Abstract
Background
While many genetic epidemiology and biomarker studies have been conducted to examine associations of genetic variants and circulating proteins with cardiovascular disease and risk factors; there has been little study of gene expression or transcriptomics. Quantitative differences in the abundance of transcripts has been demonstrated in malignancies, but gene expression from a large community-based cohort examining risk of cardiovascular disease has never been reported.
Methods and Results
Based on preliminary microarray data and previously suggested genes from the literature, we measured expression of 48 genes by high-throughput quantitative reverse transcription PCR (qRT-PCR) in 1,846 participants of the Framingham Offspring cohort from RNA derived from isolated platelets and leukocytes. A multivariable stepwise regression model was used to assess clinical correlates of quantitative RNA expression. For specific inflammatory platelet-derived transcripts including ICAM1, IFNG, IL1R1, IL6, MPO, COX2, TNF, TLR2 and TLR4, there were significant associations with higher body mass index (BMI). Compared with platelets, fewer leukocyte-derived transcripts were associated with BMI or other cardiovascular risk factors. Select transcripts were found to be highly heritable, including GPIBA and COX1. Almost uniformly, heritable transcripts were not those associated with BMI.
Conclusions
Inflammatory transcripts derived from platelets, particularly those part of the NFκB pathway, are associated with BMI while others are heritable. This is the first study, using a large community-based cohort, to demonstrate clinical correlates of gene expression and is consistent with the hypothesis that specific peripheral blood transcripts play a role in the pathogenesis of coronary heart disease and its risk factors.
Keywords: genes, epidemiology, leukocytes, platelets
INTRODUCTION
Large numbers of genetic epidemiology and biomarker studies have been conducted to examine associations of common genetic variants and circulating proteins with clinically apparent cardiovascular disease and associated risk factors; however, there has been relatively little study of gene expression or transcriptomics. It is believed that quantitative differences in the abundance of specific transcripts partially accounts for phenotypic differences between individuals.1 The power of gene expression analysis is based on the strong quantitative relation between DNA, RNA, and proteins and it is estimated that 85% of autosomal transcript expression is heritable.1 The most extensive descriptions using peripheral blood cells for the identification of disease state have been in oncology, which have noted the utility of gene expression for the detection of disseminated breast cancer and leukemia.2–4 However, no adequately powered study has reported the clinical correlates of gene expression in a large community-based cohort.
Advances in the fields of genomics and biomarker signatures have modified the definition of complex diseases in recent years. In the area of cardiovascular disease, the human genome-wide association studies have analyzed up to 2.5 million SNPs in the human genome in large numbers of subjects and genetically redefined many common diseases and risk factors.5–8 Conversely, large gene expression profiling studies have not yet been reported from epidemiological cohorts focused on risk factors for atherothrombotic disease or obesity. As compared to oncological studies, for obesity or heart disease, transcriptomics data is far more limited and studies are much smaller in size. While there are no large studies showing changes in transcript expression in obesity, aortic samples have been analyzed for gene expression from patients with abdominal aortic aneurysm and arterial occlusive disease9, 10 and gene expression profiling has been shown to predict cardiomyopathy etiology in heart failure.11
Researchers interested in studying the relation of gene expression to obesity and cardiovascular disease in the community must study peripheral blood cells. This is because, unlike oncological tissue, the RNA of direct interest (i.e. vessel wall, fat tissue, and heart) is not readily available. In smaller studies12, 13 and healthy populations,1 peripheral blood cells have been studied to define vascular disease. Gene expression has been associated with HDL concentrations,1 myocardial infarction,13 and sickle cell disease14 with an emphasis on the identification of transcripts mirroring the intense inflammatory nature of these pathophysiological conditions. In the present study, we hypothesized that cardiovascular risk factors influences inter-individual variability in expression of targeted transcripts from isolated circulating blood cells. Isolated platelets and leukocytes were chosen because preliminary data from our group and others demonstrated that these RNA sources are reflective of atherothrombotic disease. Additionally, they provide complementary cellular and genetic information which may be important as comparative data of RNA derived from blood suggests that the primary determinant of gene expression may be the specific source of RNA.15 While these data provide the first large scale study of gene expression and risk factors, specifically obesity, in a community based cohort; a limitation of this study is the cross sectional design and future prospective studies will be needed to determine the extent to which transcript profiles can predict the development cardiovascular disease or if modification of BMI leads to changes in transcript expression. Importantly, unlike plasma or serum based markers which are easily obtained and stored, RNA is only recently being isolated in larger cohorts and it will be some time until such results are available.
METHODS
Study sample
We examined participants of the Framingham Heart Study Offspring cohort who attended Exam 8 (April 2005–January 2008). The Framingham Heart Study was initiated in 1948 to investigate risk factors for cardiovascular disease in the community.16, 17 Starting in 1971, the Framingham Offspring study enrolled 5,124 offspring of the Original cohort and the spouses of the offspring.17 Participants of the Framingham Offspring study cohort are examined at the Heart Study clinic approximately every four to eight years. During these visits, participants undergo a targeted medical history, physical examination, and laboratory assessment of cardiovascular risk factors. The Framingham Study is reviewed by the Boston University Medical Center Institutional Review Board and all participants gave written informed consent. A total of 1,846 participants were included. Participants examined between 01/27/2006 – 07/05/2006 were not included (n=824) due to a gap in funding for the gene expression portion of the study. In addition, 4 samples were not included due to technical problems during cell isolation and 15 samples were lost during RNA isolation.
Risk Factor Assessment
At each routine clinic visit, participants underwent physical examination, 12-lead ECG, anthropometry, and laboratory assessment of vascular risk factors. Participants with systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg (mean reading of 2 readings taken by an examining physician)or receiving medication for the treatment of hypertension were defined as having hypertension. Plasma glucose and total cholesterol were measured. Diabetes mellitus was defined (throughout the study period) as fasting plasma glucose ≥126 mg/dL, a nonfasting glucose of ≥200 mg/dL, or treatment with either insulin or hypoglycemic agents. Participants were considered to be current smokers if they smoked on average at least 1 cigarette per day during the year before examination.
Selection of genes
The original hypothesis of this project (NHLBI 5R01HL087201) was that increased activation of thrombotic and related inflammatory pathways specifically mediated by NFκB is a pro-atherothrombotic phenotype that can be assayed noninvasively via study of gene expression of NFκB dependent receptors and enzymes in circulating platelets and leukocytes. We postulated that the expression of these genes is influenced by both environmental factors and by genetic variation, and would be related to incident coronary heart disease (CHD) longitudinally (Table 1, Supplemental Table 1). Several of these genes were chosen from pilot data examining isolated platelet and leukocyte microarray and qRT-PCR gene expression from patients with cardiovascular disease. In addition, select genes from previously published studies that associated cardiovascular risk factors with gene expression (derived from a variety of cellular sources) were included (Supplemental Table 1).
