Gametophytes of the fern Pteris vittata can accumulate and tolerate more than 1% of their dry weight as arsenic. The authors provide evidence that the ACR3 arsenic transporter protein plays an important role in tolerance to high levels of arsenic by transporting arsenic into the vacuole.
Abstract
The fern Pteris vittata tolerates and hyperaccumulates exceptionally high levels of the toxic metalloid arsenic, and this trait appears unique to the Pteridaceae. Once taken up by the root, arsenate is reduced to arsenite as it is transported to the lamina of the frond, where it is stored in cells as free arsenite. Here, we describe the isolation and characterization of two P. vittata genes, ACR3 and ACR3;1, which encode proteins similar to the ACR3 arsenite effluxer of yeast. Pv ACR3 is able to rescue the arsenic-sensitive phenotypes of yeast deficient for ACR3. ACR3 transcripts are upregulated by arsenic in sporophyte roots and gametophytes, tissues that directly contact soil, whereas ACR3;1 expression is unaffected by arsenic. Knocking down the expression of ACR3, but not ACR3;1, in the gametophyte results in an arsenite-sensitive phenotype, indicating that ACR3 plays a necessary role in arsenic tolerance in the gametophyte. We show that ACR3 localizes to the vacuolar membrane in gametophytes, indicating that it likely effluxes arsenite into the vacuole for sequestration. Whereas single-copy ACR3 genes are present in moss, lycophytes, other ferns, and gymnosperms, none are present in angiosperms. The duplication of ACR3 in P. vittata and the loss of ACR3 in angiosperms may explain arsenic tolerance in this unusual group of ferns while precluding the same trait in angiosperms.
INTRODUCTION
Arsenic is a naturally occurring metalloid that is toxic to most organisms. It is found in soils and ground water in many regions of the world, including the United States (reviewed in Nordstrom, 2002). Chronic exposure to arsenic ingested from drinking water may lead to lung, bladder, and kidney cancers in humans (reviewed in Smith and Hira-Smith, 2004; Rahman et al., 2009). While arsenic contamination is a significant environmental problem in many regions of the world, arsenic also has beneficial uses in medicine, especially in the treatment of acute leukemia and parasitic diseases (Hermine et al., 2004; Lallemand-Breitenbach et al., 2008; Tatham et al., 2008).
The fern Pteris vittata, along with other members of the Pteridaceae, are unusual because they tolerate and hyperaccumulate extremely high levels of arsenic (Ma et al., 2001; Francesconi et al., 2002; Visoottiviseth et al., 2002; Zhao et al., 2002; Raab et al., 2004; Srivastava et al., 2006; Wang et al., 2006, 2007), with arsenic making up >1% of the dry weight of a P. vittata frond. Given its unique ability to tolerate and hyperaccumulate high levels of arsenic, P. vittata is an excellent system for studying the mechanisms responsible for arsenic metabolism, toxicity, and resistance in a multicellular organism and thus offers potential benefits for both medicine and the remediation of arsenic-contaminated soils (Kertulis-Tartar et al., 2006; Gonzaga et al., 2008; Shelmerdine et al., 2009).
Arsenic exists in two main forms in the environment, as arsenate (AsV) or arsenite (AsIII), depending on the redox potential of the environment (Cullen and Reimer, 1989). Arsenate, a phosphate analog, is the predominant form of arsenic in aerated soils. When P. vittata is grown in soils containing arsenate, arsenic is taken up by the root and rapidly transported to the frond (Chen et al., 2005), with up to 25 times more arsenic accumulating in the fronds than in the roots (Tu and Ma, 2005). Within the frond, arsenic is localized to upper and lower epidermal cells and trichomes, where it is likely stored in the vacuoles of these cells (Lombi et al., 2002; Li et al., 2005). Whereas 62.5% of the arsenic in the root is arsenate when plants are grown in the presence of arsenate (Su et al., 2008), >95% of the arsenic transported in the xylem sap is arsenite (Su et al., 2008), and arsenic is stored as arsenite in the fronds (reviewed in Zhao et al., 2008). This suggests that the roots are the main site of arsenate reduction to arsenite and that arsenite is preferentially loaded into the xylem.
