Abstract
BACKGROUND
Non-small-cell lung cancer (NSCLC) is the major determinant of overall cancer mortality worldwide. Despite progress in molecular research current treatments offer limited benefits. Since NSCLC generates early metastasis and this behavior requires great cell motility, herein we assessed the potential value of CFL1 gene (main member of the invasion/metastasis pathway) as a prognostic and predictive NSCLC biomarker.
METHODS
Meta-data analysis of tumor tissue microarray was applied to examine expression of CFL1 in archival lung cancer samples from 111 patients and investigated its clinicopathologic significance. The robustness of our finding was validated using another independent data set. Finally, we assayed in vitro the role of CFL1 levels in tumor invasiveness and drug resistance using six human NSCLC cell lines with different basal degree of CFL1 gene expression.
RESULTS
CFL1 levels in biopsies discriminate between good and bad prognosis within early tumor stage (IA, IB and IIA/B), where high CFL1 levels are correlated with lower overall survival rate (P<0.0001). Biomarker performance was further analyzed by immunohistochemistry, hazard ratio (P<0.001) and receiver-operating characteristic (ROC) curve (area=0.787; P<0.001). High CFL1 mRNA levels and protein content are positive correlated with cellular invasiveness (determined by Matrigel Invasion Chamber System) and resistance (two-fold increase in drug GI50 value) against a list of 22 alkylating agents. Hierarchical clustering analysis of CFL1 gene network had the same robustness to stratified NSCLC patients.
CONCLUSIONS
Our study indicates that CFL1 gene and its functional gene network can be used as prognostic biomarker for NSCLC and could also guide chemotherapeutic interventions.
Keywords: prognosis, biomarker, lung cancer, NSCLC, cofilin, CFL1 expression, drug resistance
Lung cancer accounts for 1.3 million deaths annually (World Health Organization) of which 85% are of non-small-cell lung cancer patients (NSCLC). These patients present an average survival rate of 10 months and only 15% survive for five years.1 Currently, prognosis of NSCLC patients is done by considering patient performance status and tumor staging.2,3 However, accumulating data4 have shown that these have unsatisfactory power in predicting patient outcome or in guiding physicians on the best course of action for each patient. A novel prognostic method for early-stage NSCLC patients can potentially increase survival rates by indicating those in need of more aggressive treatment.5
Lung cancers in particular show poor prognosis because of their ability to generate early metastasis within the lungs and then in distant organs. This behavior requires great cell motility, which is performed by several proteins that act on the actin cytoskeleton by regulating cycles of polymerization and depolymerization of actin filaments, which in turn generates cell motion.
One of the main proteins in charge of cell motility is cofilin (CFL1, cofilin-1; non-muscle isoform; Gene ID: 1072),6 which is regulated by factors such as phosphorylation, pH, binding of phosphoinositides, and subcellular compartmentalization. In a recent study we have found that cofilin mediates apoptosis in response to oxidative stress, which is a novel regulatory role described for this protein.7 The role of the cofilin pathway in cell mobility has been shown extensively.8 Its activation occurs locally and in response to EGFR signaling in chemotaxis.9 High cofilin activity has been correlated with breast cancer invasion and metastasis,10,11 where it is essential for directional sensing,12 and with epithelial - mesenchymal transition, a process that is involved in the regulation of cell migration, adhesion and invasion, suggesting the acquisition of a invasive phenotype.13 Thus, we raised the hypothesis that cofilin amount in NSCLC could provide relevant information about tumor’s aggressiveness and therefore be used as a prognostic marker.
Herein, we assessed the potential prognostic value of CFL1 as a NSCLC biomarker. To assay that, we used three different experimental approaches: the first one based on the correlation of gene expression levels and patient outcome using meta analysis of clinical data from a large, homogeneous, well-defined collection of samples from NSCLC cohorts; a second one based on the analysis of in vitro data obtained with six different human NSCLC cell lines; and a third one in which we constructed a network-based model of CFL1 gene and analyzed the role of each network component on the cellular resistance profile to different chemotherapeutic drugs.
MATERIALS & METHODS
Cohort studies and data analysis
Patients, tumor samples and microarray datasets
For NSCLC cohort analysis we accessed a large well-defined collection of lung cancer samples with expression data and relevant clinical and pathologic information on 111 patients (testing cohort), from core biopsies of patients’ tumor. The data was obtained from GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/; Series GSE3141) and the Duke Institute for Genome Sciences & Policy website (http://data.cgt.duke.edu/oncogene.php). Gene array data is available on Affymetrix U133 Plus 2.0 GeneChip.14 To test the reproducibility of the data we assessed a second, independent microarray data set (validation cohort), which is available on different microarray platform (Affymetrix HG_U95Av2 GeneChip).15 The validation cohort comprises microarray data from 86 tumor biopsies obtained from sequential patients seen at the University of Michigan Hospital for stage I or stage III lung adenocarcinomas. All gene array data of the validation cohort are available at http://dot.ped.med.umich.edu:2000/ourimage/pub/Lung/index.html.
Survival data analysis
Standard Kaplan–Meier mortality curves and their significance levels were generated for clusters of patients using SPSS software (SPSS for Windows, release 14.0.0. SPSS Inc., Chicago, IL). The survival curves are compared using the log-rank test and patients are clustered according to either biomarker expression level or NCSLC stage grouping.4,16
Cox multivariable regression analysis
Multivariate Cox proportional hazards regression models were used to test the independent contribution of each variable on mortality. Graphical assessment was used to assess the Cox model’s proportional hazard assumption. Results of multivariate analysis were summarized by calculating hazard ratios (HR) and corresponding 95% confidence intervals (CI).
Biomarker accuracy
The area under the receiver-operating characteristic (ROC) curve was used to evaluate the biomarker’s ability in discriminating patients who survived and those who died. An optimal cut-off value was obtained considering the combination of highest sensitivity and specificity.
In vitro assays
Immunohistochemical staining
Paraffin-embedded sections of lung samples from 20 patients with NSCLC (classified according to World Health Organization criteria) were obtained as archival specimens from the Department of Pathology at the São João Batista Hospital in Criciúma, SC, Brazil. Hematoxylin–eosin (H&E)–stained slides of lung tissue were examined by a national board–certified pathologist. Selected areas of lung cancer and corresponding benign samples were sectioned into 3 µm slices, and immunohistochemical staining was performed according to the standard avidin-biotin immunoperoxidase complex technique. Rabbit polyclonal anti-human cofilin-1 antibody (Abcam®) (1 µg/mL) was used as the primary antibody. The brownish-color was considered to be evidence of a positive expression of cofilin-1 in the tumor cells. Unstained red blood cells and labeled macrophages were considered, respectively, as negative and positive internal controls. The Helsinki Declaration of Human Rights was strictly observed when performing these experiments.