Table 1.
Gene List, Gene Function, and Criteria for Selection
| Gene Symbol | Gene Name | NFkappaB |
|---|---|---|
| 18S | Eukaryotic ribosamal protein 18S | NO |
| ACTB | actin, beta | NO |
| ALOX5 | arachidonate 5-lipoxygenase | YES |
| APP | amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease) | NO |
| B2M | beta-2-microglobulin | YES |
| CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | YES |
| CCL5 | chemokine (C-C motif) ligand 5 (RANTES) | YES |
| CD163 | CD163 molecule | NO |
| CD36 | CD36 molecule (thrombospondin receptor) | NO |
| CD40 | CD40 molecule, TNF receptor superfamily member 5 | YES |
| CD40LG | CD40 ligand (TNF superfamily, member 5, hyper-IgM syndrome) | YES |
| CD69 | CD69 molecule | YES |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | NO |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | NO |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | NO |
| GP1BA | glycoprotein Ib (platelet), alpha polypeptide | NO |
| GSTP1 | glutathione S-transferase pi | YES |
| ICAM1 | intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | NO |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | NO |
| IFNG | interferon, gamma | YES |
| IL1R1 | interleukin 1 receptor, type I | NO |
| IL6 | interleukin 6 (interferon, beta 2) | YES |
| IL6R | interleukin 6 receptor | NO |
| ITGA2B | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | NO |
| MIF | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | NO |
| MPO | myeloperoxidase | NO |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | YES |
| OR10J1 | olfactory receptor, family 10, subfamily J, member 1 | YES |
| P2RY12 | purinergic receptor P2Y, G-protein coupled, 12 | NO |
| PF4 | platelet factor 4 (chemokine (C-X-C motif) ligand 4) | NO |
| PLA2G7 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | NO |
| PTGER2 | prostaglandin E receptor 2 (subtype EP2), 53kDa | NO |
| PTGS1 | prostaglandin-endoperoxide synthase 1 (COX1) | NO |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (COX2) | YES |
| PTPN1 | protein tyrosine phosphatase, non-receptor type 1 | YES |
| PTPRN2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | NO |
| S100A9 | S100 calcium binding protein A9 (calgranulin B) | NO |
| SELENBP1 | selenium binding protein 1 | NO |
| SELP | selectin P (granule membrane protein 140kDa, antigen CD62) | YES |
| SERPINB9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | NO |
| TLR2 | toll-like receptor 2 | YES |
| TLR4 | toll-like receptor 4 | NO |
| TNF | tumor necrosis factor (TNF superfamily, member 2) | ? NO |
| TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | NO |
| TNFRSF1B | tumor necrosis factor receptor superfamily, member 1B | YES |
Laboratory analyses
Samples were processed and assayed (including RT-PCR) within two months of completion of Offspring Cohort Participant visits. All cell isolation took place on site at the Framingham Heart Study. Further processes and analyses were at the Boston University School of Medicine.
Cell isolation
Citrated venous blood (35 mL) was collected from the participants. Blood samples were centrifuged (80 g) and platelet rich plasma (PRP) was collected. An equal volume of platelet washing buffer (PWB), apyrase and hirudin were added to the PRP in 0.05 U/mL and 0.08 U/mL concentrations, respectively. Prostaglandin E1 (PGE1) was added to the PRP at a final concentration of 0.3 μM. The PRP was then centrifuged at 450 g to remove the plasma. The platelet pellet was resuspended with PWB and filtered through a 5 μm syringe adaptable filter to remove white blood cell (WBC) contaminants. Additional analyses by the lab using flow cytometry, confocal microscopy, and RNA assessment has confirmed less than 1/50,000 leukocyte contamination of the platelet preparation.
The platelets were pelleted by centrifugation (450 g) and lysed with RNA lysis solution (RLT; Qiagen, Germantown, MD). From the remaining blood, the buffy coat was transferred into a ficoll separation tube called CPT (Becton Dickinson, Franklin Lakes, NJ) and centrifuged at 1,700 g for 20 minutes at room temperature. The peripheral mononuclear cells (PBMC) were transferred into a clean tube, washed with phosphate buffer saline (PBS), pelleted by centrifugation and lysed with RLT solution. Platelet and PBMC lysates were kept at −80°C for further RNA isolations.
RNA Isolation
Total RNA was isolated in both platelets and PBMCs using RNeasy Mini Kits (Qiagen, Germantown, MD). The RNA isolation was completed using an automated purification instrument, QIAcube (Qiagen, Germantown, MD). Following the isolation, the concentration of the isolated RNA was measured using a ND-1000 NanoDrop Spectrophotometer (NanoDrop, Wilmington, DE). The RNA samples were stored at −80°C until later cDNA conversion.
cDNA conversion
Isolated RNA samples, both platelets and PMBCs, were converted to cDNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA) in a PTC-200 Peltier Thermal Cycler (MJ Research, Watertown, MA). The RNA was converted to cDNA using the following conditions: 25°C for 10 min, 37°C for 120 min, 85°C for 5 sec and 4°C until further processing or storage. cDNA samples were kept at −80°C for further PCR analysis.
Pre-amplification
Prior to PCR reactions, the cDNA samples were pre-amplified using Taqman PreAmp Master Mix (Applied Biosystems, Foster City, CA). The PreAmp Master Mix and 0.2x TaqMan assays were added to each cDNA sample and the pre-amplification protocol was as follows: 50°C for 2 min, 95°C for 10 min, 95°C for 15 sec and 60°C for 1 min (these last two steps were repeated for 14 cycles), 4°C until further processing or storage. The pre-amplified cDNA samples were kept overnight at 4°C.
Real-Time PCR
Quantitative Real-Time PCR reactions (qRT-PCR were performed with a high throughput RT-PCR instrument (BioMark; Fluidigm, San Francisco, CA). Pre-amplified cDNA samples were mixed with TaqMan Universal Master Mix (Applied Biosystems, Foster City, CA) and Sample Loading Reagent (Fluidigm, San Francisco, CA) and pipetted into sample inlets of the DynamicArray 48.48 chips (Fluidigm, San Francisco, CA). TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) were diluted 1:2 times with Assay Loading Reagent (Fluidigm, San Francisco, CA) and then pipetted into the assay inlets of the DynamicArray. The DynamicArray was placed into the NanoFlex controller to distribute the assays and samples into the reaction wells of the chip through microfluidic delivery. All qRT-PCR reactions were performed in the BioMark Real-Time PCR system using the following protocol: 2 min at 50°C, 10 min at 95°C, 15 sec at 95°C and 1 min at 60°C for 30 cycles.
