Abstract
Objectives
We sought to construct and partially characterize complementary DNA (cDNA) libraries prepared from the middle ear mucosa (MEM) of chinchillas to better understand pathogenic aspects of infection and inflammation, particularly with respect to leukotriene biogenesis and response.
Methods
Chinchilla MEM was harvested from controls and after middle ear inoculation with nontypeable Haemophilus influenzae. RNA was extracted to generate cDNA libraries. Randomly selected clones were subjected to sequence analysis to characterize the libraries and to provide DNA sequence for phylogenetic analyses. Reverse transcription–polymerase chain reaction of the RNA pools was used to generate cDNA sequences corresponding to genes associated with leukotriene biosynthesis and metabolism.
Results
Sequence analysis of 921 randomly selected clones from the uninfected MEM cDNA library produced approximately 250,000 nucleotides of almost entirely novel sequence data. Searches of the GenBank database with the Basic Local Alignment Search Tool provided for identification of 515 unique genes expressed in the MEM and not previously described in chinchillas. In almost all cases, the chinchilla cDNA sequences displayed much greater homology to human or other primate genes than with rodent species. Genes associated with leukotriene metabolism were present in both normal and infected MEM.
Conclusions
Based on both phylogenetic comparisons and gene expression similarities with humans, chinchilla MEM appears to be an excellent model for the study of middle ear inflammation and infection. The higher degree of sequence similarity between chinchillas and humans compared to chinchillas and rodents was unexpected. The cDNA libraries from normal and infected chinchilla MEM will serve as useful molecular tools in the study of otitis media and should yield important information with respect to middle ear pathogenesis.
Keywords: cDNA library, chinchilla, leukotriene, molecular phylogeny, otitis media
INTRODUCTION
Infectious and inflammatory diseases of the middle ear, collectively referred to as otitis media (OM), represent one of the most common clinical entities requiring medical attention. Acute OM (AOM) is the most common diagnosis in pediatric patients who visit physicians for illness in the United States1 and is responsible for an estimated 5 million annual episodes in the United States alone, at a cost of $3 to $6 billion.2 Chronic OM with effusion (OME) is associated with AOM, is a leading cause of hearing loss in children, and can lead to speech, language, and developmental delays.
One of the most robust animal models for the study of middle ear disease has been the chinchilla. However, efforts to utilize this animal model have been hampered by the lack of molecular tools to carry out the needed investigations. A primary specific aim of this study was to begin to develop a more robust set of data to allow the development of further molecular tools to study AOM and chronic OME in this animal model. The experiments detailed in this manuscript represent the initial efforts of a long-term project to characterize middle ear mucosa (MEM) gene expression patterns in this animal model in both the normal and diseased states. A comprehensive gene expression profile is being created, along with gene sequencing, which will allow investigators to utilize this animal model to further investigate middle ear pathogenesis on a molecular level.
Leukotriene (LT)–mediated inflammatory pathways have been demonstrated to be important in the pathogenesis of AOM and OME.3,4 LTB4 has been linked to the development of OME. The cysteine LTs (CysLTs) LTC4, LTD4, and LTE4 regulate vascular permeability and promote mucosal edema — important events in AOM. These CysLTs have also been associated with regulation of mucin production and mucociliary transport. These crucial processes in the pathogenesis of both AOM and chronic OME have been shown in our laboratories to be fundamental in bacterial clearance and development of chronic middle ear diseases.5,6 As such, mediators important in the regulation of these specific LTs, which have been demonstrated in previous studies to influence OM pathogenesis, were a focus of this investigation. The production of LTs follows two divergent pathways. LTA4 can be converted to LTB4 or LTC4. LTA4 hydrolase regulates the production of LTA4, whereas LTC4 synthase regulates the production of LTC4. LTC4 is then further converted to LTD4 and finally LTE4. Each of these LTs also has epithelial receptors that, when bound by the specific LT, allow for the cellular response to the LT to take place. For the CysLTs these receptors are CysLT1R and CysLT2R, and for LTB4 the receptor is BLT1. A primary aim of this investigation was to assess the expression of the important LTs and their receptors in infected and uninfected chinchilla MEM, as well as the expression of enzymatic control in this pathway and the epithelial receptors that allow these LTs to elicit their cellular responses.
MATERIALS AND METHODS
Specimen Acquisition
The MEM tissue specimens used in this study were obtained from young adult (6 to 10 months old), mixed-breed chinchillas weighing between 400 and 600 g that were free of middle ear disease, obtained as culls from the fur industry (McClenahan Chinchilla Ranch, New Wilmington, Pennsylvania). A total of 12 chinchillas, 6 each for the 2 clinical conditions, were used to perform the experiments described.
