Abstract
The methylated-CpG island recovery assay (MIRA) exploits the intrinsic specificity and the high affinity of a methylated CpG-binding protein complex (MBD2B and MBD3L1) to methylated CpG dinucleotides in genomic DNA. The MIRA approach works on double-stranded DNA and does not depend on the application of methylation-sensitive restriction enzymes. It can be performed on a few hundred nanograms of genomic DNA. Recently, the MIRA technique has been used to profile DNA methylation patterns at a resolution of 100 base pairs along the entire genome of normal human B-lymphocytes. The MIRA method is compatible with microarray and next generation DNA sequencing approaches. We describe the principles and details of this method applied for methylation profiling of genomes containing methylated CpG sequences.
1. Introduction
DNA methylation profiling has become an important technology in many areas of epigenetics research. Methylation profiling approaches are aimed at investigating DNA methylation patterns either across the entire genome and at different levels of resolution, or they are used to study methylation patterns at specific areas of interest, for example at promoters and other regulatory elements. Many such techniques have been reviewed (1) and some are covered in different sections of this issue. One ultimate goal is to measure the methylation status at every potential methylation site thus covering over 28 million CpG dinucleotides in the human genome. Aside from repetitive DNA sequences, this has recently been accomplished by employing sodium bisulfite conversion and high-throughput DNA sequencing (2). However, this approach still is very expensive and requires substantial computational resources. This extraordinary level of detail often is not required for many studies aimed at investigating DNA methylation in unique cell types or tissues. Often, it is sufficient to analyze DNA methylation at lower resolution, i.e. at ~ 100 bp, or to focus on specific areas of interest, for example specific chromosomal segments (3) or sequences of regulatory potential, e.g. CpG islands (4, 5). In our laboratories, we have developed a technology that provides a similar level of resolution as chromatin immunoprecipitation coupled to microarrays or next generation sequencing (ChIP-chip or ChIP-seq). This technique is called the methylated-CpG island recovery assay (MIRA) (4, 6, 7).
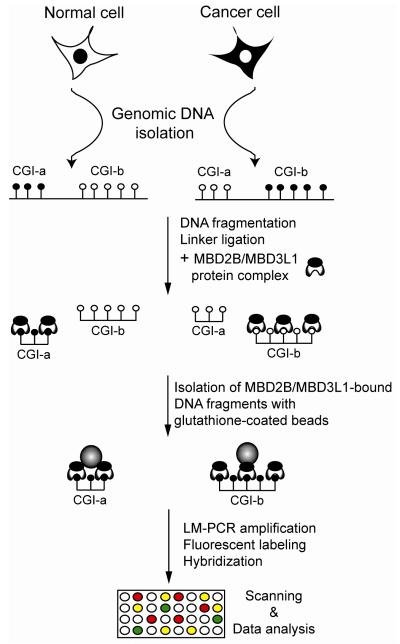
A small family of mammalian proteins contains a domain that mediates binding of the protein to double-stranded CpG-methylated DNA, the methylated-CpG binding domain (MBD) family, which is comprised of MECP2, MBD1, MBD2, MBD3, and MBD4 (8). MBD2B, the shorter protein isoform of MBD2, possesses the highest affinity to methylated DNA among the MBD proteins (9) and can heterodimerize with other MBD proteins via its C-terminal domain. MBD3L1 is also a member of the MBD family, is a homologue of MBD2 and MBD3, has no DNA binding domain itself, but can be a binding partner of MBD2, via heterodimer formation (10). The MBD2B/MBD3L1 heterodimer has higher affinity to methylated DNA than MBD2B alone (4, 6). In the MIRA procedure, the fragmentized genomic DNA is incubated with purified GST-MBD2B and His-MBD3L1 proteins. The high affinity MBD2B/MBD3L1 complex is formed and binds to the methylated genomic DNA templates. The methylated genomic DNA fragments are easily recovered from the binding reaction by using glutathionecoated magnetic beads (Fig. 1). The selectively isolated methylated DNA fraction is amplified, labeled and analyzed on different microarray platforms or with high throughput DNA sequencing on the Illumina Genome Analyzer (Fig. 2, and unpublished data).
Figure 1. MIRA-assisted genomic DNA methylation analysis.
