Abstract
The role of NF-κB in the expression of inflammatory genes and its participation in the overall inflammatory process of chronic diseases and acute tissue injury are well-established. We and others have demonstrated a critical involvement of poly(ADP-ribose) polymerase (PARP)-1 during inflammation, in part, through its relationship with NF-κB. However, the mechanism by which PARP-1 affects NF-κB activation has been elusive. Here, we show that PARP-1 inhibition by gene knockout, knockdown, or pharmacological blockade prevented p65-NF-κB nuclear translocation in smooth muscle cells (SMCs) upon TLR4 stimulation, NF-κB DNA-binding activity, and subsequent iNOS and ICAM-1 expression. Such defects were reversed by reconstitution of PARP-1 expression. PARP-1 was dispensable for LPS-induced I-κBα phosphorylation and subsequent degradation but was required for p65-NF-κB phosphorylation. A perinuclear p65-NF-κB localization in LPS-treated PARP-1−/− cells was associated with an export rather an import defect. Indeed, while PARP-1 deficiency did not alter expression of importin α3 and α4 and their cytosolic localization, the cytosolic levels of exportin (Crm)-1 were increased. Crm1 inhibition promoted p65-NF-κB nuclear accumulation as well as reversed LPS-induced p65-NF-κB phosphorylation and iNOS and ICAM-1 expression. Interestingly, p65-NF-κB poly(ADP-ribosyl)ation decreased its interaction with Crm1 in vitro. Pharmacological inhibition of PARP-1 increased p65-NF-κB-Crm1 interaction in LPS-treated SMCs. These results suggest that p65-NF-κB poly(ADP-ribosyl)ation may be a critical determinant for the interaction with Crm1 and its nuclear retention upon TLR4 stimulation. These results provide novel insights into the mechanism by which PARP-1 promotes NF-κB nuclear retention, which ultimately can influence NF-κB-dependent gene regulation.
Introduction
The role of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammation has been investigated intensely in the context of its direct participation, by way of its catalytic activity in cellular responses to DNA-damaging agents, including oxidative stress (1). In a number of pathological conditions that involve massive DNA damage, the excessive activation of PARP-1 depletes cellular stores of both NAD and its precursor ATP, leading to irreversible cytotoxicity and potentially, cell death (2–4). We recently showed that PARP-1 plays important roles in allergic asthma and atherosclerosis (5–7). An emerging role for this protein, however, is the ability of PARP-1 to participate, directly or indirectly, in the regulation of a number of inflammatory genes, especially those mediated by NF-κB (reviewed in (8). NF-κB is a pleiotropic transcription factor that plays a critical role in the regulation of the expression of multiple genes involved in inflammatory responses, including inducible nitric oxide synthase (iNOS), and adhesion molecules (9). NF-κB binds to the promoter regions of target genes as a dimer of two Rel family proteins, most frequently p50 and p65 (9, 10). In quiescent cells, NF-κB is sequestered in the cytoplasm as a result of its interaction with members of the IκB family of proteins, which includes I-κBα and IκBβ. I-κBα is phosphorylated, polyubiquitinated, and degraded by the 26S proteasome in response to cell stimulation, resulting in the release of the nuclear localization signal of NF-κB and its subsequent translocation to the nucleus (9).
Interestingly, while p65 NF-κB nuclear translocation in TNF-treated smooth muscle cells (SMCs) was sufficient for the expression of VCAM-1, we recently demonstrated that PARP-1 is required for expression of ICAM-1 (11). The expression of ICAM-1 was associated with a transient interaction between PARP-1 and p65 NF-κB when examined in COS-7 cells and in the airway epithelial cell line, A549. We (5, 7, 11, 12) and others (13, 14) have reported that NF-κB nuclear translocation requires PARP-1 expression as assessed by electrophoretic mobility shift assay (EMSA) upon TLR4 stimulation by LPS treatment. The defect in the nuclear translocation of NF-κB culminated in the severe reduction in the expression of NF-κB-targeted genes such as iNOS, MCP-1, COX-2, and adhesion molecules.
The nuclear localization signal (NLS) embedded within NF-κB binds to importins and thus promotes nuclear translocation of the transcription factor by allowing its passage through the nuclear pore complex (15). The import of p65 NF-κB to the nucleus upon stimulation has been attributed predominantly to importin α3 and importin α4 (15). Additionally, NF-κB has been reported to interact with exportins, which through a multifactor complex can transport the transcription factor from the nucleus to the cytosol (16). Crm1, an important member of the nuclear export machinery, recognizes and export proteins containing a leucine-rich nuclear export signal (NES) (17). Crm1 binds cooperatively with RanGTP and the cargo to form a trimeric complex NES-RanGTP-Crm1, which then exports through the nuclear pore complexes.
The mechanism(s) by which PARP-1, a nuclear enzyme, influences cytosolic events leading to the release of NF-κB from the cytosol and translocation to the nucleus are not understood. Thus, the goals of the present study were to determine the mechanism(s) by which PARP-1 participates in the retention of NF-κB in the nucleus upon TLR4 stimulation by LPS.
Materials and Methods
Animals, isolation of SMCs and fibroblasts, and treatment protocols
Mice were bred in a specific pathogen-free facility at LSUHSC, New Orleans, LA, and allowed unlimited access to sterilized chow and water. Maintenance, experimental protocols, and procedures were all approved by the LSUHSC Animal Care & Use Committee. C57BL/6 wild-type (WT) mice were purchased from Jackson Laboratories (Bar Harbor, ME). The generation of C57BL/6 PARP-1−/− mice was previously described (6). All animals were genotyped by PCR.
Isolation and assessment of purity of SMCs preparations were conducted as described (11). SMCs were used between passages 4 and 7. Mouse embryonic fibroblasts (MEF) were isolated as described (18). 293T cells and SMCs were maintained in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Prior to treatment, SMCs at 50–80% confluence were starved by incubation in DMEM/F12 with 0.5% cell culture-tested BSA (Sigma-Aldrich) for 18 h. Cells were treated with 1μg/mL LPS (AXXORA, San Diego, CA) for the indicated times in the absence or presence 50 μM of the PARP-1 inhibitors NU1025 (Santa Cruz Biotechnology, Santa Cruz, CA) or 1 mM 3-aminobenzamide (Santa Cruz Biotechnology). In some experiments, cells were treated with 1 mM H2O2 for the indicated time intervals before processing for immunofluorescence. For microscopy, cells were incubated in CO2-independent medium.
Total, cytoplasmic and nuclear extract preparations and immunoblot analysis
After treatment with the indicated agents, cells were washed with ice-cold PBS followed by centrifugation. For total protein extracts, cell pellets were incubated for 15 min on ice in lysis buffer supplemented with proteases and phosphatase inhibitors as previously described (19). The preparation of cytoplasmic and nuclear extracts was performed using a commercial Kit (Active Motif, Carlsbad, CA) according to manufacturer’s instructions. A portion (15 μg protein) of each lysate was then fractionated by SDS-PAGE on a 4 to 20% gradient gel, and the separated proteins were transferred to a nitrocellulose filter. The filter was stained with Ponceau S to confirm equal loading and transfer of samples, and was then probed with antibodies to IκBα, ICAM-1, VCAM, iNOS, Crm1, p65 NF-κB, PARG, or actin (Santa Cruz Biotechnology), PARP-1 (BD-Pharmingen, San Jose, CA), importin α3 (Sigma), importin α4 (Imgenex, San Diego, CA), GST (Novus Biologicals, Littleton, CO) as well as antibodies to the phosphorylated form (serine residues 32 and 36) of I-κBα (p-I-κBα) (Cell Signaling Technology, Danvers, MA), or of p65 NF-κB at serine 276, serine 529, or serine 536. Immune complexes were detected with appropriate secondary antibodies and chemiluminescence reagents (PerkinElmer Life Science Inc., Boston, MA).
