Abstract
Alternatively spliced β-neurexins (β-NRXs) and neuroligins (NLs) are thought to have distinct extracellular binding affinities, potentially providing a β-NRX/NL synaptic recognition code. We have utilized surface plasmon resonance to measure binding affinities between all sixty combinations of alternatively spliced ectodomains of β-NRXs 1–3 and NLs 1–3. Binding was observed for all β-NRX/NL pairs. The presence of the NL1 B splice insertion lowers β-NRX binding affinity by ~2-fold, while β-NRX splice insertion 4 has small effects that do not synergize with NL splicing. New structures of glycosylated β-NRXs 1 and 2 containing splice insertion 4 reveal that the insertion forms a new β-strand that replaces the β10-strand, leaving the NL binding site intact. This helps to explain the limited effect of splice insert 4 on NRX/NL binding affinities. These results provide new structural insights and quantitative binding information to help determine whether and how splice isoform choice plays a role in β-NRX/NL mediated synaptic recognition.
INTRODUCTION
In vertebrates, with nervous systems comprising billions of neurons with trillions of connections between them (Cowan et al., 2000), the construction of functional circuits requires appropriate specification of a large number of neural connections. However, the molecular mechanisms underlying this extraordinary specificity remain unclear. Pre-synaptic neurexins (NRXs) bind to post-synaptic neuroligins (NLs) to form complexes that span the synapse and are thought to play important roles in synaptic specification (Sudhof, 2008). Both NRX and NL gene transcripts can be spliced alternatively, yielding diverse ectodomains. It has been suggested that this diversification may provide a mechanism to help specify either the formation (Chih et al., 2004; Graf et al., 2004; Nam and Chen, 2005; Scheiffele et al., 2000) or validation (Chubykin et al., 2007; Ko et al., 2009b; Missler et al., 2003; Varoqueaux et al., 2006) of synaptic connections. The importance of NRX/NL interactions to neural function is highlighted by the association of mutations in both NRX and NL genes with autism spectrum disorders (Garber, 2007; Geschwind and Levitt, 2007; Jamain et al., 2003; Laumonnier et al., 2004; Moretti et al., 2006; Sudhof, 2008; Szatmari et al., 2007; Tabuchi et al., 2007).
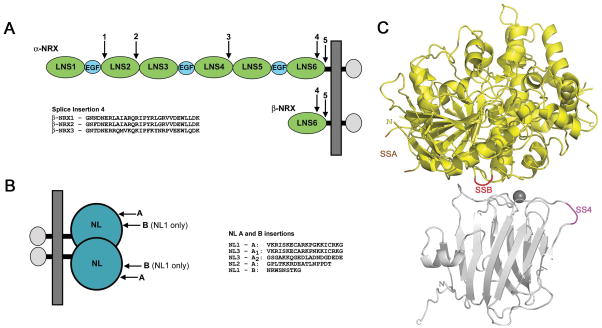
Vertebrate genomes encode three NRX genes, each of which can be transcribed from two alternative promoters resulting in the longer α-NRX and the shorter β-NRX forms (Figure 1A) (Ushkaryov et al., 1992). Both α- and β-NRXs are class I transmembrane proteins with C-terminal regions in common, but with distinct N-terminal ectodomain sequences (Ushkaryov et al., 1994). The extracellular part of α-NRXs consists of an α-specific signal peptide, which is removed from the mature protein, and three repeats, each containing an EGF-like domain flanked by two LNS (laminin, neurexin, sex-hormone-binding globulin) domains, A and B (Figure 1A). The shorter β-NRXs contain their own unique signal peptide followed by a β-NRX-specific sequence, and a single LNS B domain. The membrane proximal LNS B ectodomain region, transmembrane and cytoplasmic regions are identical in both α and β isoforms (Figure 1A). In addition to this diversification of gene products by promoter choice, both α- and β-NRXs undergo extensive alternative splicing. α-NRXs have five alternative splice sites, SS1 through SS5, resulting in thousands of possible different α-NRX protein isoforms, whereas β-NRXs can be alternatively spliced at two sites, SS4 and SS5, resulting in 4 different isoforms for β-NRXs 1 and 2 and up to 32 for β-NRX3 which has 16 variants at SS5 (Rowen et al., 2002; Tabuchi and Sudhof, 2002; Ushkaryov and Sudhof, 1993). The SS5 splice site is in the “stalk” region that connects the β-NRX LNS domain to the transmembrane segment, thus the SS4 site is the only locus of splice form diversification within the LNS domain that binds NLs. Alternative splicing of β-NRXs results in the presence or absence of “insertion sequences”. Thus, for β-NRXs 1, 2, and 3, six alternatively spliced LNS domains in total can be produced: those lacking an insertion at SS4 (referred to here as the Δ4 isoform, e.g. β-NRX1Δ4) and those containing an insertion sequence at SS4 (referred to as the +4 isoform, e.g. β-NRX1+4). These six different β-NRX gene products, including their splice site positions and insertion sequences (Figure 1A), are highly conserved across mammalian genomes, suggesting that the function of the different genes and splice isoforms serve conserved biological roles (Ullrich et al., 1995; Ushkaryov et al., 1994). The SS4 insert sequences of β-NRX1 and β-NRX2 differ at only one amino acid position, but the β-NRX3 SS4 insert differs more substantially (Figure 1A and Figure S5). Crystallographic studies of LNS domains from NRXs have revealed a 14-stranded β-sandwich structure similar to those of serum amyloid P protein and some lectins (Koehnke et al., 2008b; Rudenko et al., 1999; Sheckler et al., 2006; Shen et al., 2008).
Figure 1. NRX and NL alternative splicing.
(A) NRXs are transcribed from two independent promoters for each gene resulting in the longer α-NRXs and shorter β-NRXs. α-NRXs contain three repeats, each of which consists of an EGF domain flanked by two LNS domains. α-NRX and β-NRXs share the LNS 6 domain, a transmembrane region and a short cytoplasmic domain. The arrows indicate the alternative splicing sites in each molecule. The amino acids sequences for the insert at splice site 4 of each β-NRX are also shown. (B) NLs 1, 2, and 3 contain an extracellular domain, which forms homodimers, a stalk region, transmembrane and cytoplasmic domains. All three NLs undergo splicing at site A and NL1 only can also be spliced at site B, resulting in 10 different NL variants. The amino acid sequences for the inserts introduced as a result of splicing are shown. (C) Structure of a β-NRX1Δ4/NL1Δ complex with β-NRX1Δ4 shown in silver and NL1Δ in yellow (PDB ID 3B3Q) (Chen et al., 2008). The SS4 insertion point for β-NRXs is shaded in magenta, the SSA insertion point for NLs is shaded in orange and the SSB insertion point for NL1 in red.
NLs in vertebrates are encoded by five genes, NL1–5, although NLs 4 and 5 are more distant relatives (Bolliger et al., 2001). Like their NRX binding partners, each NL is subject to insertion-type alternative splicing which leads to the production of diverse ectodomains. All three NLs studied here (NLs 1–3) contain a splice site at position A, while NL1 alone contains an additional splice site at position B (Figure 1B). In total, the gene products of NLs 1–3 encompass a set of ten distinct isoforms. The three NL genes show a high level of conservation between genes and among vertebrate species (Ichtchenko et al., 1996). Crystallographic and biophysical characterization of NL proteins show that NLs adopt a homodimeric form similar to acetylcholinesterase, where a tight dimer interface is formed through a four-helix bundle near the C-terminus of the esterase-like domain (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007; Koehnke et al., 2008a).
A single NL dimer binds two β-NRX molecules in a Ca2+-dependent manner ((Comoletti et al., 2003; Comoletti et al., 2006), and see below). As demonstrated by recent structures of β-NRX/NL complexes (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007), a calcium ion that shares ligation between β-NRX and NL partners plays an important role in complex formation. Although several crystal structures for β-NRX/NL complexes are available (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007), each structure represents a complex between the Δ-isoforms of both binding partners. Therefore, while the positions of the splice insertion sites relative to the β-NRX/NL interface have been determined (Figure 1C), the structural role of the insertion sequences remains unclear.
Several recent studies (Boucard et al., 2005; Chih et al., 2006; Graf et al., 2006; Reissner et al., 2008) investigated the effects of alternative splicing on β-NRX/NL binding, hemisynapse formation, and neuronal function in cell culture. Results from these studies are partly contradictory: some results indicate that splicing at site 4 in β-NRX1 and at site B in NL1 jointly regulate selectivity, but other results fail to support this conclusion. It was shown that the presence of the 30-residue SS4 insert in β-NRX1 reduced binding to cells expressing NL1B (NL1 that contains an insert at site B), while maintaining binding to cells expressing NL1Δ (NL1 without any insert), or to NL2 (which lacks a B splice insertion) (Chih et al., 2006). However, results of a similar assay suggested that alternative splicing had no discernable effect on the ability of soluble β-NRXs to bind to NL-expressing cells, although in the same study the authors found splice-dependent binding differences in pull-down assays (Boucard et al., 2005). Further, it has been reported that N-linked glycosylation of the B splice insert is critical for regulation of the interaction of NL1 with NRX, since mutation of the glycosylation site in the B insert ablates its reported function in attenuating NRX1 binding and glutamatergic synapse induction (Chih et al., 2006; Comoletti et al., 2003). The presence or absence of an insert at the A site of NLs did not affect binding of NL to β-NRX in these cell binding studies (Boucard et al., 2005; Chih et al., 2006).