A schematic diagram of the basic MIRA approach is shown. For DNA fragmentation, we generally use sonication. Methylated DNA molecules are enriched in the MIRA procedure by binding to the MBD2B/MBD3L1 complex. In the microarray approach, input and MIRA-enriched (methylated) fractions are linker-ligated, amplified and labeled with different dyes, mixed, and hybridized to the slides, and the relative enrichment factors between different cell types are determined. Alternatively, MIRA-enriched DNA from two cell types can be compared directly. Open and closed circles represent methylated and unmethylated CpG dinucleotides, respectively; CGI, CpG island. In the high throughput sequencing approach (see Figure 2), the MIRA-enriched fractions are amplified in the Illumina cluster station after ligation of adaptors, and the ends are sequenced. Sequence reads are aligned to the genome.
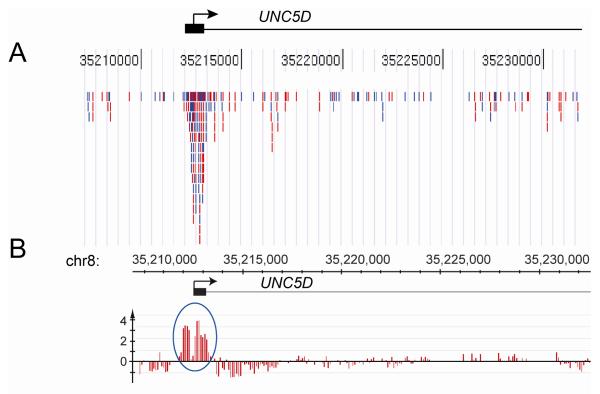
Figure 2. MIRA sequencing.
A. In the MIRA-seq approach, the methylated fragments of the genome are enriched by the MIRA technique and are sequenced using the Illumina Genome Analyzer. DNA obtained from a stage I lung squamous cell carcinoma was sonicated and the methylated fragments were collected by MIRA. Illumina adaptors were ligated, and the DNA analyzed on the Illumina Genome Analyzer. Using five lanes, ~15 million sequence reads were created and ~8 million uniquely mapped DNA sequence reads were aligned to the human genome. 1,254 of ~28,000 CpG islands had 30 or more mapped reads suggesting that they are methylated. The Figure shows an example of the sequence tags for the UNC5D gene aligned to the UCSC Genome Browser. The blue and red bars represent sequence reads of the forward and reverse strand, respectively. The peak maps to the CpG island (black box) near the 5′ end of the UNC5D gene.
B. A NimbleGen tiling array for chromosome 8 was used to analyze the DNA of the same lung tumor. The peak maps to the CpG island (black box) near the 5′ end of the UNC5D gene.
2. Experimental design
Depending on the ultimate goal of the analysis, two different experimental designs have generally been used for MIRA-assisted methylome characterization. a) When methylation profile analysis along entire chromosomes or segments of chromosomes is the aim, the MIRA-enriched fraction is hybridized against the input sample, similar as in ChIP-chip experiments. This design provides the opportunity to map the continuous methylation profile, i.e. the actual methylation landscape over megabase stretches of DNA. b) For methylation marker discovery studies, for example, simple co-hybridization of the MIRA-enriched tumor sample with the MIRA-enriched matching normal sample can pinpoint directly any cancer-associated methylation changes of interest.
3.1. Expression and purification of recombinant proteins
It is critical that high quality MBD proteins are used in the binding reaction. GST-tagged MBD2B and His-tagged MBD3L1 proteins can be expressed and purified from bacterial cells. Neither one of the two proteins tends to form inclusion bodies after overexpression in bacteria, which makes their affinity purification relatively easy. Buffer components and other reagents used in the MIRA procedure must be molecular biology-grade fine chemicals. Recombinant plasmids for bacterial expression of both MBD proteins are available upon request.
