucoma models (Table 10).6–12 Among the different microarray studies, some common themes were shared. The immune response genes were upregulated in the Morrison rat model,8 laser-induced monkey or rat model,7,11 and DBA2J mice.6 In the present study, complement component genes were enriched in glaucomatous RGCs confirming in situ hybridization observations by Stevens et al.27 A decrease in neurofilament genes has been reported in the monkey and the rat model of glaucoma7–9,11 and in our study, mRNA levels for Nefh, Nefm, and Nefl were decreased specifically in RGCs in response to elevated IOP. In addition, other structural genes vital for the survival or development of neuronal structures (Gja1, Snap25, Cit, Nrep, Gabrg2, and Vdac3) were among the most downregulated (Table 5). Genes vital for neuronal structure are affected by increased IOP, and these changes may contribute to RGC vulnerability in glaucoma or altered cellular functions that precede RGC death. Because of the detection ability of microarrays, some genes with small changes in expression may not have been detected in our microarray experiment. For example, Thy1 was not significantly changed in our microarray analysis of glaucomatous RGCs. We have reported that Thy1 mRNA levels were decreased approximately twofold in glaucomatous RGCs isolated by LCM followed by RT-PCR.20 Because RT-PCR has been shown to be more sensitive than microarrays,28 some genes with small changes in expression may not be detected by microarray experiments.
Table 10.
Comparison of Our Study with Previous Microarray Studies Using Whole Retina Glaucoma Model
| Our Study | Miyahara et al.7 | Ahmed et al.8 | Piri et al.9 | Steele et al.6 | Naskar et al.10 | Yang et al.11 | Walsh et al.12 | ||
|---|---|---|---|---|---|---|---|---|---|
| Species | Brown Norway rat | Cynomolgus monkey | Brown Norway rat | Wistar rat | DBA2J mouse | Rat | Wistar rat | C57 Mice | |
| Glaucoma model | Saline injection | Laser-induced glaucoma | Saline injection | Optic nerve transsection | 8 months after IOP elevation | Hereditary high IOP | Laser-induced glaucoma | Optic nerve transsection | Saline injection |
| Samples | RGC | Whole retina | Whole retina | Whole retina | Whole retina | Whole retina | Whole retina | Whole retina | |
| Array* | Affymetrix rat 230.2 | UniGEM Human ver 2.0 | Affymetrix rat U34A | Aligent rat G4130A | Affymetrix mouse 430.2 | MWG Biotech 10K chip | Affymetrix rat 230.2 | Affymetrix mouse 430.2 | |
| Statistic setting | >2-fold change | >1.7-fold change | >2-fold change | >2-fold change | >1.8-fold change | >3-fold change | >2-fold change | >1.5-fold change | |
| Number of upregulated genes | 330 | 45 | 74 | 85 | 32 | 75 | 347 | 212 | 50 |
| Number of downregulated genes | 575 | 17 | 7 | 21 | 36 | 45 | 196 | 480 | 140 |
| Major upregulated genes | Apoptosis genes (Table 4), Complement component genes (C1qa, C1qb, C3) | Immune response genes, glia activation genes, ceruloplasmin | Immune response genes, glia activation genes | Apoptotic genes, ceruloplasmin | Glial activation, immune response genes, signaling, gene expression | Glial activation genes | Early phase: immune and stress response genes. Late phase: genes related to protein synthesis | Early phase: genes related to cell death | Saa1, Saa2 |
| Major downregulated genes | Pro-survival genes (Cpg15, Nrg1, Ywhaz, Ywhab, Grb2, Nfl, Stmn2, Map2k1, Nell2, Vsnl1), neurogenesis genes (Gja1, Snap25, Cit, Nrep, Gabrg2 and Vdac3) | Neurofilament genes | Neurofilament genes | Neurofilament genes, Neuritin (Cpg15) | Multiple crystal genes | RGC-enriched genes; neurofilament genes | |||
Affymetrix, Santa Clara, CA; Agilent, Palo Alto, CA; UniGem, Incyte Genomics, Palo Alto, CA; MWG Biotech, High Point, NC.
In the present study, apoptosis and oxidative stress–related genes (Angptl4, Ednrb, Pnkp, Gpx1, Ca3, RT1-Aw2, Nek6, and Stat3) were among the most upregulated (Table 4). RGC death in glaucoma has been characterized as involving the apoptotic pathway of cell death.2,4,5 In addition, in the GSEA analysis, the caspase cascade was significantly enriched in glaucomatous RGCs, and one of the genes that contributed significantly to this enrichment was caspase 3. Also, growing evidence supports the possibility that oxidative stress is involved om glaucomatous neurodegeneration in different subcellular compartments of RGCs.29 Our findings support the view that both apoptosis and oxidative damage contribute to glaucomatous RGC death.