Cell culture and western blot immunoassay
The human NSCLC cell lines were obtained from NCI-Frederick Cancer DCTD tumor/cell line repository, and grown in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 2 mM L-Glutamine at 37°C in 5% CO2 in air. Exponentially growing cells were washed twice with PBS and resuspended in lysis buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1% Triton, 1 mM Na3VO4 and protease inhibitors. After sonication, 30 µg of protein was electrophoresed on 4–12% Bis-Tris NuPage gels (Invitrogen), transferred to PVDF membranes (Immobilon P, Millipore) and blocked with 5% milk. The following antibodies were used for Western blot immunoassay: rabbit polyclonal anti-cofilin (1:1,000), rabbit polyclonal anti-actin (1:2,000) (Cytoskeleton, Denver, CO. USA). Horseradish peroxidase-linked secondary antibody (1:10,000) was from DakoCytomation®. Bands were visualized by chemiluminescence using the ECL Detection kit from Amersham Biosciences and exposure of X-ray film. Quantification of band was done with ImageJ 1.36b software (NIH, USA).
Drug citotoxicity
Drugs GI50 was determined as described elsewhere. Briefly, exponential growing NSCLC cell lines were treated with different concentrations of drugs (cisplatin, carboplatin, 5-fluorouracil, hydroxyurea and taxol) (Sigma®). After 72 h, the medium was removed and cells were fixed with cold 10% TCA for 1 h at 4°C. Plates were washed five times with distilled water and left to dry at room temperature. Cells were stained with 0.4% of sulforhodamine B (SRB) (Sigma®) (w/v) in 1% acetic acid (v/v) at room temperature for 20 min. SRB was removed and the plates washed five times with 1% acetic acid before air-drying. Bound dye was solubilized with 10 mM unbuffered Tris-base solution and plates were left on a plate shaker for at least 10 min. Absorbance was measured in a 96-well plate reader (VERSAmax, Molecular Devices) at 492 nm. The growth inhibition (GI50) was calculated according to the concentration-response curve. The mean of three independent experiments for each condition run in triplicates is plotted.
Cell Migration and Invasion Assays
In vitro migration and invasion assays were performed using the BioCoat™ Matrigel™ Invasion Chamber System (BD Bioscience®). Briefly, Matrigel inserts were rehydrate in RPMI medium and cells (2.5 × 104 cells) were seeded at each 24 well chamber. The chemoattractant (medium RPMI with 10% of SFB) were added to the lower wells and the movement of cells through the 8.0 µm pore size Tran-swell cell culture inserts (Falcon), either un-coated (migration) or Matrigel coated (invasion), were determined after 22 h of incubation at 37°C in a humidified incubator with 5% CO2 atmosphere. At the end of the assay, cells were removed from the top side of the insert using a cotton swab. Cells that penetrated to the underside surfaces of the inserts were fixed and stained with HEMA 3 staining kit (Fisher Scientific) and counted under the microscope. Data is expressed as the percent invasion through the Matrigel relative to the migration though the un-coated membrane, and expressed as invasion index. The mean of three high power fields for each condition run in triplicates is plotted.
Bioinformatics analysis
Microarray data from NCI-60 cancer cell panel
Transcript expression profiles of the six human NSCLC cell lines were obtained from the NCI-60 human tumor cell line anticancer drug screen (http://discover.nci.nih.gov/datasetsNature2000.jsp). To test the reproducibility of the data we assessed a second, independent microarray data set available at http://discover.nci.nih.gov/cellminer/home.do (RMA normalized Affymetrix HG-U133A/B data set). This second microarray platform comprises the human transcriptome and consistently identifies gene probes (e.g. it follows approved gene IDs from the HGNC nomenclature committee – http://www.genenames.org/), allowing the proper identification of CFL1 partners in the biological network analysis.
The drug database
For drug panel activity analysis we considered those compounds listed in the “mechanism of action” drug activity database, NCI Developmental Therapeutics Program (http://discover.nci.nih.gov/datasetsNature2000.jsp) This panel consists of 118 compounds whose mechanisms of action are putatively classified: i) alkylating agents; ii) topoisomerase I inhibitor; iii) topoisomerase II inhibitor; iv) DNA/RNA antimetabolites (DNA binder, DNA incorporation, antifols, ribonucleotide reductase inhibitor, DNA synthesis inhibitor, RNA synthesis inhibitor); v) antimitotics; and vi) others (protein synthesis inhibitor; HSP90 binder or unknown). Drug activity against the NCSLC cell lines is expressed by 50% growth inhibition doses (GI50; also known as IC50) and the entire GI50 dataset is available at http://dtp.nci.nih.gov/dtpstandard/cancerscreeningdata/index.jsp.
CFL1 chemotherapeutic drug resistance/sensitivity data analysis
The relation between the activity of the drug dataset (i.e. 118 standard chemotherapy agents) and CFL1 expression levels was estimated by Spearman correlation analysis in SPSS software (SPSS for Windows, release 14.0.0. SPSS Inc., Chicago, IL). Positive correlations occurred when relatively high levels of gene expression were found in relatively sensitive cell lines. Negative correlations occurred when relatively high levels of gene expression were found in resistant cell lines. Therefore, P values < 0.05 indicate a significant negative correlation (resistance) and P > 0.95 indicate a significant positive correlation (sensitivity). Due to multiple comparisons, only drug categories showing reproducible results were considered for further analysis (i.e. consistent results among the drugs of a given class).
