Abstract
The distal 3′UTR of prothrombin mRNA exhibits significant sequence heterogeneity reflecting an inexact 3′ cleavage/polyadenylation reaction. This same region encompasses a single-nucleotide polymorphism that enhances the normal post-transcriptional processing of nascent prothrombin transcripts. Both observations indicate the importance of 3′UTR structures to physiologically relevant properties of prothrombin mRNA. Using a HepG2-based model system, we mapped both the primary structures of reporter mRNAs containing the prothrombin 3′UTR, as well as the secondary structures of common, informative 3′UTR processing variants. A chromatographic method was subsequently employed to assess the effects of structural heterogeneities on the binding of candidate trans-acting regulatory factors. We observed that prothrombin 3′UTRs are constitutively polyadenylated at seven or more positions, and can fold into at least two distinct stem-loop conformations. These alternate structures expose/sequester a consensus binding site for hnRNP-I/PTB-1, a trans-acting factor with post-transcriptional regulatory properties. hnRNP-I/PTB-1 exhibits different affinities for the alternate 3′UTR secondary structures in vitro, predicting a corresponding regulatory role in vivo. These analyses demonstrate a critical link between the structure of the prothrombin 3′UTR and its normal function, providing a basis for further investigations into the molecular pathophysiology of naturally occurring polymorphisms within this region.
Keywords: prothrombin, mRNA, 3′UTR, mRNA processing
INTRODUCTION
The activated form of prothrombin (F.II) plays a pivotal role in physiologically important thrombotic processes. Thrombin promotes blood clotting by converting fibrinogen to fibrin; by activating coagulation factors V, VIII, and XIII; by stimulating endothelial cell release of von Willebrand factor, PAI-1, endothelin, and tissue factor; and by inducing platelet aggregation[1]. The hemostatic activities of thrombin are balanced by its participation in anticoagulant processes, including thombomodulin-dependent activation of protein C, and by its stimulated production of vasoregulatory factors such as tPA, prostacyclin, and NO.
Recent studies have suggested that post-transcriptional mechanisms play a critical--but previously unrecognized--role in regulating steady-state plasma prothrombin levels. Several groups have focused on the functional consequences of a naturally occurring G→A transition at position 20210, the nominal site at which F.II pre-mRNAs undergo 3′ cleavage/polyadenylation. Two studies have demonstrated that the G20210A polymorphism enhances the efficiency of 3′-end formation for prothrombin pre-mRNAs[2, 3], while a third report has concluded that the single-nucleotide exchange additionally prolongs the cytoplasmic half-life of mature F.II mRNA[4]. The clinical consequences of post-transcriptionally dysregulated prothrombin mRNA are highly significant: heterozygotes for the G20210A mutation exhibit plasma prothrombin levels that are approximately 25% higher than normal[5–7], with corresponding increases in risks for venous thromboembolism[5, 8], cerebral vein thrombosis[9, 10], and other vaso-occlusive events[11, 12]. The incidences of thromboses in individuals with other genetic thrombophilias, including factor V Leiden and protein C deficiency, are also elevated in prothrombin G20210A heterozygotes[13].
The pathological properties of G20210A-polymorphic mRNAs are likely to be mediated through altered interactions between the terminal 3′UTR and trans-acting factors that are critical to normal post-transcriptional processes. We[14] and others[15] have noted that this same region of 3′UTR also exhibits significant sequence heterogeneity resulting from the utilization of several alternate 3′-cleavage/polyadenylation sites. These observations have led us to speculate that the accuracy of 3′-end processing, too, might impact the regulation of F.II mRNA. Precedent literature endorses the principle that structural alterations in this region can create or ablate structural determinants with subsequent, highly consequential effects on mRNA function[16, 17]. A description of mechanistically relevant structures within the F.II 3′UTR, as well as trans-acting effector factors that bind to them, would provide important insights into constitutive processes affecting prothrombin gene expression, and enhance our understanding of the molecular pathophysiology of G20210A-mutant prothrombin mRNA.
The current manuscript formally tests for structural heterogeneities within the F.II 3′UTR that affect its post-transcriptional regulation. Using reporter genes that are stably expressed in cell-homologous HepG2 cells, we quantitate a significant variability in the accuracy of 3′ cleavage/polyadenylation for 3′UTRs derived from both wild-type and G20210A prothrombin gene sequences. Subsequent mRNA mapping analyses demonstrate that the different 3′-terminal isotypes also assume distinct secondary structures. We validate the assembly of two such high-order mRNA conformations, and note that they differentially incorporate a consensus binding site for hnRNP-I/PTB-1, a trans-acting factor that regulates the post-transcriptional processing of other, unrelated pre-mRNAs. Finally, we show that hnRNP-I/PTB-1 exhibits different binding affinities for the two conformationally distinct prothrombin 3′UTRs. These studies demonstrate the capacity of a specific 3′UTR structure to attract functional trans-acting factors, and indicate how this event can be subverted by modest changes in the sequence of the prothrombin 3′UTR.
DESIGN AND METHODS
Cell culture
Parental hepatocellular HepG2 cells were maintained in MEM supplemented with Earle’s salts, 10% FBS, 1.0 mM pyruvate, 1.4 g/L bicarbonate, 0.1 mM nonessential amino acids, 4 mM L-glutamine, and penicillin/streptomycin. Subclones expressing the tetracycline trans-activator (tTA) fusion protein[18] were generated by G418 selection of cells transfected with pTetOff (Clontech). Clonal tTA activity was established by Dual-luciferase analysis (Promega) using cells that had been transiently cotransfected with a TRE-linked firefly luciferase gene (pTRE-luc; Clontech) and a tTA-indifferent control renilla luciferase gene (pRL-SV40; Promega). A single clone exhibiting high tTA activity was used for subsequent subcloning.