A minor portion (<5%) of AsIII is coordinated by thiol groups in P. vittata (Lombi et al., 2002; Wang et al., 2002; Zhang et al., 2002; Webb et al., 2003; Zhao et al., 2003; Pickering et al., 2006; Singh and Ma, 2006), and this is limited to a cylindrical sheath, 40- to 50-μm thick, immediately surrounding the arsenic observed in the veins of the vascular tissue of the fronds (Pickering et al., 2006). However, this coordination is lost as the arsenic is stored in the fronds, primarily as arsenite. The accumulation of free arsenite in the P. vittata frond contrasts with that observed in other plants that are able to resist only low levels of arsenic, such as Brassica juncea and Arabidopsis thaliana. In these species, arsenate taken up by the root is reduced to arsenite and permanently coordinated to the thiolate ligands of phytochelatins (glutathione oligomers), where it remains sequestered in the root (Ha et al., 1999; Pickering et al., 2000; Dhankher et al., 2002). These differences in the chemical speciation and distribution of arsenic in P. vittata and other angiosperms suggest that P. vittata and its relatives have evolved a mechanism for tolerating, transporting, and accumulating arsenic that is unique in plants.
P. vittata, like other homosporous ferns, releases millions of haploid spores that generate free-living, autotrophic, haploid gametophytes capable of producing both eggs and motile sperm. Each gametophyte is small (<3 mm), lacks vascular tissue, and naturally grows on the surface of soils, yet the gametophytes can also be cultured aseptically under well-defined and controlled conditions (Gumaelius et al., 2004). These properties make the P. vittata gametophytes particularly useful as an experimental system for the study of arsenic hyperaccumulation. Like the sporophyte, arsenate-grown P. vittata gametophytes tolerate high levels of arsenic (30 mM arsenate) and accumulate 2 to 3% of their dry weight as arsenic (Gumaelius et al., 2004), with 95% in the form of arsenite (Ellis et al., 2006). Similar to what is observed in the bulk of the sporophyte tissues, thiol coordination of arsenite is not observed in the gametophyte. Furthermore, high resolution x-ray absorption spectroscopy in vivo imaging suggests that arsenite is localized primarily to the vacuoles of gametophyte cells, although some regions of the gametophyte show no significant arsenite accumulation, including the rhizoids and the base of the prothallus where rhizoids arise (Pickering et al., 2006).
Several genes involved in arsenic metabolism have been characterized in angiosperms and in rice (Oryza sativa) in particular, as rice grain can accumulate arsenic at concentrations that could potentially harm humans (Williams et al., 2007; Zhu et al., 2008). Arsenate is taken up by phosphate transporters in the root (Meharg and Macnair, 1992; Meharg and Hartley-Whitaker, 2002; Quaghebeur and Rengel, 2004; Catarecha et al., 2007) and then rapidly reduced to arsenite. This reduction is, at least in part, facilitated by a dual function CDC25 phosphatase/ACR2 arsenate reductase in Arabidopsis (Landrieu et al., 2004a, 2004b; Sorrell et al., 2004; Bleeker et al., 2006; Dhankher et al., 2006). CDC25-like arsenate reductases have also been characterized in rice (Duan et al., 2007), velvetgrass (Holcus lanatus), and P. vittata (Bleeker et al., 2006; Ellis et al., 2006). However, the P. vittata enzyme lacks the phosphatase activity found in the Arabidopsis, rice, and H. lanatus enzyme.