Before chinchilla middle ear inoculation, or MEM harvest in the case of the uninfected cohort, all ears were examined by pneumatic otoscopy. Abnormal findings on evaluation would have required elimination of the animal from the study. One cohort of animals was used to harvest normal MEM, and the second cohort was infected with a low-passage clinical isolate (PittDD) of nontypeable Haemophilus influenzae (NTHi). The animals in the normal control group were evaluated and sacrificed immediately after their appropriate acclimation period. Before tissue harvest, chinchillas were anesthetized via intramuscular injection of 0.1 mL of a solution of ketamine hydrochloride 100 mg/mL, xylazine hydrochloride 30 mg/mL, and acepromazine maleate 5 mg/mL. Deep anesthesia was confirmed by the abolishment of the eye-blink reflex, followed by euthanasia via intracardiac injection of 2 mg pentobarbital sodium (Abbott Laboratories, North Chicago, Illinois) as approved by the Panel on Euthanasia of the American Veterinary Medical Association. The temporal bone, including the tympanic membrane and middle ear cavity, was removed on both sides. The tympanic membrane and middle ear cavity were examined closely to ensure that there was no evidence of inflammation or infection. The animals in the NTHi group were inoculated 3 days before MEM harvest. Anesthesia was induced as described above followed by bilateral transbullar injection of 0.1 mL of 105 colony-forming units per milliliter of NTHi suspension by means of a 0.5-inch, 27-gauge needle attached to a 1-mL syringe.
The pathogen PittDD is a low-passage, ampicillin-sensitive NTHi isolate that was obtained from a child with OME at Children's Hospital of Pittsburgh.7 PittDD was initially cultured on chocolate agar and then subcultured once in brain-heart infusion broth (Becton Dickinson, Sparks, Maryland) supplemented with 10 μg/mL hemin (Fisher Scientific, Pittsburgh, Pennsylvania), 2 μg/mL nicotinamide adenine dinucleotide (Sigma, St Louis, Missouri), and 20 μg/mL thiamine hydrochloride (Sigma) and grown at 37°C in a humidified 5% carbon dioxide atmosphere before the preparation of freezes. For all subsequent studies, one of the initial freezes was thawed and used for culture.
RNA Harvest
Total RNA was prepared from chinchilla MEM specimens that were collected in 500 μL RNAlater (Ambion, Inc, Austin, Texas) with the TRIzol reagent (Invitrogen, Inc, Carlsbad, California). The tissue samples were disrupted with Lysing Matrix D (Bio101, Qbiogene, Inc, Carlsbad, California) in a FastPrep automatic extractor (Bio101) at setting No. 6 for 25 seconds. The lysis matrix was removed by centrifugation, and the resulting TRIzol-containing samples were processed according to the manufacturer's instructions by the addition of 50 μL trichloromethane (chloroform) per 750 μL sample. The organic and aqueous phases were separated by centrifugation, the upper aqueous phase was saved, and the total RNA was recovered by 2-propanol precipitation. RNA quality utilizing the Agilent bioanalyzer with the RNA 6000nanochip (Santa Clara, California) was used to assess the 18s-to-28s ratio and indicated that the RNA was of high quality. DNA contamination was removed by DNAse treatment with Ambion Turbo DNAse. Purified RNA was stored at –80°C before use in complementary DNA (cDNA) library construction or reverse transcription–polymerase chain reaction (RT-PCR)–based assay.
Preparation of Double-Stranded cDNA and cDNA Library Construction
Double-stranded cDNA was synthesized from the DNAsed MEM RNA by means of the Super Smart PCR cDNA synthesis kit (Clontech, Mountain View, California) according to the manufacturer's recommendations. The PCR-amplified double-stranded cDNA was then blunt-end–repaired with T4 DNA polymerase and Klenow enzyme and dephosphorylated with calf intestinal alkaline phosphatase to make the DNA suitable for use in the Topo cloning system (Invitrogen). The prepared cDNA was then ligated into the pCR4-Blunt-Topo vector according to the manufacturer's recommendations. In test ligations, the proper vector/insert ratio was calibrated to achieve the highest possible ligation and transformation efficiencies. Transformed TOP10 Escherichia coli cells were then plated on LB agar containing 50 μg/mL kanamycin sulfate on 22 × 22 cm culture dishes and incubated for 16 hours at 37°C. For each library, clones were collected with the Q Bot, a 3-axis multitasking/multifunctional robot (Genetix, New Milton, England), into 384-well culture plates containing 8% glycerol and 100 μg/mL ampicillin in LB broth. The libraries then were replicated and stored at –80°C at different locations for safety. One of the replicas was designated as the working library and used for all further manipulations.
Characterization of Chinchilla cDNA MEM Libraries
Randomly selected clones from the normal chinchilla MEM cDNA library were characterized. For each clone analyzed, the recombinant plasmid was isolated from its respective E coli clone with the Macherey-Nagel NucleoSpin Robot 96 (Macherey-Nagel GmbH, Düren, Germany) plasmid preparation kit on a Biomek FX liquid-handling robot (Beckman, Fullerton, California). The isolated plasmid DNA was then digested with Eco R1 and subjected to electrophoresis through a 1.2% agarose slab gel to determine the insert size. To assess the complexity of the normal MEM chinchilla cDNA library, we sequenced the randomly selected clones from both forward and reverse directions using a pair of Beckman CEQ8000 automated capillary gel sequencers.
Creating Primers for LT Pathway Gene Expression
Because no chinchilla DNA sequence was known, to facilitate amplification of chinchilla transcripts corresponding to the genes associated with LT biosynthesis and response, primers were designed to conserved regions of the orthologous genes (identified by aligning the sequences from guinea pig, mouse, rat, and human). A series of forward and reverse primers were designed for the cysteine-associated LT-1 receptor gene (CysLT1R), the cysteine-associated LT-2 receptor gene (CysLT2R), the LTA4 hydrolase gene (LTA4H), the BLT1 receptor gene (BLT1R), and the LTC4 synthase gene (LTC4S; Table 1).
TABLE 1.