Plasmid construction
All plasmids were derived from parental TRE-βWT, comprising a 3.3-kb fragment of DNA, containing the intact human (h) β-globin gene and contiguous 3′-flanking region, inserted into the polylinker SacII-ClaI site of pTRE2 (Clontech)[19]. TRE-βF.IIG was generated from TRE-βWT by a two-step splice-overlap-extension/polymerase-chain-reaction[20, 21] using DNA fragments corresponding to (i) the native 31-nt F.II 5′UTR, (ii) the native hβ-globin coding region and included introns, and (iii) the native 100-nt F.II 3′UTR and 63 nts of contiguous 3′-flanking region. TRE-βF.IIA was similarly constructed to contain a G→A substitution at 3′UTR position 20210. Plasmids used for stable transfections were modified to contain a hygromycin-resistance gene derived from the 1.5-kb XhoI fragment of pTRE2hyg (Clontech). Double-stable cells were generated by selecting TRE-βF.IIG and -βF.IIA-transfected tTA-expressing HepG2 cells with 0.6 μg/ml hygromycin. Subclones selected for study encompassed a two-log range of steady-state βF.II mRNA expression levels, allowing artifacts of saturation kinetics, if present, to be recognized.
3′-cleavage site
Approximately 750 ng total RNA purified from βF.II-expressing HepG2 cells (Pure-link, Invitrogen) was RT-PCR amplified (One-Step, Invitrogen) in the presence of reverse (5′GGATCCGAGCTCT203′) and forward oligomers (5′GTGCTGGTCTGTGTGCTGG3′). The forward oligomer is positioned within the β-globin coding region and does not amplify endogenous, full-length prothrombin mRNAs. Reaction products were ligated into pCR4-Topo, and chemically competent cells transformed (Top-10, Invitrogen). Plasmid DNAs prepared from individual colonies were sequenced by the Nucleic Acid/Protein Core Facility at The Children’s Hospital of Philadelphia.
RNA secondary-structure mapping
DNA templates for in vitro transcription of F.IIA, F.IIG, or F.IIG+2 3′UTRs, each containing an 18-nt poly(A) tail, were directionally inserted into the XhoI-BglII polylinker site of pSP72. BglII-linearized plasmids were transcribed, and RNAs subsequently 5′-end labeled with [γ-32P]ATP using Maxiscript and Kinase Max kits, respectively (Ambion). [32P]-labeled RNAs were digested with RNase A (100, 10, 1.0, and 0.1 ng/ml), RNase T1 (100, 10, 1.0, and 0.1 mU/μl), or RNase V1 (10, 1.0, 0.1, and 0.01 mU/μl) as directed by the supplier (Ambion). RNases A and T1 exhibit cleavage specificities for pyrimidine bases and guanosines within single-stranded regions of RNA, respectively, while RNase V1 cleaves 3′ to bases within double-stranded regions of RNA. Reaction products were resolved on a 33 × 40 cm 6%:8M acrylamide:urea gel, in parallel with a migration-control ‘ladder’ generated by alkaline hydrolysis of [32P]-labeled transcripts.
Protein identification
Affinity-enrichment: Molar equivalents of 5′-biotinylated ssDNAs were liganded to streptavidin-coated agarose beads and incubated with HepG2 cell extract as previously described[19]. Retained factors were resolved on a precast 4–12% gradient Bis-tris SDS-PAGE gel (Invitrogen). Mass spectroscopy: Tryptic digests of excised gel slices were resolved by TOF-TOF analysis (University of Pennsylvania Proteomics Core Facility); protein identities were deduced from MS-Fit analysis of peptide fragments using the NCBInr database.
Data analyses
Individual panels are derived from single autoradiographs; irrelevant lanes have been omitted to preserve clarity. Gamma levels were adjusted, when required, to facilitate analysis.
RESULTS
Generation of a cell-homologous model system
Post-transcriptional events that affect eukaryotic mRNAs can be highly cell-type specific[22, 23]. Consequently, we elected to characterize naturally occurring features of the F.II 3′UTR in hepatocellular carcinoma HepG2 cells, which express liver-restricted factors[24] and recapitulate characteristic hepatocellular processes[25]. Derivative HepG2 cells were engineered to constitutively express β-globin reporter genes containing the complete prothrombin 3′UTR and contiguous 3′-flanking region (Fig 1A). The reporter genes contain either a G or an A at F.II position 20210, reproducing the 3′UTR sequences of wild-type and G20210A-polymorphic sequences, respectively. Steady-state levels of the encoded βF.IIG and βF.IIA mRNAs were determined in 17 and 18 subclones, respectively, using an RNase protection method (Fig 1B), and representative βF.IIG and βF.IIA clones selected for further analyses.
Fig 1. Generation of HepG2 cells that constitutively express allelic F.II reporter genes.
Chimeric TRE-βF.IIG and -βF.IIA DNAs comprise elements of the human prothrombin and β-globin genes (black and gray, respectively) linked to a TRE promoter (crosshatched). The two genes are identical except for a G→A substitution at position 20210. (B) Isolation of HepG2 subclones that express βF.IIG and βF.IIA mRNAs. Total RNA from representative hygromycin-resistant clones expressing βF.IIG and βF.IIA mRNAs was analyzed by RNase protection using [32P]-labeled β-globin and control β-actin RNA probes. Representative autoradiographs are illustrated. PC = positive control β-globin mRNA.
