Carbohydrates in cells play very important roles in a wide range of biological processes, and impact health and disease.[1] Consequently, interference with the recognition and processing of carbohydrates is a strategy for drug development that is gaining favour. Examples of drugs that are already in the clinic include amylase inhibitors for the reduction of blood glucose levels and sialidase inhibitors as anti-influenza drugs.[2] A newer class of molecules currently under development are those that stabilise otherwise unstable mutant forms of lysosomal glycosidases, and thereby chaperone them to their lysosomal location and bypass proteasomal degradation. These pharmacological chaperones are typically inhibitors, but can be used to rescue deficient glycosidase activity in lysosomes and thereby provide a potential treatment for this class of storage disorders.[3] The glycosidase inhibitors employed in such approaches are typically high affinity transition-state analogues, but such compounds often do not have high linkage specificity for the specific target glycosidase since they typically do not contain components of the “aglycone”. Further, very high affinities are not desirable in this application since potent inhibitors would not be released upon reaching the lysosome. Alternatively, noncleavable substrate analogues can be used as competitive inhibitors for both glycosidases and carbohydrate-binding proteins. Although typically less potent, such reagents are generally more specific. Some of the best such analogues are thioglycosides, wherein a sulfur atom replaces the intersugar glycosidic oxygen of the normal substrate. These are generally good mimics of the natural substrate, but are recalcitrant to cleavage by essentially all glycosidases.[4]
While the synthesis of such thioglycosides can be achieved by standard synthetic organic approaches, a particularly attractive and speedy approach for the assembly of collections of thioglycosides of different types from a set of donors and acceptors involves the use of thioglycoligase technology.[5] Thioglycoligases are mutant enzymes derived from retaining glycosidases in which the acid/base carboxylic acid residue has been replaced by an amino acid that has no negative charge (Scheme 1). When presented with substrates that bear a good leaving group, such as dinitrophenol or fluoride, the engineered retaining glycosidases rapidly form a covalent glycosyl–enzyme intermediate but, due to the absence of a general base catalyst, the rates of turnover of the intermediate through hydrolysis or transglycosylation to an alcohol acceptor are extremely low. However, turnover of the intermediates occurs efficiently with acceptors that bear a suitably positioned thiol, since the thiol group is much more nucleophilic and requires no general base catalytic assistance. Consequently, good yields of product are typically obtained.
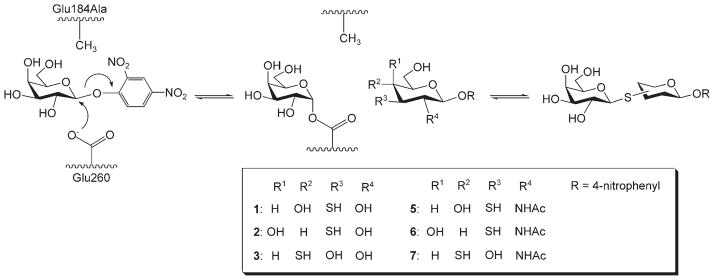
Scheme 1.
Scheme of the generation of galactosyl thio-linked derivatives catalyzed by a thioglycoligase from BgaX (BgaX-Glu184Ala) with Gal-DNP (4) and various thiosugar acceptors.
In this manuscript we describe the use of a thioglycoligase derived from the Xanthomonas manihotis β-galactosidase (BgaX)[6] to generate a small collection of five galactosyl thio-β-glycosides. These analogues were each screened as potential inhibitors of five β-galactosidases that represent the three major families of these enzymes (GH2, GH35 and GH42), and which included the GH35 human lysosomal β-galactosidase (hLyBga) that is responsible for catabolism of gangliosides. Deficiencies arising from mutations in hLyBga lead to the neurological disorders GM1-gangliosidosis and Morquio B. syndrome,[7] for which pharmacological chaperone therapies could be useful. One analogue screened proved to be a reasonably potent (Ki = 8 μM) inhibitor of the human enzyme.