Statistical analysis
Descriptive statistics are displayed as mean (SD) for continuous variables and percentage (count) for categorical variables. Multivariable linear regression models for gene expression data as cycle threshold (Ct) values were fitted using stepwise selection with alpha=0.001 to enter and to stay in the model; sex, age and “housekeeping-gene-expression-value” were forced in. Candidates for entry (all assessed at the same exam when RNA was collected) were body mass index, heart rate, smoking status, total cholesterol, HDL cholesterol, triglycerides, systolic blood pressure, diastolic blood pressure, glucose level, diabetes, coronary heart disease, lipid-lowering therapy, hormone replacement therapy, anti-hypertensive therapy, and regular aspirin use (at least 3x per week).
To display selected results, we grouped participants by obesity status (BMI < 25, ≥25 and <30, ≥30 kg/m2), sex, heart disease status or age groups (below 60, 60 to 69, 70 and above) and calculated east-squares means (LSMs) for expression Ct values for each gene adjustting for sex, age and housekeeping expression level. LSMs represent the predicted values for each group when adjustment variables are held fixed at sample mean values. We graphed differences in predicted Ct values (interest group – reference group) ordered by decreasing value for display.
Hierarchical clustering of genes was carried out on “delta Ct” values with the SAS procedure VARCLUS19 using principal components with clustering criterion “maximum eigenvalue = 0.80” for splitting clusters. When choosing a cluster to split, the cluster with the largest second eigenvalue is selected provided that its second eigenvalue exceeds the criterion.
For heritability estimation, we used variance-component models fitted with SOLAR (Sequential Oligogenic Linkage Analysis Routine)separately for each gene from each specimen source.18 Except for heritability estimation, statistical analyses were performed using SAS v9.1 with significance criterion alpha=0.001 (two-sided).19
To examine gene expression jointly in leukocytes and platelets, we estimated partial correlations accounting for sex, age and housekeeping expression levels. We carried out power calculations using Cohen’s approach in the setting of multiple regression analyses. 20 We held fixed the following values: sample size N=1857 and significance level α=0.001 and we calculated the values of true correlations between two variables necessary to provide power = 1−β= 0.80, 0.90 and 0.95. We found that ρ=0.096 yields power ≥ 0.80, ρ=0.107 yields power ≥0.90 and ρ=0.115 yields power ≥ 0.95. Thus, we had excellent power to detect very modest correlations.
RESULTS
Transcript Expression Level
Genes (n=48) were selected based on the criteria discussed above including; preliminary microarray data in cardiovascular patients, inflammatory genes of interest to the Framingham investigators, and transcripts previously associated with coronary disease with the acknowledgement that many were derived from other peripheral blood sources (Table 1, Supplemental Table 1). Of these 48 genes, 4 genes were selected to be used as reference genes (constitutively expressed) in order to define appropriate “housekeeping genes”; i.e. stability, across a large population of individuals.
General expression levels for the 48 genes are shown in Supplemental Table 2 for platelets and leukocytes for all genes for the raw Ct values. After normalization with the housekeeping genes (see below), expression levels (delta Ct values) are shown in Supplemental Table 2 for platelets and leukocytes. A direct comparison of platelet and leukocyte gene expression levels (Supplemental Table 2) demonstrated that there was only partial homology for the levels of gene expression between platelets and leukocytes, particularly for the raw Ct values. For the raw Ct values, other than the housekeeping genes, only SELENBP1, PF4, GP1BA, ITGA2B, P2RY12, SELP, PTGS1, CD36, CCL5, GSTP1, and MIF were significantly correlated.
Housekeeping Genes
Four reference genes were chosen to be used as housekeeping genes in the normalization of the qRT-PCR results. Because there is no literature available validating a suitable housekeeping gene in a large epidemiological sample using peripheral blood cells, RNA expression from four general reference genes were measured; beta actin (ACTB), beta-2-microglobulin (B2M), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and eukaryotic ribosomal protein 18S (18S). For both platelets and leukocytes, all but 18S were highly correlated (Supplemental Figures 1a and 1b); therefore, in subsequent analyses, the mean of ACTB, B2M, and GAPDH for each individual was used to standardize gene expression.
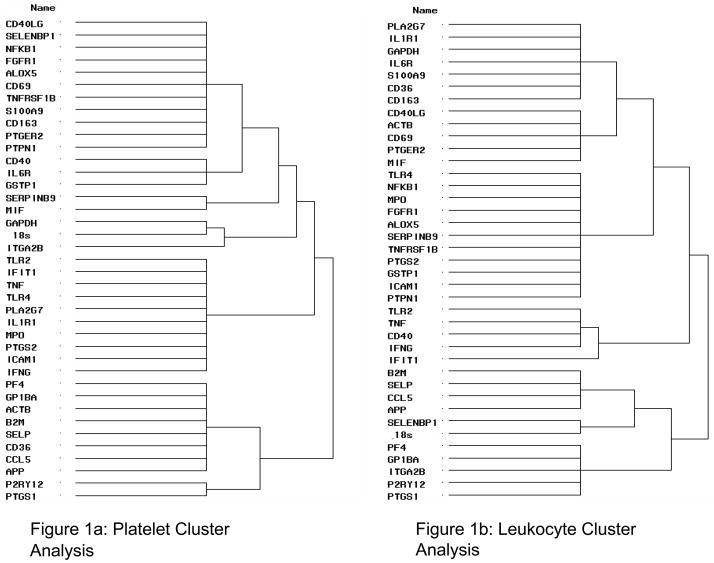
Cluster Analysis
To determine if specific genes have related expression patterns (co-expressed), cluster analysis was performed. Six genes with low expression levels were removed prior to cluster analysis – CCL2, FCER1A, IL6, OR10J1, PTPRN2 and TNFRSF11B. Cluster analysis of gene expression was carried out using delta CT values. Clustering by principal components method with a criterion of maximum eigenvalue > 0.80 was used. For platelets, the housekeeping genes fell in two clusters and many of the NFκB-dependent genes fell in a distinct cluster (Figure 1a). Results obtained from clustering of platelet expression data by multiple methods led to similar clusters with all three methods producing 3 large clusters each with 7–12 members. Clustering of leukocyte expression data by the principal components method produced 8 clusters with 1–11 members (Figure 1b). Clusters seen for leukocyte gene expression were notably distinct as compared to those seen with platelet derived transcripts.
Figure 1. Platelet and Leukocyte Cluster Analysis.
Related expression patterns for platelets (Figure 1a) and leukocytes (Figure 1b) using cluster analysis. Cluster analysis of gene expression was carried out using delta CT values. Clustering by principal components method with criterion max. eigenvalue > 0.80 was used.