Construction of the network-based model of CFL1 interaction partners
Experimental evidences of protein-protein interactions were obtained from STRING database (http://string.embl.de/).17 STRING integrates different curated, public databases containing information on direct and indirect functional protein-protein associations. We retrieved all proteins described in that database inferred by experimental evidences and that directly interact with CFL1 (cofilin-1; non-muscle isoform; Ensembl Peptide ID: ENSP00000309629). The final network was drawn using spring model algorithm and then handled in Medusa software.18
CFL1 gene partner analysis
Microarray data of NSCLC cell lines were crossed against GI50 values of 118 standard chemotherapy agents in order to estimate drug sensitivity/resistance profile according to the expression levels of CFL1 gene partners (i.e. all genes identified in the network-based model of CFL1 interaction partners). The statistical analysis follows the original method described in the National Cancer Institute’s drug discovery program.19
Clustering analysis and expression profile of CFL1 gene network
The strategy to assess the functional status of tumor samples based on gene expression network profiles have been previously described.20,21 Two-way hierarchical clustering analysis was performed with CLUSTER 3.0 software package using the complete linkage clustering option.22 For visualization purposes, the gene expression values were median-centered and normalized. The results were processed and visualized in TREEVIEW software23. The color intensity was set to the log2 ratio of the microarray signal. Probes of all genes listed in the CFL1 gene network could be retrieved from the microarray platform (i.e. the cohort study – its corresponding gene expression database – is provided on Human Genome U133-Plus 2.0 Array).
RESULTS
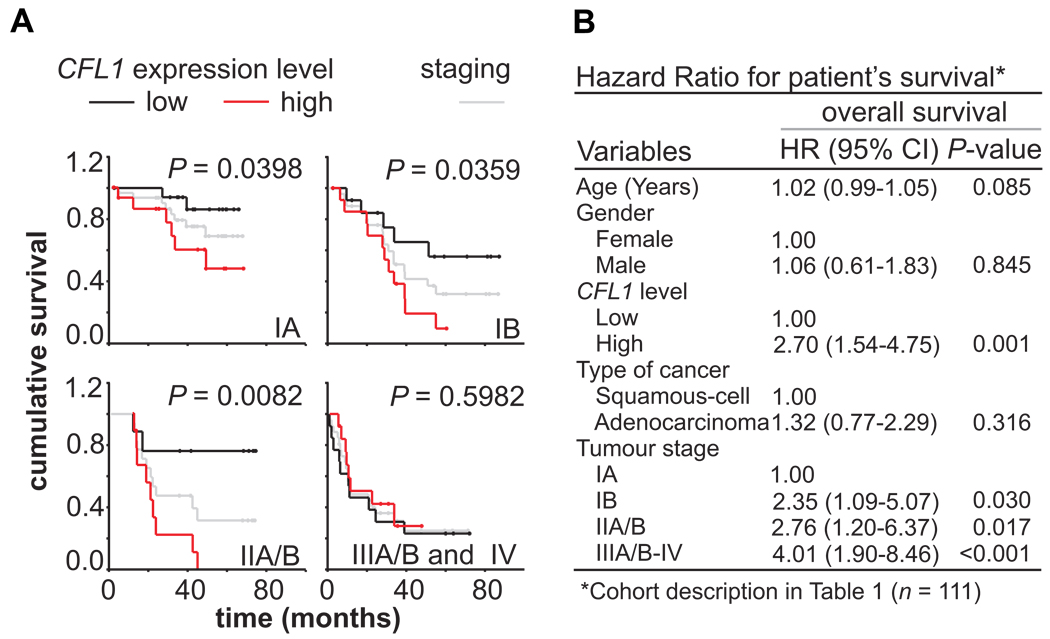
Kaplan-Meier estimates of patient cumulative survival by time (months) according to the expression level of CFL1 showed that when patients are grouped by CFL1 gene expression (upper fifth vs. lower fifth of transcript abundance levels), the expression levels can be used to discriminate patients in early disease stages (IA, IB, IIA, and IIB) between good or bad outcome (Fig. 1A; based on meta-data analysis). Data on microarray gene expression and patient information such as age, sex, cancer histological type, and NSCLC staging were considered (cohort description can be found in Table 1). Cox multivariate regression revealed that lower CFL1 expression was significantly associated with a high overall survival (hazard ratio for high risk vs. low risk, 2.7; 95% C.I., 1.5 to 4.7, P = 0.001) (Fig. 1B).
FIGURE 1. Prognostic value of CFL1 mRNA levels in NSCLC patients.
(A) Meta-analysis of cohort data grouped according to the International Staging System for Lung Cancer and CFL1 gene expression level (i.e. upper-fifth vs. lower-fifth), and plotted as survival probabilities using Kaplan-Meier method. Black lines represent patients with low CFL1 expression; red lines with high CFL1 expression. Differences in survival rates were assessed with the log-rank test. Gray lines represent all patients according to tumor staging. P values lower than 0.05 were considered significant. (B) Cox multivariable regression analysis to estimate hazard ratios for cohort clinical covariates and CFL1 expression. Hazard ratios indicate that patients with high CFL1 expression level presented poor outcome.
Table 1.
Clinical Characteristics of the Original and Validation Cohorts.