Thioglycoligase reactions were performed by using the p-nitrophenyl glycosides of the thiosugar acceptors, since the presence of the nitrophenyl aglycone improves binding of the acceptor sugar to the thioglycoligase and simplifies reaction monitoring. Coupling of galactose to the thiosugar analogues of glucose and galactose was carried out by using an equimolar ratio of donor (2,4-dinitrophenyl β-D-galactopyranoside, Gal-DNP, 4) to the thiosugar acceptors (3SGlc-pNP, 1; 3SGal-pNP, 2; 4SGlc-pNP, 3) in the presence of 13 μg of BgaX-Glu184Ala per μmole of thiosugar acceptor. As shown in Table 1, the disaccharide products were isolated in yields that were around 80% consistent with earlier findings.[6] In contrast, when their 2-deoxy-2-acetamido counterparts were used as acceptors (3SGlcNAc-pNP, 5; 3SGalNAc-pNP, 6; 4SGlcNAc-pNP, 7) an equimolar ratio of donor to the thiosugar acceptors was insufficient for reaction completion, even with increased enzyme loadings (36 μg BgaX-Glu183Ala per μmole thiosugar acceptor). Presumably the bulkier 2-acetamido group was not well accommodated in the +1 site of BgaX-Glu185Ala. By increasing the amount of donor sugar to twice that of the thiosugar acceptors the thiodisaccharides formed with 3SGlcNAc-pNP (5) and 4SGlcNAc-pNP (7) were obtained in acceptable to good yields of 47% and 79%, respectively (Table 1). However, useful coupling of 3SGalNAc-pNP (6) could not be effected, since only a trace amount of disaccharide was formed. This is consistent with earlier studies of the aglycone specificity of BgaX.[8] Products from the thioglycoligase reactions were acetylated with Ac2O/pyridine, purified by silica gel chromatography and their structures confirmed by one- and two-dimensional 1H and 13C NMR analyses (see Experimental Section). The regioselective formation of only thioglycosidic bonds by BgaX-Glu184Ala was consistent with previous results on thioglycoligases.[6, 9]
Table 1.
Summary of donor and acceptor sugars used with BgaX-Glu184Ala and products formed in the respective reactions.
| Donor | Acceptors | Products | Yield [%][a] |
|---|---|---|---|
| Gal-DNP(4) | 3SGlc-pNP (1) | Gal-β-S-1,3-Glc-pNP (9) | 85 |
| 3SGal-pNP (2) | Gal-β-S-1,3-Gal-pNP (12) | 83 | |
| 4SGlc-pNP (3) | Gal-β-S-1,4-Glc-pNP (13) | 80 | |
| 3SGlcNAc-pNP (5) | Gal-β-S-1,3-GlcNAc-pNP (10) | 79 | |
| 3SGalNAc-pNP (6) | no product | – | |
| 4SGlcNAc-pNP (7) | Gal-β-S-1,4-GlcNAc-pNP (11) | 47 |
Isolated yield after silica gel column chromatography.
While our primary interest was in finding specific inhibitors of the human lysosomal β-galactosidase to serve as chaperones, it was also of interest to test the set of thiodisaccharides as inhibitors of a range of β-galactosidases that represent the three major families of such enzymes. This would give us some measure of the specificities of the inhibitor–enzyme combinations. Consequently, we prepared samples of a family GH2 β-galactosidase: the E. coli lacz β-galactosidase (LacZ), and two other GH35 β-galactosidases with which we had worked previously—the wild-type X. manihotis β-galactosidase (BgaX) and the Bacillus circulans β-galactosidase (BgaC).[10] In order to study a family GH42 enzyme it was necessary to first clone and express one such protein. The GH42 β-galactosidase from Bacillus subtilis (LacA) was chosen since it has recently been identified as a β-galactosidase with no β-glucosidase, β-mannosidase or β-xylosidase activity.[11] Thus, the gene (yvfN) encoding LacA was cloned into E. coli based on the genome sequence of B. subtilis subsp. subtilis strain 168,[12] and the recombinant LacA with a His6 tag at its C terminus was purified by using Ni–NTA affinity chromatography. By evaluating two other enzymes from within the same family as the human galactosidase it should be possible to see whether the specificities observed are inherent to that fold, while sampling enzymes from the other two major β-galactosidase families might provide insight into cross-family specificity.