Stepwise Regression Analysis
Clinical characteristics for the 1,857 participants are listed in Table 2 and divided according to BMI categories. By multiple regression analysis, most genes show little variation in CT values beyond that accounted by housekeeping genes, i.e. most gene expression levels were highly correlated with the housekeeping genes, but there are some factors that added to model R-squared particularly body mass index (BMI). A summary of the regression analysis (Table 3) demonstrates select associations with specific genes for platelets or leukocytes. Notably, there is little overall homology between these associations when comparing platelets to leukocytes. For both platelet and leukocyte gene expression, associations for most cardiovascular risk findings other than sex and age, are absent. Notably present, however, is BMI and, to a lesser extent, diabetes (Type II) and HDL levels (Table 3).
Table 2.
Characteristics of Framingham Offspring Study sample participants by BMI
| BMI Group | BMI < 25 | 25 ≤ BMI < 30 | BMI ≥ 30 | |||
|---|---|---|---|---|---|---|
| Sample Size, N (%) | 508 (27%) | 764 (41%) | 585 (32%) | |||
| Variable | Mean | Std Dev | Mean | Std Dev | Mean | Std Dev |
| Female sex | 355 | 46% | 363 | 50% | 301 | 50% |
| Age (years) | 67 | 9 | 67 | 9 | 66 | 8 |
| BMI (kg/m2) | 22.6 | 1.8 | 27.5 | 1.5 | 34.5 | 4.2 |
| Waist (inches), N=1837 | 34.2 | 3.1 | 39.7 | 2.9 | 45.9 | 4.4 |
| Weight (pounds) | 137 | 20 | 172 | 23 | 212 | 34 |
| Height (inches) | 65 | 4 | 66 | 4 | 66 | 4 |
| Lipid Treatment | 163 | 47% | 354 | 50% | 301 | 50% |
| Total Chol (mg/100 ml) | 191 | 36 | 184 | 38 | 180 | 38 |
| HDL Chol (mg/100 ml) | 68 | 21 | 56 | 16 | 51 | 15 |
| Triglyceride (mg/100 ml) | 93 | 51 | 117 | 64 | 135 | 78 |
| Anti-Hypertensive Treatment | 168 | 47% | 396 | 50% | 355 | 49% |
| SBP (mm Hg) | 125 | 17 | 130 | 18 | 129 | 16 |
| DBP (mm Hg) | 71 | 10 | 74 | 10 | 74 | 10 |
| Heart Rate (beats per minute) | 62 | 10 | 61 | 10 | 64 | 11 |
| Glucose (mg/dL) | 100 | 19 | 106 | 22 | 114 | 29 |
| Diabetes | 36 | 26% | 93 | 33% | 153 | 44% |
| Prevalent CHD | 41 | 27% | 95 | 33% | 68 | 32% |
| Aspirin (3 week) | 211 | 49% | 346 | 50% | 269 | 50% |
| Current HRT | 50 | 30% | 36 | 21% | 25 | 20% |
| Smoker | 54 | 31% | 62 | 27% | 47 | 27% |
N = 1857, except waist (N=1837)
Table 3.
Relations of gene expression with demographic and clinical variables (p < 0.001 listed associations)
| Platelet Expression | Leukocyte Expression | |||||
|---|---|---|---|---|---|---|
| Gene | sex/age | chd/bmi | all other | sex/age | chd/bmi | all other |
| 18s | ||||||
| ACTB | ||||||
| ALOX5 | s | HDL | a | |||
| APP | ||||||
| B2M | ||||||
| CCL2 | a | s | bmi | |||
| CCL5 | ||||||
| CD163 | a | |||||
| CD36 | ||||||
| CD40 | s | HDL | s | |||
| CD40LG | s | |||||
| CD69 | s | |||||
| FCER1A | s | bmi | lipidrx | |||
| FGFR1 | s | |||||
| GAPDH | bmi | |||||
| GP1BA | ||||||
| GSTP1 | s/a | |||||
| ICAM1 | s/a | bmi | ||||
| IFIT1 | s | HDL | ||||
| IFNG | s/a | bmi | a | |||
| IL1R1 | s/a | bmi | s | |||
| IL6 | s | bmi | ||||
| IL6R | s | |||||
| ITGA2B | ||||||
| MIF | ||||||
| MPO | a | bmi | trig | s | ||
| NFKB1 | s | |||||
| OR10J1 | s | |||||
| P2RY12 | tot chol | |||||
| PF4 | s | |||||
| PLA2G7 | a | bmi | a | |||
| PTGER2 | s/a | a | ||||
| PTGS1 | a | |||||
| PTGS2 | a | bmi | ||||
| PTPN1 | s/ | |||||
| PTPRN2 | diabetes | |||||
| S100A9 | a | bmi | ||||
| SELENBP1 | s | chd/bmi | smoking | trig | ||
| SELP | ||||||
| SERPINB9 | s | |||||
| TLR2 | s/a | bmi | a | |||
| TLR4 | a | a | ||||
| TNF | s | HR/HDL | ||||
| TNFRSF11B | ||||||
| TNFRSF1B | s/a | |||||
Age, Sex, and Gene Expression
As seen in Table 3, expression of many genes was associated with age and sex. To further understand these associations, the delta Ct value for each gene for platelets and leukocytes was compared to age or sex. As seen in Supplemental Figure 2a, platelet gene expression was nearly uniformly increased for subjects over the age of 70 years, particularly as compared to subjects between the ages of 60–69 or younger than 60 years. This is contrasted to the age-related associations seen with leukocytes (Supplementary Figure 2b); many of the genes appeared to have decreased expression with increasing age. With regard to sex, most platelet-derived genes were more highly expressed in women (Supplementary Figure 3a) and this increase in gene expression in females was also seen but to a lesser extent in leukocytes (Supplementary Figure 3b). Notably, MPO was the most highly expressed in men in both platelet and leukocyte transcripts.