The GPCRs are a diverse family of transmembrane signaling proteins. They transduce a wide variety of cellular stimuli, and a large number of the drugs in current use exert their effects through GPCRs.30 In addition, multiple endogenous ligands are thought to act through GPCR pathways. Our data show an unexpected finding in glaucomatous RGCs of significant downregulation of the GPCR pathway. In addition, we found decreased expression of several adenylate cyclase genes and cAMP-dependent protein kinase (PKA) genes in RGCs from eyes with elevated IOP. It has been shown that GPCRs modulate neuronal signaling via specific G proteins in the central nervous system, including structures of the eye.31 In the present study, the decreased message levels of GPCR and G proteins in RGCs in glaucoma may lead to a decrease in adenylate cyclase–regulated cAMP signaling, which is critical for RGC survival.32,33
Alterations in neurotrophins and neurotrophin receptors have been implicated in RGC death in glaucoma with a decrease in retinal levels of both the levels of BDNF and in the Trkb receptors.34 Our observation that the GPCR pathway is downregulated prompts consideration of another intriguing possibility. Transactivation of the Trkb receptor through a GPCR-mediated, non-neurotrophin mechanism has been described,35 and one possibility is that downregulation of the GPCR pathway has the potential to cause a further decrease in Trkb signaling in RGCs in glaucoma.
In summary, in our work that elevated IOP led to complex changes in RGC gene expression in a rat model of glaucoma. A critical finding of this study is that multiple cellular pathways were changed in RGCs in response to elevated IOP. The expression of genes involved in apoptotic pathways (Caspase pathway, Fas pathway, JNK pathway) was increased, and the genes involved in the GPCR, 14-3-3 pathway, and PI3K pathway were decreased. By contrast, gene expression of the prosurvival JAK/STAT pathway was increased, suggesting the activation of endogenous neuroprotective mechanisms. We also found decreased expression of many prosurvival genes that are normally expressed in RGCs. Thus, gene expression changes led to an alteration of the balance between prosurvival and prodeath genes and pathways in glaucomatous RGCs. We speculate that the decreased expression of prosurvival genes contributes to RGC death in glaucoma. These findings warrant further investigation of the roles of these genes in glaucomatous RGC death.
Supplementary Material
Acknowledgments
The authors thank the NIH Neuroscience Microarray Consortium for gene microarray hybridization and scanning.
Footnotes
Supported by the Massachusetts Lions Eye Research Fund, National Eye Institute Grant R01-EY13399, the Cannistraro Fund, the Margolis fund, and National Eye Institute Core Facility Grant EY014104.
Disclosure: D.Y. Wang, None; A. Ray, None; K. Rodgers, None; C. Ergorul, None; B.T. Hyman, None; W. Huang, None; C.L. Grosskreutz, None
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol 2006;90:262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farkas RH, Grosskreutz CL. Apoptosis, neuroprotection, and retinal ganglion cell death: an overview. Int Ophthalmol Clin 2001;41:111–130 [DOI] [PubMed] [Google Scholar]
- 3.Huang W, Dobberfuhl A, Filippopoulos T, et al. Transcriptional up-regulation and activation of initiating caspases in experimental glaucoma. Am J Pathol 2005;167:673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Valenzuela E, Shareef S, Walsh J, Sharma SC. Programmed cell death of retinal ganglion cells during experimental glaucoma. Exp Eye Res 1995;61:33–44 [DOI] [PubMed] [Google Scholar]
- 5.Wax MB, Tezel G, Edward PD. Clinical and ocular histopathological findings in a patient with normal-pressure glaucoma. Arch Ophthalmol 1998;116:993–1001 [DOI] [PubMed] [Google Scholar]
- 6.Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci 2006;47:977–985 [DOI] [PubMed] [Google Scholar]
- 7.Miyahara T, Kikuchi T, Akimoto M, Kurokawa T, Shibuki H, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomolgus monkey. Invest Ophthalmol Vis Sci 2003;44:4347–4356 [DOI] [PubMed] [Google Scholar]
- 8.Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci 2004;45:1247–1258 [DOI] [PubMed] [Google Scholar]