| Characteristic |
CFL-1 Expression |
P value | |
|---|---|---|---|
| High | Low | ||
| Testing Cohort (n = 111) | 55 (49%) | 56 (51%) | |
| Age (years) | 64.6 ± 9.6 | 64.9 ± 9.7 | 0.842 |
| Gender | |||
| Male | 30 (54%) | 33 (59%) | 0.784 |
| Female | 25 (46%) | 23 (41%) | |
| Tumor type | |||
| Adenocarcinoma | 28 (51%) | 30 (54%) | 0.928 |
| Squamous-cell | 27 (49%) | 26 (46%) | |
| Tumor TNM Stage | |||
| Ia | 20 (36%) | 20 (36%) | 0.999 |
| Ib | 13 (24%) | 14 (25%) | |
| II | 9 (16%) | 9 (16%) | |
| III–IV | 13 (24%) | 13 (23%) | |
| Validation Cohort (n = 86) | 43 (50%) | 43 (50%) | |
| Age (years) | 62.3 ± 8.8 | 65.1 ± 10.7 | 0.187 |
| Gender | |||
| Male | 21 (49%) | 14 (33%) | 0.198 |
| Female | 22 (51%) | 29 (67%) | |
| Tumor type/differentiation | |||
| Adenocarcinoma/well | 12 (28%) | 12 (28%) | 0.964 |
| Adenocarcinoma/ moderate | 21 (49%) | 20 (47%) | |
| Adenocarcinoma/ poor | 10 (23%) | 11 (26%) | |
| Tumor TNM Stage | |||
| I | 34 (79%) | 33 (77%) | 0.999 |
| III | 9 (21%) | 10 (23%) | |
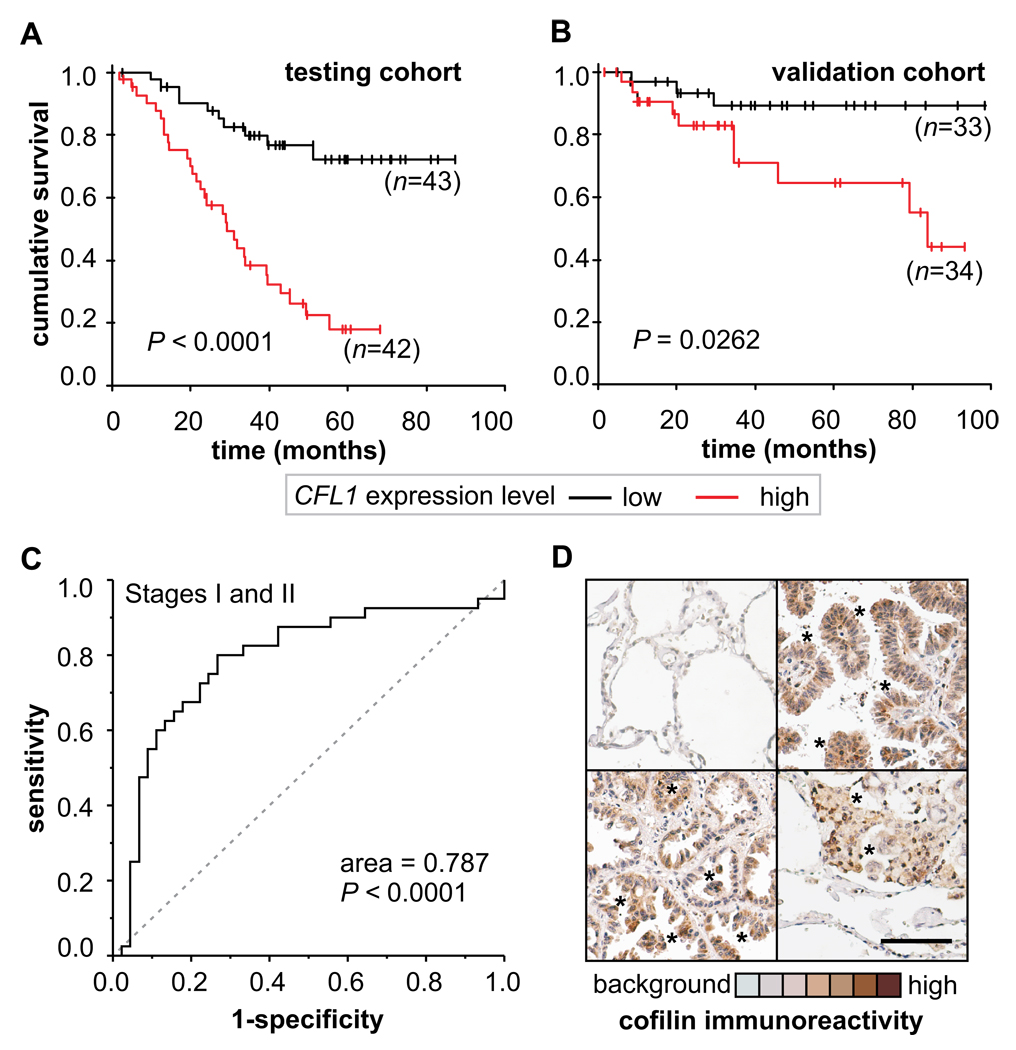
Analysis of 85 patients with disease stages I or II (the testing cohort), revealed that patients with high CFL1 expression (n=42) had an overall survival rate shorter than those with low CFL1 expression (n=43) (Fig. 2A). To test the robustness of this finding, we analyzed a second, independent data set of 67 patients in early stages (the validation cohort) (Fig. 2B). Our meta-analysis showed that high CFL1 levels are associated with shorter overall survival in both cohorts. ROC curve analysis showed that CFL1 sensitivity/specificity is high enough to indicate patient’s outcome of those with early disease stages (area under ROC curve = 0.787) (Fig. 2C). Immunohistochemical stains revealed an increased cofilin immunocontent within the neoplasic tissue (Fig. 2E). The data presented in Figure 2 suggests that CFL1 levels can be used to indicate patient outcome.
FIGURE 2. Biomarker performance in early stage NSCLC patients.
(A) Kaplan Meier plot are shown for patients in stages I and II (n=85) in the original cohort (testing cohort) stratified by CFL1 expression level and (B) in an independent cohort (validation cohort) obtained from a different set of published NSCLC microarray data (n=67). (C) Biomarker performance estimated by Receiver Operating Characteristic (ROC) analysis. (D) Representative immunohistochemical (IHC) analysis of cofilin immunocontent in tumor biopsies. Healthy human alveolar tissue obtained from tumor margins is mostly negative to cofilin IHC staining (upper left). High staining for cofilin is found within the neoplasic lung cells (asterisks). Original magnification ×200; scale bar = 100 µM.