Studies of the effect of NRX and NL splice isotype on intermolecular NRX/NL binding interactions have also yielded contradictory results. A recent study (Comoletti et al., 2006) used surface plasmon resonance (SPR) measurements to determine the effect of the SS4 insertion sequence on NL binding. Results from this SPR study, which reported only relative binding strengths, suggested that while both the Δ4 and +4 splice forms bound NL1Δ, β-NRX1+4 binding was attenuated. In another study (Boucard et al., 2005), the authors concluded from pull-down studies that binding of β-NRX1+4 to NL1B is abolished. However, analogous pull-down assays performed by Reissner and colleagues indicate that NL1B is capable of binding to β-NRX1+4 (Reissner et al., 2008). A structural rationale for the effects of splice insertion 4 on β-NRX binding to NLs was also explored in recent crystallographic studies of a bacterially produced β-NRX1+4 ectodomain (Shen et al., 2008). In this molecule, residues of the splice insertion form a helix extending from the connection between β-strands 10 and 11. This helix protrudes from the NL binding face of the protein, adopting a conformation that would be expected to sterically inhibit NL binding (Shen et al., 2008). This result has been interpreted as supportive of the affinity-regulation model for alternative splicing. In contrast, we have reported based on NMR measurements that splice insertion 4 of a bacterially produced β-NRX1+4 is unstructured in solution (Koehnke et al., 2008b)
To gain understanding of β-NRX/NL interactions, and to provide a detailed quantitative description of the energetic effects of alternative splicing, we have used SPR to determine the kinetic rates (ka, kd) and the binding affinities (KDs) for the interaction of all pair-wise combinations of β-NRXs 1–3 and NLs 1-3. Although previous reports have used SPR to measure the relative strength of a small subset of β-NRX/NL interactions, here we employ a global approach to provide quantitative analysis of the binding of the two splice isoforms (+4 and Δ4) of the LNS domains from each of the three β-NRXs with the ten splice isoforms of NLs 1, 2, and 3. Our results show that all NL family members, whether or not they carry insert A or B, can bind to all β-NRX isoforms whether they contain or lack splice insertion 4. The presence of insert B in NL1 weakens the binding with β-NRXs, yet the changes in KD are moderate (< 2.4-fold in all instances). On the other hand, the presence of splice insert 4 weakens binding of β-NRXs 1 and 2 but strengthens binding of β-NRX3 with NLs. NL3 exhibits ~2-250-fold weaker binding than NL1 or NL2, irrespective of splice isoform.
We interpret these results based on a new structure of a glycosylated β-NRX3Δ4 which is nearly identical to non-glycosylated β-NRXs, and two new crystal structures of splice insert-containing glycosylated β-NRX ectodomains, β-NRX1+4 and β-NRX2+4. Distinct from the structure of non-glycosylated β-NRX1+4 (Shen et al., 2008), our new structures show that the SS4 insertion sequence interacts with glycan and part of the insertion sequence forms a “new” β-strand that replaces the “old” β10 strand in the LNS domain β-sandwich. The displaced β10 strand and another segment of splice insertion 4 form a new structure that protrudes from the β-NRX face most distal from the NL binding site, while the NL binding surface remains largely intact. The position of the new structure formed by elements from the displaced β10 strand, on a β-NRX face distal from the NL binding site, suggests, together with the SPR results, that insertion sequences at β-NRX SS4 may function in a role other than to modulate binding affinity for NLs.
RESULTS
All of the sixteen proteins used in this work were produced in a mammalian expression system as detailed in the Experimental Procedures section (Figure S1). Analytical ultracentrifugation (AUC) studies show that each NL forms tight constitutive dimers independent of the presence of splice insertions and all six β-NRX isoforms were monomeric regardless of splice isoform (data not shown), in agreement with prior results (Comoletti et al., 2003; Comoletti et al., 2006; Koehnke et al., 2008a; Koehnke et al., 2008b).
SPR binding experiments
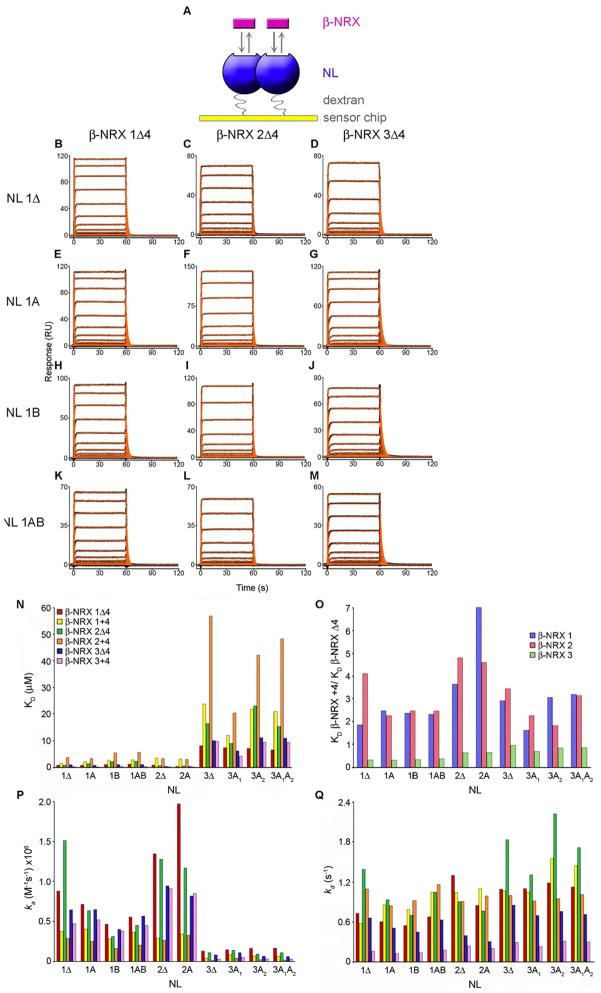
SPR binding assays have been used previously to investigate the binding properties of β-NRX to NL (Arac et al., 2007; Boucard et al., 2005; Comoletti et al., 2003; Comoletti et al., 2006). In our experiments we have optimized the assay design to obtain accurate binding parameters for each interaction as outlined in Figure 2A and discussed in the supplemental text. Figures 2B–M and S2–3 show results typical of the SPR binding assays reported here for the interaction of each of the six β-NRX isoforms with each of the ten NL isoforms. The SPR traces for all β-NRXs reported here indicate that binding to the immobilized NLs was specific and concentration dependent. Triplicate injections at the same β-NRX concentration superimpose with each other, demonstrating that (1) the assay is reproducible, (2) nonspecific binding has been eliminated, and (3) the NL surfaces do not lose binding activity as a result of the regeneration conditions. The experimental binding signals of each of the sixty pair-wise β-NRX/NL interactions, which are illustrated in black, overlay well with the red traces that represent the fit to a 1:1 binding model. The complete matrix of binding affinities (Figure 2N, Table 1), shows a significant spread with KDs for the strongest binders ~0.25μM compared to the weakest interactions at ~60μM, demonstrating a substantial range in affinities for β-NRX/NL ectodomain interactions of ~250-fold for the set of proteins tested here. The complete set of ka, kd and KD values are listed in Table S1. To help assess the contribution of β-NRX and NL splice insertion sequences to the affinities of β-NRX/NL interactions, we calculated fold-changes in KD, relative to Δ-isoforms, for each pair-wise interaction. The measured KDs for each splice insert-containing β-NRX/NL complex were normalized against the KDs for the corresponding insert-less β-NRXΔ4/NLΔ complex (Table 1, blue quadrant). Similarly, to isolate the effects of splice inserts for β-NRXs (Table S2) or NLs (Table S3), we normalized KDs against those measured for the corresponding complex with Δ-isoform β-NRXs or NLs, respectively.
Figure 2. SPR binding analysis of β-NRXs 1–3 splice variants to NLs 1–3 isoforms.
(A) In the SPR binding assay, NL was immobilized to the sensor chip surface (ligand) and increasing concentrations of β-NRX were injected in the solution phase (analyte). Two molecules of β-NRX bind to each dimeric NL. (B–M) β-NRXΔ4 isoforms binding over sensor chip surfaces immobilized with NL isoforms 1Δ, 1A, 1B and 1AB at a concentration range of 8.0-0.039 μM with the exception of panels B and C, where the highest concentration tested was 4 μM. Black traces show the experimental data and red traces show the fit to a 1:1 model with a step to account for mass transport. The KDs, along with fold change analyses, are listed in Tables 1, S2-S3, and the kinetic rates are shown in Table S1. (N) KD values determined by SPR binding analysis for the interaction of the two isoforms of each β-NRX 1, 2 and 3, with the ten different NL isoforms, containing or lacking splice inserts A and for NL1 splice insert B. The values for this plot are listed in Table 1. (O) The KD values of β-NRX+4 interacting with a given NL, normalized against the KDs of the same NL binding to β-NRXΔ4. The fold changes for each β-NRX+4 binding to a certain NL compared to the binding of β-NRXΔ4 are shown in Table S2. (P) A plot of the association rates (ka) and (Q) a plot of the dissociation rates (kd) for β-NRX/NL interactions. The values in this plot are shown in Table S1. The association and dissociation rates were used to calculate the KDs in (N) using the relationship KD=kd/ka.