Conventional RT-PCR
RNA was extracted from cells using standard methods and cDNA was generated using reverse transcriptase III (Invitrogen, Carlsbad, CA). Oligonucleotide primers to specifically amplify a fragment of ICAM-1, iNOS, or β-actin were purchased from Integrated DNA Technologies. The specific primers were as follows: ICAM-1: forward primer, 5′-TCC TAA AAT GAC CTG CAG ACG - 3′; reverse primer, 5′- AGT TTT ATG GCC TCC TCC TGA - 3′; iNOS: forward primer, 5′- CAG CTG GGC TGT ACA AAC CTT-3′; reverse primer, 5′ CAT TGG AAG TGA AGC GTT TCG -3′; and β-actin: forward primer, 5′-ACC GTG AAA AGA TGA CCC AGA TC -3′; reverse primer, 5′-TAG TTT CAT GGA TGC CAC AGG -3′. The amplification program was as follows: 3 min at 95 °C, 30 sec at 95 °C, 45 sec at 60 °C, and 45 sec. at 72 °C. The cycle numbers were optimized for each primer pair. The PCR products were then incubated for 15 min at 72 °C. The resulting PCR products were subjected to electrophoresis in a 2%-agarose gel and stained with ethidium bromide.
Construction of the GST-p65 NF-κB, p65 NF-κB-EYFP and PARP-1-EYFP plasmids, and adenoviral vector expressing PARP-1-EYFP, transduction, and transfections
Construction of the p65 NF-κB-EYFP expression vector was described elsewhere (11). For the construction of the GST-p65 NF-κB expression vector, the p65 NF-κB cDNA was cloned into a Gateway entry vector pENTR/SD/D-TOPO (Invitrogen). The accuracy of the construct was verified by sequence analysis. The expression clone was generated by performing an LR recombination reaction between the entry clone and the destination vector GST-pDEST27 (Invitrogen).
SMCs were transiently transfected with p65 NF-κB-EYFP using the Mirus TransIT-LT1 transfection reagent (Mirus, Madison, WI) according to the manufacturer’s instructions. After starvation, SMCs were cultured in CO2-independent medium (Invitrogen) and treated with LPS as described earlier (11). Subcellular localization of p65 NF-κB was monitored throughout the treatment using a Carl Zeiss inverted fluorescence microscope. The PARP-1-EYFP expression vector was described previously (20). All sequences were confirmed by sequencing. Knockdown of PARP-1 (sc-29438-V, Santa Cruz Biotechnology) or PARG (sc-152026-V, Santa Cruz Biotechnology) in wild type SMCs was achieved using lentiviral vectors encoding the respective targeting shRNA, according to the manufacturer’s specifications and instructions, before treatment with the indicated reagents.
Pulldown assay, EMSA, and Luciferase Assay
293T cells were transiently transfected with the GST-p65 NF-κB vector using Lipofectamine LTX (Invitrogen) and were treated with 1μg/mL LPS (AXXORA, San Diego, CA) for the indicated times. GST-p65 NF-κB was pulled down using Glutathione Sepharose™ 4 Fast Flow (GE Healthcare, Piscataway, NJ). EMSA analysis of DNA-binding activity in prepared nuclear extracts was performed as described (21). MEF were transiently transfected with a total of 5 μg of the RapidReporter pRR-High-NFκB plasmid (Active Motif) using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s instructions. After treatment, cells were lysed (20 μl of Lysis Buffer per well) then incubated for 30 minutes at room temperature. Substrate was added to extracts according to the manufacturer’s instructions and measured on a tristar LB 941 luminometer (Berthold Technologies, USA).
Poly(ADP-ribosyl)ation in Vitro
Purified recombinant GST-p65 or untagged p65 (Active Motif, Carlsbad, CA) NF-κB and Purified PARP were incubated in a mixture (25 μl) containing reaction buffer (100 mM Tris-HCl, 1 mM DTT, 10 mM MgCl2), sonicated salmon sperm DNA and 2 mM NAD (sigma) for 30 min at 37 °C. The reaction was either terminated by the addition of an equal volume of SDS sample buffer and heating at 95 °C for 5 min or subjected to pulldown with GST beads after addition of recombinant Crm1 protein. Samples were then subjected to immunoblot analysis with antibodies to poly(ADP-ribose) (PAR) (AXXORA, San Diego, CA), p65 NF-κB, or GST.
Indirect immunofluorescence
Cells grown on chamber slides were washed in PBS three times after the indicated treatment, and then fixed with 3.7% paraformaldehyde for 20 min at room temperature. After permeabilization in 0.05% Triton X-100 in PBS for 5 min, cells were incubated with primary antibodies for 1 h at room temperature. Antibody–antigen complexes were detected with Alexa-546-conjugated secondary antibody for 1 h at room temperature. The cells were thoroughly washed after each incubation and then counterstained with DAPI. Cells were examined under a fluorescence microscope (Leica) using a 40× objective lens.
Results
PARP-1 is required for LPS-induced translocation of p65 NF-κB to nuclei of SMCs and expression of iNOS and ICAM-1
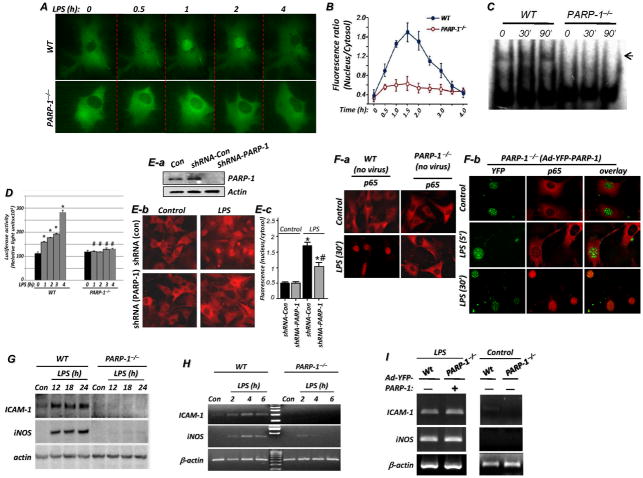
Using both live imaging of SMCs transfected with YFP-tagged p65 NF-κB (Fig. 1A) and immunofluorescence with antibodies to p65 NF-κB (Fig. 1F-a), we showed that PARP-1 gene deletion severely reduced the ability of p65 NF-κB to translocate to the nuclei of primary SMCs upon TLR4 stimulation by LPS, confirming earlier reports from our laboratory (7, 11, 12) and that of others (13, 14). Fig. 1B provides a quantitative assessment of NF-κB subcellular localization with additional time points. Treatment of SMCs with LPS resulted in a marked increase in the DNA-binding activity of NF-κB as assessed by EMSA, and this effect was completely blocked in LPS-treated PARP-1−/− SMCs (Fig. 1C). PARP-1 expression appeared to be required for the expression of ICAM-1 and iNOS as well as other NF-κB-dependent genes in cells other than SMCs. Indeed, expression of ICAM-1 (see supplemental Fig. S1) and iNOS (7, 22) was blocked in LPS-treated PARP-1−/− macrophages. Furthermore, in MEF transiently transfected with the RapidReporter pRR-High-NF-κB plasmid, treatment with LPS induced a pronounced increase in luciferase activity; such increase was completely blocked in the PARP-1−/− counterparts (Fig. 1D), which is consistent with published reports (5, 14, 23). PARP-1 knockdown with a lentiviral vector encoding a shRNA targeting mouse PARP-1 (Fig. 1E-a) significantly reduced nuclear localization of p65 NF-κB upon LPS treatment of wild type SMCs (Fig. 1E-b-c). The impairment of p65 NF-κB nuclear translocation in LPS-treated PARP-1−/− cells (Fig. 1F-a) was fully reversed upon reconstitution of PARP-1 expression using adenovirus gene transfer (Ad-YFP-PARP-1) as assessed by immunofluorescence with antibodies to p65 NF-κB (Fig. 1F-b). PARP-1 gene deletion markedly reduced expression of iNOS and ICAM-1 at the protein (Fig. 1G) and mRNA levels (Fig. 1H). The expression of iNOS and ICAM-1 was fully restored by adenoviral-mediated reconstitution of PARP-1 expression (Fig. 1I), confirming the specificity and validity of our observations.