PRIMER SEQUENCES USED TO AMPLIFY AND CHARACTERIZE CHINCHILLA GENES IN LIPOXYGENASE PATHWAY
| Gene | Orientation | Sequence |
|---|---|---|
| CysLT2R | Reverse | GAG TGC CTG GAT CCT CTG TG |
| CysLT2R | Forward | GGT CAG TGC CTT CCT GTG AG |
| CysLT1R | Reverse-1 | TTC ATG CCA TAT CAT TAT TCA AC |
| CysLT1R | Forward | TGG ACA AAG AAT GCT TTC TAA |
| CysLT1R | Reverse-2 | GTA GGC TTC TTT GGC AAT GGC |
| LTA4 hydrolase | Reverse | ATG CCA GAG GTC GTG GAT ACC |
| LTA4 hydrolase | Forward | GCT CAA GGT GAA AGA GTA AAG |
| BLT1R | Reverse-1 | ATG CGT GCA AAC ACT ACA TCT |
| BLT1R | Reverse-2 | GCC TTC CTG AGC AGC AGC |
| BLT1R | Forward | GAC ACC TCC GAG CCA GTG |
| LTC4R synthase | Reverse | GCC RAC TTG TCC CTA GAG |
| LTC4R synthase | Forward | GGA GCC ATC TGA AGA GCG |
CysLT — cysteine leukotriene; LT — leukotriene.
Transcriptional Activity of Chinchilla LT Receptor Genes and Characterization of Transcript Sequence
RNA extracted from the MEM of both uninfected and NTHi-infected chinchillas were used in RT-PCR–based assays to assess transcriptional activity of the LT genes. It should be noted that the PCR products and the cDNA library were made from the same total RNA preparation from the same animals with different aliquots. After DNAse treatment, the chinchilla MEM total RNA specimens were tested for residue genomic DNA contamination by PCR, by use of primer sets for both the CysLT1R gene and for the 28th exon of the chinchilla's von Willebrand factor gene8 (GenBank accession number, AJ238385; forward TGGTAGTGCCCCCTACAGAA; reverse TGAGSTCAAGGTAAGCATGA). The latter served as a positive control. All RNA samples were established as being free of DNA. The DNA-free RNA was then converted to cDNA, which was then subjected to PCR. The amplification products were analyzed on a 1.5% aga-rose gel stained with ethidium bromide and imaged with a Kodak IS440 digital imaging station.
The nucleotide sequence of cDNA generated from the CysLT1R primers was determined on a Beckman CEQ8000 automated capillary DNA sequencer. The generated additional sequence data for the CysLT1R gene, the original 3' primer, and a newly designed 5' primer (GTAGGCTTCTTTGGCAATGGC) that was homologous to the putative translational start site were utilized. This amplification product was column-purified with the QIAquick Spin PCR purification kit (Qiagen, Inc, Valencia, California) and visualized on agarose gel with mass ladder standards (Invitrogen). The new nucleotide sequence was then determined on the automated capillary DNA sequencer.
The unweighted pair group method with arithmetic mean method of phylogenetic tree building, available in the Mega version 2.1 software package (Molecular Evolutionary Genetics Analysis, Tempe, Arizona), was used for all phylogenetic analyses. This program constructs a phylogenetic tree using a unitless relative difference scale calculated as a Kimura gamma distance using algorithms and formulas previously described.9
RESULTS
Characterization of Chinchilla MEM cDNA Libraries
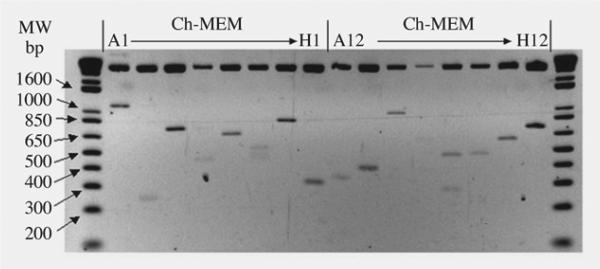
This partial characterization of the chinchilla MEM cDNA libraries revealed that more than 87% of randomly selected clones contained an insert with an average length of 680 ± 180 bp (Fig 1). The uninfected MEM chinchilla cDNA library contained 14,976 clones, of which 921 have been randomly selected for sequence analysis. Each clone produced average read lengths of at least 500 bases from both ends; thus, for longer clones, primer walking was used to close the gaps. The collected sequence information represents approximately 250,000 nucleotides and is available at the CGS website (www.centerforgenomicsciences.org) or as an electronic text file. These sequence data are almost entirely novel and represent the largest sequencing project to date for the chinchilla.
Fig 1.
Insert length of randomly selected clones from uninfected cDNA library. MW — molecular weight markers; Ch-MEM — chinchilla middle ear mucosa.
Homology searches were performed for all clones by use of various BLAST (Basic Local Alignment Search Tool) algorithms against the GenBank database. The proposed identities of the sequenced gene fragments are summarized in Table 2; only the species with the greatest homology to the chinchilla sequence is shown in each case. Interestingly, the chinchilla sequences consistently displayed their greatest homology to the primate gene, as opposed to the rodent orthologs. Homology was detected to numerous rodent species; however, the level of identity was almost always lower by several percentage points than that observed in the human homologs.
TABLE 2.