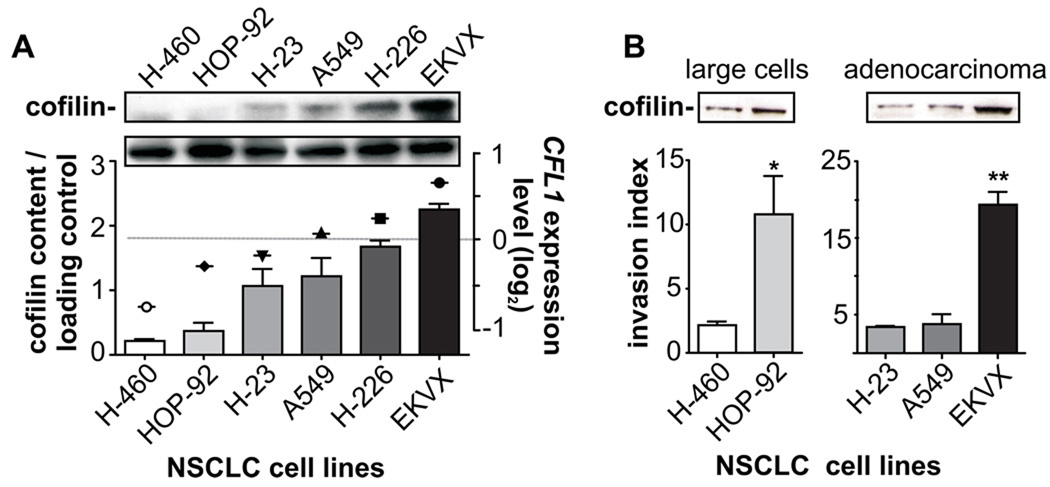
We also asked whether CFL1 levels could provide additional insights into the pathophysiology of NSCLC, predicting tumor aggressiveness and/or chemotherapy response. To do that, we used NSCLC data from the US National Cancer Institute in vitro anticancer drug screen (NCI60 cancer panel).19 Six human cell lines of the three major histological types of NSCLC, namely adenocarcinomas cells (H-23, A549, EKVX), squamous cells carcinomas (H-226), and large cells carcinomas (H-460, HOP-92) were analyzed. Relative levels of CFL1 gene expression obtained by microarray are presented in Figure 3A (symbols) and match the amount of cofilin protein evaluated here (Fig. 3A; bars). Then, using the BD BioCoat™ Matrigel™ Invasion System (to assess the tumor’s metastatic potential), we found that different histological types expressing higher CFL1 levels presented higher invasion indexes, which indicates a more aggressive invasiveness behavior (Fig. 3B) (*P < 0.02, Mann Whitney test; **P < 0.0001, One-way ANOVA).
FIGURE 3. Cofilin immunocontent correlates with tumor invasiveness and resistance against alkylating drugs.
Six human NSCLC cell lines composed of adenocarcinomas (H-23, A549, EKVX), large cells (H-460, HOP-92) and squamous-cells carcinomas (H-226) from the NCI-60 panel were selected based on different levels of CFL1 gene expression (http://discover.nci.nih.gov/datasetsNature2000.jsp) to establish the role of CFL1 in tumor aggressiveness, evaluated by assays of cell invasion and drug resistance. (A) Western blot analysis shows that the pattern of CFL1 mRNA (symbols) matches with the level of cofilin immunocontent (bars). (B) Invasion index was obtained by determining the movement of cells through an 8.0 µm pore size, either uncoated (migration) or matrigel-coated (invasion), attracted by a chemotactic gradient of serum. The mean of four fields for each condition in quadruplicates is plotted. *P < 0.02 (Mann Whitney test); **P < 0.0001 (One-way ANOVA).
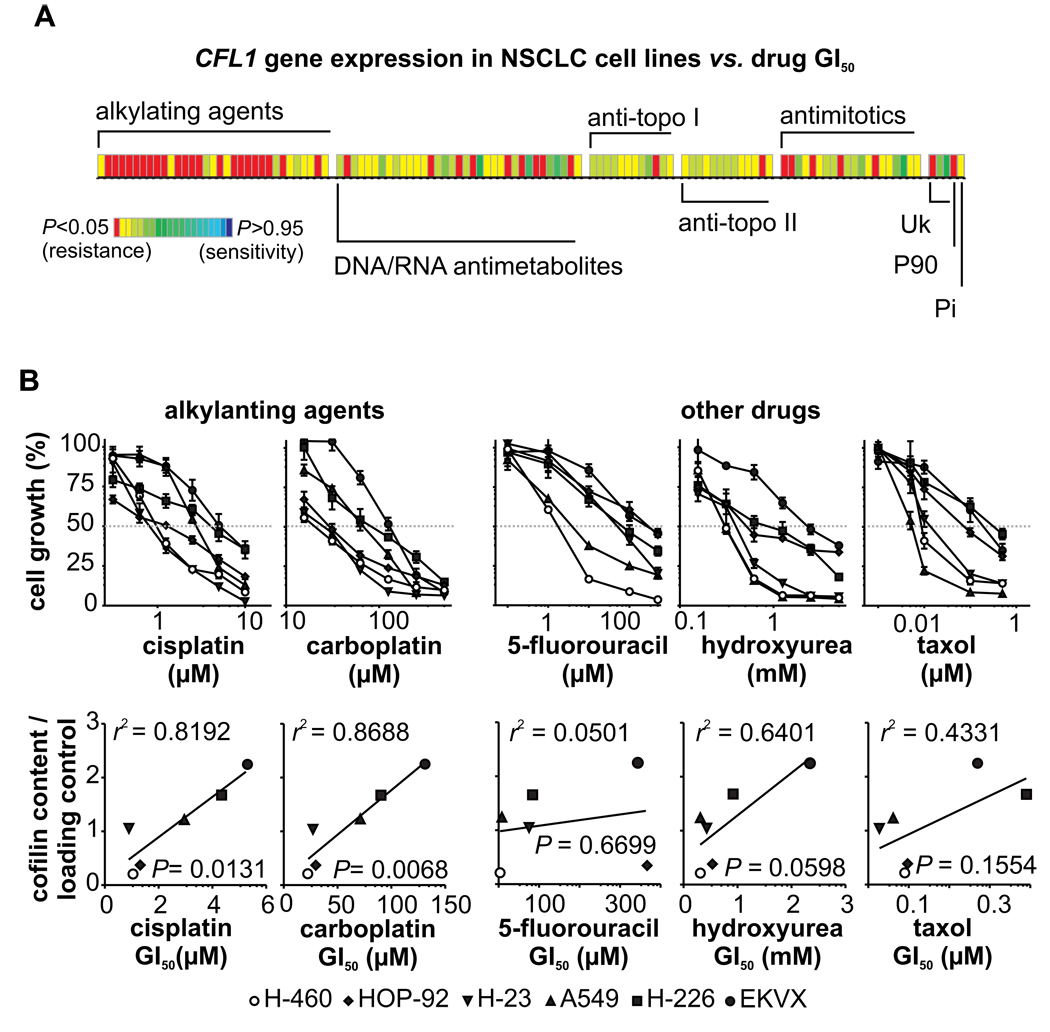
In addition to this higher invasiveness potential, analysis of microarray data of the six cell lines and respective GI50 values of 118 standard chemotherapy agents (from the NCI60 drug discovery pipeline) revealed that high levels of CFL1 mRNA is also correlated with resistance against different anticancer drugs – mainly alkylating agents (Fig. 4A; meta-analysis) (for a list of all correlated alkylating drugs see Table 2). Exposure of the cell lines to different concentrations of selected chemotherapy drugs (namely cisplatin, carboplatin, 5-fluorouracil, hydroxyurea, and taxol) revealed significant correlations between cofilin immunocontent and resistance to cisplatin and carboplatin, the two alkylating agents tested (Fig. 4B; in vitro analysis).