As a first step, kinetic parameters for hydrolysis of the substrate, either Gal-DNP (4) for hLyBgal or p-nitrophenyl β-D-gal-actopyranoside (Gal-pNP, 8) for other β-galactosidases, were measured, and the following Km values were determined: BgaC (pH 7) 0.01 mM; BgaX (pH 7) 0.12 mM; hLyBgal (pH 4.5) 0.45 mM; LacZ (pH 7) 4.1 mM and LacA (pH 7) 4.3 mM. The Ki values for each inhibitor with each enzyme (for which inhibition was observed) were then determined by measuring reaction rates at substrate concentrations that ranged from 0.5 × Km to 2 × Km for each enzyme at each of a series of inhibitor concentrations. The hydrolysis of thiodisaccharides by the β-galactosidases under study was not detectable under the reaction conditions employed for kinetic studies. The results are presented in Table 2.
Table 2.
The Ki values of thiodisaccharides with various β-galactosidases.
| Thiodisaccharides | BgaX | BgaC | Ki [μM][a] LacA | hLyBga | LacZ |
|---|---|---|---|---|---|
| Gal-β-S-1,3-Glc-pNP (9) | 700 | n.i.[b] | n.i.[b] | 8 | 9 |
| Gal-β-S-1,3-Gal-pNP (12) | 1200 | n.i.[b] | n.i.[b] | 350 | 51 |
| Gal-β-S-1,4-Glc-pNP (13) | 1300 | n.i.[b] | n.i.[b] | 1700 | 150 |
| Gal-β-S-1,3-GlcNAc-pNP (10) | n.i.[b] | n.i.[b] | n.i.[b] | 2600 | n.i.[b] |
| Gal-β-S-1,4-GlcNAc-pNP (11) | n.i.[b] | n.i.[b] | n.i.[b] | 2500 | n.i.[b] |
Each Ki value was determined by using substrate concentrations that ranged from 0.5 × Km to 2 × Km for the corresponding enzymes.
Inhibition was not observed at a 2 mM concentration of thiodisaccharide.
BgaX, the parent enzyme of the thioglycoligase used in this study, was not inhibited efficiently by any of the thiodisacchar-ides, Gal-β-S-1,3-Glc-pNP (9) was the best inhibitor with a Ki value of 700 μM. The better inhibition potency of 9 than of other thiodisaccharides in this study is reasonable because its O-linked counterpart, Gal-β-1,3-Glc, was the best acceptor for BgaX in the earlier assessment of the aglycone specificity of this enzyme.[10] Indeed, it is somewhat surprising that it is not a better inhibitor of this enzyme, but perhaps fortunate since, had it been a good inhibitor, then its synthesis by the thioglycoligase variant might have been compromised by serious product inhibition—this was not the case. The lack of inhibition by Gal-β-S-1,3-GlcNAc-pNP (10) was somewhat surprising since that glycosidic linkage is commonly found in glycoconjugates and glycoproteins that are the probable natural substrates of BgaX.[13] It is, however, consistent with the greater difficulty encountered in getting the thioglycoligase reactions to work with hexosaminide substrates. None of the thiodisaccharides inhibited BgaC, even at concentrations of up to 2 mM, so these were not studied further. Likewise, inhibition of the GH42 LacA β-galactosidase was not seen with any of the analogues under these same conditions. However, the lack of inhibition of LacA is not so surprising since most GH42 β-galactosidases best cleave β-1,4-galactobiose with only trace activity towards lactose.[11] They have presumably evolved to degrade galactans or galactooligosaccharides of plant origin.