Obesity and Gene Expression
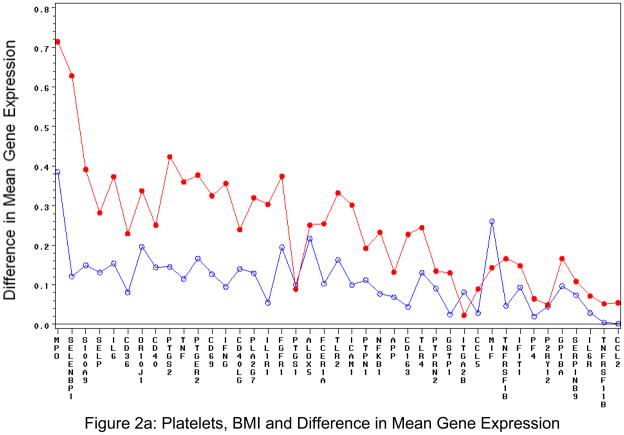
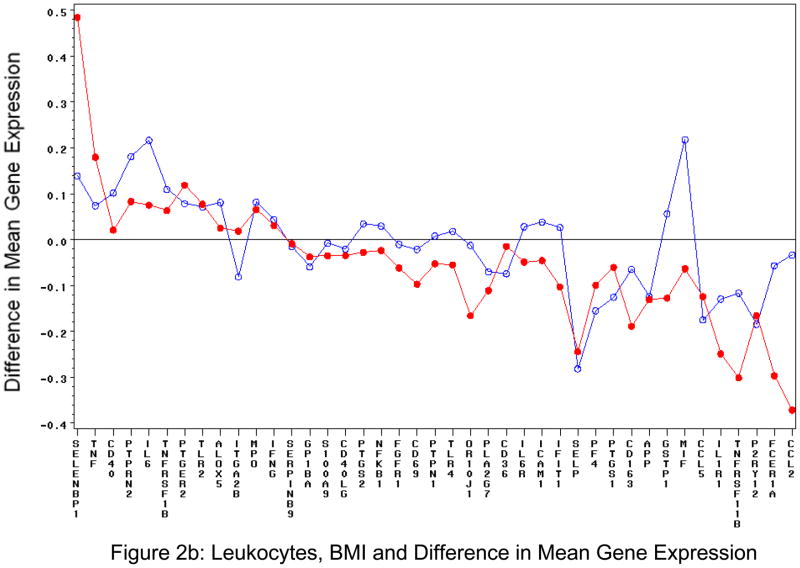
Of all the clinical variables studied other than age and sex, the most commonly associated variable with gene expression was body mass index (BMI; Table 3). For the least squares means plots, the following was significant at the p<0.005 level for BMI and platelet transcripts: MPO, SELENBP1, GAPDH, S100A9, IL6, PTGS2, IFNG, PLA2G7, IL1R1, TLR2, ICAM1. For BMI and leukocytes: FCER1A and CCL2. For both platelet and leukocytes, there were both increased and decreased gene expression associated with BMI, specifically for BMI>30 kg/m2 (Figures 2a and 2b).
Figure 2. BMI and Difference in Mean Gene Expression in Isolated Circulating Cells.
Platelet (Figure 2a) and leukocyte (Figure 2b). The blue line (open circle) is difference in predicted delta CT values between overweight (BMI>25) and normal weight. Red line (closed circle) is difference in predicted delta CT values between obese (BMI>30)and normal weight. Delta CT is gene-specific CT – housekeeping CT.
Because of these findings, we also ran models with interactions of BMI with sex and age and examined the interaction of gene expression with BMI and waist circumference after adjusting for sex, age, and housekeeping gene expression. Each interaction test was conducted with a separate model to avoid collinearity. None of the interactions [(age group, sex) × (BMI category, waist category)] yielded p < 0.001 for any gene in either specimen source (Supplemental Figures 4a and 4b). This finding is not surprising as waist and BMI are highly correlated (r=0.889 crude, partial r=0.896 accounting for sex and age).
Coronary Heart Disease and Gene Expression
Expression of several platelet derived transcripts were borderline associated with prevalent cardiovascular disease including: S100A9, ALOX5, and NFKB1 with only expression of SELENBP1 statistically significant. (P<0.001; Supplemental Figure 5a) There were no significant associations for prevalent cardiovascular disease with gene expression from leukocytes (Supplemental Figure 5b).
Heritability
While a significant portion of variation in the transcriptome must be assumed to be due to environmental influences, genetic factors also have been reported to regulate gene expression.1 As seen in Table 4 (significant associations are shaded), several of the platelet and leukocyte genes demonstrated moderate to substantial heritability. Unexpectedly, heritability of gene expression was only partially homologous for platelets and leukocytes. Highly heritable gene expression (estimates H2 > 0.30) were found in lymphocytes for GP1BA, PTGS1 and GSTP1; similarly, in platelets for GP1BA and CD36. None of the highly heritable expressed genes were shown to have association with cardiovascular risk factors or prevalence of cardiac disease. However, it is noted that GP1BA and CD36 were associated with elevated triglyceride levels.
Table 4.
Heritability Estimates for Gene Expression in Platelets and Leukocytes. Shading=significant heritability.
| Platelet Expression | Leukocyte Expression | |||||
|---|---|---|---|---|---|---|
| Gene | H2r | Std. Error | p value | H2r | Std. Error | p value |
| 18s | 0.070 | 0.083 | 0.20 | 0.09 | 0.092 | 0.16 |
| ACTB | 0.138 | 0.101 | 0.08 | 0.14 | 0.096 | 0.066 |
| ALOX5 | 0.000 | 0.50 | 0.10 | 0.094 | 0.13 | |
| APP | 0.229 | 0.097 | 0.009 | 0.13 | 0.101 | 0.10 |
| B2M | 0.208 | 0.097 | 0.015 | 0.14 | 0.097 | 0.074 |
| CCL2 | 0.125 | 0.091 | 0.080 | 0.18 | 0.088 | 0.019 |
| CCL5 | 0.278 | 0.096 | 0.002 | 0.15 | 0.099 | 0.065 |
| CD163 | 0.100 | 0.092 | 0.13 | 0.08 | 0.091 | 0.20 |
| CD36 | 0.385 | 0.103 | 0.0001 | 0.13 | 0.092 | 0.067 |
| CD40 | 0.000 | 0.50 | 0.00 | 0.50 | ||
| CD40LG | 0.047 | 0.094 | 0.31 | 0.22 | 0.106 | 0.017 |
| CD69 | 0.077 | 0.088 | 0.19 | 0.26 | 0.099 | 0.003 |
| FCER1A | 0.000 | 0.50 | 0.08 | 0.093 | 0.20 | |
| FGFR1 | 0.008 | 0.101 | 0.47 | 0.25 | 0.093 | 0.003 |
| GAPDH | 0.094 | 0.098 | 0.17 | 0.00 | 0.50 | |
| GP1BA | 0.492 | 0.083 | 0.00 | 0.39 | 0.089 | <0.001 |
| GSTP1 | 0.203 | 0.092 | 0.011 | 0.38 | 0.098 | <0.001 |
| ICAM1 | 0.000 | 0.50 | 0.07 | 0.081 | 0.20 | |
| IFIT1 | 0.016 | 0.093 | 0.43 | 0.26 | 0.105 | 0.005 |
| IFNG | 0.015 | 0.087 | 0.43 | 0.15 | 0.084 | 0.032 |
| IL1R1 | 0.042 | 0.087 | 0.31 | 0.14 | 0.093 | 0.063 |
| IL6 | 0.091 | 0.093 | 0.16 | 0.09 | 0.094 | 0.16 |
| IL6R | 0.000 | 0.50 | 0.16 | 0.095 | 0.038 | |
| ITGA2B | 0.242 | 0.102 | 0.008 | 0.26 | 0.093 | 0.002 |
| MIF | 0.023 | 0.082 | 0.39 | 0.13 | 0.085 | 0.060 |
| MPO | 0.082 | 0.091 | 0.18 | 0.09 | 0.086 | 0.15 |
| NFKB1 | 0.046 | 0.102 | 0.33 | 0.08 | 0.090 | 0.20 |
| OR10J1 | 0.000 | 0.50 | 0.00 | 0.50 | ||
| P2RY12 | 0.039 | 0.084 | 0.32 | 0.13 | 0.094 | 0.071 |
| PF4 | 0.176 | 0.088 | 0.022 | 0.01 | 0.093 | 0.45 |
| PLA2G7 | 0.017 | 0.095 | 0.43 | 0.09 | 0.092 | 0.15 |
| PTGER2 | 0.031 | 0.096 | 0.37 | 0.13 | 0.092 | 0.070 |
| PTGS1 | 0.208 | 0.097 | 0.015 | 0.38 | 0.099 | <0.001 |
| PTGS2 | 0.000 | 0.50 | 0.21 | 0.086 | 0.007 | |
| PTPN1 | 0.040 | 0.097 | 0.34 | 0.11 | 0.091 | 0.11 |
| PTPRN2 | 0.162 | 0.093 | 0.038 | 0.10 | 0.093 | 0.14 |
| S100A9 | 0.094 | 0.097 | 0.16 | 0.04 | 0.092 | 0.31 |
| SELENBP1 | 0.237 | 0.092 | 0.003 | 0.18 | 0.087 | 0.015 |
| SELP | 0.264 | 0.098 | 0.003 | 0.24 | 0.109 | 0.012 |
| SERPINB9 | 0.042 | 0.093 | 0.33 | 0.09 | 0.092 | 0.16 |
| TLR2 | 0.000 | 0.50 | 0.00 | 0.50 | ||
| TLR4 | 0.000 | 0.50 | 0.16 | 0.090 | 0.037 | |
| TNF | 0.000 | 0.50 | 0.21 | 0.094 | 0.011 | |