Prothrombin 3′UTRs exhibit reproducible heterogeneity in the position of 3′ cleavage/polyadenylation
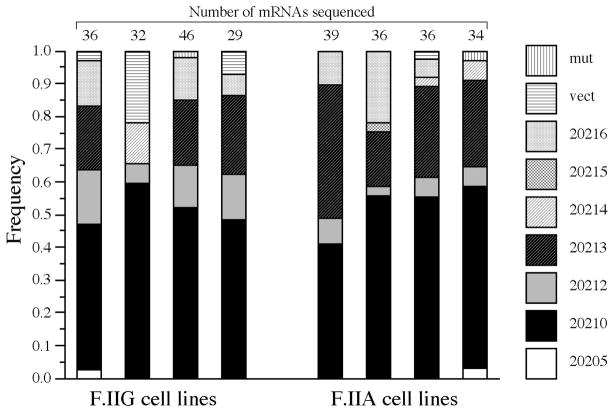
Previous analyses of reporter mRNAs expressed in HepG2 cells[15] and full-length F.II mRNAs expressed in primary hepatocytes[14] have noted inexact targeting of the 3′-cleavage/polyadenylation reaction. We specified both the locations of these 3′ processing sites, as well as their relative strengths, by performing 3′-end analyses on 3′UTRs from more than 120 βF.IIG mRNAs expressed in four independent subclones. Approximately one-half of the wild-type 3′UTRs were polyadenylated at position 20210, while another one-third were polyadenylated at downstream positions 20212 and 20213 (Fig 2, left). Four other native 3′-cleavage sites (at positions 20205, 20214, 20215, and 20216) were used less frequently. A parallel analysis of 3′UTRs from G20210A-containing βF.IIA mRNAs expressed in each of four additional subclones revealed a similar pattern of 3′-cleavage heterogeneity (Fig 2, right). These quantitative data reveal an unexpectedly high variability in the primary structures of 3′UTRs that are preserved in both wild-type and G20210A-polymorphic prothrombin mRNAs.
Fig 2. Heterogeneity in F.II mRNA 3′-end formation.
Constitutively expressed βF.IIG or βF.IIA mRNAs from representative HepG2 cell lines were subjected to 3′ cleavage-site analysis. The frequency of polyadenylation at each of eight sites is indicated. mRNAs terminating at sites encoded by vector sequence (vect) or specifying mutant sequence (mut) are indicated. F.IIG 3′UTRs that undergo 3′-cleavage/polyadenylation at positions 20210G and 20211A produce identical sequences, as do F.IIA 3′UTRs that are processed at positions 20209C, 20210A, and 20211A. In both instances, transcripts are categorized as 20210-terminal mRNAs.
The position of 3′-cleavage dictates the secondary structure of the F.II 3′UTR
While the properties of prothrombin mRNA may be affected by the location of 3′-end processing and/or the identity of the 20210 nucleotide, we considered the possibility that secondary-structure configurations of the 3′UTR, specified by variations in its nucleotide sequence, might also subserve important regulatory functions. As these structures are not reliably predicted by in silico methods, we proceeded to map the high-order conformations of informative 3′-variable and 20210-polymorphic 3′UTRs using an enzyme-based method (Fig 3A).
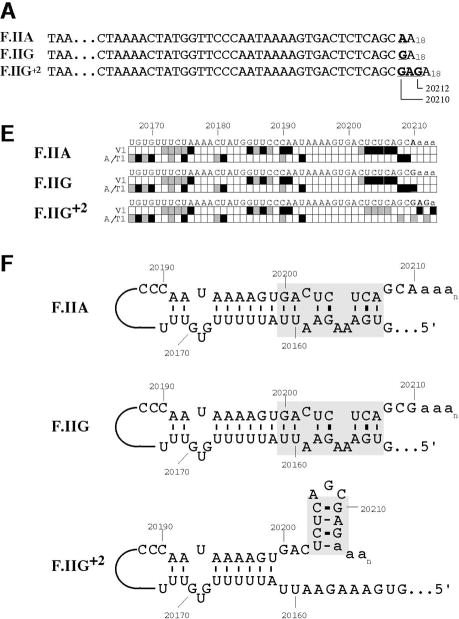
Fig 3. Secondary structures of F.II 3′UTRs with different 3′-processing sites.