Much more interesting results were obtained, fortunately, with the human lysosomal β-galactosidase, with Gal-β-S-1,3-Glc-pNP (9), which exhibited potent inhibition of the human enzyme (Ki = 8 μM). Interestingly, simply inverting the stereo- chemistry of the C-4 carbon of the reducing-end sugar had severe deleterious effects upon binding; Gal-β-S-1,3-Gal-pNP (12) bound 40-times less tightly than 10. Similarly, changing the glycosidic linkage from a β-S-1,3-linkage to a β-S-1,4-linkage was also highly deleterious; Gal-β-S-1,4-Gal-pNP (13) bound 200-times weaker than 9. The 2-acetamido sugar-containing analogues (10 and 11) showed only trace inhibitory activity. These results are somewhat surprising given that the natural substrate of this enzyme has a Gal-β-1,3-GalNAc linkage at its cleavage position. One might have therefore expected that the best inhibitor of the set studied would be the Gal-β-S-1,3-GlcNAc derivative (given that the GalNAc derivative could not be made). In fact, a gluco-derivative was preferred, but not one with an acetamide substituent. This result was somewhat reminiscent of the finding of an unexpectedly strong inhibition of a cellulase by an “incorrectly” linked thioglycoside. In that case an alpha rather than beta linkage was formed; this class of molecules have been dubbed “by-pass” inhibitors due to their binding mode.[14]
A similar set of data was obtained with LacZ, and the same order of affinities was observed. There was no a priori reason why these two enzymes should have such similar profiles. The family 2 and 35 enzymes have the same (β/α)8 structural fold, however, so do the enzymes from family 42, and of course the other enzymes from GH35, all of which are members of clan GH-A.[15] It will be of interest to see whether this similarity extends to other inhibitor classes as this study evolves; if this were the case, the E. coli enzyme might serve as a less expensive model system for inhibitor development and structural analyses for the human enzyme. Indeed, structural studies of inhibitor binding to LacZ are available.[16]
In summary, an efficient inhibitor of both hLyBga and LacZ was identified through screening of a small collection of thioglycosides created by a thioglycoligase derived from BgaX. The best inhibitor identified was of reasonable potency for a first lead and, interestingly, was not exactly the structure that would have been predicted as the best inhibitor based upon substrate specificities. This unexpected inhibitory activity might arise from the subtle differences in bond lengths and bond angles, plus hydrogen-bonding abilities of the thioglycoside bond compared to the O-glycosidic bond. Hopefully, future structural analyses will reveal the basis for this. However, this result opens the interesting possibility of finding novel and unpredicted inhibitors of enzymes of interest through this relatively simple strategy of library generation in which aglycone-diverse thioglycosides are created by thioglycoligases. From a modest set of thiosugar acceptors it is reasonable to envisage the assembly of a substantial library of thiodisaccharides for testing as inhibitors of glycosidases or carbohydrate-binding proteins of interest.
Experimental Section
Materials and general analysis
All chemicals were obtained from Sigma unless otherwise specified. Compounds 4SGlc-pNP (3) and 4SGlcNAc-pNP (7) were synthesized according to literature procedures.[5] The synthetic details of the preparation of 3SGlc-pNP (1), 3SGal-pNP (2), 3SGlcNAc-pNP (5) and 3SGalNAc-pNP (6) will be published elsewhere. All 1H and 13C NMR spectra were recorded at 400 MHz by using a Bruker AV-400 spectrometer. Mass spectra of thiodisaccharides were recorded by using a PE-Sciex API 300 triple quadrupole mass spectrometer (Sciex, Thornhill, ON, Canada) equipped with an electrospray ionization ion source. TLC was performed on aluminium-backed sheets of silica gel 60F254 (Merck) of thickness 0.2 mm. The plates were visualized by using UV light (254 nm) and/or by exposure to sulfuric acid (10 %) in methanol followed by charring.
Gene cloning and protein purification
The gene (yvfN) encoding the β-galactosidase from B. subtilis was amplified with PCR by using 1 μM of each primer (Bsu_lacA_fw: 5′-CACCATGTCAAAGCTT-GAAAAAACGCACGTAAC-3′, and Bsu_lacA_rv: 5′-ATGTGTGTTTACGA-CAATTCTCACTTC-3′), the four dNTPs (0.2 mM each), B. subtilis genomic DNA (50 ng) from The American Type Culture Collection, and Pwo polymerase (2.5 unit; Roche) in 1× Pwo polymerase buffer (50 μL). Twenty-five PCR cycles (45 s at 94°C, 30 s at 55°C and 90 s at 72 °C) were performed in a thermal cycler (Perkin–Elmer, GeneAmp PCR System 2400). The PCR product was sub-cloned into pET101 by using Directional TOPO Expression Kit™ (Invitrogen). The resulting plasmid was designated as pET101-Bsu-LacA and was used for the expression of LacA. The recombinant LacA was purified with nickel-nitrilotriacetate affinity chromatography. The precursor form of hLyBga was purified as previously described by Zhang et al.[17] Other bacterial β-galactosidases and BgaX-Glu184Ala were purified as described previously.[6,10, 18]
Kinetic analysis of β-galactosidases
All kinetic studies were performed at 30°C, in sodium-acetate buffer (100 mM; pH 4.5) for hLyBga, and in phosphate buffer (100 mM; pH 7.0) for other β-galactosidases. Enzyme (20 μL) was added to buffer (100 μL) containing either Gal-DNP (4) for hLyBga or Gal-pNP (8) for other bacterial β-galactosidases. The release of the nitrophenols was monitored at 400 nm by using a microplate reader (SPECTRAMax plus, Molecular Devices Corporation). The values of Km and Vmax were determined by fitting the initial velocity curves to the Michaelis–Menten equation by using nonlinear regression with the program GraFit (Erithacus Software Ltd., Staines, UK). For inhibition kinetics, the assay buffers contained a fixed concentration of substrate and varying amounts of inhibitors. The experiments were repeated at different substrate concentrations (0.5 × Km, 1 × Km and 2 × Km of each enzyme). In a Dixon plot of 1/rate as a function of inhibitor concentration for each substrate concentration a line given by 1/Vmax intersected the other lines at an inhibitor concentration equal to −Ki.