FIGURE 4. CFL1 mRNA and protein levels vs. drug sensitivity/resistance profile.
(A) Microarray meta-data of the cell lines are crossed against GI50 values of 118 standard chemotherapy agents (from NCI-60 drug discovery pipeline). P values have been color coded according to the scale shown; P < 0.05 indicates a significant negative correlation (resistance) while P > 0.95 indicates a significant positive correlation (sensitivity). The major mechanism of drug action is shown (the term “alkylating agents” is used broadly to include platinating agents; Uk: unknown; P90: hsp90 binder; Pi: protein synthesis inhibitor). Each column within the matrix represents the Spearman correlation between gene expression and toxicity of an individual drug. (B) In vitro validation of the citotoxicity for selected drugs assayed by the sulforhodamine B (SRB) method (upper plots). The obtained drug GI50 values were correlated with cofilin immunocontent (lower plots).
Table 2.
List of alkylating agents for which CFL1 mRNA levels are biomarker1 for drug resistance.
| Class2 | Drugs | Rs | P-value |
|---|---|---|---|
| A2 | Porfiromycin | 0.771 | 0.036 |
| A6 | Carmustine (BCNU) | 1.000 | 0.000 |
| A6 | Chlorozotocin | 0.943 | 0.002 |
| A6 | Clomesone | 0.943 | 0.002 |
| A6 | Lomustine (CCNU) | 0.771 | 0.036 |
| A6 | Mitozolamide | 0.943 | 0.002 |
| A6 | PCNU | 0.943 | 0.002 |
| A6 | Semustine (MeCCNU) | 0.886 | 0.009 |
| A7 | Asaley | 0.771 | 0.036 |
| A7 | Carboplatin | 0.829 | 0.021 |
| A7 | Chlorambucil | 0.829 | 0.021 |
| A7 | Cisplatin | 0.829 | 0.021 |
| A7 | Cyclodisone | 0.943 | 0.002 |
| A7 | Hepsulfam | 0.771 | 0.036 |
| A7 | Iproplatin | 1.000 | 0.000 |
| A7 | Mechlorethamine | 0.943 | 0.002 |
| A7 | Melphalan | 0.771 | 0.036 |
| A7 | Piperazine mustard | 0.943 | 0.002 |
| A7 | Piperazinedione | 0.771 | 0.036 |
| A7 | Spiromustine | 0.886 | 0.009 |
| A7 | Uracil mustard | 0.829 | 0.021 |
| A7 | Yoshi-864 | 0.771 | 0.036 |
Meta-analysis data of chemotherapeutic drugs from a panel of 33 alkylating agents (from Fig.4A) tested for positive correlation (resistance) between drug GI50 (µM) and the pattern of CFL1 gene expression in six human NSCLC cell lines (A549, EKVX, HOP-92, NCI-H226, NCI-H23, NCI-H460) obtained from the NCI-60 cell panel.
Mechanism of action codes: A2= alkylating at N-2 position of guanine; A6= alkylating at O-6 position of guanine; A7= alkylating at N-7 position of guanine.
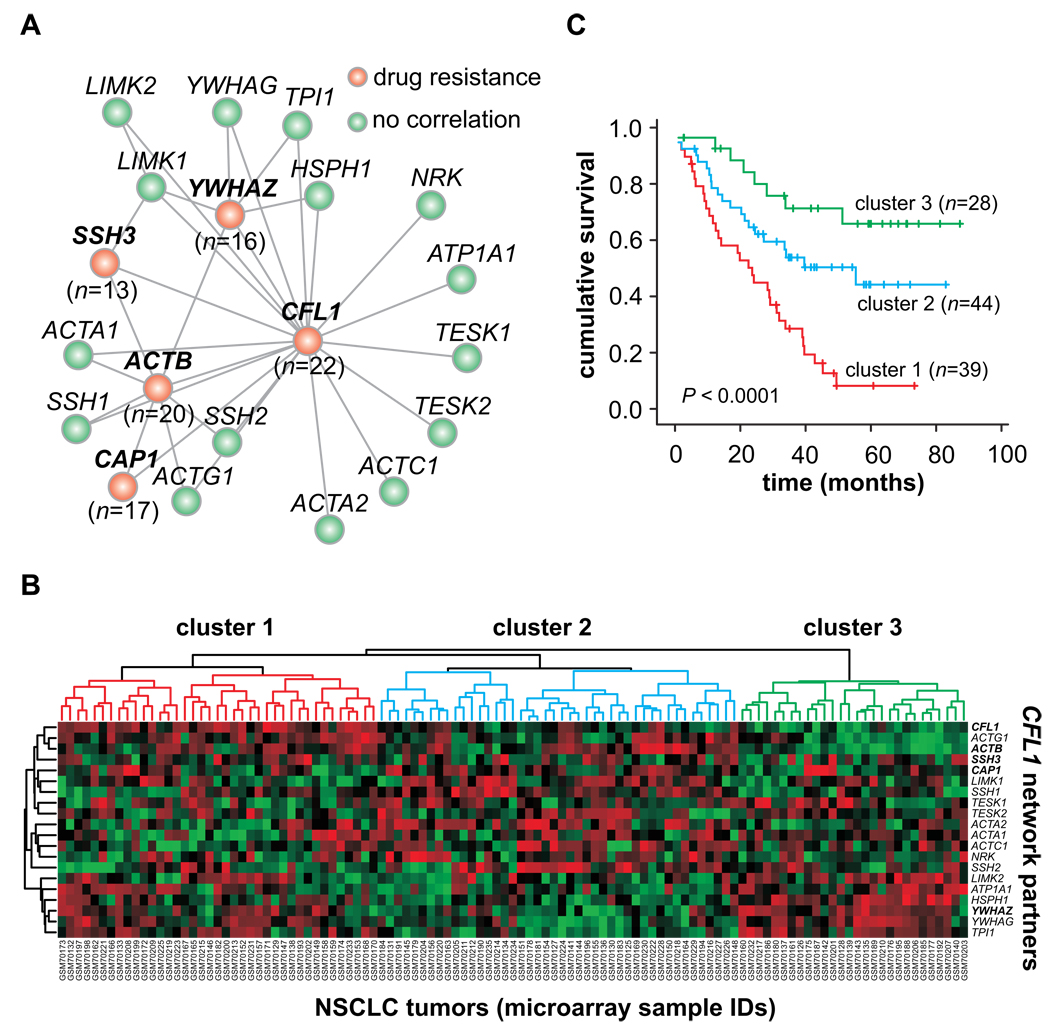
Using the same approach on drug resistance, we evaluated the resistance profile against alkylating agents of each gene product that interacts directly with CFL1. Four cofilin’s partners (CAP1, ACTB, SSH3, YWHAZ genes) show a resistance profile similar to cofilin, suggesting that a functional network is correlated with this tumor phenotype. These results are presented as network-based model of the cofilin biological pathway (Fig. 5A, red nodes), where nodes represent gene products and connecting lines indicate physical and/or functional associations according to experimental data (http://string.embl.de/).