Table 1.
Effect of the β-NRX SS4 insert and NL A/B inserts on the β-NRX/NL binding KDs
| KD(μM) | Fold-change compared to | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-NRX 1Δ4 | β-NRX 2Δ4 | β-NRX 3Δ4 | β-NRX 1+4 | β-NRX 2+4 | β-NRX 3+4 | β-NRX 1Δ4/NLΔ | β-NRX 2Δ4/NLΔ | β-NRX 3Δ4/NLΔ | β-NRX 1+4/NLΔ | β-NRX 2+4/NLΔ | β-NRX 3+4/NLΔ | |
| NL1Δ | 0.828(2) | 0.917(5) | 1.02(1) | 1.54 (1) | 3.77(3) | 0.3320(1) | 1.00 | 1.00 | 1.00 | 1.86 | 4.11 | 0.33 |
| NL2Δ | 0.964(2) | 0.7055(8) | 0.418(2) | 3.520(9) | 3.388(7) | 0.262(5) | 1.00 | 1.00 | 1.00 | 3.65 | 4.80 | 0.63 |
| NL3Δ | 8.17(3) | 16.48(3) | 10.1(3) | 23.8(3) | 56.8(3) | 9.73(1) | 1.00 | 1.00 | 1.00 | 2.91 | 3.45 | 0.96 |
| NL1A | 0.845(3) | 1.466(7) | 0.782(3) | 2.096(4) | 3.317(7) | 0.2460(4) | 1.02 | 1.60 | 0.77 | 2.53 | 3.62 | 0.24 |
| NL1B | 1.16 (1) | 2.21(1) | 1.10(8) | 2.754(8) | 5.48(2) | 0.3650(8) | 1.40 | 2.41 | 1.08 | 3.33 | 5.98 | 0.36 |
| NL1AB | 1.21 (6) | 2.29 (5) | 1.10(7) | 2.82(1) | 5.66(2) | 0.393(1) | 1.46 | 2.50 | 1.08 | 3.41 | 6.17 | 0.39 |
| NL2A | 0.430(5) | 0.65 (1) | 0.369(8) | 3.168(9) | 3.013(8) | 0.239 (3) | 0.45 | 0.92 | 0.88 | 3.29 | 4.27 | 0.57 |
| NL3A1 | 7.45(3) | 9.05(8) | 6.26(3) | 12.0(5) | 20.5(1) | 4.32(1) | 0.91 | 0.55 | 0.62 | 1.47 | 1.24 | 0.43 |
| NL3A2 | 7.15(5) | 23(3) | 11.2(1) | 21.9(1) | 42.2(8) | 9.50(2) | 0.88 | 1.40 | 1.11 | 2.68 | 2.56 | 0.94 |
| NL3A1A2 | 6.57(3) | 15.34(9) | 10.9(8) | 21.0(1) | 48(1) | 9.39(1) | 0.80 | 0.93 | 1.08 | 2.57 | 2.91 | 0.93 |
The left side of the table shows KD values for the interactions of isoforms of NLs 1, 2 and 3 with isoforms of β-NRXs 1, 2 and 3. The blue quadrant shows the KDs for interactions where both NLs and β-NRXs lack a spice insert (A or B for NLs and SS4 for β-NRXs). The green quadrant displays the KDs for NLs that contain inserts A or B with β-NRXs that lack splice insert 4. The orange quadrant describes the binding of NLs that lack a splice insert with β-NRXs containing insert 4, and the purple quadrant shows the KDs for the interaction of both insert-containing NLs and β-NRXs. The right side of the table shows all the KD values normalized against the same NL/β-NRX pair that lacks both inserts, which are in the left blue quadrant. The number in brackets represents the error of the fit in the last digit.
NL3 binds β-NRXs weakly compared to NL1 and NL2
As can be seen in Figure 2N and Table 1, the strengths of β-NRX/NL interactions vary somewhat depending on β-NRX subtype (e.g., NL1Δ/β-NRXΔ4 ranges in KD from 0.83 to 1.02 μM), but the most significant differences are associated with NL subtype. Whereas interactions of all three β-NRXΔ subtypes are on the order of 1μM KD for NL1 and NL2, interactions with NL3 are at least an order of magnitude weaker. Figure 2N shows that the ~10-fold lesser strength of β-NRX interaction with NL3 is generally preserved in character irrespective of the splice isoform of either binding partner. Thus, for interactions between NLΔs and β-NRX+4s (Table 1, orange quadrant), or interactions of insert-containing NLs with either β-NRXΔ4s (Table 1, green quadrant) or β-NRX+4s (Table 1, purple quadrant), the most significant contributor to differential affinity is the NL subtype, as NL3 binds to identical β-NRXs with ~4–40 fold reduced affinity compared to NL1 or NL2. Unlike the binding differences observed for NLs with respect to subtype, the three Δ-isoform β-NRX subtypes yield affinities that varied by less than 2.3-fold for each of the NLΔs and less than 3.2-fold for interactions with each insert-containing NL.
Splice insert 4 weakens β-NRX1 and 2 binding, but strengthens β-NRX3 binding to NLs
Figure 2O shows that the presence of the SS4 insert in β-NRX1+4 led to KDs for binding to NL that were ~2–7-fold higher than for interactions with the β-NRX1Δ4 variant, demonstrating that the SS4 insert exhibits an inhibitory effect on β-NRX1/NL interactions. Similarly, for β-NRX2+4 KDs are increased between 1.8 and 4.8-fold from those for β-NRX2Δ4 (Figure 2O, Table S2). Notably, the strength of interaction is decreased for the +4 isoforms of β-NRXs 1 and 2 for all NLs, irrespective of the NL subtype or splice form. These findings are consistent with the results recently reported by Reissner and colleagues (Reissner et al., 2008), where both isoforms of β-NRX1 could precipitate NL1 from brain lysates, although to a smaller extent if the SS4 insert was present.
In contrast to the uniform inhibition of NL interaction observed for β-NRXs 1 or 2 when the SS4 insert was included, the effect on NL binding affinity was opposite for β-NRX3 (Figure 2O). The β-NRX3+4 isoform exhibited greater affinity for all NLs than β-NRX3Δ4, with KDs for β-NRX3+4/NL interactions ~0.3–0.9 of the corresponding β-NRX3Δ4/NL interactions. Notably, whereas the SS4 insertion sequences are identical except for one conserved substitution between β-NRX1 and β-NRX2, the β-NRX3 SS4 insert differs more substantially with fourteen amino acid substitutions (Figure 1A and Figure S5). These differences are likely to underlie the different effects of the SS4 insert in β-NRX3. Kinetic analysis of these SPR data revealed that the dissociation rates for complexes involving β-NRX3+4 were slower than for β-NRX3Δ4, leading to more stable interactions (Figure 2P–Q, Table S1).
NL splice isoform choice gives rise to relatively small changes in affinity for β-NRXs
To gain a better understanding of the effects of NL splice inserts A and B on the binding affinities of interactions with β-NRXs, the KDs for all β-NRX/NL interactions were normalized against the KDs of the same β-NRX binding to the corresponding NLΔ (Table S3).
The presence of the B insertion sequence in NL1 increased the KDs for all the β-NRXs tested, indicating that the inhibitory effect of the B insert is maintained for all β-NRX family members, regardless of the presence of the SS4. The relative changes in KD for each β-NRX, compared to the KD for NL1Δ, were 2.4-fold or less, providing further support for the idea that, although the presence of the B insert can lead to weaker interactions, NL1B can still maintain strong binding to each of the β-NRXs, even in the presence of the SS4 insert.
Inclusion of the A insert sequence in NL1 also gave rise to small effects, leading to KDs for β-NRXs that were ~0.7–1.6 of the corresponding KDs for NL1Δ. NL2A had slightly increased affinities for all β-NRXs, with KDs ~0.4–0.9 of the corresponding KDs for NL2Δ. NL3, all isoforms of which bind β-NRXs more weakly than NL1 and NL2, has insert sequences A1 and A2, which can alternatively be included together or separately at the A splice insertion site. Inclusion of the A1 insertion led to enhancement of β-NRX binding (KD changes ~0.4–0.9); inclusion of A2 led to changes of affinity that depended on the β-NRX partner (KD changes ~0.7–1.4). Inclusion of the concatenated A1A2 insert also yielded a small range of changes in KD relative to NL3Δ (~0.9–1.1). These lesser effects of A inserts as compared to the B insert on binding affinity are consistent with the close proximity of the NL1 B insertion site to the β-NRX binding surface and the distal nature of the A insertion site, which is ~31Å from the nearest β-NRX atom in the complex crystal structures (Figure 1C) (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007).