Fig. 1. Translocation of p65 NF-κB is defective in PARP-1−/− SMCs in response to LPS treatment and is reversed by reconstitution of PARP-1 expression.
(A) SMCs derived from WT or PARP-1−/− mice were transfected with the YFP-p65 NF-κB plasmid. Cells were then cultured in CO2-independent medium and incubated on a temperature-controlled inverted fluorescence microscope. LPS (1 μg/ml) was added to the culture chambers and images were then taken at the times indicated. (B) p65 NF-κB nuclear translocation was quantified and assessed as the ratios between nuclear and cytosolic fluorescence in at least 10 cells and expressed as mean ± SEM; *, difference from LPS-treated WT cells at a respective time point, p < 0.01. (C) WT or PARP-1−/− SMCs were treated with LPS for 30 or 90 min or left untreated. Nuclear extracts were then prepared and subjected to EMSA with a radioactively labeled κB sequence; arrow indicates NF-κB-DNA complex. (D) WT or PARP-1−/− MEF transiently transfected with the RapidReporter pRR-High-NFκB plasmid were treated with LPS for the indicated time intervals after which luciferase activity was assessed; Data (luciferase activity) are expressed in relative light units and are means ± SD of triplicate values from representative experiments; *difference from untreated cells, P < 0.05; #difference from cells treated with LPS at the respective time point, P < 0.05. (E) PARP-1 knockdown by shRNA reduces p65 NF-κB translocation in LPS-treated SMCs. (E-a) WT SMCs were transduced with lentiviral particles encoding control shRNA (shRNA-Con) or a shRNA targeting PARP-1 (shRNA-PARP-1); expression of PARP-1 and actin was assessed by immunoblot analysis and compared to that in uninfected cells. Cells transduced with either virus were treated with LPS for30 min after which p65 NF-κB nuclear localization was examined by immunofluorescence (E-b) and quantified (E-c). *, difference from untreated cells; #, difference from LPS-treated cells transduced with shRNA-Con virus; p < 0.01. (F) The defective p65 NF-κB translocation in PARP-1−/− SMCs in response to LPS treatment, confirmed by immunofluorescence (F-a), is fully reversed by reconstitution of WT PARP-1 expression (F-b). WT and PARP-1−/− SMCs were infected with the adenoviral vector expressing an YFP-WT PARP-1 (Ad-YFP-PARP-1) or left uninfected. Thirty-six hours later, cells were treated with 1 μg/ml LPS or left untreated (Con) for the indicated time periods. NF-κB nuclear translocation was assessed by immunofluorescence (IF) with antibodies to p65 NF-κB. PARP-1 was visualized with YFP. (G–I) Expression of iNOS and ICAM-1 is blocked in LPS-treated PARP-1−/− SMCs and fully restored upon reconstitution of PARP-1 expression. WT or PARP-1−/− SMCs were treated with LPS for the indicated time intervals after which protein extracts or total RNA were prepared and subjected to immunoblot analysis with antibodies to mouse iNOS, ICAM-1, or actin (G) or subjected to RT-PCR using primers specific to mouse iNOS, ICAM-1, or β-actin with amplicons analyzed by agarose electrophoresis (H). (I) WT SMCs or Ad-YFP-PARP-1-infected PARP-1 −/− SMCs were treated with LPS for 3 h after which total RNA was prepared and subjected to RT-PCR using primers specific to mouse iNOS, ICAM-1, or β-actin; the right panels represent expression of iNOS or ICAM-1 mRNA in untreated WT or PARP-1−/− cells.
PARP-1 expression is dispensable for I-κBα phosphorylation and subsequent degradation but is required for p65 NF-κB phosphorylation in response to TLR4 stimulation
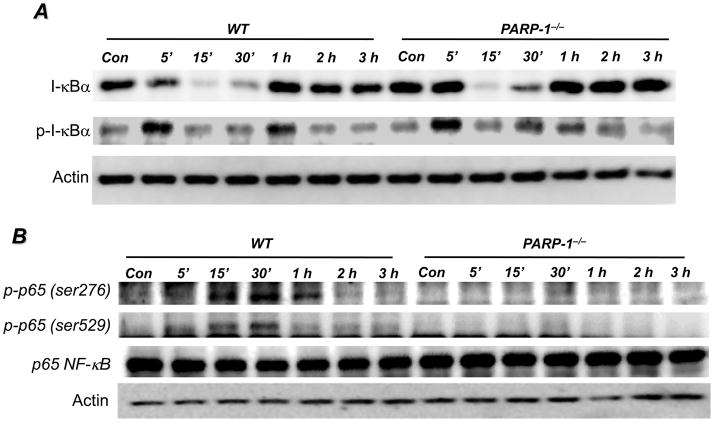
We next investigated whether PARP-1 gene deletion prevented NF-κB nuclear translocation in response to LPS by affecting the critical events leading to its activation and consequent nuclear translocation as well as its subsequent post-translocation phosphorylation. Surprisingly, I-κBα phosphorylation and subsequent degradation were almost identical in both LPS-treated WT and PARP-1−/− SMCs (Fig. 2A) with the clear preservation of the transient nature of I-κBα phosphorylation and degradation. These results strongly suggest that the NF-κB signal transduction machinery post-TLR4 stimulation is intact in PARP-1−/− cells and that the defect may reside in the actual translocation of the transcription factor to the nucleus rather than its association with I-κBα.
Fig. 2. Effects of PARP-1 gene deletion on phosphorylation and degradation of I-κBα and phosphorylation of p65 NF-κB in SMCs upon LPS treatment.
WT or PARP-1−/− SMCs were treated with 1 μg/ml LPS for different time intervals or left untreated (Con). (A) Proteins extracts were subjected to immunoblot analysis with antibodies to I-κBα or phosphor-I-κBα at Ser32/Ser36. (B) The same extracts were subjected to immunoblot analysis with antibodies to phospho-p65 NF-κB at Ser276 or Ser529, total p65 NF-κB, or actin.