Preparation of thiodisaccharides synthesized with BgaX-Glu184Ala
All thioglycoligase reactions were carried out at room temperature in sodium phosphate buffer (3–5 mL, 100 mM, pH 7.0). Thiosugar acceptors (~9 mg) were dissolved in DMF (200 μL), and then Gal-DNP (4; 1 or 2 equiv) was added as donor. BgaX-Glu184Ala (0.3 or 0.9 mg) was added and the mixture was incubated. Reactions were monitored by using TLC. Upon completion, the reaction mixtures were subjected to a C18 SEP PAK cartridge (Waters) to remove free sugars, enzyme and salts, and then the solvent was evaporated under reduced pressure. Transfer products were purified by flash chromatography (EtOAc/MeOH/H2O 17:2:1→7:2:1) by using silica gel 60 (230±400 mesh), and reaction yields were determined by weighing the isolated products. The purified compounds were acetylated in pyridine and Ac2O. The acetylated thiodisaccharides, which were purified by using flash chromatography (EtOAc/hexanes 1:1) were subjected to ESI-MS and 1H and 13C NMR spectroscopy.
4-Nitrophenyl (2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)-(1→ 3)-S-2,4,6-tri-O-acetyl-β-D-galactopyranoside (per-O-acetylated 9)
1H NMR (400 MHz, CDCl3): δ = 8.21 (d, 2H; Ar-H), 7.07 (d, 2 H; Ar- H), 5.45 (m, 3 H), 5.14 (m, 2 H), 5.01 (dd, 1 H; J =10.0, 3.4 Hz), 4.65 (d, 1 H; J =10.1 Hz), 4.23–4.04 (m, 5H), 3.93 (t, 1 H; J =6.4 Hz), 3.26 (dd, 1H; J =11.4, 3.1 Hz), 2.18–1.99 (7 s, 21H; 7Ac). 13C NMR (100 MHz, CDCl3): δ = 20.7, 20.8, 20.8, 20.9, 20.9, 21.0, 47.3, 61.2, 62.3, 67.1, 67.3, 70.5, 72.0, 74.6, 74.7, 84.8, 99.7, 116.8, 126.0, 143.4, 161.5, 169.1, 169.7, 169.8, 170.2, 170.4, 170.5, 170.6; ESI-MS: calcd for [C32H39NO19S + Na] + 773.2; found: m/z 773.2.
4-Nitrophenyl (2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)-(1→ 3)-S-2,4,6-tri-O-acetyl-β-D-glucopyranoside (per-O-acetylated 12)
1H NMR (400 MHz, CDCl3): δ = 8.20 (d, 2H; Ar-H), 7.08 (d, 2 H; Ar-H), 5.42 (d, 1H; J =3.2 Hz), 5.35 (dd, 1 H; J =10.4, 7.4 Hz), 5.12 (m, 2H), 5.01 (m, 2H), 4.68 (d, 1 H; 10.0 Hz), 4.22 (m, 3 H), 4.04 (dd, 1 H; J = 11.0, 6.8 Hz), 3.93 (m, 2 H), 3.12 (t, 1H; J =10.6 Hz). 2.16–1.98 (7 s, 21H; 7Ac). 13C NMR (100 MHz, CDCl3): δ=20.7, 20.8, 20.9, 20.9, 21.0, 21.2, 49.9, 61.4, 62.5, 66.7, 67.1, 67.4, 71.9, 72.4, 74.4, 75.1, 77.4, 84.8, 99.5, 116.7, 126.0, 143.4, 161.4, 168.6, 169.5, 169.6, 170.3, 170.4, 170.7; ESI-MS: calcd for [C32H39NO19S + Na] + 773.2; found: m/z: 773.2.