| TNFRSF11B | 0.054 | 0.099 | 0.29 | 0.07 | 0.076 | 0.19 |
| TNFRSF1B | 0.007 | 0.097 | 0.47 | 0.08 | 0.088 | 0.17 |
DISCUSSION
In the current community-based sample, we found that expression of specific genes from isolated peripheral blood cells is associated with coronary heart disease and selected risk factors, particularly obesity, and is also heritable. Based on preliminary microarray data and previously suggested genes from the literature, expression of 48 genes was measured in 1,846 participants of the Framingham Offspring cohort from RNA derived from isolated platelets and leukocytes. Specific inflammatory platelet-derived transcripts including ICAM1, IFNG, IL1R1, IL6, MPO, and TLR2 were significantly associated with higher body mass index. Compared with platelets, fewer leukocyte-derived transcripts were associated with obesity or other cardiovascular risk factors. Select transcripts were found to be highly heritable, including GPIBA and COX1. Almost uniformly, heritable transcripts were not those associated with obesity or a history of heart disease. These data showed that inflammatory transcripts derived from platelets, particularly those part of the NFκB pathway (See Figure 4-Summary), were associated with obesity and coronary heart disease while other distinct transcripts, many known to be related to platelet function, were heritable.
In addition to examining the relations between cardiovascular risk factors and gene expression, this study begins to answer some more fundamental questions about the approach to measuring transcripts in a large community-based group. The first question was the correct use of a gene for normalization (“housekeeping” gene). Four reference genes were chosen to be used as housekeeping genes in the normalization of the qRT-PCR results. Currently, there is no literature available validating a suitable housekeeping gene in a large epidemiological sample using peripheral blood cells. Therefore, data was generated and analyzed from all participants examining four potential housekeeping genes. For both platelets and leukocytes, all but 18S were highly correlated (Supplementary Figures 1a and 1b) establishing that the mean of ACTB, B2M, and GAPDH for each individual would be suitable to standardize gene expression.
Another important question is whether all sources of blood derived RNA are the same or if a specific source of RNA better reflects differences between individuals. The distinct results for the platelet and leukocyte derived RNA as well as the cluster analysis suggest that the source is very important. Currently, whole blood RNA derivation is simpler and less costly than leukocyte or platelet isolation and, thus, is being widely used. The conventional assumption has been that any source of RNA from the blood is equivalent to detect clinically meaningful differences. However, it has been shown that whole blood methods do not provide the same data as isolated leukocytes15 and the most predominant mRNA in whole blood are residual mRNAs associated with erythrocytes or reticulocytes.15 When comparing the two methods, the primary determinant of gene expression was shown to be the source of RNA and not the disease state.15 Heritability has been established for transcripts derived from blood but this large study specifically utilized peripheral blood lymphocytes.1 Only further studies will define the appropriate source of RNA for a particular phenotype and ease of preparation will also factor into the debate.
Sex differences seen in this large human cohort are consistent with smaller gene expression animal studies that suggest that sex-biased gene expression is widespread across organisms and genomes21. Sex-biased genes have been shown to have rapidly evolved and may be labile in their pattern of expression.21 It has also been suggested that expression of sex-biased genes and the selective forces that influence this form of phenotypic diversity are underappreciated.21 In monkeys, there are major differences in gene regulation exist between males and females in vascular aging.22 Differences in genes regulating vascular structure involves sex differences in vascular stiffness that develop with aging and are programmed at an early stage in development. Therefore, while sex linked genes are known to influence gene expression, a fundamental question is whether sex is associated with non X- or Y chromosome-linked gene expression. In this community cohort, the data would suggest that expression of specific genes varies among the two sexes. Although not all statistically significant, nearly all of the genes measured from platelets had higher expression in women as compared to men (Supplementary Figure 3a). This was seen to a lesser extent in leukocytes (Supplementary Figure 3b). Of note, the expression of MPO was significantly higher in men compared to women in both platelets and leukocytes.
Another unknown question is whether advancing age is associated with alterations in gene expression. The data from this community based sample would suggest that, particularly from platelet derived genes, expression of inflammatory genes is enhanced with increasing age (Supplementary Figure 2a). However, this result is not as clearly seen when examining leukocyte derived gene expression (Supplementary Figure 2b) where almost half of the genes appeared to be decreased in participants over the age of 70 years. Thus, the influence of age on gene expression varies depending on cellular source evaluated.