3.1.1. Expression of MBD2B and MBD3L1 proteins
Transformation of BL21 (DE3)-competent cells (Epicurian Coli Competent Cells; Stratagene; Santa Clara, CA) with the recombinant protein-expressing plasmids can be performed by any chemical- or electro-transformation protocols. GST-MBD2B transformed cells are plated onto ampicillin-containing plates, while His-MBD3L1 transformed cells are plated onto kanamycin-containing plates, respectively. After overnight incubation at 37°C, 10-20 well-developed bacterial colonies are inoculated into 50 ml of the appropriate antibiotic-containing LB bacterial growth media and grown at 37°C until the OD reaches 0.6 at a fixed wavelength of 600 nm. Protein overexpression is induced by addition of 50 μl of 100 mM IPTG solution (Sigma; St. Louis, MO) and cells are allowed to grow for an additional 4 to 6 h at 37°C. Induced bacterial cultures are collected into a 50 ml tube and centrifuged at 3500g for 10 min at 4°C. Bacterial cells can be stored at −80°C for several months, or proceed immediately for protein purification.
3.1.2. Purification of the overexpressed proteins
The first part of the purification procedure is the same for both proteins but later the protocol diverges.
3.1.2.1. Suspend the bacterial pellet in 10 ml of buffer (10mM Tris–HCl, pH 7.8, 150 mM NaCl, 0.1 mM EDTA) containing 100 μg/ml lysozyme and 0.1 mM PMSF (Sigma; St. Louis, MO).
3.1.2.2. Induce lysis of bacterial cells by addition of 1 ml of 10% (w/v) N-lauroylsarcosine (Sigma; St. Louis, MO) solution.
3.1.2.3. Sonicate the bacterial lysate until it clears up.
3.1.2.4. Add 1 ml Triton X-100 (Sigma; St. Louis, MO) solution to the lysate to promote the solubilization of the overexpressed proteins. Vortex the lysate for at least 20 sec.
3.1.2.5. To separate the insoluble material from the lysate, centrifuge at 3500g for 10 min and transfer the supernatant into a new tube.
From this step forward, the two protocols diverge, GST-tagged MBD2B is affinity purified with Glutathione Sepharose 4B beads (GE Healthcare; Uppsala, Sweden), while His-tagged MBD3L1 can be isolated with Ni-NTA agarose beads (Novagen; Darmstadt, Germany).
3.1.3. Affinity purification of MBD2B protein
3.1.3.1. Add 0.1ml of Glutathione Sepharose 4B beads (50% slurry) to 12 ml cleared lysate and mix gently by rotating at 4°C for 45 min.
3.1.3.2. Pellet the beads at 1000g for 1 min and wash the pellet with 10 ml of buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.5) containing 0.1% (v/v) Triton X-100 by inverting the tube several times.
3.1.3.3. Repeat the previous step two more times.
3.1.3.4. Elute the GST-tagged MBD2B protein from the beads with 1 ml of elution buffer (50 mM Tris-HCl, pH 8.5, 150 mM NaCl, 20 mM reduced glutathione; Sigma; St. Louis, MO) and 0.1% (v/v) Triton X-100 at 4°C for 4 h on a rotating platform.
3.1.3.5. The eluted GST-MBD2B protein should be dialyzed against 2 liters of phosphate-buffered saline (PBS) in the cold-room for 5 h and then overnight against protein dialysis buffer consisting of 50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM β-mercaptoethanol and 50% (v/v) glycerol. After dialysis, purified MBD2B protein can be kept at −20°C for 6 months.
3.1.3.6. Check the protein concentration on a 10% SDS-PAGE gel using BSA controls.
3.2. Affinity purification of MBD3L1 protein
3.2.1. Add 0.1 ml of Ni-NTA agarose beads (50% slurry) to 12 ml cleared lysate and mix gently by rotating at 4°C for 45 min.
3.2.2. Pellet the beads at 1000g for 1 min and wash the pellet with 10 ml of buffer (50 mM NaH2PO4, pH adjusted to 8.0 with 1 M NaOH, 300 mM NaCl, 20 mM imidazole) containing 0.1% (v/v) Triton X-100 by inverting the tube several times.
3.2.3. Repeat the previous step two more times.
3.2.4. Elute the His-tagged MBD3L1 protein from the beads with 1 ml of elution buffer (50 mM NaH2PO4, pH adjusted to 8.0 with 1 M NaOH, 300 mM NaCl, 250 mM imidazole) and 0.1% (v/v) Triton X-100 at 4°C for 4 h on a rotating platform.
3.2.5. The eluted His-MBD3L1 protein should be dialyzed against 2 l of PBS in the cold-room for 5 h and then overnight against protein dialysis buffer consisting of 50 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM β-mercaptoethanol and 50% (v/v) glycerol. After dialysis, purified MBD3L1 protein can be kept at −20°C for 6 months.