NUCLEOTIDE AND AMINO ACID HOMOLOGIES DISPLAYED BY SEQUENCED CHINCHILLA cDNA CLONES
| Clone | Homology | Species | % Identity (Nucleotide) | % Identity (Amino Acids) | % Similarity (Amino Acids) |
|---|---|---|---|---|---|
| Ch-MEM-N-lCll | Potassium channel protein, TWIK-2 | Guinea pig | 85 | ||
| Ch-MEM-N-1123 | Seven transmembrane helix receptor | Human | 91 | ||
| Ch-MEM-N-2A1 | Selenoprotein K | Human | 90 | 93 | 94 |
| Ch-MEM-N-2113 | Proto-oncogene Wnt-5A | Human | 91 | ||
| Ch-MEM-N-3H1 | Leukotriene A4 hydrolase | Guinea pig | 89 | 90 | 99 |
| Ch-MEM-N-4C2 | v-crk Sarcoma virus CT10 oncogene | Human | 96 | ||
| Ch-MEM-N-4D20 | Heparin-binding growth factor binding proteins | Human | 82 | ||
| Ch-MEM-N-5B9 | Nicotinamide adenine dinucleotide phosphate–dependent isocitrate dehydrogenase | Human | 83 | 70 | 84 |
| Ch-MEM-N-9A5 | Tymosin beta 4 | Human | 89 | 84 | 91 |
| Ch-MEM-N-13F1 | Melanoma metastasis clone D | Human | 82 | ||
| Ch-MEM-N-14A16 | SPARC-like 1 (mast9, hevin) | Human | 84 | ||
| Ch-MEM-N-18B6 | Spinal cord–derived growth factor β | Human | 82 | ||
| Ch-MEM-N-17F3 | Goliath protein | Human | 91 | 95 | 96 |
| Ch-MEM-N-19E10 | Matrix G1a protein | Human | 80 | ||
| Ch-MEM-N-18D3 | Laminin B2 chain | Rhesus monkey | 86 | ||
| Ch-MEM-N-11C19 | Plastin 3 (T isoform) | Human | 89 | 78 | 88 |
| Ch-MEM-N-12B16 | Lysosomal-associated protein transmembrane 4α | Human | 93 | 94 | 96 |
| Ch-MEM-N-15E21 | Ribosomal protein | Human | 91 | 92 | 96 |
| Ch-MEM-N-17K9 | Similar to CGI-45 protein | Human | 88 | ||
| Ch-MEM-N-15B3 | Lung and nasal epithelium carcinoma-associated protein | Human | 80 | ||
| Ch-MEM-N-13B19 | Sorting nexin16 | Human | 86 | 84 | 89 |
| Ch-MEM-N-16J24 | Chimerin 1 | Human | 83 | 87 | 92 |
The 921 clones sequenced represent 515 unique genes according to a BLAST search utilizing the top 2 hits from each sequence BLASTed. This finding would indicate that in this library, as a whole, there exist approximately 8,400 genes, suggesting that its complexity is quite high.
Transcriptional Activity of LT Receptor and Biogenesis Genes in Chinchilla MEM
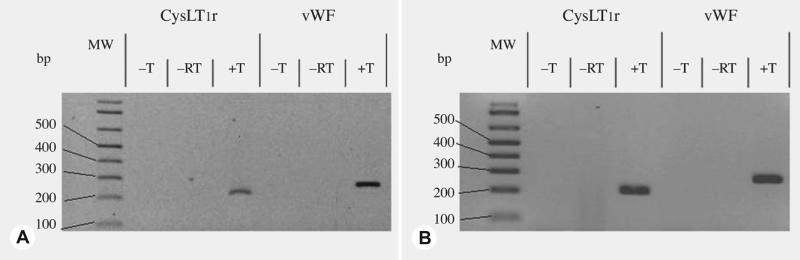
Primers were designed to support amplification of 1) the CysLT1R gene, which encodes a G protein–coupled receptor that binds the CysLTs (LTC4, LTD4, and LTE4) and is the target of the anti-inflammatory drug montelukast sodium; and 2) a control gene, von Willebrand factor. The primers were designed to yield RT-PCR amplicons using chinchilla messenger RNA (mRNA) of 211 and 266 bp, respectively. Figure 2 demonstrates the expected band for CysLT1R at 211 bp, indicating that the CysLT1R gene is transcriptionally active in uninfected and infected middle ear mucosa.
Fig 2.
Images of agarose gels (1.5%) stained with EtBr. Expression of cysteine leukotriene (CysLT1R) and von Willebrand factor (vWF) genes in A) normal chinchilla middle ear mucosa (MEM) and B) Haemophilus influenzae–infected MEM. –T — no template RNA/DNA added to polymerase chain reaction (PCR); –RT — PCR performed without prior reverse transcriptase step in presence of template RNA; +T — template RNA plus reverse transcription (RT) before PCR. +T reaction using CysLT1R-specific primers from both normal and infected MEM yield predicted 211 bp PCR product, and +T reactions for vWF gene–specific primers yielded expected 266 bp PCR product.