The biological consequences of cardiac risk factors have been previously assessed using gene expression analysis in smaller sample sizes. Cigarette smoke exposure has been shown to alter the expression of 90 genes from peripheral blood leukocytes23 and variability in expression was demonstrated to be more important in smokers than in nonsmokers.24, 25 Diabetes was associated with reduced expression of genes encoding key enzymes for oxidative metabolism and mitochondrial function.26 In addition, inflammatory receptors including TLR2 and ILIR were associated with increased BMI. The inflammatory modulators ICAM1, IFNG, IL6, and MPO were also associated with BMI. For both platelets and leukocytes, gene expression was associated both increased and decreased BMI. No cardiovascular risk factors were associated with leukocyte gene expression although TNF was associated with HDL levels. Importantly, as compared to BMI, waist circumference did not add additional or different specific gene expression findings in this study (Supplemental Figures 4a and 4b). This observation is not surprising as waist and BMI are highly correlated in this population. In addition, platelet derived expression of SELENBP1 was associated with a history of cardiac disease. Little is known about SELENBP1 although it is responsible for selenium binding in enzymes, many of which have been previously correlated with the development of cardiac disease.27
While a significant portion of variation in gene expression must be assumed to be due to environmental influences, genetic factors also influence gene expression. Although completely distinct in population, methodology, and fundamental analysis (individual genes were not reported), the genome-wide transcriptional profiling of lymphocytic samples in the San Antonio Family Heart Study1 estimated that 85% of the detected expression of autosomal transcripts were significantly heritable and linkage analysis suggested greater than 1,000 cis-regulated transcripts. In our data, it is notable that the expression of specific genes that were heritable were almost uniformly not the genes associated with risk factors and, specifically, BMI. Interestingly, several transcripts known to regulate platelet function were found to heritable including GP1BA. This is important in that platelet function itself as assessed by a variety of methods has been found to be heritable but, to this date, known genetic factors have not correlated with thrombosis or platelet function.28, 29
While there have been other studies examining transcripts in blood;30–33 this study is unique in many respects. It is the first large community based study, notably in an older population, to examine gene expression and cardiovascular disease. This study reports individual genes using the more robust platform of qRT-PCR. It is strengthened by the use of two sources of RNA from isolated cells and not whole blood. However, there are several limitations to our investigation. We did not perform a complete transcriptomics survey of the participants using microarray analysis. Importantly, in the current study, genes were chosen in a targeted manner with an eye towards biological plausibility. In addition, microarray studies by nature are not hypothesis driven and may have problems with multiple testing and false-discovery particularly when examining the impact of individual genes. Additionally, arrays are often normalized to commercially available “normals” making a substantial leap in faith as to what one is controlling for. Quantitative RT-PCR relies on “housekeeping” genes but these genes themselves may be influenced by disease states. By measuring multiple housekeeping genes and utilizing only those that were internally consistent, potential error from erroneous normalization was minimized. In addition, the present study was conducted in middle-aged to elderly individuals of European-descent; the generalizability to younger individuals or those of other ethnicity/race is unknown. We did not account for multiple testing; hence, our data will need to be replicated in an external cohort.
Over the past few years, information obtained from gene expression data has been used to define and discover new mechanisms for disease and has clearly aided in diagnosis and treatment in the field of oncology. Cardiac disease is influenced by multiple risk factors including and its inherent complexity make the potentially diverse data provided from transcriptomic methods both exciting and daunting in the goals of diagnosis and treatment. While blood samples are readily available from patients and hold the promise of assisting in the understanding, treatment, and diagnosis of atherothrombotic disease, much remains unknown particularly which cell types are most appropriate for diagnostic and therapeutic use in each of these areas. In the future, transcriptomic data will be linked with additional phenotypic and genomic information and may provide a bridge in understanding the biological complexity of cardiac disease and its link to obesity.
Clinical Impact: Freedman et al.
There have been many genetic epidemiology and biomarker studies examining associations of common genetic variants (DNA) and circulating proteins with clinically apparent cardiovascular disease and associated risk factors, however; there has been relatively little study of gene expression or transcriptomics. Quantitative differences in the abundance of transcripts (mRNA) has been demonstrated in specific malignancies, but gene expression from a large community-based cohort examining cardiovascular disease or its risk factors has never been reported. In this study, we measured quantitative expression of 48 genes in 1,846 participants of the Framingham Offspring cohort from RNA derived from isolated platelets and leukocytes. Specific inflammatory platelet-derived transcripts were significantly associations with higher body mass index (BMI). Compared with platelets, fewer leukocyte-derived transcripts were associated with BMI or other cardiovascular risk factors. Select transcripts were found to be highly heritable. This study demonstrates that inflammatory transcripts derived from platelets, particularly those part inflammatory regulating pathways, are associated with BMI while other distinct transcripts, many known to be related to platelet function, are heritable. This is the first study, using a large community-based cohort, to demonstrate that quantitative gene expression is associated with risk factors, most notably BMI.
Supplementary Material
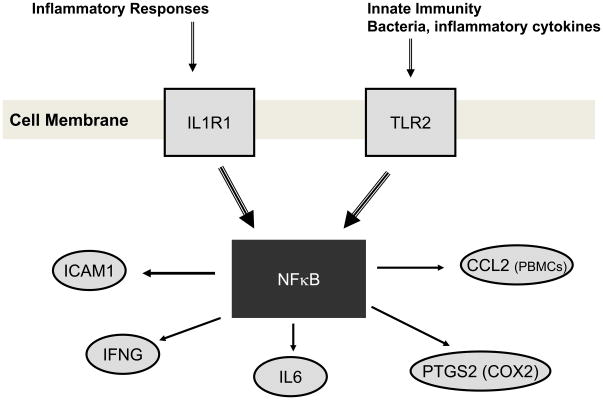
Figure 3. Summary of genes significantly associated with BMI and their role in the NFκB pathway.
Most of the genes found to be significantly associated with BMI are either known to stimulated NFκB activity or are expressed as a result of activation of this pathway. All, except for CCL2, were platelet expressed transcripts.
Acknowledgments
Source of funding
The Framingham Heart Study is supported by N01-HC 25195; the work was also supported by HL087201 (J.E.F.); HL092577-01A1, HL076784, AG028321 (EJB).
Footnotes
Authors’ contributions statement
All authors contributed to this work. There were no paid authors or writing assistants used in the preparation of the manuscript or analysis of the data.
The authors have no conflicts of interest to report.
Disclosures
None.