Affinity effects of NL and β-NRX splice insertions are largely independent of one another
The SPR measurements reported here show that the β-NRX1 SS4 insert lessens affinity for NLs in all cases (1.6–7.4 fold-increase in KD, Table S2), and similarly the NL1 B insert weakens binding to all β-NRXs tested (1.1–2.4 fold-increase in KD, Table S3). If the inhibition of binding arose from interference between the NL1 B insert and the β-NRX SS4 insert, as has been suggested (Boucard et al., 2005; Chih et al., 2006; Comoletti et al., 2006), effects of splice insert inclusion in one binding partner would be expected to be larger when the other binding partner contained an insertion sequence. However, results from our SPR experiments show that the binding inhibition, which results from inclusion of an insertion sequence, is similar whether or not the binding partner also has an insertion sequence. The inclusion of the SS4 insert in β-NRX1 increases the KD by 1.9-fold for NL1Δ as compared to ~2.3-fold increases for NL1B and NL1AB (Table S2). On the other hand, for β-NRX2, whose SS4 insert is nearly identical to that of β-NRX1, inclusion of the SS4 insert leads to a 4.1-fold increase in the KD for interaction with NL1Δ, but only ~2.5-fold for NL1B and NL1AB (Table S2). These results demonstrate that binding inhibition by the β-NRX and NL insertion sequences are independent of whether the interacting partner contains an insertion sequence.
Co-precipitation and AUC experiments are consistent with results from SPR
Pull-down experiments reported by Boucard et al. showed that only NL1 isoforms lacking splice insertion B could be precipitated by immobilized β-NRX1+4, implying that the combination of B and SS4 inserts inhibited detectable binding (Boucard et al., 2005). However, in the same work, when the orientation of this experiment was reversed such that the NLs were immobilized, binding to β-NRX1+4 was observed independent of the presence of the NL1 B insert (Boucard et al., 2005). These results are in apparent disagreement, and therefore we sought other experimental means to determine whether β-NRX1+4 and NL1B can form tight complexes as suggested by the SPR experiments presented here.
We performed co-precipitation experiments in which we coupled each of the four NL1 isoforms, which contained C-terminal 8xHis affinity tags, to Ni-NTA beads and incubated these with an excess of either β-NRX1Δ4 or β-NRX1+4. After washing, the protein retained on the beads was analyzed by SDS-PAGE (Figure S4A). These results are consistent with the SPR data showing that, irrespective of the splice form of either binding partner, β-NRX1 and NL1 can associate to form complexes.
We also performed sedimentation equilibrium analytical ultracentrifugation experiments on stoichiometric mixtures of β-NRX1Δ4 or β-NRX1+4 and NL1Δ or NL1B (Figure S4B). Each of these AUC experiments, fit with a 1:1 (1 NL dimer: 2 β-NRX molecules) binding model, revealed the formation of tight complexes with micromolar affinities, in agreement with the SPR measurements reported here (Table S4).
Splice insertion 4 mediates β-NRX strand displacement with minimal changes to NL binding site
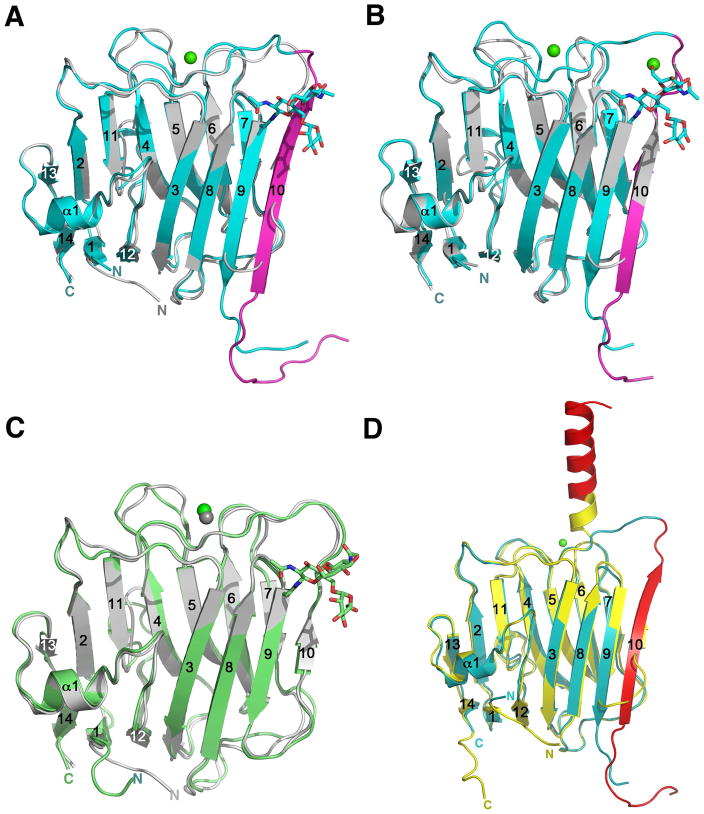
The structure of β-NRX1+4 was determined by molecular replacement using the structure of β-NRX1Δ4 (PDB ID 3BOD) as a search model. The β-NRX1+4 structure, refined at 2.7Å resolution, contains two molecules in the C2 asymmetric unit, each of which includes residues 86–197 and 210–288. The structure of β-NRX2+4 was determined by molecular replacement using the structure of β-NRX2Δ4 (PDB ID 3BOP) as the search model. The final structure, refined at 2.3Å, has one molecule in the asymmetric unit including residues 88–197 and 214–290. The RMSD between β-NRX1Δ4 and β-NRX1+4 is 0.97Å for 170 Cα atom pairs, and the RMSD between β-NRX2Δ4 and β-NRX2+4 is 1.02Å for 165 Cα atom pairs (Figure 3A, B). The structures of β-NRX1+4 and β-NRX2+4 are highly similar to each other, with an RMSD of 0.67Å for 189 Cα atom pairs, as expected from a high sequence similarity between the two β-NRXs.
Figure 3.
Structural rearrangements take place to accommodate the splice insert 4. (A) Superposition of the β-NRX1Δ4 structure (PDB ID 3BOD) in silver and the β-NRX1+4 structure in cyan. The SS4 insert is highlighted in magenta. (B) Superposition of the β-NRX2Δ4 structure (PDB ID 3BOP) in silver and the β-NRX2+4 structure in cyan. The SS4 insert is highlighted in magenta. (C) Superposition of the β-NRX1Δ4 structure (PDB ID 3BOD) in silver and the glycosylated β-NRX3Δ4 structure in green. (D) Superposition of the new β-NRX1+4 structure in cyan with the non-glycosylated β-NRX1+4 structure (PDB ID 2R1B) (Shen et al., 2008) in yellow. The SS4 is shown in red in both structures, highlighting identical sequences adopting completely different secondary structures.
Structural rearrangements take place to accommodate the SS4 insert
The overall folds of both β-NRX1+4 and β-NRX2+4 consist of 14 β strands arranged in two curved β sheets, one α helix, and a 310 helix, similar to those of previously determined structures of β-NRX LNS domains (Koehnke et al., 2008b; Rudenko et al., 1999; Shen et al., 2008) and to the glycosylated β-NRX3Δ4 structure we report here (see below). Despite conservation in the core fold, there are significant structural rearrangements to accommodate the thirty-residue SS4 insertion sequence. In the β-NRX1+4 and β-NRX2+4 structures, the segment that forms the β10 strand in the Δ4 isoform structures is displaced and forms an extension of the β9 strand/β9-β10 loop (Figures 3A, 3B, and S5A). Residues 195–200 (β-NRX1+4 numbering for clarity), which form part of the β10 strand in the Δ4 isoform structures, are disordered in the +4 isoform structures. A segment from the SS4 insert itself forms a “new” β10 strand at the edge of the convex sheet, as can be seen unambiguously in simulated annealing omit maps calculated with residues from the SS4 insert omitted (Figure S6A). The first 9 and 13 residues of the SS4 insert of the β-NRX 1+4 and 2+4 structures respectively, are disordered indicating a high degree of structural mobility in this region.
A detailed analysis of the “new” β10 strand of the +4 isoform in comparison with the “old” β10 strand in Δ4 isoform structures reveals that although the amino acid composition of the two strands is different, the hydrophobicity and polarity of residues in corresponding positions are retained to some extent (Figures S5B and S6B). For example, Val222 of the new β10 strand aligns with the position of Val194 in β-NRX1Δ4, Val223 aligns with Ile195, Asp224 with Glu196, and Trp226 with Tyr198. Upon SS4 insertion-induced structural rearrangement, a new hydrophobic cluster is formed at the N-terminal extremity of the “new” β10 strand in both of the β-NRX1+4 and β-NRX2+4 structures, which may stabilize the new fold. This cluster is composed of highly conserved residues, some of which were already in place before the strand replacement (Tyr173 and Val175 from the β8 strand), and the remaining of which were positioned to be part of this structure due to strand replacement (Trp192, Pro193, and Val194 from the “old” β10 strand, and Ile215, Pro216, and Tyr217 from the insert itself) (Figure S7). The strand replacement also places the side chain of Arg218 from the “new” β10 strand in a position to form a salt bridge with Asp190, which may provide additional stability to the new structure (Figure S7).