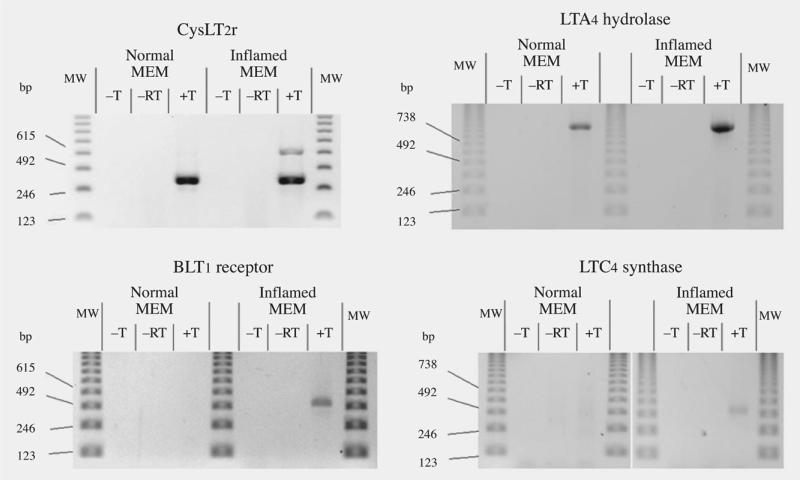
Expression of the CysLT2 receptor gene in the MEM was assessed by designing intron-spanning primers that were designed using sequence of the orthologous human gene. Amplification of a 283 bp fragment was evident from both normal and infected chinchilla MEM RNA (Fig 3). An additional band of approximately 650 bp was also observed from the NTHi-infected MEM. Using an identical experimental strategy, we demonstrated that the LTA4 hydrolase gene is also expressed in both normal and NTHi-infected chinchilla MEMs (Fig 3). The finding of expression of this important LT biosynthesis gene was corroborated by identification of a cDNA clone (Ch-MEM-N-3H1) containing this gene from our random screening of the normal MEM cDNA library.
Fig 3.
RT-PCR expression of CysLT2r, LTA4 hydrolase, BLT1R, and LTC4 synthase genes.
In contrast to the CysLT receptors, the gene for the BLT1 receptor is expressed only in the infected MEM (Fig 3) and not in normal mucosa. Similarly, the RT-PCR results show clearly that expression of the LTC4S gene is up-regulated in the infected MEM (Fig 3) compared to normal MEM.
LT Receptor and Hydrolase Gene Characterization in Chinchilla
After determining that the chinchilla genes were most highly homologous to their cognate primate genes, we designed primers based on human sequences to amplify an 841 bp fragment of the CysLT1R gene. Gel analysis of the RT-PCR product revealed an amplimer of the expected size. The sequence of this reverse transcript, detailed in Fig 4, is predicted to encode for a segment of the chinchilla CysLT1R that includes several of the extracellular domains, including a glycosylation site and several of the transmembrane domains. BLASTn and BLASTx homology searches of the GenBank nucleotide and protein databases revealed that the CysLT1R gene of the chinchilla shared 91% nucleo tide homology with human, 89% with mouse, and 88% with rat and pig, and shared 97% amino acid sequence identity with the human protein, but only 92% with the other species. Sequencing of the 283 bp RT-PCR product of the CysLT2R gene, amplified from normal MEM, yielded a sequence highly homologous to the corresponding human gene region (Fig 5).
Fig 4.
Partial cDNA sequence of CysLT1R gene (GenBank accession No. AY462138).
Fig 5.
Partial cDNA sequence of CysLT2R gene (GenBank accession No. AY462139).
The insert of clone Ch-MEM-N-3H1, randomly selected for sequence analysis from the normal chinchilla MEM cDNA library, was identified as an ortholog of the guinea pig's LTA4 hydrolase gene (Fig 6). The BLAST sequence analysis revealed that the gene fragment begins at the 3' (polyadenylated) end of LTA4 hydrolase mRNA and encompasses approximately 50% of the coding sequence of the mRNA.
Fig 6.
Complete cDNA sequence of chinchilla LTA4 hydrolase gene (GenBank accession No. AY462137).
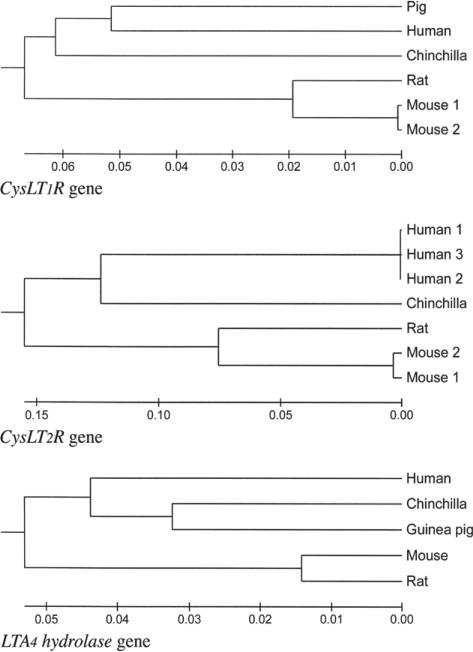
Phylogenetic tree building (Fig 7) for the LT-related genes, CysLT1R, CysLT2R, and LTA4 hydrolase, indicates that the chinchilla represents a very deep fork in the rodent lineage. Although data are available from only a limited number of rodent species, it is nevertheless clear that the chinchilla represents a more ancient lineage within the rodentia that has diverged less from the most recent common ancestor of primates and rodents than have other rodent species that are more commonly used as experimental models of human disease.
Fig 7.
Phylogenetic trees demonstrate that chinchilla CysLT1R, CysLT2R, and LTA4 hydrolase genes have generally greater similarity to humans than other rodents.