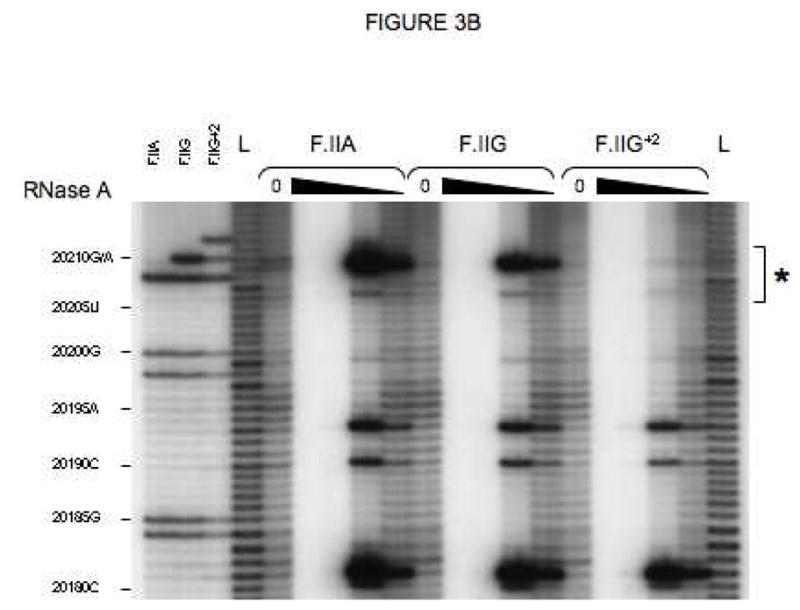
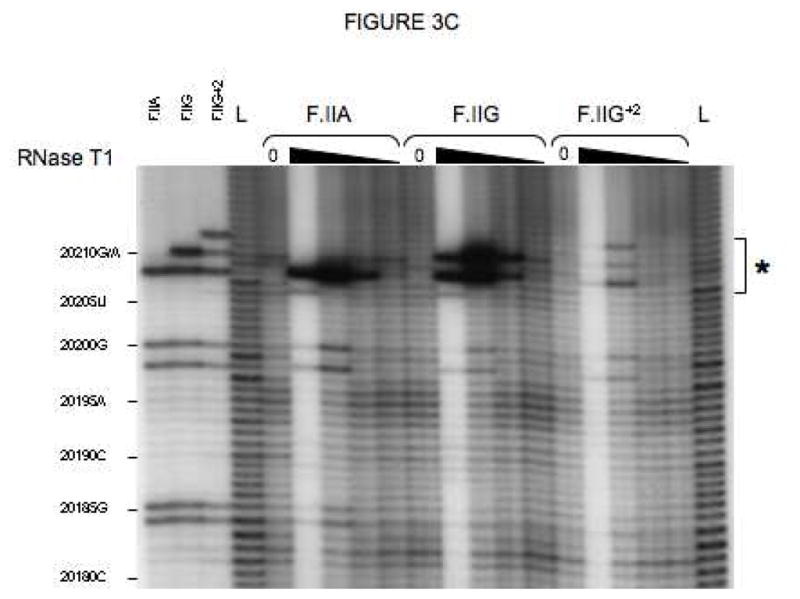
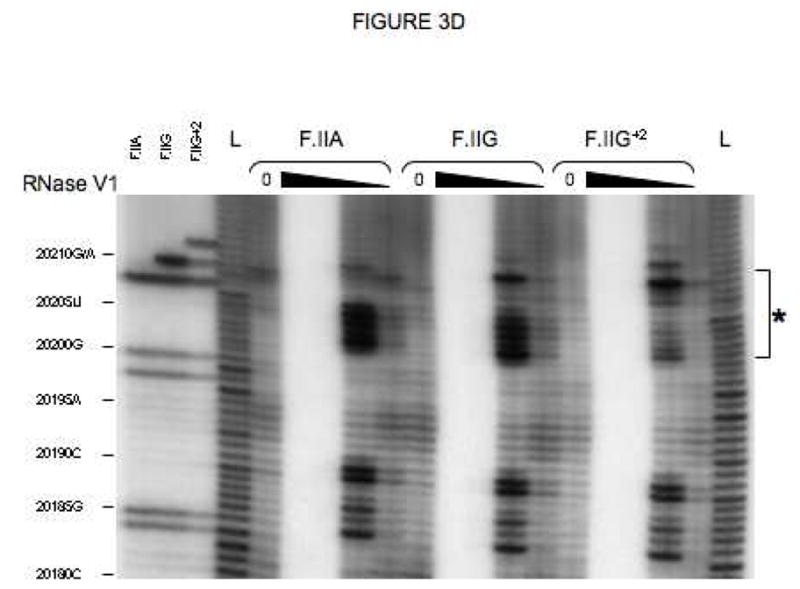
(A) Probes used for enzymatic structure-mapping studies. [32P]-labeled RNAs corresponding to three representative F.II 3′UTRs (F.IIA, F.IIG, F.IIG+2) were transcribed in vitro from the cognate DNA templates. All three RNAs contain identical polyadenylate (A)18 tails. Primary sequence differences are bolded and underlined. (B–D) Enzymatic secondary-structure mapping of F.II 3′UTRs. 5′-end [32P]-labeled F.II 3′UTRs were digested with RNase A (panel B), RNase T1 (panel C), and RNase V1 (panel D) at each of four different concentrations (wedges). Reaction products were resolved on an acrylamide-urea gel, and exposed to film. Nucleotide assignments (left) were deduced from comparison to denatured, RNase T1-treated 3′UTRs (F.IIA, F.IIG, F.IIG+2) and to an alkaline hydrolysis ladder (L). Regions exhibiting differential sensitivities to individual RNases are indicated with an asterisk. (E) 20210- and 20212-terminal F.II 3′UTRs exhibit different secondary structures. The 3′-terminal sequences of the F.IIA, F.IIG, and F.IIG+2 3′UTRs are illustrated (uppercase), along with the initial portion of the poly(A) tail (lowercase). Nucleotides exhibiting high and intermediate RNase sensitivities are indicated by black and grey boxes, respectively. (F) In silico structural mapping studies. Computational analyses were refined by constraining defined nucleotides to single- or double-stranded interactions based upon the experimental results from panels B and C. Relevant features of the predicted F.IIG and F.IIG+2 structures are illustrated. Elements of stem structures encompassing the terminal F.II 3′UTR are boxed in gray.