4-Nitrophenyl (2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)-1(4)-S-2,3,6-tri-O-acetyl-β-D-glucopyranoside (per-O-acetylated 13)
1H NMR (400 MHz, CDCl3): δ = 8.18 (d, 2H; Ar-H), 7.06 (d, 2 H; Ar-H), 5.43 (d, 1H; J =3.1 Hz), 5.27–5.20 (m, 2 H), 5.15 (d, 1 H; J =7.5 Hz), 5.11 (t, 1H; J =9.8 Hz), 5.02 (t, 1 H; J =3.3 Hz), 4.72 (d, 1 H; 9.8 Hz), 4.63 (dd, 1 H; J =12.0, 1.6 Hz), 4.43 (d, 1H; 5.4 Hz), 4.10 (m, 2H), 4.04 (m, 1 H), 3.93 (m, 1 H), 3.03 (t, 1 H; J =10.6 Hz), 2.16–1.96 (7 s, 21H; 7Ac). 13C NMR (100 MHz, CDCl3): δ=20.3, 20.4, 20.5, 20.6, 45.9, 61.8, 63.3, 67.0, 69.7, 71.5, 72.1, 74.4, 74.5, 82.1, 98.0, 116.6, 125.6, 143.1, 161.1, 169.1, 169.2, 169.8, 170.0, 170.1; ESI-MS: calcd for [C32H39NO19S + Na] + 773.2; found m/z: 773.2.
4-Nitrophenyl (2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)-(1→ 3)-S-4,6-tri-O-acetyl-2-acetamido-2-deoxy-β-D-glucopyranoside (per-O-acetylated 10)
1H NMR (400 MHz, CDCl3): δ = 8.20 (d, 2H; Ar-H), 7.04 (d, 2 H; Ar-H), 6.41 (d, 1H; J =6.0 Hz), 6.15 (d, 1H; J = 8.0), 5.49 (d, 1 H; J =4.0 Hz, NH), 5.18 (t, 1 H; J =10.0 Hz), 5.10 (dd, 1H; J =10.0, 3.2 Hz), 5.02 (t, 1 H; J =10.4, 9.6 Hz), 4.82 (d, 1 H; J = 9.6 Hz), 4.32–4.09 (m, 4 H), 3.99 (m, 3H), 3.25 (m, 1 H), 2.19–1.99 (7 s, 21H; 7Ac). 13C NMR (100 MHz, CDCl3): δ = 20.7, 20.8, 20.9, 21.0, 21.1, 23.8, 47.2, 57.6, 62.7, 63.1, 66.5, 66.6, 67.7, 71.8, 75.0, 75.1, 81.6, 97.7, 116.9, 126.0, 143.3, 161.6, 169.4, 169.8, 170.0, 170.3, 170.7, 170.9, 171.8; ESI-MS: calcd for [C32H40N2O18S + Na] + 772.2; found m/z: 772.3.
4-Nitrophenyl (2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl)-(1→ 4)-S-3,6-tri-O-acetyl-2-acetamido-2-deoxy-β-D-glucopyranoside (per-O-acetylated 11)
1H NMR (400 MHz, CDCl3): δ = 8.18 (d, 2H; Ar-H), 7.08 (d, 2 H; Ar-H), 5.86 (m, 1H), 5.45 (m, 3H), 5.10 (m, 2H), 4.74 (m, 1H), 4.64 (d, 1H; J =11.6 Hz), 4.45 (m, 2H), 4.22–3.97 (m, 4H), 3.02 (t, 1 H; J =9.6 Hz), 2.18–1.83 (7 s, 21H; 7Ac). 13C NMR (100 MHz, CDCl3): δ = 20.7, 20.8, 20.9, 20.9, 21.0, 21.0, 23.6, 46.5, 56.1, 62.0, 63.9, 67.4, 67.5, 71.9, 74.6, 74.9, 77.4, 82.6, 98.2, 116.9, 125.9, 143.3, 161.7, 169.6, 170.0, 170.2, 170.4, 170.5, 170.7, 170.8, 170.9; ESI-MS: calcd for [C32H40N2O18S + Na] + 772.2; found m/z: 772.3.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Protein Engineering Network of Centres of Excellence (PENCE) for financial support. We also acknowledge fellowship support from the Michael Smith Foundation for Health Research (Y.W.K), the Korea Research Foundation (J.H.K), and Austrian FWF (J.M.).