FIGURE 5. Prognostic and drug resistance marker of CFL1 functional gene network.
(A) Graph model of CFL1 functional gene network vs. alkylating drug sensitivity/resistance profile. Nodes represent gene products; connecting lines indicate physical and/or functional associations according to experimental data (http://string.embl.de/). Gene expression data (http://discover.nci.nih.gov/cellminer/home.do) were crossed against GI50 values of all alkylating agents identified in the resistance panel at Figure 4A. Four CFL1 network partners follow the same resistance profile (red nodes; n = number of drugs for which gene expression showed correlation). Network drawn was built using a spring model algorithm. Further detains in Methods. (B) Two-way hierarchical clustering analysis of NCSLC tumors. This panel presents the NSCLC cohort data (referred to as Testing Cohort in Table 1) arranged according to the gene expression profile of all CFL1 network components. Complete linkage clustering of tumor samples is shown in TREEVIEW format. The color intensity is relative to the log2 ratio of the microarray signal (red: positive values; green: negative values). For visualization purposes, the gene expression values were median centered and normalized using CLUSTER 3.0 software. (c) Kaplan Meier plot of the entire NSCLC cohort data (n=111), where patients are stratified according to the hierarchical clustering analysis of CFL1 functional gene network.
To further explore the role of this gene network in NCSLC patient outcome, a cluster analysis was carried out using the data-bank from testing cohort. As the microarray dataset from this cohort study was produced on Affymetrix U133 Plus 2.0 platform, all genes listed in our network could be retrieved. Complete linkage clustering of tumor samples is shown in TREEVIEW format (Fig.5B). From the Heat Map, we identified three large tumor clusters, which were then used to re-stratify the NSCLC patients according to the gene expression profile. Kaplan-Meier estimates based on this new stratification showed that the CFL1 gene network can also be used to discriminate patient’s outcomes (Fig.5C).
DISCUSSION
Although much progress has been made in reducing overall mortality rates, cancer is a major public health problem worldwide, accounting for more deaths than heart disease. Most recent epidemiological data show a notable trend in stabilization of incidence rates for all cancer and a continued decrease in the cancer death rate.1 While the decrease in death rates for colorectal, breast, and prostate cancer largely reflects improvements in early detection and treatment, the decrease in lung cancer death rates reflects manly the reduction in tobacco use.24,25
In this scenario, non-small-cell lung cancer (NSCLC) is the leading cause of deaths annually. Currently, prognosis of NSCLC patients is still based almost exclusively on the anatomical extent of disease and may have reached their limit of usefulness for predicting outcomes.4 Advances in molecular pathology underwent to the development of many candidate biomarkers with potential clinical value. However, according to the TNM tumor staging system, only few tumors are formally staged with the addition of molecular biomarker information (e.g. TNM+S; where S= serum levels of selected biomarker), which does not include lung cancers.3
Herein we are proposing the use of CFL1 gene expression levels as a prognostic and predictive NSCLC biomarker based on the following findings: i) CFL1 mRNA levels are highly sensitive and specific to discriminate between good and bad patient outcome in two independent cohort – specially in early-stage disease – where tumor with low expression of CFL1 gene are associated with high overall survival; ii) an association exists between cofilin immunocontent and tumor invasion; iii) cells with high cofilin mRNA and protein levels are resistant to alkylating drug treatment, and iv) four other genes that interact in the CFL1 pathway (named SSH3, YWHZ, CAP1 and ACTB) also demonstrate the same resistance profile.
As previously shown, to be able to generate early metastasis, tumor cells require the activity of cofilin to modulate actin cytoskeleton, generating cell mobility.9,10 Therefore, as cofilin is associated with epithelial-mesenchymal transition and tumor invasion, it stands to reason that NSCLC patients with high tumor CFL1 expression levels present low overall survival rates, even in early-stage disease. Our data obtained by in vitro experiments suggest that cofilin levels also could be used to predict tumor resistance to alkylating agents. The correlation between high levels of cofilin and alkylating drug resistance probably is the most important finding of this study, since this class of drugs is among the most effective cytotoxic agents for advanced cancer treatments and has long been the cornerstone of NSCLC management.26,27 Even though this treatment improves patient survival, the benefit is stage-dependent. Unfortunately, intrinsic or acquired resistance to alkylating agents is frequently encountered and severely limits its therapeutic potential.28 Our findings may have great impact on survival rates, as currently there is no way to predict and identify potential responders.
Although we focused our analysis on the role of CFL1 gene in alkylating drugs resistance, we also expanded the potential biological relevance of our findings by testing the role of other cofilin’s partners on tumor resistance. Doing so, we obtained a signature based on five biological related genes (members of the cofilin pathway). These genes can be used in combination to characterize the tumor resistance phenotype. This approach is consistent with other studies that have been proposing the use of gene combination to enhance biomarker robustness, which may potentially deal better with intrinsic intra and inter-sample heterogeneity29–31. For instance, Chen & cols32 have described a biomarker cluster comprising the combination of DUSP6, MMD, STAT1, ERBB3 and LCK gene expression to predict clinical outcome of NSCLC patient. This signature was obtained based on the statistical (not biological) combination of high-throughput screening of cDNA microarray probes. Likewise, other authors used the same strategy to identify low/high NSCLC risk phenotypes.14,15
In this sense, our five-gene signature emerges from a functional gene network comprising all described cofilin partners. To further explore this finding using the NSCLC cohort data, we assigned subsets of tumors (clusters) based on related expression patterns, represented in the tree structure, or dendrograms. Using all CFL1 gene network components, the hierarchical clustering analysis put together the similar network datasets, stratifying NSCLC patients in three large subgroups, whose outcomes differ in the same extent as observed for CFL1 gene alone. The effect of this strategy of assessing tumors is the distribution of biomarker task among related genes, not focused on one or several non-related ones, which can potentially reduce the effect of random fluctuations on biomarker performance. Further investigation of the molecular properties of this network should be helpful to validate these genes as prognostic and predictive markers in NSCLC, or even in other cancer types, given that CFL1 gene is widely expressed34 and more specifically in some subtypes (e.g. colorectal adenocarcinomas34,35).