Accumulating evidence indicates that after the release of NF-κB from inhibition by I-κBα, the phosphorylation of the p65 subunit at serine 276, serine 529, and serine 536 is critical for its interaction with transcriptional co-activators such as CBP/p300 and for binding to its target sites on DNA (24). We and others have reported that interaction between PARP-1, p65 NF-κB, CBP/p300 takes place upon stimulation with TNF or LPS (11, 25). Given that the TLR4-associated signal transduction appeared to be intact in PARP-1−/− cells, while expression of NF-κB-dependent genes was compromised, it became important to determine whether PARP-1 plays a role in events leading to p65 NF-κB phosphorylation. Fig. 2B shows that phosphorylation of p65 NF-κB at serine 276 and serine 529 was evident in WT SMCs but almost completely absent in PARP-1−/− cells upon TLR4 stimulation by LPS. These results clearly suggest that PARP-1 expression is necessary for events leading to the phosphorylation of p65 NF-κB. It is unclear how PARP-1 influences these phosphorylation events.
Expression of importin α3 and importin α4 and their cytosolic localization are not altered in PARP-1−/− cells
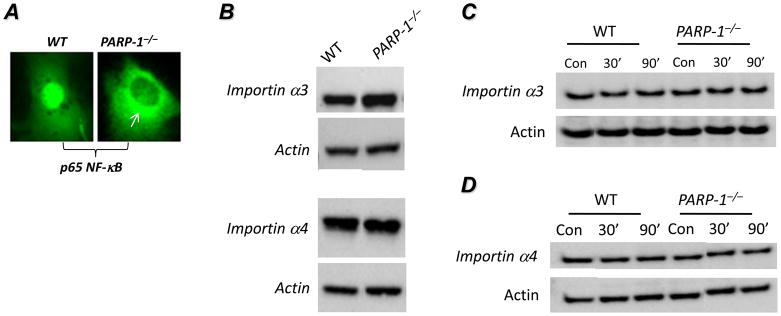
A closer examination of p65 NF-κB localization in LPS-treated PARP-1−/− cells revealed that the protein appeared to accumulate in the perinuclear area of a subpopulation (30–40%) of the examined cells (Fig. 3A), which suggested a potential defect in the nuclear import system in these cells. We surmised that the defect in p65 NF-κB nuclear translocation in response to LPS may be coupled to a defective importin system. First, we examined whether PARP-1 gene deletion exerted any effect on expression of importin α3 and importin α4. Fig. 3B shows that the expression levels of importin α3 and importin α4 were not affected by PARP-1 gene deletion in SMCs. Given that interaction between p65 NF-κB and importins takes place in the cytosol after release from I-κBα upon stimulation, we examined whether the levels of cytosolic importins were different between WT and PARP-1−/− cells upon stimulation with LPS. The levels of cytosolic importin α3 (Fig. 3C) and importin α4 (Fig. 3D) were not different between WT and PARP-1−/− cells suggesting that the defect in p65 NF-κB nuclear translocation in LPS-treated PARP-1−/− cells resides upstream of importins.
Fig. 3. Effect of PARP-1 gene deletion on expression and cytosolic localization of importins in SMCs upon LPS treatment.
(A) High magnification image of nuclear and perinuclear localization of YFP-p65 NF-κB in LPS-treated WT and PARP-1−/− SMCs, respectively. (B) Total protein extracts, prepared from WT or PARP-1−/− SMCs, were subjected to immunoblot analysis with antibodies to importin α3, importin α4, or actin. WT or PARP-1−/− SMCs were treated with 1 μg/ml LPS for 30 or 90 min or left untreated (Con), after which cytosolic fractions were prepared. The resulting extracts were subjected to immunoblot analysis with antibodies to importin α3 (C), importin α4 (D), or actin.
Inhibition of Crm1 by leptomycin B (LMB) promotes accumulation of p65 NF-κB in nuclei of PARP-1−/− cells, and reverses expression of NF-κB-dependent genes upon TLR4 stimulation
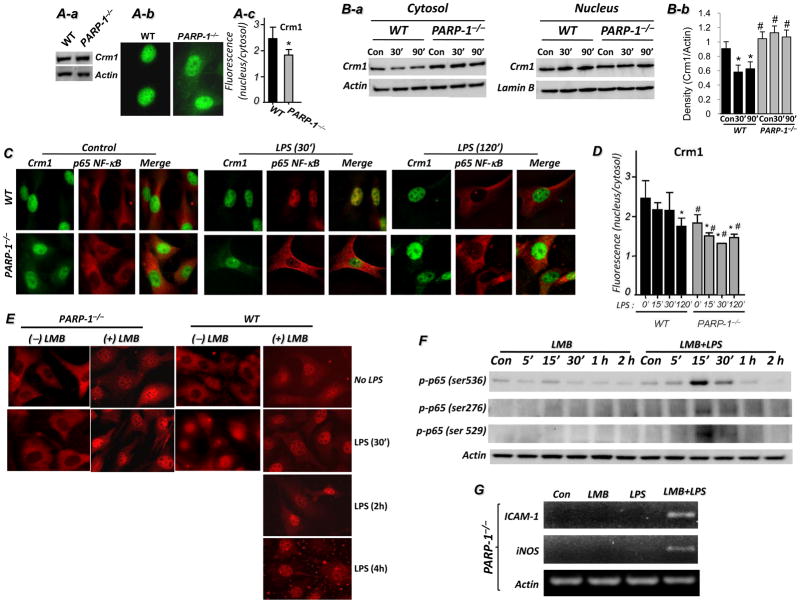
Since the importin system was not affected by PARP-1 gene deletion, we reasoned that elevated export activity resulted in NF-κB-nuclear accumulation upon TLR4 stimulation in PARP-1−/− cells. Accordingly, we tested the hypothesis that NF-κB was not detected in the nucleus because it was rapidly exported by Crm1-associated export machinery. PARP-1 gene deletion did not alter total Crm1 expression (Fig. 4A-a) but was associated with an increase in cytosolic Crm1 in control cells (Fig. 4A-b-c). This increase in cytosolic Crm1 was verified by cell fractionation followed by immunoblot analysis (Fig. 4B). LPS treatment promoted a decrease in cytosolic Crm1 in WT cells. In sharp contrast, the levels of cytosolic Crm1 were relatively unchanged in LPS-treated PARP-1−/− cells (Fig. 4B). Interestingly, no major changes in nuclear Crm1 was observed in both WT and PARP-1−/− cells suggesting that the level of the translocated Crm1 to the cytosol was minor compared to that in the nucleus. A quantitative assessment of the data is presented in Fig. 4B-b. The persistence of Crm1 in the cytosol of control and LPS-treated PARP-1−/− SMCs was verified by immunofluorescence (Fig. 4C–D). These results suggest that PARP-1 may play a role in the dynamics of Crm1 trafficking.
Fig. 4. Effect of PARP-1 gene deletion on expression and cytosolic localization of Crm1 and effects of Crm1 inhibition on p65 NF-κB nuclear accumulation and expression of iNOS and ICAM-1 in LPS-treated SMCs.
(A) WT or PARP-1−/− SMCs were subjected to protein extracts followed by immunoblot analysis with antibodies to Crm1 or actin or subjected to immunofluorescence with antibodies to Crm1. (B) The same cytosolic fractions described in Fig. 3C were subjected to immunoblot analysis with antibodies to Crm1 and actin. (C) WT or PARP-1−/− SMCs cultured on chamber slides were treated with 1 μg/ml LPS for 30 or 120 min or left untreated (Con), after which cells were fixed and subjected to immunofluorescence with antibodies to Crm1 (Alexa Fluo 488, green) and p65 NF-κB (Alexa Fluo 594, red). (D) Crm1 cellular distribution was quantified as described for p65 NF-κB in Fig. 1C; *, difference from respective untreated cells; #, difference from WT cells at the respective treatment time; p < 0.01. (E) WT or PARP-1−/− SMC were pretreated with LMB or left untreated after which they were treated with 1 μg/ml LPS for the indicated time intervals. Cells were then fixed and subjected to immunofluorescence with antibodies to p65 NF-κB. Inhibition of Crm1 by leptomycin B (LMB) promotes accumulation of p65 NF-κB in nuclei of PARP-1−/− cells and expression of iNOS and ICAM-1 in LPS-treated PARP-1−/− SMCs. WT or PARP-1−/− SMCs, pretreated with LMB or left untreated, were exposed to 1 μg/ml LPS for different time intervals or left untreated (Con). Proteins extracts were prepared and subjected to immunoblot analysis with antibodies to phospho-p65 NF-κB at ser276, ser529, or Ser536, or actin (F). (G) Cells were treated as in (F) but for 3 h, after which total RNA was extracted and subjected to RT-PCR analysis with primers to iNOS, ICAM-1, and β-actin.