3.2.6. Check the protein concentration on a 10% SDS-PAGE gel using BSA controls.
Note: Recombinant MBD2B and MBD3L1 proteins can be purchased from Active Motif (Carlsbad, CA) as part of the MethylCollector™ Ultra kit.
3.3. Functional testing of the affinity-purified recombinant proteins
After the purification of the two proteins, it is advisable to test their activity in a binding reaction. For this purpose, a pilot MIRA reaction (with 1 μg of MBD2B and 1 μg of MBD3L1 proteins) can be set up with 100 ng of fragmented genomic DNA template. Check the selectivity of the system by monitoring the amplification of known methylation-positive and methylation-negative controls in an endpoint PCR (see section 5.3).
4. Template preparation for MIRA-assisted methylation profiling
4.1. Genomic DNA preparation
High molecular weight genomic DNA can be isolated from cells or tissues by routine proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation or with any commercially available genomic DNA purification kit.
4.2. DNA fragmentation
Before setting up a MIRA binding reaction, genomic DNA must be fragmented. Application of enzymes that cut outside of CpG islands is advised, e.g. MseI (5′-TTAA-3′), Tsp509I (5′-AATT-3′) or CviQI (5′-GTAC-3′). Proper fragmentation can also be achieved by sonication, which is the preferred method as it lacks a significant sequence bias. Ideally, the resulting DNA fragment size should be between 300 and 500 bps. After digestion or sonication, DNA should be purified by phenol/chloroform extraction and precipitation or through the use of a DNA purification column.
4.3. Linker ligation
Since a typical MIRA reaction yields only several nanograms of methylation-enriched DNA, an amplification step generally is introduced to create substantial amounts of DNA (amplicon) for the subsequent microarray procedure. Ligation-mediated PCR (LM-PCR) can be one tool for achieving this goal. Unidirectional linker ligation to the ends is a prerequisite of LM-PCR. Depending on how the genomic DNA was fragmented, appropriate linker ligation must be performed.
4.3.1. Linker ligation onto restriction enzyme-cut fragments
MseI cutting produces sticky ends that make the appropriate adaptor ligation easy and efficient. Double stranded unidirectional linkers (adaptors) are created from the annealing of two complementary oligonucleotides (Short: 5′- TAGAATTCAGATCTCCCG-3′ and Long: 5′- GCGGTGACCCGGGAGATCTGAATTC - 3′) and are ligated to the ends.
Set up the following ligation reaction and incubate overnight at 16°C: 5 μg of MseI cut DNA, 5 μl of 10x NEB ligation buffer, 5 μl of 20 μM adaptor, 1 μl of 400 U/μl T4 DNA ligase (New England Biolabs, NEB; Ipswich, MA); then add H2O to a total volume of 50 μl. Linker-ligated DNA should be purified by phenol/chloroform extraction and ethanol precipitation or through use of a DNA purification column. Suspend the precipitated DNA in 20-30 μl of TE buffer.
4.3.2. Adaptor ligation onto sonicated DNA
Sonication creates DNA fragments with heterogeneous ends; therefore, an end-treatment/ end-polishing step must be introduced before the blunt-ended linker ligation.
Set up the following T4 DNA polymerase reaction and incubate at 12°C for 30 min: 5 μg of sonicated DNA, 5 μl of NEB buffer 2, 0.5μl of 5 mg/ml BSA, 0.5 μl of 10 mM dNTP, 0.5 μl of T4 DNA polymerase (NEB); add H2O to a total volume of 50 μl.
Linker ligated DNA must be purified by phenol/chloroform extraction and precipitation or through use of a DNA purification column. The purified and blunt-ended DNA can be used in the subsequent ligation reaction. Double-stranded adaptors are created from the annealing of the two oligonucleotides (Short linker oligo: 5′-GAATTCAGATCTCCCG-3′ and Long linker oligo: 5′-GCGGTGACCCGGGAGATCTGAATTC - 3′) and are ligated to the ends.