Our findings have clear implications for NSCLC management and therapy, as CFL1 expression levels can be used to indicate which patients should receive a more aggressive therapy in an attempt to reverse the poor prognosis. Because CFL1 expression levels also correlate with drug resistance, our findings can also be used to decide the best course of action for each patient, representing a contribution into translational medicine for treating NSCLC. In the adjuvant setting, for example, cisplatin-based chemotherapy constitutes the standard first-line treatment for patients with early stage and good performance status.33 Since CFL1 expression appears to be a marker of resistance to platinum agents, patients whose tumors harbor high levels of CFL1 would benefit from a different treatment modality. In these cases, possible trials to test alternative adjuvant regimens would be based on the combination of other drugs commonly used in NSCLC (e.g. gencitabine, docetaxel and vinorelbine) or EGFR-targeted monoclonal antibodies. The combination of EGFR inhibitors with first-line chemotherapy is currently under evaluation and efforts have been made to identify subgroups of NSCLC patients that respond to these agents.36–38 The refinement of patient stratification with the use of CFL1 gene signature provides the opportunity to design a prospective, large-scale, randomized clinical trial that would evaluate these ideas.
Acknowledgments
Financial support: Brazilian MCT/CNPq Universal funds (479860/2006-8) and MCT/CNPq INCT-TM (573671/2008-7) funds and federal funds from the National Cancer Institute, National Institutes of Health (NO1-CO-12400). We would like to thank Dr Marcia Triunfol at Publicase for editing the article and for manuscript comments.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:1–25. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Potti A, Mukherjee S, Petersen R, et al. A Genomic Strategy to Refine Prognosis in Early-Stage Non-Small-Cell Lung Cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 3.Greene FL, Page DL, Fleming ID, Fritz A, Balch CM, Haller DG, Morrow M. AJCC Cancer Staging Manual. 6.ed. New York: Springer; 2002. [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Kaminski N, Krupsky M. Gene expression patterns, prognostic and diagnostic markers, and lung cancer biology. Chest. 2004;125:111S–115S. doi: 10.1378/chest.125.5_suppl.111s-a. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh M, Song X, Mouneimne G, et al. Cofilin Promotes Actin Polymerization and Defines the Direction of Cell Motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 7.Klamt F, Zdanov S, Levine RL, Pariser A, Zhang Y, Zhang B, et al. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol. 2009;11:1241–1247. doi: 10.1038/ncb1968. DOI: 10.1038/ncb1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rheenen J, Song X, van Roosmalen W, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–1259. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 12.Mouneimne G, DesMarais V, Sidani M, Scemes E, Wang W, Song X, et al. Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr Biol. 2006;16:2193–2205. doi: 10.1016/j.cub.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 14.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 15.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 16.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 17.von Mering C, Jensen LJ, Kuhn M, et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 19.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 20.Castro MAA, Mombach JCM, de Almeida RMC, Moreira JCF. Impaired expression of NER gene network in sporadic solid tumors. Nucleic Acids Res. 2007;35:1859–1867. doi: 10.1093/nar/gkm061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro MAA, Rybarczyk Filho JL, Dalmolin RJS, et al. ViaComplex: software for landscape analysis of gene expression networks in genomic context. Bioinformatics. 2009;25:1468–1469. doi: 10.1093/bioinformatics/btp246. [DOI] [PubMed] [Google Scholar]
- 22.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 23.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 24.Ravdin PM, Cronin KA, Howlader N, et al. The Decrease in Breast-Cancer Incidence in 2003 in the United States. N Engl J Med. 2007;356:1670–1674. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 25.Jemal A, Clegg LX, Ward E, et al. Annual report to the nation on the status of cancer, 1975–2001, with a special feature regarding survival. Cancer. 2004;101:3–27. doi: 10.1002/cncr.20288. [DOI] [PubMed] [Google Scholar]
- 26.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 27.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 28.Imaizumi M. Postoperative adjuvant cisplatin, vindesine, plus uracil-tegafur chemotherapy increased survival of patients with completely resected p-stage I non-small cell lung cancer. Lung Cancer. 2005;49:85–94. doi: 10.1016/j.lungcan.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 29.Castro MAA, Onsten TGH, Moreira JCF, de Almeida RMC. Chromosome aberrations in solid tumors have a stochastic nature. Mutat Res. 2006;600:150–164. doi: 10.1016/j.mrfmmm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen HY, Yu SL, Chen CH, et al. A Five-Gene Signature and Clinical Outcome in Non-Small-Cell Lung Cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 33.IALT. Cisplatin-Based Adjuvant Chemotherapy in Patients with Completely Resected Non-Small-Cell Lung Cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 34.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. BioGPS – biogps.gnf.org. [Google Scholar]
- 36.Linardou H, Dahabreh IJ, Bafaloukos D, Kosmidis P, Murray S. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC. Nat Rev Clin Oncol. 2009;6:352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- 37.Hirsch FR. The role of genetic testing in the prediction of response to EGFR inhibitors in NSCLC. Oncogene. 2009;28 Suppl 1:S1–S3. doi: 10.1038/onc.2009.195. [DOI] [PubMed] [Google Scholar]
- 38.Rosell R, Robinet G, Szczesna A, Ramlau R, Constenla M, Mennecier BC, et al. Randomized phase II study of cetuximab plus cisplatin/vinorelbine compared with cisplatin/vinorelbine alone as first-line therapy in EGFR-expressing advanced non-small-cell lung cancer. Ann Oncol. 2008;19:362–369. doi: 10.1093/annonc/mdm474. [DOI] [PubMed] [Google Scholar]