The conformational impact of 3′-end heterogeneity was investigated by comparing the secondary structures of βF.II 3′UTRs that terminate at positions 20210 and 20212 (F.IIG and F.IIG+2, respectively; Fig 3A). The F.IIG and F.IIG+2 3′UTRs displayed strikingly different sensitivities to both single and double strand-specific RNases in the region surrounding nt 20210 (Figs 3B–D and 3E). Formal structures for both 3′UTRs were subsequently predicted by in silico analyses using the mfold server[26, 27]. Linear F.IIG and F.IIG+2 query sequences were refined by constraining individual nucleotides to single- or double-strand interactions corresponding to their experimentally determined RNase sensitivities. As anticipated, thermodynamically favored structures for F.IIG and F.IIG+2 3′UTRs explicitly differ distal to position 20200 (Fig 3F). The 3′-terminal region of the F.IIG 3′UTR anneals with an upstream U-rich sequence to form a weak, discontinuous stem structure, while the corresponding region of F.IIG+2 3′UTR assembles a stronger local stem-loop motif. The RNase sensitivities of both 3′UTRs were identical upstream of position 20200 (Figs 3B–D, and data not shown), suggesting that functional differences between mRNAs with distinct 3′-terminal processing sites can be attributed to elements positioned near nt 20210.
The structural impact of the G→A polymorphism at position 20210 was similarly determined by comparing the RNase sensitivities of the corresponding F.II 3′UTRs (F.IIG and F.IIA; Fig 3A). Surprisingly, nucleotide identity at this position does not affect the secondary-structure conformation of the F.II 3′UTR. Both the F.IIG and F.IIA 3′UTRs display similar sensitivities to single and double strand-specific RNases (Figs 3B–D and 3E), predicting homologous stem structures (Fig 3F). This result suggests that differential regulation of wild-type and G20210A-polymorphic prothrombin mRNAs is likely to be independent of post-transcriptional processes that distinguish between prothrombin 3′UTRs on the basis of their secondary structural conformations.
Trans-acting factors differentially bind to conformationally distinct F.II 3′UTRs
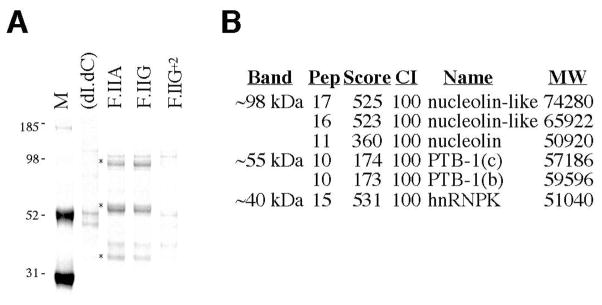
The significant heterogeneities in both the primary and secondary structures of the F.II 3′UTRs suggests a potential impact upon one or more critical post-transcriptional processes. Specifically, we considered the possibility that the productive interaction of regulatory transacting factors might depend upon the exact structural configuration of the F.II 3′UTR. To test this hypothesis, we conducted affinity chromatographic analyses using probes corresponding to F.IIG and F.IIA 3′UTRs (both terminating at position 20210) and the F.IIG+2 3′UTR (terminating at position 20212). Three Coomassie-stained bands were observed to associate with the 20210-terminal F.IIG and F.IIA probes, but not with the 20212-terminal F.IIG+2 probe. These factors were subsequently identified using a mass spectrometric method (Figs 4A and 4B). A ~98 kDa band comprised a structural isoform of nucleolin, a ubiquitous polyfunctional RNA-binding protein that participates in regulating the stabilities and translational efficiencies of a heterogeneous group of mRNAs[19, 28, 29]. A second ~40 kDa band contained hnRNP-K, another RNA-binding regulatory factor that enhances efficient splicing and translation of eukaryotic mRNAs[30, 31]. A third, ~55 kDa band comprised two structural isoforms of a polyfunctional mRNA-binding factor, polypyrimidine tract-binding protein-1 (PTB-1/hnRNP-I), an assignment that was subsequently confirmed by western transfer analysis of affinity-enriched HepG2 extract (not shown). The observation that three distinct factors, each with known mRNA-regulatory functions, exhibit structural isoform-specific binding to the F.II 3′UTR is consistent with their proposed role in the post-transcriptional regulation of prothrombin mRNA.
Fig 4. Differential binding of cytoplasmic factors to 20210- and 20212-terminal F.II 3′UTRs.
(A) Chromatographic analysis of HepG2 extract. Molar equivalents of streptavidin-ligated ssDNAs corresponding to F.IIG, F.IIA, and F.IIG+2 3′UTRs were incubated with HepG2 cell extract, and adherent proteins resolved by SDS-PAGE. Proteins that differentially bind 20210- and 20212-terminal F.II 3′UTR probes are indicated (asterisks). Agarose-liganded poly(dI.dC) ssDNA was assessed as a specificity control. M=protein size standards (kDa). (B) Mass spectrometric analysis of 3′ cleavage site-dependent binding factors. Tryptic digests of gel slices containing the three bands of interest were resolved by TOF-TOF analysis. For each identity, the peptide species count (Pep), protein score (Score), and confidence of identification (CI) are indicated. Score and CI values that are >70 and >95, respectively, are considered highly significant; other identities not meeting either criteria have been excluded from the figure to preserve clarity. MW=calculated molecular weight (Da).