Structural effects of glycosylation
To assess the structural effects of glycosylation independent of the SS4 insertion, we also determined the structure of glycosylated β-NRX3Δ4 at 2.0Å resolution. The structure is highly similar to that of bacterially produced non-glycosylated β-NRX ectodomains, with 0.70Å RMSD between the glycosylated β-NRX3Δ4 and the non-glycosylated β-NRX1Δ4 for 174 corresponding Cα atom pairs (Figure 3C). The structure of a glycosylated β-NRX1Δ4, determined in complex with NL1Δ (Arac et al., 2007), also reveals a high-level of similarity to the non-glycosylated β-NRX1Δ4 (Koehnke et al., 2008b) with an RMSD of 0.62Å for all Cα atom pairs. Therefore, it can be concluded that glycosylation does not induce significant structural rearrangements in β-NRXs lacking the splice insertion 4.
In β-NRXs containing the splice insertion 4, however, N-glycosylation at a conserved asparagine residue (Asn184 in β-NRX1, Asn186 in β-NRX2, and Asn182 in β-NRX3) partially accounts for stabilization of the ordered part of the SS4 insert. In both of the new β-NRX1+4 and β-NRX2+4 structures, a mannose moiety linked to a N-acetyl-D-glucosamine (GlcNAG), which is covalently attached to Asn184, stacks against the indole ring of Trp226 from the SS4 insert. In the glycosylated β-NRX3Δ4 structure, Tyr196 (equivalent to Tyr198 in β-NRX1) makes similar, although less extensive, van der Waals interactions with the mannose moiety of the carbohydrate chain covalently attached to Asn182 (Asn184 in β-NRX1) (Figure S6B). Because Trp226 makes more extensive hydrophobic interactions with the mannose than Tyr196 does, the presence of a tryptophan residue in this position may be more energetically favorable than a tyrosine residue, when both residues are available in the presence of the SS4 insert.
Ca2+ binding in the splice insert 4-containing β-NRX structures
Due to the presence of a calcium chelator, citrate, and low pH in the crystallization solution, no bound Ca2+ ions were seen in the β-NRX1+4 structure. Our analysis of Ca2+ binding is therefore based on the β-NRX2+4 structure. For convenience in comparing with the β-NRX1Δ4/NL1Δ structures, β-NRX1+4 numberings are used.
Electron density is observed for two Ca2+ ions in the β-NRX2+4 structure, one of which is at the same site seen in previously reported β-NRX structures (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007; Koehnke et al., 2008b; Shen et al., 2008) and in one of the three β-NRX3Δ4 molecules in the asymmetric unit of the β-NRX3Δ4 structure reported here. In the presence of the SS4 insert, the octahedral coordination of this Ca2+ ion remains largely unchanged. Side chain oxygen atoms of Asp137 and Asn238 and main chain carbonyl oxygen atoms of Val154 and Ile236 serve as ligands, along with two well-ordered water molecules, water 1 and water 2 (Figure S8A). These two water molecules are also present in the β-NRX1Δ4/NL1Δ complex structure (Chen et al., 2008). In our previous β-NRX1Δ4 structure, only water 1 is present, while the sixth ligand is fortuitously contributed by the Glu249 side chain from a symmetry-related molecule (Koehnke et al., 2008b), overlapping the position of water 2 (Figure S8A). In the β-NRX1Δ4/NL1Δ complex structure, water 2 mediates the Ca2+-dependent interaction between NL1 and β-NRX1Δ4 by hydrogen bonding to the main chain carbonyl oxygen of Gln395 and the side chain carboxyl of Glu397 from NL1 (Figure S8A).
The second “new” Ca2+ ion in the β-NRX2+4 structure is coordinated by a side chain carboxylate from Gln233, and main chain carbonyl oxygen atoms from Asp229 and Gly231 (Figure S8B). Presumably, this new binding site can only be present in splice isoforms containing the SS4 insertion sequence, because the Ca2+ ion is coordinated by residues from the insert. The biological relevance of this “new” Ca2+ binding site is unclear. From a purely structural standpoint, Ca2+ binding at this site is an additional element that is likely to stabilize the “new” fold, after β10 strand replacement.
DISCUSSION
The results of the systematic β-NRX/NL affinity measurements presented here show that, among the substantial set of proteins we studied, all NLs can bind to all β-NRXs, irrespective of the splice isoform of either partner. As for classical cadherins (Katsamba et al., 2009), there is no evidence for a “binary code” which would require that some pairs of NLs and β-NRXs fail to bind to one another. Rather, although splice isoform choice alters KDs by as much as a factor of 7.4, binding affinity for all β-NRX/NL pairs remains within a range likely to be consistent with productive cell-adhesive interactions. Moreover, the effects on KDs of the NL and β-NRX splice inserts appear to act independently of one another leading to a graded set of affinities. This finding is in contrast to the results of previous experiments which suggested that the combination of β-NRX insert 4 and NL1 insert B collaborate to inhibit binding (Boucard et al., 2005; Chih et al., 2006), but supports other studies which failed to find such effect (Graf et al., 2006; Reissner et al., 2008).
Effects of gene identity and splice inserts on β-NRX/NL interaction affinities
Some affinity effects can be rationalized in light of the atomic-level structures of β-NRXs, NLs, and complexes between them. The β-NRX1Δ4/NL1Δ complex structures (Arac et al., 2007; Chen et al., 2008) show that the insert points for both splice insert B of NL1 and splice insert 4 of β-NRX1 are at the “edges” of the interaction interface (Figure 1C). The presence of splice inserts at either of these sites could potentially alter β-NRX/NL interaction, although our measurements indicate that the effects are modest.
In all cases, irrespective of the subtype or splice isoform of the β-NRX binding partner, the presence of NL1 insert B reduced affinity, albeit by only up to a factor of 2.4. On the other hand, inclusion of the A inserts in NLs had very small effects on affinities, consistent with the distal nature of the A splice site in the complex structures (Figure 1C) (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007).
The similar NL binding affinities of the three Δ4 isoform β-NRXs are not surprising since all are expected to bind to NLs via a conserved canonical interface in which Ca2+ ions are jointly coordinated between the two proteins (Arac et al., 2007; Chen et al., 2008; Fabrichny et al., 2007). The presence of splice insertion 4 in β-NRXs has somewhat larger effects than NL inserts, but the character of these effects depends on the β-NRX subtype. The +4 isoforms of β-NRX1 and β-NRX2 have lower affinity for NLs, irrespective of NL subtype or splice isoform, with KDs increased by 1.6–7.4 fold as compared to their respective Δ4 isoform proteins. By contrast, inclusion of the SS4 insert in β-NRX3 had the opposite effect, increasing affinity for all NLs, leading to KDs that were decreased to ~90-30% of those for the corresponding NLs with β-NRX3Δ4. This difference is consistent with divergence of the β-NRX3 SS4 insert sequence as compared to those of β-NRXs 1 and 2, which are far more similar (Figure 1A and Figure S5).
The binding affinities for each β-NRX/NL pair appear to vary by a maximum of about sevenfold as a consequence of the splice isoforms of the component proteins. It is likely that β-NRX/NL complexes of various splice isoform combination will exist as either stable or transient complexes in vivo. The single biggest effect on affinities is NL subtype dependent and is observed for NL3 where KDs are an order of magnitude or more larger than for NLs 1 and 2, independent of splice isoform. The single largest KD we observe is for the complex formed between NL3Δ and β-NRX2+4 although all β-NRX splice variants exhibit reduced affinities to all variants of NL3. Thus, in this case it is the gene identity of NL rather than the nature of the insertion that is the primary determinant of variations in binding affinities.
Structural changes of β-NRXs induced by SS4
The structural studies presented here provide an explanation of the effects on NL binding affinity of β-NRX SS4 insertions that is not evident from earlier structures of non-glycosylated proteins. Structures of the glycosylated β-NRX1+4 and β-NRX2+4 ectodomains each depict nearly identical changes from insert-less β-NRXΔ4 structures. Most notably, regions from the SS4 insertion sequence displace the β10 strand of the Δ4 isoform structure, producing new molecular surfaces along the length of the LNS domain edge defined by the new β10 strand, but leaving the NL binding site essentially unaffected.
Recently, Shen and colleagues determined the structure of a β-NRX1+4 expressed in bacteria (Shen et al., 2008). In their structure, while the first 15 residues of the SS4 insert are disordered, the last 15 residues from SS4, 216–230, form an α helix protruding from the LNS domain surface near the NL binding site in a conformation that would clearly interfere with NL binding (Figure 3D). The helical configuration of SS4 seen in this structure may be due to crystal packing, as the SS4 residues interact with residues from the convex sheet of a neighboring symmetry mate. Indeed, our previous NMR studies of β-NRX1+4, also produced in bacteria, indicate that the SS4 insertion of the non-glycosylated protein is unstructured in solution (Koehnke et al., 2008b). The structural data presented here indicate that β-NRXs, when post-translationally glycosylated as they are in vivo, undergo a significant and defined structural rearrangement to accommodate the SS4 insertion sequence, in a manner entirely different from that seen previously for a bacterially expressed β-NRX1+4.