Set up the following ligation reaction and incubate overnight at 16°C: 10-20 μl of end-treated DNA, 5 μl of 10x NEB ligation buffer, 5 μl of 20 μM adaptor, 1 μl of 400 U/μl T4 DNA ligase NEB; then add H2O to a total volume of 50 μl. Linker-ligated DNA must be purified by phenol/chloroform extraction and ethanol precipitation or through use of a DNA purification column. Pick up the precipitated DNA in 20-30 μl of TE buffer and measure the concentration of the linker-ligated DNA by using a spectrophotometer.
5. MIRA-binding reaction and amplicon generation
MIRA is routinely carried out with several hundred nanograms of fragmented genomic DNA, generally 250 to 500 ng. To form a high affinity MBD2B/MBD3L1 protein complex, a pre-incubation step precedes the actual DNA binding reaction. The protein complex-captured methylation-enriched fraction is purified from the reaction and is LM-PCR amplified. MIRA can also be used with unamplified DNA but much more DNA is needed as starting material. Approximately 25 μg of input DNA have been used successfully without amplification.
5.1. MIRA binding reaction
5.1.1. Set up the following binding reaction in a 1.5 ml Eppendorf tube:
40 μl of 10 x MIRA buffer (100 mM Tris-HCl, pH 7.9, 500 mM NaCl, 10 mM DTT, 100 mM MgCl2, 1.0% (v/v) Triton X-100).
10 μl of 50 ng/μl of JM110 DNA (sonicated bacterial DNA).
1 μg of purified GST-MBD2B.
1 μg of purified His-MBD3L1.
Add H2O to a final volume of 350 μl.
5.1.2. Mix by pipetting and preincubate at 4°C for 20 min on a rotating platform.
5.1.3. Add 250 ng of linker-ligated DNA in 50 μl. (The final volume is now 400 μl).
5.1.4. Incubate the binding reaction at 4°C at least for 4 h (or overnight) on a rotating platform.
5.1.5. Add 10 μl of preblocked MagneGST beads (Promega; Madison, WI) and incubate at 4°C for 45 min on a rotating platform.
5.1.6. Retrieve MagneGST beads carrying the enriched methylated DNA fraction. Use the magnetic stand to capture the beads, and carefully remove the supernatant with a pipette.
5.1.7. Add 800 μl of MIRA wash buffer (10mM Tris-HCl, pH 7.5, 700 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 0.1% (v/v) Triton X-100) into the tube and invert 4 to 5 times.
5.1.8. Retrieve beads (methyl-CpG-rich fraction) by using the magnetic stand and carefully decant supernatant.
5.1.9. Repeat the previous two steps two more times.
5.1.10. Elute and purify the methylated-CpG-enriched fraction from the MagneGST beads with QIAquick PCR purification kit (Qiagen; Valencia, CA) according to the company’s protocol. Briefly, after the third wash of the beads with 700 mM NaCl containing buffer, directly add 400 μl of Quagen “binding buffer” onto the beads, vortex and load the sample onto the provided affinity column. The next steps are carried out exactly as described in the company’s purification protocol.
5.1.11. Elute the methyl-CpG-rich fraction from the column with 50 μl of H2O.
5.1.12. Reduce the volume of the eluted fraction to ~20μl in a Speed Vac concentrator.
5.2. LM-PCR amplification
The amplification should be performed in such a way that cycling is stopped right after the linear phase of amplification. The easiest way of monitoring the amplification is to perform the PCR in a real-time thermocycler. Adding SYBR green dye into the PCR does not interfere with any of the subsequent procedures.
5.2.1. Set up the following amplification reaction in a PCR tube:
20 μl of eluted fraction from the step 5.1.12.
5 μl of 10 x PCR buffer.
5 μl of 2.5 mM dNTPs.
2 μl of 25 μM long LM-PCR linker oligonucleotide.
1 μl 5 U/μl Taq polymerase.
2 μl SYBR green [1:667 dilution of SYBR green (Roche)].
15 μl of H2O.
Run the LM-PCR under the following condition: 2 min at 72°C, 3 min at 95°C, 40 cycles of [30 sec at 94°C; 1 min 60°C; 5 min 72°C], followed by 5 min at 72°C and hold at 4°C.
Stop the LM-PCR after the liner phase of amplification (usually this takes 12-13 cycles). Purify amplicons with QIAquick PCR purification kits and determine the concentration by using a spectrophotometer.