PTB-1 exhibits sequence- and 3′ cleavage isotype-specific binding to the 3′UTR
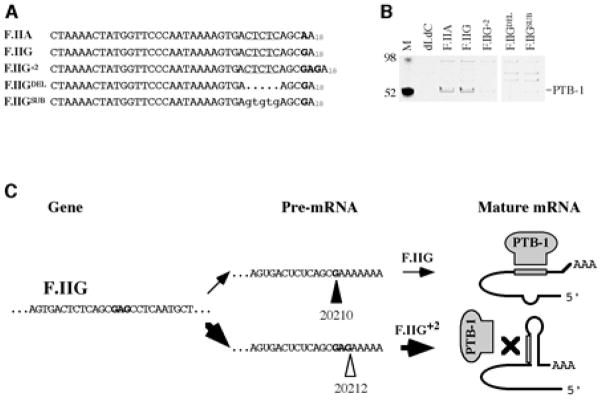
We elected to focus on the potential regulatory properties of PTB-1 because of the positioning of a CUCUC pentanucleotide, encompassing residues 20202–20206, within the structurally variable region of its 3′UTR. This sequence reproduces all four binding domain-specific RNA recognition consensi for PTB-1 determined by NMR spectroscopy[32], and is highly similar to an optimized UCUU binding site for PTB-1 determined using an iterative selection method[33]. As PTB-1 is known to exhibit binding specificity for both ribonucleic and deoxyribonucleic polypyrimidine sequences, we investigated the PTB-1-binding characteristics of the F.II 3′UTR using informative single-stranded DNA probes (Fig 5A). Cytosolic PTB-1 was retained by probes corresponding to the F.IIG and F.IIA 3′UTRs, but not to related probes containing deletion (F.IIGDEL) or purine substitution (F.IIGSUB) of the candidate CUCUC sequence (Fig 5B). Importantly, PTB-1 also failed to bind to a F.IIG+2 probe that retained the pentanucleotide target, validating our previous mapping studies indicating that the CUCUC target is sequestered in strong secondary structure in 20212-terminal 3′UTRs, but relatively weak secondary structure in 20210-terminal 3′UTRs (Fig 3). These results implicate the pyrimidine pentanucleotide as a functional binding site for PTB-1 and, moreover, suggest that its steric availability is highly sensitive to the specific secondary-structure conformation of the F.II 3′UTR.
Fig 5. Primary and secondary structural requirements for PTB-1 binding.
(A) Probes used for affinity chromatography. ssDNAs were designed to correspond to polyadenylated F.IIA, F.IIG, and F.IIG+2 3′UTRs, or to F.IIG 3′UTRs containing deletion (F.IIGDEL) or purine substitution (F.IIGSUB) of the PTB-1 consensus binding sequence. (B) PTB-1 binding affinity is affected by the site of 3′-end processing. Cytoplasmic factors adhering to specific F.II probes were resolved by SDS-PAGE; the position of PTB-1 is indicated. An oligo(dI.dC) probe was assessed in parallel as a negative control. M=protein size standards (kDa). (C) A model for secondary structure-dependent binding of PTB-1 to the F.II 3′UTR. See text for discussion. Positions of 3′-terminal cleavage/polyadenylation at positions 20210 and 20212 are emphasized by closed and open wedges, respectively. The PTB-1 consensus binding sequence is illustrated as a grey bar.
Based upon this data, we propose that 3′-terminal variants of F.II mRNA assume at least two structurally distinct conformations that differentially interact with trans-acting factors, affecting their post-transcriptional regulation (Fig 5C). Some 3′-cleavage isotypes--like the 20210-terminal F.IIG and F.IIA 3′UTRs--assume conformations that permit PTB-1 to access its functional binding site. Other 3′-cleavage isotypes--including the 20212-terminal F.IIG+2 3′UTR--form alternate conformations that restrict PTB-1 access. This model accounts for the F.II 3′UTR conformation-specific binding properties of PTB-1, and predicts that polymorphisms facilitating its interaction with this region of F.II mRNA will dysregulate normal expression of the cognate prothrombin protein.
DISCUSSION
The importance of regulated prothrombin expression to normal physiology is illustrated by the hemorrhagic or thrombophilic phenotypes of individuals who under- or overexpress this critical coagulation factor, respectively. Recent analyses emphasize the critical contributions of regulatory events that act post-transcriptionally to specify the efficiency of 3′-end formation[2, 3, 34] and/or the stability[4] of F.II mRNA. The current work extends this theme by defining constitutive structural heterogeneities in the F.II 3′UTR that appear to contribute to the efficiency of related post-transcriptional processes.
The molecular basis for the constitutive, highly variable 3′-end processing of F.II mRNA is not known, but is likely to reflect a relative weakness in cis-acting regulatory sequences that mediate cleavage-site recognition[2, 35]. The functional consequence of 3′-end variability, however, seems likely to reflect corresponding alterations in high-order mRNA structure. Specifically, 3′-terminal heterogeneities (Fig 2) foster conformationally distinct 3′UTRs (Fig 3) that differentially interact with one or more factors subserving critical post-transcriptional regulatory processes (Figs 4 and 5). This model is consistent with an abundant literature describing mRNA-specific cis-acting structures and trans-acting factors that participate in the post-transcriptional regulation of heterologous mRNAs[36–38], including effects on mRNA processing[39, 40], polyadenylation[39], and/or stability[41, 42].