The coincidence of Crm1 and p65 NF-κB in the cytosolic compartments of LPS-treated PARP-1−/− cells is suggestive of a potential link between PARP-1 gene deletion and the defect in NF-κB nuclear translocation. To further investigate the relationship between PARP-1, p65 NF-κB, and Crm1 and to establish that the export system is affected by PARP-1 gene deletion, we hypothesized that inhibition of Crm1 would allow p65 NF-κB nuclear accumulation in LPS-treated PARP-1−/− cells. Indeed, Crm1 inhibition allowed p65 NF-κB nuclear accumulation in PARP-1−/− cells without LPS treatment, similar to that observed in WT cells (Fig. 4E). LPS treatment appeared to increase the nuclear retention of p65 NF-κB with almost a complete absence of the protein in the cytosolic compartment in PARP-1−/− cells (Fig. 4E). These results suggest that nuclear accumulation of p65 NF-κB in PARP-1−/− cells in response to TLR4 stimulation results from enhanced Crm1-associated export of the transcription factor from the nucleus.
The nuclear retention of p65 NF-κB in LMB-treated PARP-1−/− cells coincided with a robust increase in its phosphorylation state at ser-276, ser-529, and ser-536 but only upon LPS treatment, suggesting a potential transcriptional activity (Fig. 4F). An important question to be addressed was whether such nuclear accumulation of p65 NF-κB and concomitant phosphorylation at ser-536 in LPS and LMB-treated PARP-1−/− cells culminated in expression of NF-κB-driven genes such as iNOS or ICAM-1. Fig. 4G shows that, indeed, inhibition of Crm1 restored LPS-stimulated iNOS or ICAM-1 expression in PARP-1−/− cells. Together, these data strongly suggest that PARP-1 promotes the nuclear function of NF-κB by preventing its export from the nucleus by Crm1.
Poly(ADP-ribosyl)ation of p65 NF-κB by PARP-1 decreases its interaction with Crm1
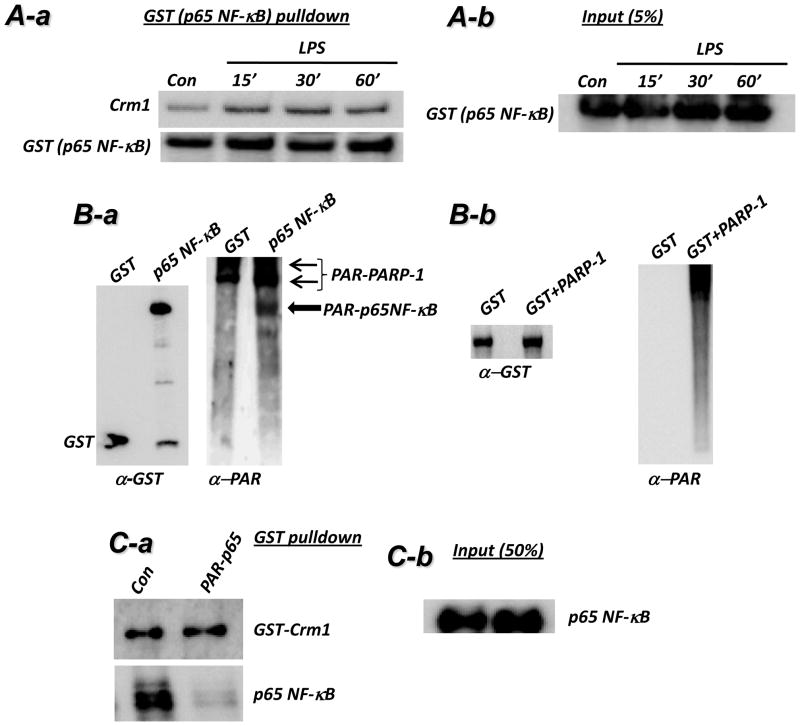
We next investigated the potential mechanism by which PARP-1 influences the relationship between Crm1 and p65 NF-κB. Fig. 5A confirms the interaction between Crm1 and p65 NF-κB and the dynamic nature of such an interaction upon LPS stimulation in 293T cells. Using an in vitro poly(ADP-ribosyl)ation system (26), we showed that p65 NF-κB can be modified by PARP-1 (Fig. 5B), confirming the report by Kameoka et al. (27). We next examined whether PARP-1-mediated modification of p65 NF-κB would affect the interaction between the transcription factor and Crm1 in vitro. Fig. 5C shows that poly(ADP-ribosyl)ation of p65 NF-κB almost completely abolished the interaction between the two proteins. These observations are consistent with the hypothesis that poly(ADP-ribosyl)ation of p65 NF-κB inhibits its association with Crm1 and hence attenuates nuclear export of the transcription factor.
Fig. 5. Interaction between Crm1 and p65 NF-κB is blocked by PARP-1-mediated modification of p65 NF-κB.
(A-a) 293T cells, transfected with GST-p65 NF-κB, were treated with 1 μg/ml LPS for 15, 30, or 60 min or left untreated (Con). Protein extracts were then prepared and were subjected to a pulldown assay with GST-beads. The resulting precipitates were subjected to immunoblot analysis with antibodies to Crm1 and GST. (A-b) A 5% portion of total protein was used as input control (right panel). (B-a) GST-p65 NF-κB protein purified from transiently transfected 293T was incubated with or without recombinant PARP-1 in a poly(ADP-ribosyl)ation reaction containing NAD and activated DNA for 15 min. The reactions were terminated with sample buffer and subjected to immunoblot analysis with antibodies to poly(ADP-ribose) (α-PAR) or GST. (B-b) Purified GST was subjected to a poly(ADP-ribosyl)ation reaction in the absence or presence of recombinant PARP-1 followed by immunoblot analysis with antibodies to either GST (left panel) or PAR (right panel). (C-a) p65 NF-κB protein (not tagged with GST) was incubated in a poly(ADP-ribosyl)ation reaction mixture with or without recombinant PARP-1 (not tagged with GST) for 30 min (saturated reaction). Recombinant GST-Crm1was then added to the reaction for 15 min. The mixture was then subjected to a pulldown assay with glutathione beads followed by immunoblot analysis with antibodies to GST or p65 NF-κB. (C-b) A portion equal to 50% of p65 NF-κB added to the poly(ADP-ribosyl)ion reaction was subjected to immunoblot analysis with antibodies to p65 NF-κB.