References
- 1.a) Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kolter T, Sandhoff K. Biochim Biophys Acta Biomembr. 2006;1758:2057–2079. doi: 10.1016/j.bbamem.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.a) Scott LJ, Spencer CM. Drugs. 2000;59:521–549. doi: 10.2165/00003495-200059030-00012. [DOI] [PubMed] [Google Scholar]; b) De Clercq E. Nat Rev Drug Discovery. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Cohen FE, Kelly JW. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]; b) Tropak MB, Reid SP, Guiral M, Withers SG, Mahuran D. J Biol Chem. 2004;279:13478–13487. doi: 10.1074/jbc.M308523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Aguilera B, Fernandez-Mayoralas A. J Org Chem. 1998;63:2719–2723. doi: 10.1021/jo971784a. [DOI] [PubMed] [Google Scholar]; b) Driguez H. ChemBioChem. 2001;2:311–318. doi: 10.1002/1439-7633(20010504)2:5<311::AID-CBIC311>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]; c) Rich JR, Bundle DR. Org Lett. 2004;6:897–900. doi: 10.1021/ol036460p. [DOI] [PubMed] [Google Scholar]
- 5.Jahn M, Marles J, Warren RAJ, Withers SG. Angew Chem. 2003;115:366–368. doi: 10.1002/anie.200390114. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2003;42:352–354. [Google Scholar]
- 6.Kim YW, Chen H, Kim JH, Withers SG. FEBS Lett. 2006;580:4377–4381. doi: 10.1016/j.febslet.2006.06.095. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, Sakuraba H, Oshima A. Beta-Galactosidase Deficiency (Beta-Galactosidosis): GM1 Gangliosidosis and Morquio B Disease. McGraw–Hill; New York: 1995. pp. 2787–2823. [Google Scholar]
- 8.Blanchard JE, Withers SG. Chem Biol. 2001;8:627–633. doi: 10.1016/s1074-5521(01)00038-2. [DOI] [PubMed] [Google Scholar]
- 9.a) Stick RV, Stubbs KA. Tetrahedron: Asymmetry. 2005;16:321–335. [Google Scholar]; b) Mullegger J, Jahn M, Chen HM, Warren RAJ, Withers SG. Protein Eng Des Sel. 2005;18:33–40. doi: 10.1093/protein/gzi003. [DOI] [PubMed] [Google Scholar]; c) Hancock SM, Vaughan MD, Withers SG. Curr Opin Chem Biol. 2006;10:509–519. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard JE, Gal L, He S, Foisy J, Warren RAJ, Withers SG. Carbohydr Res. 2001;333:7–17. doi: 10.1016/s0008-6215(01)00108-2. [DOI] [PubMed] [Google Scholar]
- 11.Shipkowski S, Brenchley JE. Appl Environ Microbiol. 2006;72:7730–7738. doi: 10.1128/AEM.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 13.Taron CH, Benner JS, Hornstra LJ, Guthrie EP. Glycobiology. 1995;5:603–610. doi: 10.1093/glycob/5.6.603. [DOI] [PubMed] [Google Scholar]
- 14.Fort S, Varrot A, Schulein M, Cottaz S, Driguez H, Davies GJ. Chem-BioChem. 2001;2:319–325. doi: 10.1002/1439-7633(20010504)2:5<319::AID-CBIC319>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B, Bairoch A. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juers DH, Heightman TD, Vasella A, McCarter JD, Mackenzie L, Withers SG, Matthews BW. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, McCarter JD, Okamura-Oho Y, Yaghi F, Hinek A, Withers SG, Callahan JW. Biochem J. 1994;304:281–288. doi: 10.1042/bj3040281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juers DH, Heightman TD, Vasella A, McCarter JD, Mackenzie L, Withers SG, Matthews BW. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]