The concept of heterogeneous 3′-end processing as a post-transcriptional regulatory mechanism has attracted particular interest over the last decade. A large proportion--perhaps half--of all mammalian mRNAs exhibit variability in 3′-end formation[43–45]. This heterogeneity results both from the use of alternate polyadenylation sites, as well as from inexact cleavage of pre-mRNAs downstream of individual polyadenylation sites. The former process--in which two or more alternative polyadenylation sites are in competition--plays an unambiguous role in the regulation of gene expression following the activation of both macrophages[17] and T-cells[16]. Our manuscript extends this theme by suggesting that the functional characteristics of mRNAs may be similarly affected by constitutive variations in the position of the 3′-cleavage reaction. We speculate that targeting of 3′ cleavage/polyadenylation may, in fact, be a dynamic process that is affected by genetic and/or environmental conditions. Such a mechanism would account for the lack of agreement regarding the magnitude of F.II mRNA 3′-processing heterogeneity among investigators using different model systems and experimental conditions.
Importantly, our analyses of eight independent cultured cell lines indicate that the site-specificity of the F.II 3′-cleavage/polyadenylation reaction is indifferent to the presence of a G or an A at position 20210 (Fig 2). While a larger data set might reveal subtle differences in the rates at which the polymorphic mRNAs utilize the panoply of available processing sites, we doubt that any such allele-specific biases would be either biologically or physiologically meaningful. Our data does not fully exclude an alternate model in which low-frequency 3′-processing isotypes (e.g., those that are polyadenylated at positions 20214–20216) assume functionally distinct conformations when the nucleotide at position 20210 is changed from a G to an A. A full accounting of this possibility will require comparison of the 14 secondary structures resulting from the interaction of two position-20210 polymorphisms with each of seven distinct F.II 3′UTR cleavage isotypes.
Our affinity chromatographic studies clearly identify three mRNA-binding factors--each with known mRNA regulatory properties--that differentially bind to F.II 3′UTRs terminating at positions 20210 and 20212 (Fig 4). These results, which are consistent with demonstrated differences in the secondary structures of the two processing isoforms (Fig 3), are likely to reflect differences in the thermodynamic stabilities of the corresponding stem structures enveloping the proposed PTB-1 binding site. While we suspect that the factors assemble into a single functional complex, we cannot exclude the possibility that they compete for one or more overlapping RNA target sequences. Regardless, all three factors would have been overlooked by comparative gel-mobility shift studies utilizing 20210-terminal, but not 20212-terminal probes[4].
Among the three factors that display mRNA cleavage site-dependent binding, PTB-1 merits special attention. This mRNA-binding protein interacts with the 3′UTRs of both human[46] and viral[47] mRNAs, and is reported to play critical roles in directing both the site[48] and the efficiency[49] of 3′-end formation. PTB-1 is also reported to affect mRNA function, impacting the translation of viral RNA[50]. In each case, PTB-1 is believed to remodel high-order mRNA structure, utilizing its four independent RNA-binding domains to bring distant, discontinuous sections of mRNA into proximity by looping out intervening sequence[32]. Results from our studies raise the possibly that PTB-1 similarly remodels the secondary structure of F.II pre-mRNA to facilitate its processing, or alters the structure of mature cytoplasmic F.II mRNA to enhance its translation, consistent with demonstrations that both processes play a critical role in regulating prothrombin gene expression. Both processes would also be consistent with the known functions of the two other factors (nucleolin and hnRNP-K) that exhibit cleavage site-dependent binding to the F.II 3′UTR.
Acknowledgments
This work was supported in part by the University Research Foundation (University of Pennsylvania) and NIH awards R01-HL082754 and -HL061399.
Abbreviations
- F.II
prothrombin
- PTB-1
polypyrimidine tract binding protein 1
Footnotes
CONFLICT-OF-INTEREST STATEMENT
The authors do not have either financial or personal relationships with other people or organizations that could bias their work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 2.Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nature Genetics. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 3.Ceelie H, Spaargaren-van Riel CC, Bertina RM, Vos HL. G20210A is a functional mutation in the prothrombin gene; effect on protein levels and 3′-end formation. J Thromb Haemost. 2004;2:119–127. doi: 10.1111/j.1538-7836.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 4.Carter AM, Sachchithananthan M, Stasinopoulos S, Maurer F, Medcalf RL. Prothrombin G20210A is a bifunctional gene polymorphism. Thromb Haemost. 2002;87:846–853. [PubMed] [Google Scholar]
- 5.Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–3703. [PubMed] [Google Scholar]
- 6.Franco RF, Trip MD, ten Cate H, van den Ende A, Prins MH, Kastelein JJ, Reitsma PH. The 20210 G->A mutation in the 3′-untranslated region of the prothrombin gene and the risk for arterial thrombotic disease. Brit J Heme. 1999;104:50–54. doi: 10.1046/j.1365-2141.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 7.Soria JM, Almasy L, Souto JC, Tirado I, Borell M, Mateo J, Slifer S, Stone W, Blangero J, Fontcuberta J. Linkage analysis demonstrates that the prothrombotic G20210A mutation jointly influences plasma prothrombin levels and risk of thrombosis. Blood. 2000;95:2780–2785. [PubMed] [Google Scholar]
- 8.Hillarp A, Zoller B, Svensson PJ, Dahlback B. The 20210 A allele of the prothrombin gene is a common risk factor among Swedish outpatients with verified deep venous thrombosis. Thromb Haemost. 1997;78:990–992. [PubMed] [Google Scholar]
- 9.Reuner KH, Ruf A, Grau A, Rickmann H, Stolz E, Juttler E, Druschky KF, Patscheke H. Prothrombin gene G20210->A transition is a risk factor for cerebral venous thrombosis. Stroke. 1998;29:1765–1769. doi: 10.1161/01.str.29.9.1765. [DOI] [PubMed] [Google Scholar]