In-press papers
There are no in press papers
References
- 1.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 2.Martin KJ, Graner E, Li Y, Price LM, Kritzman BM, Fournier MV, Rhei E, Pardee AB. High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc Natl Acad Sci U S A. 2001;98:2646–2651. doi: 10.1073/pnas.041622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greiner TC. mRNA microarray analysis in lymphoma and leukemia. Cancer Treat Res. 2004;121:1–12. doi: 10.1007/1-4020-7920-6_1. [DOI] [PubMed] [Google Scholar]
- 4.Zent CS, Zhan F, Schichman SA, Bumm KH, Lin P, Chen JB, Shaughnessy JD. The distinct gene expression profiles of chronic lymphocytic leukemia and multiple myeloma suggest different anti-apoptotic mechanisms but predict only some differences in phenotype. Leuk Res. 2003;27:765–774. doi: 10.1016/s0145-2126(03)00015-8. [DOI] [PubMed] [Google Scholar]
- 5.Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin EJ, Dupuis J, Larson MG, Lunetta KL, Booth SL, Govindaraju DR, Kathiresan S, Keaney JF, Jr, Keyes MJ, Lin JP, Meigs JB, Robins SJ, Rong J, Schnabel R, Vita JA, Wang TJ, Wilson PW, Wolf PA, Vasan RS. Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med Genet. 2007;8 (Suppl 1):S11. doi: 10.1186/1471-2350-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 8.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong PJ, Johanning JM, Calton WC, Jr, Delatore JR, Franklin DP, Han DC, Carey DJ, Elmore JR. Differential gene expression in human abdominal aorta: aneurysmal versus occlusive disease. J Vasc Surg. 2002;35:346–355. doi: 10.1067/mva.2002.121071. [DOI] [PubMed] [Google Scholar]
- 10.Seo D, Wang T, Dressman H, Herderick EE, Iversen ES, Dong C, Vata K, Milano CA, Rigat F, Pittman J, Nevins JR, West M, Goldschmidt-Clermont PJ. Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- 11.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, Garcia JG, Hare JM. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110:3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. [DOI] [PubMed] [Google Scholar]
- 12.Healy AM, Pickard MD, Pradhan AD, Wang Y, Chen Z, Croce K, Sakuma M, Shi C, Zago AC, Garasic J, Damokosh AI, Dowie TL, Poisson L, Lillie J, Libby P, Ridker PM, Simon DI. Platelet expression profiling and clinical validation of myeloid-related protein-14 as a novel determinant of cardiovascular events. Circulation. 2006;113:2278–2284. doi: 10.1161/CIRCULATIONAHA.105.607333. [DOI] [PubMed] [Google Scholar]
- 13.Wettinger SB, Doggen CJ, Spek CA, Rosendaal FR, Reitsma PH. High throughput mRNA profiling highlights associations between myocardial infarction and aberrant expression of inflammatory molecules in blood cells. Blood. 2005;105:2000–2006. doi: 10.1182/blood-2004-08-3283. [DOI] [PubMed] [Google Scholar]
- 14.Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, Raghavachari N, Beigel JH, Shelhamer JH, Danner RL, Gladwin MT. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood. 2004;104:270–280. doi: 10.1182/blood-2003-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, Fay A, MacMillan S, Laramie J, Xiao W, Moldawer LL, Cobb JP, Laudanski K, Miller-Graziano CL, Maier RV, Schoenfeld D, Davis RW, Tompkins RG. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 16.Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci. 1963;107:539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 17.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 18.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SAS Institute Inc. SAS/STATR 9.1 User’s Guide. Cary, NC: SAS Institute Inc; 2004. [Google Scholar]
- 20.Cohen J, editor. Statistical Power Analysis for the Behavioral Sciences, Revised Edition. New York, NY: Academic Press; 1988. [Google Scholar]
- 21.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 22.Qiu H, Tian B, Resuello RG, Natividad FF, Peppas A, Shen YT, Vatner DE, Vatner SF, Depre C. Sex-specific regulation of gene expression in the aging monkey aorta. Physiol Genomics. 2007;29:169–180. doi: 10.1152/physiolgenomics.00229.2006. [DOI] [PubMed] [Google Scholar]
- 23.Ryder MI, Hyun W, Loomer P, Haqq C. Alteration of gene expression profiles of peripheral mononuclear blood cells by tobacco smoke: implications for periodontal diseases. Oral Microbiol Immunol. 2004;19:39–49. doi: 10.1046/j.0902-0055.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang XL, Mahaney MC. Genotype-specific effects of smoking on risk of CHD. Lancet. 2001;358:87–88. doi: 10.1016/s0140-6736(01)05377-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang XL, Rainwater DL, VandeBerg JF, Mitchell BD, Mahaney MC. Genetic contributions to plasma total antioxidant activity. Arterioscler Thromb Vasc Biol. 2001;21:1190–1195. doi: 10.1161/hq0701.092146. [DOI] [PubMed] [Google Scholar]
- 26.Patti ME. Gene expression in humans with diabetes and prediabetes: what have we learned about diabetes pathophysiology? Curr Opin Clin Nutr Metab Care. 2004;7:383–390. doi: 10.1097/01.mco.0000134359.23288.72. [DOI] [PubMed] [Google Scholar]
- 27.Rayman MP. Selenoproteins and human health: Insights from epidemiological data. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Frelinger AL, 3rd, Furman MI, Linden MD, Li Y, Fox ML, Barnard MR, Michelson AD. Residual arachidonic acid-induced platelet activation via an adenosine diphosphate-dependent but cyclooxygenase-1- and cyclooxygenase-2-independent pathway: a 700-patient study of aspirin resistance. Circulation. 2006;113:2888–2896. doi: 10.1161/CIRCULATIONAHA.105.596627. [DOI] [PubMed] [Google Scholar]
- 29.O’Donnell CJ, Larson MG, Feng D, Sutherland PA, Lindpaintner K, Myers RH, D’Agostino RA, Levy D, Tofler GH. Genetic and environmental contributions to platelet aggregation: the Framingham heart study. Circulation. 2001;103:3051–3056. doi: 10.1161/01.cir.103.25.3051. [DOI] [PubMed] [Google Scholar]
- 30.Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr, Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu H, Goldin E, Baird AE. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- 31.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radich JP, Mao M, Stepaniants S, Biery M, Castle J, Ward T, Schimmack G, Kobayashi S, Carleton M, Lampe J, Linsley PS. Individual-specific variation of gene expression in peripheral blood leukocytes. Genomics. 2004;83:980–988. doi: 10.1016/j.ygeno.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Dempsey AA, Stamatiou D, Marshall KW, Liew CC. Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects. Atherosclerosis. 2007;191:63–72. doi: 10.1016/j.atherosclerosis.2006.05.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.