- 9.Piri N, Kwong JM, Song M, Elashoff D, Caprioli J. Gene expression changes in the retina following optic nerve transection. Mol Vis 2006;12:1660–1673 [PubMed] [Google Scholar]
- 10.Naskar R, Thanos S. Retinal gene profiling in a hereditary rodent model of elevated intraocular pressure. Mol Vis 2006;12:1199–1210 [PubMed] [Google Scholar]
- 11.Yang Z, Quigley HA, Pease ME, et al. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci 2007;48:5539–5548 [DOI] [PubMed] [Google Scholar]
- 12.Walsh MM, Yi H, Friedman J, et al. Gene and protein expression pilot profiling and biomarkers in an experimental mouse model of hypertensive glaucoma. Exp Biol Med (Maywood) 2009;234(8):918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science 1996;274:998–1001 [DOI] [PubMed] [Google Scholar]
- 14.Battle TE, Frank DA. The role of STATs in apoptosis. Curr Mol Med 2002;2:381–392 [DOI] [PubMed] [Google Scholar]
- 15.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem 2007;282:20059–20063 [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Li H, Liu MG, et al. STAT3 activation protects retinal ganglion cell layer neurons in response to stress. Exp Eye Res 2008;86:991–997 [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Tang H, Jin X, Jia G, Hsieh JT. p53 regulates Stat3 phosphorylation and DNA binding activity in human prostate cancer cells expressing constitutively active Stat3. Oncogene 2002;21:3082–3088 [DOI] [PubMed] [Google Scholar]
- 18.Morrison JC, Moore CG, Deppmeier LM, Gold BG, Meshul CK, Johnson EC. A rat model of chronic pressure-induced optic nerve damage. Exp Eye Res 1997;64:85–96 [DOI] [PubMed] [Google Scholar]
- 19.Fileta JB, Huang W, Kwon GP, et al. Efficient estimation of retinal ganglion cell number: a stereological approach. J Neurosci Methods 2008;170:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W, Fileta J, Guo Y, Grosskreutz CL. Downregulation of Thy1 in retinal ganglion cells in experimental glaucoma. Curr Eye Res 2006;31:265–271 [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 2001;98:31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–273 [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Cen LP, Choy KW, et al. JAK/STAT pathway mediates retinal ganglion cell survival after acute ocular hypertension but not under normal conditions. Exp Eye Res 2007;85:684–695 [DOI] [PubMed] [Google Scholar]
- 25.Yao F, Yu F, Gong L, Taube D, Rao DD, MacKenzie RG. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. J Neurosci Methods 2005;143:95–106 [DOI] [PubMed] [Google Scholar]
- 26.Ohyama H, Zhang X, Kohno Y, et al. Laser capture microdissection-generated target sample for high-density oligonucleotide array hybridization. BioTechniques 2000;29:530–536 [DOI] [PubMed] [Google Scholar]
- 27.Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007;131:1164–1178 [DOI] [PubMed] [Google Scholar]
- 28.Park WD, Stegall MD. A meta-analysis of kidney microarray datasets: investigation of cytokine gene detection and correlation with rt-PCR and detection thresholds. BMC Genomics 2007;8:88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tezel G. Oxidative stress in glaucomatous neurodegeneration: mechanisms and consequences. Prog Retin Eye Res 2006;25:490–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maudsley S, Martin B, Luttrell LM. G protein-coupled receptor signaling complexity in neuronal tissue: implications for novel therapeutics. Curr Alzheimer Res 2007;4:3–19 [DOI] [PubMed] [Google Scholar]
- 31.Savinainen JR, Laitinen JT. Detection of cannabinoid CB1, adenosine A1, muscarinic acetylcholine, and GABA(B) receptor-dependent G protein activity in transducin-deactivated membranes and autoradiography sections of rat retina. Cell Mol Neurobiol 2004;24:243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron 1995;15:805–819 [DOI] [PubMed] [Google Scholar]
- 33.Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron 2002;33:689–702 [DOI] [PubMed] [Google Scholar]
- 34.Johnson EC, Guo Y, Cepurna WO, Morrison JC. Neurotrophin roles in retinal ganglion cell survival: lessons from rat glaucoma models. Exp Eye Res 2009;88:808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee FS, Rajagopal R, Kim AH, Chang PC, Chao MV. Activation of Trk neurotrophin receptor signaling by pituitary adenylate cyclase-activating polypeptides. J Biol Chem 2002;277:9096–9102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.