DISCUSSION
Chinchilla Model for Investigation of OM
The anatomic advantages of the chinchilla, including a large middle ear space and a readily visible tympanum, have made the chinchilla the most widely used animal model for the study of OM. In addition, the chinchilla demonstrates infectious susceptibility and inflammatory responses to most of the human middle ear pathogens. From a bacteriologic susceptibility standpoint, OM is readily induced in the chinchilla after a transbullar injection of NTHi or Streptococcus pneumoniae, but the chinchilla is not naturally susceptible to OM, unlike other rodent animal models such as the rabbit, the guinea pig,10 and the rat.11 The chinchilla model also has been extensively used for in vitro experimentation.5,6,12 The large middle ear space provides ample tissue for harvest and cell culture, and in vitro studies have demonstrated that the MEMs of the chinchilla and humans behave in a similar fashion with regard to inflammatory cytokine production and responses, mucin secretion, and mucin gene regulation.5,6,13 Thus, the chinchilla faithfully replicates numerous important anatomic and pathophysiologic characters of human OM.
The results of this investigation, examining the expression of elements of the LT pathways of the chinchilla MEM under naive and infected conditions, demonstrate that these key genes of the human inflammatory response are also expressed in the chinchilla and that infection with human middle ear pathogens can up-regulate expression and result in the production of additional transcripts from LT pathway genes. In addition, this data set provided a consistent observation across all genes examined, from both DNA homology and protein identity standpoints, is that the chinchilla is more closely related to primates than it is to the common experimental rodent models of the rat and mouse (Fig 7). Although debate exists regarding the evolution of rodents and the chinchilla with respect to primates, these findings corroborate earlier work from our laboratories examining mucin gene sequences, which also demonstrated greater homology between chinchilla and human sequences than between chinchilla and other rodent species.14 This finding of similarity of the chinchilla and human genetic sequences in this current work is important, because it provides evidence against what might have been an intuitive assumption that there would be significantly greater genetic differences between humans and chinchillas than between humans and other rodent models. These data, coupled with the historical experience in using the chinchilla model for OM research and the similarities in physiologic and pathophysiologic behavior of the chinchilla middle ear and its mucosa to that of humans, suggest that this model continues to have robust elements supporting its use in OM research. However, in conducting research that involves molecular techniques, these advantages have been somewhat offset by the lack of molecular reagents available for experimentation with this model. The creation of cDNA libraries, as outlined in this report, and gene sequencing from these libraries, which has begun in our laboratories (Table 2), will allow significant expansion of the molecular tools available for this model and further facilitate OM experimentation. This current investigation, although at present only a partial investigation of the cDNA libraries generated, produced a large volume of unique sequence data for the chinchilla, identifying 515 unique genes. After identification of genes with existing human homologs (Table 2), the focus of this investigation was narrowed to LTs, which are known to be important in OM. However, other genes identified in this project also play roles in human epithelial mucosa and will be the subject of future investigations.
LTs and OM
Leukotrienes have a variety of effects and have received recent attention as to their role in the pathogenesis of OM. Previous studies have shown high levels of LTB4 in bacteria- and virus-positive middle ear effusion obtained from children with AOM, and higher concentrations of LTB4 were associated with patients who then developed persistent and chronic OM.3 Similarly, the CysLTs are known to potentiate important effects on respiratory epithelial cells that are related to the pathogenesis of OM, including mucin production and mucociliary transport.4
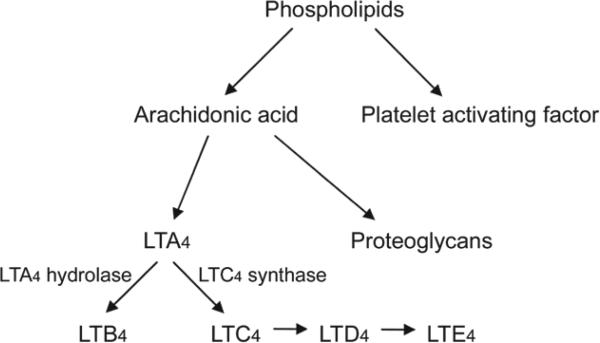
LTA4 hydrolase is a key enzyme in LTB4 production that branches away from the synthesis path leading to the production of the CysLTs (LTC4, LTD4, and LTE4; Fig 84,15). LTA4 can be processed to LTB4 by LTA4 hydrolase or to LTC4 by LTC4 synthase. LTB4 is the ligand of BLT1 receptors; LTC4 and its products are the ligands of the CysLT1 and CysLT2 receptors. The presence of the LTA4 hydrolase sequence in the cDNA library prepared from normal chinchilla MEM messenger RNA (mRNA) proves that the gene is actively transcribed in this tissue. Specific investigation of MEM using RTPCR also demonstrated LTA4 hydrolase gene expression in normal MEM and MEM infected with NTHi (Fig 3). These findings increase the likelihood that the production of LTB4 from LTA4 via the LTA4 hydrolase pathway is important in MEM and OM. Use of corticosteroids in AOM has been proposed as a potential adjuvant therapy to improve recovery because of the potential of steroids to nonspecifically block production of LTB4. In a recent trial, however, the use of corticosteroids did not demonstrate a reduction in LTB4 levels in middle ear fluid samples.4 The findings from the present study will allow for more specific investigations of this pathway in the chinchilla model, will provide means to investigate LTA4 hydrolase gene activity, and may provide for more specific regulation of this pathway in OM. The finding that gene expression of the LTB4 receptor, BLT1, is expressed only in the infected MEM and is not expressed in normal mucosa (Fig 3) also demonstrates the importance of increased understanding of these pathways on a molecular level, as the regulation of BLT1R expression may provide possibilities in modulating the effects of LTB4 in MEM.