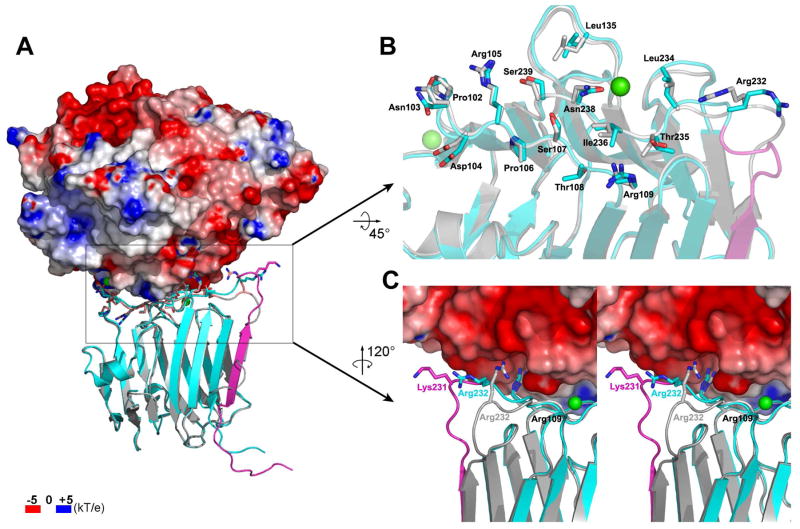
Which of these structures corresponds to a relevant biological conformation? Several lines of evidence shed light on this question: Glycosylation clearly favors the strand-displaced conformation. In vivo, Asn184 (β-NRX1 numbering) should be constitutively glycosylated, as we found for the mammalian cell-expressed proteins described here. Further, this site is conserved in vertebrate β-NRXs, arguing for its functional importance. The 30 amino acid SS4 insertion sequence is itself highly conserved both among NRX1-2 genes and across species, and this sequence conservation can be at least partly explained by constraints imposed by the structural requirements of strand displacement. In particular, as mentioned previously, the stabilization of the new structures involves many residues from the SS4 insert itself (residues 215–217 for the hydrophobic cluster, Arg218 which forms a salt bridge with Asp190, and residue Trp226 for stacking against the mannose). The most compelling evidence favoring the strand-displaced conformation comes from the effect of SS4 inserts on binding to NLs. The SS4 insert-helix of the bacterial β-NRX1+4 structure protrudes from the β-NRX surface like a stiff arm that would certainly clash with a bound NL (Figure 3D), suggesting that binding of the +4 isoform might be completely or significantly inhibited, depending on the rigidity and stability of the protruding helix. However, in early biophysical studies (Koehnke et al., 2008b), and in the comprehensive SPR binding analyses reported here, the presence or absence of the SS4 insert makes only small differences (~2–7 fold in KD) for β-NRX/NL interactions. This observed binding behavior is consistent with the glycosylated β-NRX1+4 structures we report, for which superpositions on the extant β-NRX1Δ4/NL1Δ complex structures (Arac et al., 2007; Chen et al., 2008) reveal no significant clashes, and most of the NL binding surface (~90% surface area) remains unchanged from the Δ4 isoform structures (Figure 4). Moreover, in accordance with the high structural conservation of the NL binding surface in the glycosylated β-NRX +4 isoforms we report (in particular the Ca2+ binding site and the charged residues Arg109, Arg232, Asp104, and Arg105), the surface electrostatic potential of the NL binding region is conserved upon SS4 insertion and remains complementary to the NL surface electrostatic potential pattern (Figure S9).
Figure 4.
Structural effects of the SS4 insertion on the NL binding site. (A) The β-NRX1+4 structure (cyan) is superposed onto the β-NRX1Δ4 (silver) from the β-NRX1Δ4/NL1Δ complex structure (PDB ID 3BIW). The SS4 insert is shown in magenta in the β-NRX1+4 structure. The electrostatic potential surface is shown for the NL1 molecule in the complex with the indicated color scale. (B) Close-up view of the NL binding interface of β-NRX1Δ4 with the β-NRX1+4 structure superposed. The β-NRX1Δ4 interface residues with a buried surface area greater than 5Å2 in the complex structure with NL1 and the same residues in β-NRX1+4 are represented. The colorcoding is the same as in (A). (C) Close-up view of the part of the β10–β11 loop that is the most variable upon SS4 insertion.
Possible conformations of the SS4 insert in β-NRX/NL complexes
The only significant structural difference induced by the presence of the SS4 insert in the region of the NL interface is localized at a part of the β10–β11 loop which carries Arg232. Arg232 forms a salt bridge with Asp387 from NL1 in the β-NRX1Δ4/NL1Δ complex structures (Arac et al., 2007; Chen et al., 2008) (Figure 4). This salt bridge is one of only two intermolecular ion pairs within the β-NRX/NL interface, and is the only one in the immediate vicinity of SS4. Given the immediate proximity of Arg232 relative to the SS4 insertion sequence, the presence of SS4 is likely to change the configuration of Arg232 in complexes with NLs (Figure 4C), which in principle could lead to disruption of this salt bridge interaction. Clearly, further structural studies would be needed to determine the atomic details of the actual conformation of the SS4 insert and the β10-β11 loop in a bona fide β-NRX+4/NL structure. Nevertheless, the NL region facing that variable region of β-NRXs forms a large and clearly electronegative groove that can accommodate the likely structural changes arising from the SS4 insertion (Figure 4).
Conformational differences in the Arg232 region, however, cannot explain the opposite effects on binding of splice insertions in β-NRX3 versus β-NRXs 1 and 2, because this region is well conserved between them (Figure S5). On the other hand, the β-NRX3 SS4 sequences differ substantially from those of β-NRX1 and β-NRX2, which are nearly identical (Figure 1A). Although these SS4 insert residues are localized outside of the NL binding site, we cannot exclude the possibility that they have a long range effect on NL binding affinity and that they play a role in the difference observed between β-NRX3 and β-NRXs 1 and 2. In any case, without atomic-level structural data, we cannot predict with certainty whether a similar strand displacement would be induced in β-NRX3 by its SS4 insert. Understanding the molecular basis of the slightly stronger binding of insert-containing β-NRX3s must await further investigation.
Generality of splice insert-dependent strand displacement
From a purely structural perspective, the strand replacement observed for the β10 strand in the β-NRX1+4 and β-NRX2+4 structures presented here is favored not only by the different elements that stabilize the new structure, as described above, but also by the reduced structural constraints on the β10 strand and the consequent structural plasticity of that strand. The β10 strand is at the edge of the convex β-sheet and its backbone is thus hydrogen bonded to only one other strand. Moreover, its side chains are mostly solvent exposed and none of them are involved in the hydrophobic core of the β-sandwich fold. Of note, a similar β-strand replacement has been seen in the piccolo C2A domain in the presence of a 9-residue splice insert, which forms a β-strand to displace a preexisting β-strand at the edge of β-sandwich (Garcia et al., 2004). This type of secondary structure rearrangement may represent a common mechanism to realize the effects of alternative splicing.
The crystal structures available for the LNS2 and LNS4 domains of α-NRX (Sheckler et al., 2006; Shen et al., 2008) where splice insertion sites 2 and 3 (SS2 and SS3) respectively reside, show that their topology is very similar to the that of the β-NRX LNS domain. However, SS2 is located on the β8–β9 loop such that both of the adjacent strands are internal in the β-sheet topology and are hydrogen bonded to two other strands. This suggests that a similar strand replacement is unlikely at the SS2 of the α-NRX LNS2 domain, due to the strong structural constraints imposed on the two adjacent strands. In the case of SS3, which is located on the β6–β7 loop of the LNS4 domain, one of the adjacent strands, the β7 strand, is at the edge of the concave β-sheet, suggesting that strand replacement is possible, but further studies would be required to determine the precise structural outcomes of splicing at these sites (Figure S5C).
Potential biological roles for alternative splicing of β-NRXs and NLs
The biophysical measurements presented here define a matrix of β-NRX/NL ectodomain affinities for all splice isoforms of β-NRX1, 2, and 3, interacting pair-wise with all splice isoforms of NL1, 2, and 3. We find that all β-NRXs can bind to all NLs, and while there is a dependence of affinities on splice form, all affinities are in a range consistent with productive adhesion. The structures presented here of SS4 insert-containing β-NRXs provide a structural explanation for the limited effect of SS4 insertion sequences on interaction with NLs, and show that while the NL binding site remains mainly intact, other regions of the β-NRX LNS domain are more dramatically altered. Previous cell-based studies indicated that β-NRX1Δ4 binds efficiently to all four NL1 splice isoforms, while β-NRX1+4 binds poorly to isoforms of NL1 that include the B insertion (Chih et al., 2006). However, lesser effects of splice insertion 4 were found in similar experiments (Boucard et al., 2005; Graf et al., 2006). The proximity in the Δ complex structures of NL1 splice site B and β-NRX1 splice site 4 initially suggested the attractive possibility that steric hindrance between the inserts might prevent interaction in the corresponding splice insert-containing complex. However, the extensive set of SPR experiments we present here reveals maximal splice form-dependent differences in KD of only 3.4-fold for β-NRX1/NL1 interactions. It is difficult to judge whether these differences in KD could in principle specify a molecular recognition code for β-NRX/NL interactions. Classical cadherins appear to use graded affinities as an initial filtering step in cell-sorting processes (Katsamba et al., 2009). However, it is less likely that NRXs and NLs function in the same way since they are not required for synapse formation.