- 10.Martinelli I, Sacchi E, Landi G, Taioli E, Duca F, Mannucci PM. High risk of cerebral-vein thrombosis in carriers of a prothrombin-gene mutation and in users of oral contraceptives. N Engl J Med. 1998;338:1793–1797. doi: 10.1056/NEJM199806183382502. [DOI] [PubMed] [Google Scholar]
- 11.Mercier E, Quere I, Campello C, Mares P, Gris JC. The 20210A allele of the prothrombin gene is frequent in young women with unexplained spinal cord infarction. Blood. 1998;92:1840–1841. [PubMed] [Google Scholar]
- 12.Kupferminc MJ, Peri H, Zwang E, Yaron Y, Wolman I, Eldor A. High prevalence of the prothrombin gene mutation in women with intrauterine growth retardation, abruptio placentae, and second trimester loss. Acta Obstet Gynecol Scand. 2000;79:963–967. [PubMed] [Google Scholar]
- 13.Zoller B, Garcia de Frutos P, Hillarp A, Dahlback B. Thrombophilia as a multigenic disease. Haematologica. 1999;84:59–70. [PubMed] [Google Scholar]
- 14.Pollak ES, Lam H-S, Russell JE. The G20210A mutation does not affect the stability of prothrombin mRNA in vivo. Blood. 2002;100:359–362. doi: 10.1182/blood-2002-02-0412. [DOI] [PubMed] [Google Scholar]
- 15.von Ahsen N, Oellerich M. The intronic prothrombin 19911A>G polymorphism influences splicing efficiency and modulates effects of the 20210G>A polymorphism on mRNA amount and expression in a stable reporter gene assay system. Blood. 2004;103:586–593. doi: 10.1182/blood-2003-02-0419. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CG. Proliferating cells express mRNAs with shortened 3′UTRs and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shell SA, Hesse C, Morris SM, Milcarek C. Elevated levels of 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 18.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Xu X-S, Russell JE. A nucleolin-binding 3′UTR element stabilizes [beta]-globin mRNA in vivo. Mol Cell Biol. 2006;26:2419–2429. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell JE, Liebhaber SA. The stability of human beta-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood. 1996;87:5314–5323. [PubMed] [Google Scholar]
- 21.Yu J, Russell JE. Structural and functional analysis of an mRNP complex that mediates the high stability of human [beta]-globin mRNA. Mol Cell Biol. 2001;21:5879–5888. doi: 10.1128/MCB.21.17.5879-5888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong J, Ji X, Liebhaber SA. Mechanistic analysis of human [alpha]-globin mRNA stabilization. Blood. 2001;98:437a. [Google Scholar]
- 23.Weiss IM, Liebhaber SA. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fair DS, Bahnak BR. Human hepatoma cells secrete single chain factor X, prothrombin, and antithrombin III. Blood. 1984;64:194–204. [PubMed] [Google Scholar]
- 25.Huang MN, Hung HL, Stanfield-Oakley SA, High KA. Characterization of the human blood coagulation factor X promoter. J Biol Chem. 1992;267:15440–15446. [PubMed] [Google Scholar]
- 26.Mathews DH, Sabina J, Zucker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 27.Zucker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J Biol Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- 29.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 30.Ostareck-Lederer A, Ostareck DH, Vans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear riboprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 32.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymound L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH-T. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 33.Perez I, Lin C-H, McAffee JG, Patton JG. Mutation of PTB-1 binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–768. [PMC free article] [PubMed] [Google Scholar]
- 34.Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert A-S, Gehring NH, Neu-Yilik G, Bork P, Keller W, MW, Hentze MW, Kulozik AE. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danckwardt S, Gehring NH, Neu-Yilik G, Hundsdoerfer P, Pforsich M, Frede U, Hentze MW, Kulozik AE. The prothrombin 3′-end formation signal reveals a unique architecture that is sensitive to thrombophilic gain-of-function mutations. Blood. 2004;104:428–435. doi: 10.1182/blood-2003-08-2894. [DOI] [PubMed] [Google Scholar]
- 36.Pandey NB, Marzluff WF. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klausner RD, Rouault TA, Harford JB. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 38.Zilka A, Garlapati S, Dahan E, Yaolsky V, Shapira M. Developmental regulation of heat shock protein 83 in Leishmania. 3′ processing and mRNA stability control transcript abundance, and translation is directed by a determinant in the 3′-untranslated region. J Biol Chem. 2001;276:47922–47929. doi: 10.1074/jbc.M108271200. [DOI] [PubMed] [Google Scholar]
- 39.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 40.Bilenoglu O, Basak AN, Russell JE. A 3′UTR mutation affects [beta]-globin expression without altering the stability of its fully-processed mRNA. Br J Haematol. 2002;119:1106–1114. doi: 10.1046/j.1365-2141.2002.03989.x. [DOI] [PubMed] [Google Scholar]
- 41.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 42.Holcik M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3′UTR RNA-protein complexes sharing cis- and trans-components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pauws E, van Kampen AHC, van de Graaf SAR, de Vijlder JJM, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian B, Hu J, Zhang H, Lutz C. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Research. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosinski PA, Laughlin J, Singh K, Covey LR. A complex containing polypyrimidine tract-binding protein is involved in regulating the stability of CD40 ligand (CD154) mRNA. J Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- 47.De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. Translation elongation factor-1 alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 48.Le Sommer C, Lesimple M, Mereau A, Menoret S, Allo M-R, Hardy S. PTB regulates the processing of a 3′-terminal exon by repressing both splicing and polyadenylation. Mol Cell Biol. 2005;25:9595–9607. doi: 10.1128/MCB.25.21.9595-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proodfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]