Pharmacological inhibition of PARP-1 reduces p65 NF-κB nuclear retention and subsequent expression of ICAM-1 and iNOS in response to TLR4 stimulation, in part, by promoting persistence of p65 NF-κB-Crm1 interaction
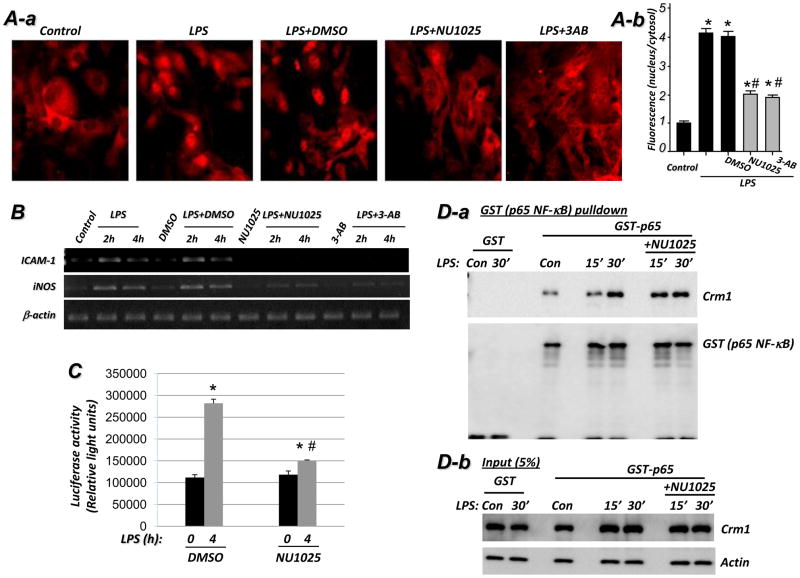
While the involvement of the PARP-1 enzymatic activity in the expression of NF-κB-dependent genes and interaction with related transcription factors remains controversial, its involvement in NF-κB nuclear translocation upon TLR4 stimulation is completely unknown. Our data predict that inhibition of PARP1 enzymatic activity should promote the p65 NF-κB-Crm1 interaction and thereby enhance Crm1-mediated nuclear export of the transcription factor. Figure 6A shows that inhibition of PARP-1 by either a non-competitive (NU1025) or a competitive (3-aminobenzamide) inhibitor significantly reduced p65 NF-κB nuclear accumulation in LPS-treated SMCs. Such reduction in nuclear translocation correlated with a decrease in p65 NF-κB phosphorylation upon pharmacological inhibition of PARP-1 (see supplemental Fig. S2). In agreement with these findings, expression of iNOS and ICAM-1 in LPS-stimulated SMCs was markedly reduced by the PARP-1 inhibitors (Figure 6B). Accordingly, these results demonstrate that the enzymatic activity of PARP-1 is required for a full nuclear translocation of p65 NF-κB and subsequent expression of iNOS and ICAM-1. The requirement of PARP-1 enzymatic activity for NF-κB-dependent gene expression was verified by a luciferase activity assay using wild type MEF that were transiently transfected with the RapidReporter pRR-High-NF-κB plasmid (Fig. 6C). Whether the requirement for PARP-1 enzymatic activity applies to all NF-κB stimuli and all cell types is not fully established.
Fig. 6. Pharmacological inhibition of PARP-1 reduces p65 NF-κB nuclear retention and subsequent expression of ICAM-1 and iNOS in response to TLR4 stimulation, in part, by promoting persistence of p65 NF-κB-Crm1 interaction.
(A) PARP-1 inhibitors reduce LPS-induced p65 NF-κB nuclear translocation. WT SMCs were treated with LPS in the presence or absence of the non-competitive (NU1025) or the competitive (3-AB) PARP-1 inhibitor for 30 min after which p65 NF-κB nuclear localization was examined by immunofluorescence (A-a) and quantified (A-b) as described in Fig. 1C. *, difference from untreated cells; #, difference from LPS-treated cells; p < 0.01. (B) WT SMCs were treated as in (A) but for 2 or 4 h after which total RNA were prepared and subjected to RT-PCR using primers specific to mouse iNOS, ICAM-1, or β-actin with amplicons analyzed by agarose electrophoresis. (C) WT MEF transiently transfected with the RapidReporter pRR-High-NF-κB plasmid were treated with LPS for 4 h in the presence or absence of NU1025 after which luciferase activity was assessed; Data (luciferase activity) are expressed in relative light units and are means ± SD of triplicate values from representative experiments; *difference from untreated cells, P < 0.05; #difference from cells treated with LPS alone, P < 0.05. (D-a) 293T cells, transfected with GST-p65 NF-κB, were treated with LPS for 15 or 30 min or left untreated (Con) in the absence or presence of NU1025. Protein extracts were then prepared and were subjected to a pulldown assay with GST-beads. The resulting precipitates were subjected to immunoblot analysis with antibodies to Crm1 and GST. (D-b) A 5% portion of total protein was used as input control.
Inhibition of PARP-1 activity can also be achieved by reducing expression of PARG, the PAR-hydrolysing enzyme through an increase in the population of automodified PARP-1 molecules (28, 29). Indeed, poly(ADP-ribosyl)ation activity was markedly reduced in cells that were subjected to PARG knockdown with a lentiviral vector encoding PARG-targeting shRNA (Supplemental data S3-A) upon treatment with the DNA damage-inducing agent H2O2 (Supplemental data S3-B). The decreased in PARP-1 activity in PARG-depleted cell extracts was confirmed using an in vitro poly(ADP-ribosyl)ation assay (Supplemental data S3-C). PARG knockdown significantly decreased p65 NF-κB retention in SMC upon LPS treatment (S3-D), which provides additional support for the involvement of PARP-1 activity in the nuclear retention of the transcription factor.
We next investigated the potential effect of pharmacological inhibition of PARP-1 on the interaction between Crm1 and p65 NF-κB. Figure 6D shows that the interaction of p65 NF-κB was persistent in cells treated with LPS in the presence of the PARP-1 inhibitor NU1025 compared to that observed in cells treated with LPS alone. These results strongly suggest that PARP-1 enzymatic activity is required for an efficient nuclear retention of p65 NF-κB upon TLR4 stimulation and confirm our in vitro data.
Overall, these results strongly suggest that the potential key regulatory mechanism of NF-κB nuclear retention may reside in the modification of p65 NF-κB by poly(ADP-ribosyl)ation, which prevents its interaction with Crm1, rendering the transcription factor resistant to export and promoting its retention in the nucleus.
Discussion
The mechanisms by which PARP-1, a nuclear protein, affects the activation of NF-κB in the cytosol and its nuclear translocation are not understood. The results of the present study shed light on these mechanisms. Our initial prediction was that the importin system was defective in PARP-1-deficient cells given the accumulation of NF-κB in the perinuclear compartment of the cells. However, our results showed that PARP-1 exerts a decisive effect on the interaction between p65 NF-κB and the exportin protein Crm1. In the absence of PARP-1, the interaction between the transcription factor and Crm1 increases such that little to no p65 NF-κB was detectable in the nuclei of cells upon TLR4 stimulation. Such an enhanced interaction culminates in the down-regulation of NF-κB-dependent genes despite the presence of functional TLR4-dependent signal transduction. The specificity of the effect of PARP-1 gene deletion on NF-κB nuclear translocation/retention was supported by the finding that a re-establishment of PARP-1 expression completely reversed the ability of the transcription factor to be retained within the nuclei of LPS-treated cells and to promote the expression of the NF-κB target genes ICAM-1 and iNOS.