Fig 8.
Leukocytes are known to express both CysLT1 and CysLT2 receptors. Middle ear mucosa leukocytes have been demonstrated in numerous histologic studies examining OM pathogenesis; however, in the normal, uninfected MEM epithelium, very few immunocytes are present.16 Therefore, it is likely that both receptor mRNAs are also expressed by epithelial cells and thus are potential targets for pharmacologic intervention in OM disease states. The epithelial expression of the CysLT2 receptor is also supported by the observation that LTC4 is a potent mitogen for airway epithelial cells, indicating the presence and involvement of this receptor type in the epithelium.17 Although the precise localization and definition of the cell type responsible for our observed expression of CysLT1 and CysLT2 receptors need to be further investigated with quantitative histologic experiments, and these experiments are planned, this data set clearly demonstrates that these receptors are clearly present in normal and infected MEMs of the chinchilla (Fig 3). These find ings support previous data demonstrating that CysLTs are important in the pathogenesis of OM.3
Our observation of the double RT-PCR band pattern from the infected MEM (Fig 3) indicates the possibility of alternative splicing of the CysLT2R gene products in the chinchilla. Interestingly, alternative splicing of the CysLT2R mRNA has been previously reported in an in vitro system in which a cloned mouse CysLT2R gene was expressed in HEK 293 cells.18 The tissue-specific distribution of the 2 spliceoforms is quite different19-22; however, it is apparent from our results that 2 transcripts are expressed in the infected chinchilla MEM. Alternative splicing may develop in the presence of inflammation as a means to regulate the LT pathway and control cellular uptake and could provide fertile ground for investigations focused on regulation of middle ear inflammation through this pathway.
Figure 3 clearly shows that the LTC4S gene is transcribed in the infected MEM; however, the result for the normal MEM was inconclusive, with a weak, diffuse band. This requires further study and quantification, as LTC4S and production of the CysLTs are known to be important in reactive airway disease, atopy, and chronic sinusitis,15,23 and have also been implicated in AOM.3 The presence of LTC4S from a leukocyte source was expected in infected MEM, given the recruitment of leukocytes that occurs in this process. However, the possibility of epithelial expression, versus low levels of expression from eosinophils or mast cells in the normal MEM, could have important implications for understanding both allergic and infectious ME pathophysiologies.
CONCLUSIONS
Characterization of cDNA libraries from naive and NTHi-infected chinchilla MEM is providing important molecular tools for investigating the pathophysiology of OM in an important animal model. The LT pathway has been identified as having an integral role in the development of recurrent OM and OME, and the findings from this study further support the importance of a more thorough understanding of the mechanisms by which LTs contribute to OM pathophysiology. The work reported herein demonstrates that the genes encoding the LT receptors and the LT biogenesis enzymes are expressed in the chinchilla MEM under normal and diseased states. This finding will provide a platform for further investigations into the infectious and inflammatory pathways of OM that are ongoing in our laboratories. The observation that chinchillas have a close evolutionary relationship to humans is an interesting finding from this work and warrants further investigation.
Acknowledgments
Supported by National Institutes of Health grants DC00192 and DC007903 (Kerschner), DC02148, DC04173, and HRSA (Ehrlich), and DC05659 (Post) from the National Institute on Deafness and Other Communication Disorders and by the Allegheny Singer Research Institute. This study was performed in accordance with the PHS Policy on Humane Care and Use of Laboratory Animals, the NIH Guide for the Care and Use of Laboratory Animals, and the Animal Welfare Act (7 U.S.C. et seq.); the animal use protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the Allegheny-Singer Research Institute.