5.3. Efficiency evaluation of the MIRA reaction
By applying general positive and negative controls in real-time PCR or performing endpoint PCR amplification, we can check whether the MIRA reaction has increased the occurrence of the methylated DNA molecules in the captured fraction and in the subsequently created amplicons. As the TATA-binding protein (TBP) promoter CpG island is an unmethylated region in all cells, we used it as a general negative control in our assays. Finding a general positive control for methylation analysis is not trivial because every promoter-associated CpG island is unmethylated in certain cell types or at a given developmental stage. Imprinted regions are frequent targets of cancer-specific hypomethylation, which makes them not ideal as markers of hypermethylation. As a general positive control, we used the SLC25A37 gene’s CpG island located at the 3′ end region of the gene. This CpG island has been proven to be densely methylated in all cell lines and human tissues tested so far.
The following primer pairs can be used as general controls for MIRA: Positive control primers (SLC25A37): 5′-CCCCCTGGACGTCTGTAAG-3′; 5′-GGCATCTGGTAGATGACACG-3′; Tm: 58°C.
Negative control primers: 5′-CTTTCCTACGTCCAGCAAGG-3′; 5′-AATGTCACTTCCGCCAGTT-3′; Tm: 58°C.
5.3.1. Set up the following amplification reaction in a PCR tube:
1 μl of eluted fraction from step 5.1.11 or 5.1.12.
5 μl of 10x PCR buffer.
5 μl of 2.5 mM dNTPs.
2 μl of 10 μM positive or negative primer mix.
0.25 μl of 5U/μl Taq polymerase.
11.75 μl of H2O.
Run 34 cycles of PCR under the following conditions: 3 min at 95°C, 34 cycles of [20 sec at 94°C, 20 sec at 58°C, 30 sec at 72°C] and 5 min at 72°C, or set up real-time PCR containing SYBR green dye.
6. Amplicon labeling and hybridization
MIRA is compatible with different types of microarray platforms. We have successfully combined it with first generation microarrays from the UHN Microarray Centre, University of Toronto, Canada (4) and tiling arrays from Agilent Technologies (7), NimbleGen (11, 12) and Affymetrix (unpublished data). Fluorescent labeling of the amplicons and the microarray hybridizations are carried out according to the appropriate company’s suggested protocols. Image and data analysis are performed as described previously (4, 7).
7. MIRA-seq
Besides microarray analysis, MIRA is compatible with next-generation sequencing platforms, such as the Illumina Genome Analyzer (Fig. 2 and unpublished data). After MIRA and adapter ligation using Illumina linker sequences, single-stranded DNA molecules are attached to a dense lawn of primers in the flow cell. Bridge amplification will form clusters in the Illumina cluster station. Analysis is done using the standard Illumina software and data analysis pipeline.
8. Concluding remarks
The methylated-CpG island recovery assay (MIRA) is based on the high affinity of the MBD2B/MBD3L1 protein complex to CpG-methylated DNA. Its advantages are its high specificity and sensitivity requiring only a few hundred nanograms of genomic DNA for analysis. It does not require DNA denaturation and is therefore not affected by genome structures that may be refractory to efficient strand separation. The current applications of MIRA include genome-scale methylation profiling along chromosomes at a resolution of ~ 100 base pairs and the efficient identification of DNA methylation differences between normal and diseased tissues or between different developmental stages or tissue types. The method is compatible with various microarray platforms and high-throughput DNA sequencing approaches allowing flexible designs of methylation profiling strategies.
Acknowledgements
The work of the authors has been supported by NIH grants CA084469 and AG036041.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: Under a licensing agreement between City of Hope and Active Motif (Carlsbad, CA), the methylated-CpG island recovery assay (MIRA) technique was licensed to Active Motif, and the authors T.A.R. and G.P.P. are entitled to a share of the royalties received by City of Hope from sales of the licensed technology.
8. References
- 1.Rauch TA, Pfeifer GP. Methods for assessing genome-wide DNA methylation. In: Tollefsbol T, editor. Handbook of Epigenetics: The New Molecular and Medical Genetics. Elsevier; 2010. in press. [Google Scholar]
- 2.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 5.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 6.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 7.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with MBD2 and components of the NuRD complex. J Biol Chem. 2004 doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 11.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]