Several studies on the potential mechanism by which PARP-1 regulates NF-κB-mediated gene activation, including those from our laboratory, have focused on factors that are related directly to NF-κB DNA-binding to the promoter regions of target genes (5, 11, 14, 23, 25, 27, 30, 31). The requirement of PARP-1 activity for the expression of iNOS and ICAM-1 as well as other NF-κB-dependent genes has been reported by several groups (32–34). Our results also show that PARP-1 activity is crucial for efficient nuclear retention of the transcription factor. Thus, to the best of our knowledge, these results reveal a novel mechanism by which PARP-1 influences p65 NF-κB nuclear localization. However, whether PARP-1 activity is the only defining factor in NF-κB nuclear retention and expression of target genes has yet to be fully settled. In several comprehensive investigations, Hottiger’s group reported that neither enzymatic nor DNA binding activity of PARP-1 is required for NF-κB to induce its target genes; only its direct protein-protein interaction with the transcription factor is required for the specific induction of gene transcription (reviewed (8). It is noteworthy that the enzymatic activity of PARP-1 is clearly required for the transcriptional activity of other factors, such as NFAT, HES1, Sp1, and Elk1 (reviewed (35). The release of p65 NF-κB from inhibition upon phosphorylation and subsequent degradation of I-κBα does not seem to be sufficient to mediate transcription of target genes; the transcription factor must be actively retained in the nucleus for transcriptional activity to take place. PARP-1 does not appear to play a role in the cytosolic events upstream of p65 NF-κB translocation to the nucleus upon TLR4 stimulation.
The role of Crm1 in the export of nuclear factors including proteins is complex and is associated with numerous sophisticated processes (36). The interaction between Crm1 and NF-κB subunits as well as I-κBα has been well-established (37). Our results showed that inhibition of Crm1 resulted in NF-κB nuclear accumulation but did not enhance the expression of the NF-κB-dependent genes, iNOS and ICAM-1; stimulation of TLR4 was still required. Nuclear accumulation of p65 NF-κB in LPS-treated WT cells coincided with a decrease in cytosolic Crm1. Interestingly, the lack of nuclear accumulation of p65 NF-κB in LPS-treated PARP-1−/− cells correlated with the persistent cytosolic expression of Crm1. It is noteworthy that PARP-1 gene deletion did not exert any effect on the total expression of Crm1. Whether the changes in cellular localization of Crm1 upon LPS stimulation were strictly associated with p65 NF-κB translocation or were a general phenomenon related to the overall signal transduction associated with TLR4 stimulation is not clear. However, it is plausible to predict that, at least, a portion of the cytosolic Crm1 is responsible for the lack of nuclear accumulation of p65 NF-κB in LPS-treated PARP-1−/− cells. This observation is supported by the finding that Crm1 inhibition by LMB promoted the accumulation of nuclear p65 NF-κB in LPS-treated PARP-1−/− cells. According to our in vitro data, such role involved PARP-1-mediated poly(ADP-ribosyl)ation of p65 NF-κB, which reduced the interaction of the modified protein with Crm1. This observation was confirmed in LPS-treated cells using NU1025, a potent inhibitor of PARP-1 enzymatic activity. A recent report by Fukasawa’s group (38) investigating Crm1 in the context of nuclear accumulation of p53 supports the notion that this mechanism of regulation is not restricted to NF-κB alone.
PARP-1 appears to be required for the phosphorylation of p65 NF-κB at serine residues 276, 529, and 536. Although the mechanism by which PARP-1 influences p65 NF-κB phosphorylation is not clear, PARP-1 has been reported to interact with several kinases such as DNA-dependent protein kinase and ERK (39). Thus, it will be very important to determine how p65 NF-κB phosphorylation relates to the role of PARP-1-mediated regulation of this transcription factor and its interaction with Crm1. It is noteworthy that PARP-1 deficiency did not seem to affect the phosphorylation of p65 NF-κB given the observation that such event was reversed by LMB-mediated Crm1 inhibition in LPS-treated PARP-1−/− cells. This may suggest that the phosphorylation was related to a defect in nuclear retention rather than PARP-1 deficiency. What is clear and established, however, is that the phosphorylation of the transcription factor at these sites is critical for the expression of NF-κB-dependent genes (40–43). Additionally, the expression levels of I-κBα (balance between expression and degradation) also may dictate the nature of the interaction between PARP-1 and p65 NF-κB as well as with Crm1. Furthermore, the role of acetylation undoubtedly is very important and may be a critical factor in the functionality of NF-κB and its ability to drive target gene expression. As stated above, we and others recently showed an interaction between PARP-1 and p300/CBP that appears to take place immediately after the interaction of PARP-1 with NF-κB (11, 25). The consequences of the sequence of these events are far from clear.
An additional important aspect of NF-κB nuclear translocation is the importin system (44). Although the data on the role of importins in p65 NF-κB shuttling between the cytosol and the nucleus presented in this study are not extensive, however, they suggest that there is no relationship between PARP-1 and these proteins as it relates to nuclear localization of p65 NF-κB, especially given the fact that PARP-1 gene deletion exerted no effect on total or cytosolic levels of importin α3 and importin α4 upon LPS treatment. These observations are supported by the fact that Crm1 inhibition by LMB leads to accumulation of p65 NF-κB within the nucleus, which again suggests that the importin system may be intact; however, this remains to be verified. Furthermore, it was recently shown that the nucleoporin Nup214 modulates NF-κB activation (in Drosophila) and influences the relative strength and duration of NF-κB signaling responses (45). Again, whether Nup214 influences the relationship between PARP-1, NF-κB, and Crm1, remains to be studied. In conclusion, the results of the present study provide novel insights into the mechanism by which PARP-1 influences activation of TLR4-associated signal transduction leading to NF-κB activation and subsequent nuclear retention.
Supplementary Material
Acknowledgments
We would like to thank Ji-Won Park and Waleed Elsegeiny for their technical assistance.
Footnotes
This work was supported, in part, by grants HL072889, and 1P20RR18766 (overall P.I. Dr. D. Kapusta) from the NIH, from the American Cancer Society (RSG-116608) and funds from the Louisiana Cancer Research Consortium (New Orleans, LA) to H. Boulares. This work was also supported by grant 1P20 RR021970/COBRE from the National Center for Research Resources to S. Koochekpour (PI: A. Ochoa).