Although splice isoform choice has limited effects on the β-NRX/NL binding, it is possible that alternative splicing of β-NRXs and NLs may serve a primary biological role other than to modulate the affinity of their interactions. Notably, recent work in two laboratories have identified the postsynaptic cell surface receptor LRRTM2 as a receptor whose binding to presynaptic β-NRXs depends on the presence of splice insert 4 (de Wit et al., 2009; Ko et al., 2009a), although the molecular basis of this specificity remains unknown. Comparison of the strand-displaced β-NRX+4 structures with the insert-less β-NRXΔ4 structures reveals areas of difference outside of the NL interacting region. Two regions of β-NRX are substantially altered by the SS4 insertion: the LNS domain edge defined by the splice form-specific β10 strand (Figure S6A and B), and a new loop which protrudes from the “bottom” of the LNS domain that is formed by residues 192–200 from the displaced β10 strand and β9–β10 loop, followed by the N-terminal 17 residues of the SS4 insert (Figure S7). It is possible that these regions of insert-dependent structure, rather than or in addition to those near the NL binding site, may contribute to the biological role of alternative splicing, for example, to regulate recruitment of synaptic binding partners other than NLs. Lateral oligomerization following trans binding, for example of cadherins, ephrins and T-cell receptors appears to be a widely used mechanism that is exploited by adhesion molecules, and β-NRX and NL splice variation could conceivably regulate such a process as well.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Although bacterially produced NRXs have been used in prior biophysical studies, as described above, appropriate glycosylation is critical to the structure of the SS4 insert in β-NRXs. We produced molecules corresponding to the LNS domains of β-NRXs 1, 2, and 3 for each alternative splice isoform (Δ4 and +4). Protein constructs for mouse β-NRX1 (residues 86–288), rat β-NRX2 (residues 87–290), and mouse β-NRX3 (residues 1094–1297, α-NRX numbering) were cloned into a proprietary mammalian expression vector obtained from Prof. Dan Leahy (Johns Hopkins University) to produce proteins with an N-terminal 8xHis-tag. Proteins were expressed in human embryonic kidney (HEK) 293F cells by transient transfection using Polyfect (Qiagen) according to the instructions of the manufacturer. The transfected cells were kept in culture for an additional 5 days. Secreted β-NRX LNS domains were purified by Ni2+ affinity chromatography followed by ion exchange and gel filtration steps. Briefly, conditioned medium was collected and buffer-adjusted to the final concentrations of 20mM Tris (pH 8.0), 500mM NaCl, 20mM imidazole, and 3mM CaCl2. The medium was filtered through a 0.22μm filter and passed over HisTrap HP columns (GE Healthcare). The columns were washed with 20mM Tris (pH 8.0), 500mM NaCl, 20mM imidazole, and 3mM CaCl2, and the protein was eluted with 250mM imidazole. Protein concentration in the eluate was estimated by SDS-PAGE and appropriate amounts of PreScission protease (GE Healthcare) added to cleave off the 8xHis tag during overnight dialysis at 4°C against 10mM Tris (pH 8.0), 150mM NaCl, 3mM CaCl2, and 1mM DTT. Next, proteins were dialyzed at 4°C against 20mM Bis-Tris (pH 6.0), 100mM NaCl, and 3mM CaCl2 and then applied to a Mono S column (GE Healthcare) equilibrated with the same buffer. Proteins containing the splice insertion 4 bound to the MonoS column and eluted at approximately 400mM NaCl. For proteins without the splice insertion 4, the flow-through was passed over a Mono Q column equilibrated with the same buffer and the flow through collected. Finally, the protein was subjected to size exclusion chromatography (Superdex 200) in 10mM Tris (pH 8.0), 150mM NaCl, and 3mM CaCl2, and concentrated to 10mg/mL.
Similarly, the cholinesterase-like regions of the rat NL1, 2, and 3 extracellular domains, NL1 (residues 48–638), NL2 (residues 46–612), and NL3 (residues 42–633), were produced for all possible splice forms. These regions were fused to the endogenous signal sequences and a C-terminal 8xHis-tag and cloned into the mammalian expression vector pCEP4 (Invitrogen). Proteins were overexpressed in HEK293F cells. Conditioned medium that contained the secreted NLs was collected and buffer-adjusted to the final concentrations of 20mM Tris (pH 8.0), 500mM NaCl, 20mM imidazole, and 3mM CaCl2. The medium was filtered through a 0.22μm filter and passed over HisTrap HP columns (GE Healthcare). The columns were washed with 20mM Tris (pH 8.0), 500mM NaCl, 20mM imidazole, and 3mM CaCl2, and the proteins were eluted with 250mM imidazole. The proteins were dialyzed overnight at 4°C against a buffer of 20mM Bis-Tris (pH 6.0), 100mM NaCl, and 3mM CaCl2, and then were applied to a Mono S column (GE Healthcare) equilibrated with the same buffer, with the exception of NLs 3A1 and 3A1A2. All NLs were present in the flow through of the Mono S column, which along with the NLs 3A1 and 3A1A2 were passed over a Mono Q column equilibrated with the dialysis buffer and eluted using a linear gradient of 200mM to 350mM NaCl. Finally, proteins were subjected to size exclusion chromatography (Superdex 200) in 10mM Tris (pH 8.0), 150mM NaCl, and 3mM CaCl2, and concentrated to 10mg/mL.
SPR binding assays
Binding assays were performed using a Biacore T100 biosensor equipped with a Series S NTA chip (GE Healthcare). The instrument was equilibrated in HBS (HEPES-buffered saline, 10 mM HEPES, pH 7.4, 150 mM NaCl) buffer at 25°C for immobilization of NL to the sensor chip surfaces. NLs were immobilized using a modified amine-coupling chemistry method (Willard and Siderovski, 2006) that allows the capture of His-tag-fused proteins to NTA moieties followed by amine coupling to the dextran layer of the sensor chip to avoid decaying surfaces that are often associated with capture of His-tagged proteins. A 60s injection of 500 μM NiSO4 at 20 μL/min was performed over all four flow cells to load the NTA groups with Ni2+. The carboxyl moieties of the dextran layer over all four flow cells were activated using a 7 min injection of 50 mM N-hydroxysuccinimide/200 mM 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide hydrochloride (Sigma-Aldrich) at the same flow rate. NLs were diluted to a concentration of 50-25 μg/mL in HBS buffer and injected over independent flow cells at 20 μL/min, using short pulses of 6–10s, until a capture level of ~500 RU was achieved in each flow cell. To block any remaining activated carboxyl groups, 1 M ethanolamine, pH 8.5 (Sigma-Aldrich) was injected over all surfaces for 4 minutes at 20 μL/min. All four surfaces were postconditioned using three 10-second-consecutive injections of 350 mM EDTA at 100 μL/min to remove any proteins that were not covalently tethered to the sensor chip surfaces and any Ni2+ that remained bound to the NTA groups. In each experiment, three different NLs were immobilized onto flow cells 1, 3 and 4, leaving flow cell 2 vacant to serve as a reference flow cell.
Kinetic binding analysis was performed in a running buffer of 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 3 mM CaCl2, 0.005% Tween 20, and 1 mg/mL BSA at 25°C. β-NRX protein stocks were diluted in running buffer to the highest concentration as described in the figure legends for each experiment. The concentration series for each β-NRX was prepared using a two-fold serial dilution in running buffer. During a binding cycle, each sample was injected over all four surfaces for 60s at a flow rate of 50 μL/min, followed by a 60-s dissociation phase at the same flow rate, during which running buffer flows over the bound complexes to collect information about the dissociation rate of the β-NRX/NL of interest. The surfaces were regenerated using a 10-s injection of 350 mM EDTA at 100 μL/min, and at the end of each cycle buffer was injected for 60 seconds at 50 μL/min. Analyte injections were performed in order of increasing concentration for each concentration series with each series tested in triplicate from lowest to highest concentration. For every two binding cycles of β-NRX, a cycle where buffer was used as the analyte was performed, which together with the signal from the vacant surface, allows for double referencing of the binding responses (Myszka, 1999) to remove such artifacts as instrument noise and drift. At the beginning of each experiment, ten additional binding cycles utilizing buffer as the analyte were performed allowing for thermal equilibration of the instrument. These binding cycles were removed from the analysis. The data were processed and analyzed using Scrubber 2.0 (BioLogic Software, Pty., Campbell, Australia). The binding data were fit to a 1:1 binding model with a step to account for mass transport to extrapolate ka and kd. The equilibrium binding constant KD was calculated based on the relationship KD=kd/ka. In each panel the data are depicted in black and the fit to the model is shown in red.