REFERENCES
- 1.Bluestone CD, Klein JO. Otitis media, atelectasis, and eustachian tube dysfunction. In: Bluestone CD, Stool SE, Kenna MA, editors. Pediatric otolaryngology. 3rd ed Saunders; Philadelphia, Pa: 1996. pp. 388–582. [Google Scholar]
- 2.Rosenfeld RM, Casselbrant ML, Hannley MT. Implications of the AHRQ evidence report on acute otitis media. Otolaryngol Head Neck Surg. 2001;125:440–8. doi: 10.1067/mhn.2001.119326. [DOI] [PubMed] [Google Scholar]
- 3.Jung TT, Park SK, Rhee CK. Effect of inhibitors of leukotriene and/or platelet activating factor on killed H influenzae induced experimental otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2004;68:57–63. doi: 10.1016/j.ijporl.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 4.McCormick DP, Saeed K, Uchida T, et al. Middle ear fluid histamine and leukotriene B4 in acute otitis media: effect of antihistamine or corticosteroid treatment. Int J Pediatr Otorhinolaryngol. 2003;67:221–30. doi: 10.1016/s0165-5876(02)00372-5. [DOI] [PubMed] [Google Scholar]
- 5.Kerschner JE, Meyer TK, Burrows A. Chinchilla middle ear epithelial mucin gene expression in response to inflammatory cytokines. Arch Otolaryngol Head Neck Surg. 2004;130:1163–7. doi: 10.1001/archotol.130.10.1163. [DOI] [PubMed] [Google Scholar]
- 6.Kerschner JE, Meyer TK, Wohlfeill E. Middle ear epithelial mucin production in response to interleukin-1β exposure in vitro. Otolaryngol Head Neck Surg. 2003;129:128–35. doi: 10.1016/S0194-59980300532-1. [DOI] [PubMed] [Google Scholar]
- 7.Shen K, Antalis P, Gladitz J, et al. Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae. Infect Immun. 2005;73:3479–91. doi: 10.1128/IAI.73.6.3479-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huchon D, Catzeflis FM, Douzery EJ. Variance of molecular datings, evolution of rodents and the phylogenetic affinities between Ctenodactylidae and Hystricognathi. Proc Biol Sci. 2000;267:393–402. doi: 10.1098/rspb.2000.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nei M, Kumar S. Molecular evolution and phylogentics. Oxford University Press; New York, NY: 2000. p. 44. [Google Scholar]
- 10.Hejek DM, Yuan Z, Quartey MK, Giebink GS. Otitis media: the chinchilla model. In: Zak O, Sande MA, editors. Handbook of animal models of infection: experimental models in antimicrobial chemotherapy. Academic Press; San Diego, Calif: 1999. pp. 389–401. [Google Scholar]
- 11.Daniel HJ III, Fulghum RS, Brinn JE, Barrett KA. Comparative anatomy of eustachian tube and middle ear cavity in animal models for otitis media. Ann Otol Rhinol Laryngol. 1982;91:82–9. doi: 10.1177/000348948209100118. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura A, Kumazawa T, Lim DJ, Demaria TF, van Blitterswijk CA. Culture of middle ear epithelium: a review. Acta Otolaryngol Suppl. 1993;500:75–9. doi: 10.3109/00016489309126185. [DOI] [PubMed] [Google Scholar]
- 13.Samuel EA, Burrows A, Kerschner JE. Cytokine regulation of mucin secretion in a human middle ear epithelial model. Cytokine. 2008;41:38–43. doi: 10.1016/j.cyto.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerschner JE, Meyer TK, Burrows A, Ehrlich G, Post JC. Mucin gene cDNA sequence characterization in chinchilla middle ear epithelium. Int J Pediatr Otorhinolaryngol. 2006;70:1449–56. doi: 10.1016/j.ijporl.2006.03.007. [Erratum in Int J Pediatr Otorhinolaryngol 2006;70:2125-6.]
- 15.Pérez-Novo CA, Watelet JB, Claeys C, Van Cauwenberge P, Bachert C. Prostaglandin, leukotriene, and lipoxin balance in chronic rhinosinusitis with and without nasal polyposis. J Allergy Clin Immunol. 2005;115:1189–96. doi: 10.1016/j.jaci.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 16.Ichimiya I, Kawauchi H, Mogi G. Analysis of immunocompetent cells in the middle ear mucosa. Arch Otolaryngol Head Neck Surg. 1990;116:324–30. doi: 10.1001/archotol.1990.01870030088015. [DOI] [PubMed] [Google Scholar]
- 17.Leikauf GD, Claesson HE, Doupnik CA, Hybbinette S, Grafström RC. Cysteinyl leukotrienes enhance growth of human airway epithelial cells. Am J Physiol. 1990;259:L255–L261. doi: 10.1152/ajplung.1990.259.4.L255. [DOI] [PubMed] [Google Scholar]
- 18.Hui Y, Yang G, Galczenski H, et al. The murine cysteinyl leukotriene 2 (CysLT2) receptor. cDNA and genomic cloning, alternative splicing, and in vitro characterization. J Biol Chem. 2001;276:47489–95. doi: 10.1074/jbc.M107556200. [DOI] [PubMed] [Google Scholar]
- 19.Heise CE, O'Dowd BF, Figueroa DJ, et al. Characterization of the human cysteinyl leukotriene 2 receptor. J Biol Chem. 2000;275:30531–6. doi: 10.1074/jbc.M003490200. [DOI] [PubMed] [Google Scholar]
- 20.Ogasawara H, Ishii S, Yokomizo T, et al. Characterization of mouse cysteinyl leukotriene receptors mCysLT1 and mCysLT2: differential pharmacological properties and tissue dis tribution. J Biol Chem. 2002;277:18763–8. doi: 10.1074/jbc.M109447200. [DOI] [PubMed] [Google Scholar]
- 21.Sarau HM, Ames RS, Chambers J, et al. Identification, molecular cloning, expression, and characterization of a cysteinyl leukotriene receptor. Mol Pharmacol. 1999;56:657–63. doi: 10.1124/mol.56.3.657. [DOI] [PubMed] [Google Scholar]
- 22.Takasaki J, Kamohara M, Matsumoto M, et al. The molecular characterization and tissue distribution of the human cys teinyl leukotriene CysLT(2) receptor. Biochem Biophys Res Commun. 2000;274:316–22. doi: 10.1006/bbrc.2000.3140. [DOI] [PubMed] [Google Scholar]
- 23.Moissidis I, Chinoy B, Yanamandra K, et al. Association of IL-13, RANTES, and leukotriene C4 synthase gene promoter polymorphisms with asthma and/or atopy in African Americans. Genet Med. 2005;7:406–10. doi: 10.1097/01.gim.0000170994.24960.48. [DOI] [PubMed] [Google Scholar]