References
- 1.Burkle A. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays. 2001;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- 2.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiarugi A, Meli E, Calvani M, Picca R, Baronti R, Camaioni E, Costantino G, Marinozzi M, Pellegrini-Giampietro DE, Pellicciari R, Moroni F. Novel isoquinolinone-derived inhibitors of poly(ADP-ribose) polymerase-1: pharmacological characterization and neuroprotective effects in an in vitro model of cerebral ischemia. J Pharmacol Exp Ther. 2003;305:943–949. doi: 10.1124/jpet.103.048934. [DOI] [PubMed] [Google Scholar]
- 4.Nicoletti VG, Stella AM. Role of PARP under stress conditions: cell death or protection? Neurochem Res. 2003;28:187–194. doi: 10.1023/a:1022316914492. [DOI] [PubMed] [Google Scholar]
- 5.Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Gene knockout or pharmacological inhibition of poly(ADP-ribose) polymerase-1 prevents lung inflammation in a murine model of asthma. Am J Respir Cell Mol Biol. 2003;28:322–329. doi: 10.1165/rcmb.2001-0015OC. [DOI] [PubMed] [Google Scholar]
- 6.Boulares H, Zoltoski A, Kandan S, Akbulut T, Yakovlev A, Oumouna M. Correlation between decreased sensitivity of the Daudi lymphoma cells to VP-16-induced apoptosis and deficiency in DNAS1L3 expression. Biochem Biophys Res Commun. 2006;341:653–662. doi: 10.1016/j.bbrc.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Oumouna-Benachour K, Hans CP, Suzuki Y, Naura A, Datta R, Belmadani S, Fallon K, Woods C, Boulares AH. Poly(ADP-ribose) polymerase inhibition reduces atherosclerotic plaque size and promotes factors of plaque stability in apolipoprotein E-deficient mice: effects on macrophage recruitment, nuclear factor-kappaB nuclear translocation, and foam cell death. Circulation. 2007;115:2442–2450. doi: 10.1161/CIRCULATIONAHA.106.668756. [DOI] [PubMed] [Google Scholar]
- 8.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto Y, Gaynor RB. Role of the NF-kappaB pathway in the pathogenesis of human disease states. Curr Mol Med. 2001;1:287–296. doi: 10.2174/1566524013363816. [DOI] [PubMed] [Google Scholar]
- 11.Zerfaoui M, Suzuki Y, Naura AS, Hans CP, Nichols C, Boulares AH. Nuclear translocation of p65 NF-kappaB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: Differential requirement for PARP-1 expression and interaction. Cell Signal. 2008;20:186–194. doi: 10.1016/j.cellsig.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hans CP, Feng Y, Naura AS, Zerfaoui M, Rezk BM, Xia H, Kaye AD, Matrougui K, Lazartigues E, Boulares AH. Protective effects of PARP-1 knockout on dyslipidemia-induced autonomic and vascular dysfunction in ApoE mice: effects on eNOS and oxidative stress. PLoS ONE. 2009;4:e7430. doi: 10.1371/journal.pone.0007430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. Embo J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha HC. Defective transcription factor activation for proinflammatory gene expression in poly(ADP-ribose) polymerase 1-deficient glia. Proc Natl Acad Sci U S A. 2004;101:5087–5092. doi: 10.1073/pnas.0306895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagerlund R, Melen K, Cao X, Julkunen I. NF-kappaB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell Signal. 2008;20:1442–1451. doi: 10.1016/j.cellsig.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Harhaj EW, Sun SC. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol Cell Biol. 1999;19:7088–7095. doi: 10.1128/mcb.19.10.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Boulares AH, Zoltoski AJ, Yakovlev A, Xu M, Smulson ME. Roles of DNA Fragmentation Factor and Poly(ADP-ribose) Polymerase in an Amplification Phase of Tumor Necrosis Factor-induced Apoptosis. J Biol Chem. 2001;276:38185–38192. doi: 10.1074/jbc.M100629200. [DOI] [PubMed] [Google Scholar]
- 19.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 20.Belmadani S, Zerfaoui M, Boulares HA, Palen DI, Matrougui K. Microvessel vascular smooth muscle cells contribute to collagen type I deposition through ERK1/2 MAP kinase, alphavbeta3-integrin, and TGF-beta1 in response to ANG II and high glucose. Am J Physiol Heart Circ Physiol. 2008;295:H69–76. doi: 10.1152/ajpheart.00341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulares HA, Giardina C, Navarro CL, Khairallah EA, Cohen SD. Modulation of serum growth factor signal transduction in Hepa 1-6 cells by acetaminophen: an inhibition of c-myc expression, NF-kappaB activation, and Raf-1 kinase activity. Toxicol Sci. 1999;48:264–274. doi: 10.1093/toxsci/48.2.264. [DOI] [PubMed] [Google Scholar]
- 22.Naura AS, Datta R, Hans CP, Zerfaoui M, Rezk BM, Errami Y, Oumouna M, Matrougui K, Boulares AH. Reciprocal regulation of iNOS and PARP-1 during allergen-induced eosinophilia. Eur Respir J. 2009;33:252–262. doi: 10.1183/09031936.00089008. [DOI] [PubMed] [Google Scholar]
- 23.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, I, Verma M. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 25.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 26.Boulares AH, Zoltoski AJ, Contreras FJ, Yakovlev AG, Yoshihara K, Smulson ME. Regulation of DNAS1L3 Endonuclease Activity by Poly(ADP-ribosyl)ation during Etoposide-induced Apoptosis. Role of Poly(ADP-ribose) Polymerase-1 cleavage in endonuclease activation. J Biol Chem. 2002;277:372–378. doi: 10.1074/jbc.M107738200. [DOI] [PubMed] [Google Scholar]
- 27.Kameoka M, Ota K, Tetsuka T, Tanaka Y, Itaya A, Okamoto T, Yoshihara K. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. Biochem J. 2000;346(Pt 3):641–649. [PMC free article] [PubMed] [Google Scholar]
- 28.Rapizzi E, Fossati S, Moroni F, Chiarugi A. Inhibition of poly(ADP-ribose) glycohydrolase by gallotannin selectively up-regulates expression of proinflammatory genes. Mol Pharmacol. 2004;66:890–898. doi: 10.1124/mol.104.000968. [DOI] [PubMed] [Google Scholar]
- 29.Erdelyi K, Kiss A, Bakondi E, Bai P, Szabo C, Gergely P, Erdodi F, Virag L. Gallotannin inhibits the expression of chemokines and inflammatory cytokines in A549 cells. Mol Pharmacol. 2005;68:895–904. doi: 10.1124/mol.105.012518. [DOI] [PubMed] [Google Scholar]
- 30.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 31.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 32.Le Page C, Sanceau J, Drapier JC, Wietzerbin J. Inhibitors of ADP-ribosylation impair inducible nitric oxide synthase gene transcription through inhibition of NF kappa B activation. Biochem Biophys Res Commun. 1998;243:451–457. doi: 10.1006/bbrc.1998.8113. [DOI] [PubMed] [Google Scholar]
- 33.Chiarugi A. Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima H, Nagaso H, Kakui N, Ishikawa M, Hiranuma T, Hoshiko S. Critical role of the automodification of poly(ADP-ribose) polymerase-1 in nuclear factor-kappaB-dependent gene expression in primary cultured mouse glial cells. J Biol Chem. 2004;279:42774–42786. doi: 10.1074/jbc.M407923200. [DOI] [PubMed] [Google Scholar]
- 35.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 37.Shen LQ, Xu X, Lu FL, Liang HP. Nucleocytoplasmic transport of nuclear factor kappa B and its regulatory mechanisms. Progress in Biochemistry and Biophysics. 2002;29:368–371. [Google Scholar]
- 38.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 39.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 41.Yang F, Tang E, Guan K, Wang CY. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol. 2003;170:5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
- 42.Seldon MP, Silva G, Pejanovic N, Larsen R, Gregoire IP, Filipe J, Anrather J, Soares MP. Heme oxygenase-1 inhibits the expression of adhesion molecules associated with endothelial cell activation via inhibition of NF-kappaB RelA phosphorylation at serine 276. J Immunol. 2007;179:7840–7851. doi: 10.4049/jimmunol.179.11.7840. [DOI] [PubMed] [Google Scholar]
- 43.Chen LY, Pan WW, Chen M, Li JD, Liu W, Chen G, Huang S, Papadimos TJ, Pan ZK. Synergistic induction of inflammation by bacterial products lipopolysaccharide and fMLP: an important microbial pathogenic mechanism. J Immunol. 2009;182:2518–2524. doi: 10.4049/jimmunol.0713933. [DOI] [PubMed] [Google Scholar]
- 44.Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 45.Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. The nucleoporin Nup214 sequesters CRM1 at the nuclear rim and modulates NFkappaB activation in Drosophila. J Cell Sci. 2006;119:4409–4419. doi: 10.1242/jcs.03201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.