Co-precipitation experiments
For each NL, 50μg protein was immobilized to 100μL of Ni-NTA resin (Qiagen). The resin was then washed with 4mL of wash buffer (10mM Tris pH 7.4, 150mM NaCl, 3mM CaCl2), followed by incubation with 1mL wash buffer containing 20μg β-NRX. The resin was then again washed with 4mL of wash buffer, followed by elution with 200μL 2 mM EDTA. Eluates were analyzed by SDS-PAGE.
Sedimentation velocity measurements
Experiments were performed in a Beckman XL-A/I analytical ultracentrifuge (Beckman-Coulter) using two-cell centerpieces with tapered walls, 12 mm path length and sapphire windows. Proteins were dialyzed overnight at 4°C against a dialysis buffer of 10 mM Tris-HCl pH 7.4, 100 mM NaCl, 3 mM CaCl2. NL1 proteins were used at concentrations of 5 μM dimer, alone or in the presence of 5, 10, and 20 μM β-NRX1Δ4 or β-NRX 1+4. All measurements were recorded at 45000 rpm at 25°C, using interference at 660 nm at three-minute intervals between each scan, collecting a total of 340 scans for each sample. Solvent density, solvent viscosity and protein v-bar were determined using the program SednTerp (Alliance Protein Laboratories). Data analysis was performed using SedFit obtained from the National Institute of Bioimaging and Bioengineering, NIH (https://sedfitsedphat.nibib.nih.gov/software/default.aspx).
Crystallization, Data Collection and Crystallographic Analysis
Crystals of mouse β-NRX1+4 were obtained in 26% PEG1000, 0.2M Li2SO4, 0.1M phosphate-citrate, pH 4.9. The crystals were cryoprotected with the mother liquor supplemented with 25% glycerol before flash-cooling in liquid nitrogen. These crystals belong to space group C2 with cell dimensions a=85.8Å, b=59.8Å, c=105.7Å, β = 103.9° Crystals of rat β-NRX2+4 were obtained in 22% PEG3350, 0.35M CaCl2, and 1mM GSH/GSSG. The crystals were cryoprotected for freezing in liquid nitrogen in mother liquor supplemented with 35% PEG400. These crystals belonged to space group C2221 with a=51.7Å, b=102.1Å, and c=78.5Å. Crystals of mouse β-NRX3Δ4 were obtained in 27% PEG3350, 0.2M Li2SO4, and 0.1M NaAc, pH 5. The crystals were cryoprotected with the mother liquor supplemented with 25% glycerol before flash-cooling in liquid nitrogen. These crystals belong to space group P212121 with cell dimensions a=74.7Å, b=83.4Å and c=119.8Å.
Diffraction data were collected on single crystals at 100K at X4C beam line in the National Synchrotron Light Source at Brookhaven National Laboratory, and processed with the HKL program suite (Otwinowski and Minor, 1997). The structures of β-NRX1+4 and β-NRX2+4 were solved by molecular replacement with PHASER (McCoy et al., 2007) using the structures of β-NRX1Δ4 (PDB ID code: 3BOD) and β-NRX2Δ4 (PDB ID code: 3BOP) as the search models, respectively. Likewise, the structure of β-NRX3Δ4 was solved by molecular replacement using the β-NRX1Δ4 structure as the search model. Manual rebuilding was done with COOT (Emsley and Cowtan, 2004), and refinement was performed using REFMAC (Murshudov et al., 1997) implemented in the CCP4 program suite (Collaborative Computational Project, 1994). The statistics of data collection and refinement are summarized in Table 2. All molecular graphics figures were generated with the program Pymol (DeLano Scientific, LLC).
Table 2.
Data collection and refinement statistics
| β-NRX1+4 | β-NRX2+4 | β–NRX3Δ4 | |
|---|---|---|---|
| Data Collection | |||
| Space group | C2 | C2221 | P212121 |
| a, b, c (Å) | 85.8, 59.8, 105.7 | 51.7, 102.1, 78.5 | 74.7, 83.4, 119.8 |
| α, β, γ(°) | 90, 103.9, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 2.7 (2.8-2.7) | 2.35(2.43-2.35) | 2.00 (2.07-2.00) |
| Rsym | 0.090(0.325) | 0.088 (0.258) | 0.120(0.628) |
| I/σI | 19.7 (2.2) | 28.8 (3.6) | 14.6(2.3) |
| Completeness (%) | 98.9 (93.4) | 99.6 (92.9) | 99.6(99.5) |
| Redundancy | 3.6(3.0) | 5.8(3.4) | 5.9(5.4) |
| Refinement | |||
| Resolution (Å) | 20-2.7 | 20-2.35 | 20-2.0 |
| Rwork/Rfree | 0.206/0.287 | 0.19/0.252 | 0.177/0.215 |
| B-factors (Å2) | |||
| All atoms | 35.8 | 34.9 | 30.6 |
| Protein | 35.7 | 34.6 | 28.7 |
| Ca2+ | N/A | 42.4 | 61.8 |
| Water | 37.0 | 37.7 | 37.0 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.008 | 0.013 | 0.009 |
| Bond angles (°) | 1.234 | 1.555 | 1.196 |
One crystal was used per dataset. Values in parentheses are for the highest resolution shell.
Rfree = Rwork calculated using 5% of the reflection data chosen randomly and omitted from the start of refinement.
DATA DEPOSITION
Coordinates have been deposited in the protein data bank with deposition IDs 3MW2 (β-NRX1+4), 3MW3 (β-NRX2+4), and 3MW4 (β-NRX3Δ4).
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant U54 CA121852 (LS. and B.H.) and by NSF grant MCB-0918535 (B.H.). B.H. is an investigator of the Howard Hughes Medical Institute. X-ray data were collected at the X4C beamline of the National Synchrotron Light Source, Brookhaven National Laboratory; the X4 beamlines are operated by the New York Structural Biology Center. We thank Tom Jessell and Peter Scheiffele for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arac D, Boucard AA, Ozkan E, Strop P, Newell E, Sudhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Bolliger MF, Frei K, Winterhalter KH, Gloor SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem J. 2001;356:581–588. doi: 10.1042/0264-6021:3560581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu H, Shim AH, Focia PJ, He X. Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions. Nat Struct Mol Biol. 2008;15:50–56. doi: 10.1038/nsmb1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Afridi SK, Clark L, Scheiffele P. Disorder-associated mutations lead to functional inactivation of neuroligins. Hum Mol Genet. 2004;13:1471–1477. doi: 10.1093/hmg/ddh158. [DOI] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Sudhof TC. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn R, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Sudhof TC, Taylor P. Characterization of the interaction of a recombinant soluble neuroligin-1 with neurexin-1beta. J Biol Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Cowan MW, Südhof TC, Stevens CF, editors. Synapses. The John Hopkins University Press; 2000. [Google Scholar]
- de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Neuroscience. Autism’s cause may reside in abnormalities at the synapse. Science. 2007;317:190–191. doi: 10.1126/science.317.5835.190. [DOI] [PubMed] [Google Scholar]
- Garcia J, Gerber SH, Sugita S, Sudhof TC, Rizo J. A conformational switch in the Piccolo C2A domain regulated by alternative splicing. Nat Struct Mol Biol. 2004;11:45–53. doi: 10.1038/nsmb707. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26:4256–4265. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proceedings of the National Academy of Sciences. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009a;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Zhang C, Arac D, Boucard AA, Brunger AT, Sudhof TC. Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 2009b;28:3244–3255. doi: 10.1038/emboj.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Jin X, Budreck EC, Posy S, Scheiffele P, Honig B, Shapiro L. Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2. Proc Natl Acad Sci U S A. 2008a;105:1873–1878. doi: 10.1073/pnas.0711701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Jin X, Trbovic N, Katsamba PS, Brasch J, Ahlsen G, Scheiffele P, Honig B, Palmer AG, 3rd, Shapiro L. Crystal structures of beta-neurexin 1 and beta-neurexin 2 ectodomains and dynamics of splice insertion sequence 4. Structure. 2008b;16:410–421. doi: 10.1016/j.str.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6137–6142. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner C, Klose M, Fairless R, Missler M. Mutational analysis of the neurexin/neuroligin complex reveals essential and regulatory components. Proc Natl Acad Sci U S A. 2008;105:15124–15129. doi: 10.1073/pnas.0801639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, Qin S, Minx P, Wilson RK, Hood L, Graveley BR. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79:587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J. The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J Biol Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KC, Kuczynska DA, Wu IJ, Murray BH, Sheckler LR, Rudenko G. Regulation of neurexin 1beta tertiary structure and ligand binding through alternative splicing. Structure. 2008;16:422–431. doi: 10.1016/j.str.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Hata Y, Ichtchenko K, Moomaw C, Afendis S, Slaughter CA, Sudhof TC. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;269:11987–11992. [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Sudhof TC. Neurexin III alpha: extensive alternative splicing generates membrane-bound and soluble forms. Proc Natl Acad Sci U S A. 1993;90:6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Willard FS, Siderovski DP. Covalent immobilization of histidine-tagged proteins for surface plasmon resonance. Analytical Biochemistry. 2006;353:147–149. doi: 10.1016/j.ab.2006.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.