In arsenic nonhyperaccumulators, AsIII complexes with phytochelatins (AsIII-PC) (Pickering et al., 2000; Raab et al., 2004, 2005), which is ultimately sequestered in the root, likely in the vacuole. The proteins that transport AsIII-PC complexes across the tonoplast into the vacuole are unknown, although transport may be similar to that observed for Cd-phytochelatin complexes (Salt and Rauser, 1995). Whereas arsenate is the predominant chemical species of arsenic in soils, the predominant species of arsenic in aquatic environments (such as flooded rice paddies) is arsenite (Marin et al., 2003; Xu et al., 2008), a small neutral molecule that requires a different mechanism of uptake in the plant. Several recent studies in rice and Arabidopsis (reviewed in Zhao et al., 2008) provide evidence that arsenite is taken up in these plants by Nodulin26-like Intrinsic Proteins (aquaglyceroporins) (Ma et al., 2008; Kamiya et al., 2009). In rice, arsenite is then exported to the xylem by the silicon/arsenite efflux carrier Lsi2 (Ma et al., 2008).
Whereas rice grains accumulate arsenic to levels of 0.60 μg/g (Williams et al., 2007), the amount of arsenic that accumulates in P. vittata fronds is at least three orders of magnitude higher than the level in rice grain, suggesting that the mechanism of arsenic hyperaccumlation in P. vittata is likely to involve novel or additional arsenic transporters. To identify such proteins, we screened for P. vittata genes capable of complementing yeast (Saccharomyces cerevisiae) mutants deficient in the function of the arsenite effluxer Arsenical Compound Resistance3 (ACR3) gene. In yeast, ACR3 encodes an arsenite transporter that exports arsenite out of the cell into the surrounding medium (Wysocki et al., 1997; Ghosh et al., 1999), thus allowing the yeast cells to tolerate low levels of arsenic. A cDNA library was generated from arsenate-grown gametophytes (Ellis et al., 2006), and a cDNA that suppresses the arsenite sensitive phenotype of a yeast strain deficient for ACR3 was identified. Here, we report the characterization of this gene and the role it plays in arsenic tolerance and hyperaccumulation in this fern.
RESULTS
Isolation of the P. vittata ACR3 Genes
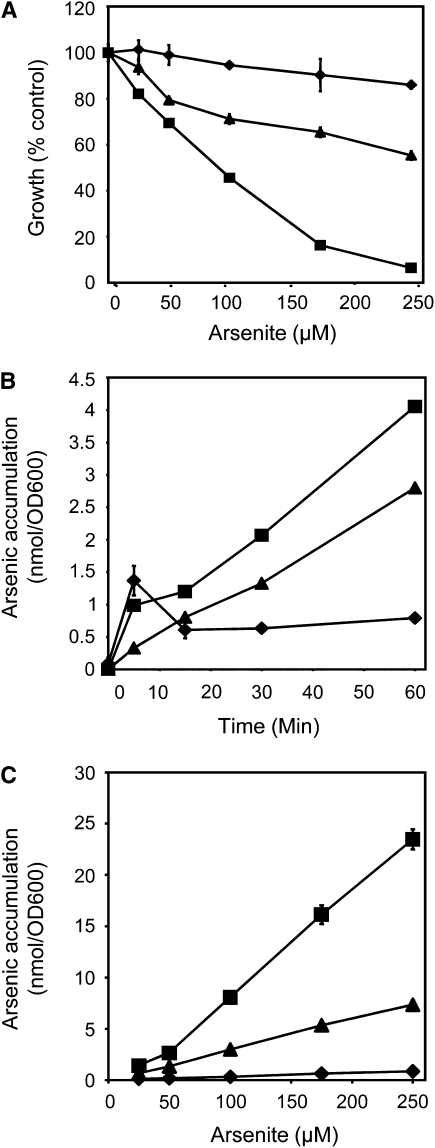
To identify a P. vittata protein that potentially functions as an arsenite efflux transporter, the Δacr3 Δycf1 arsenite-sensitive yeast strain MG102 (Ghosh et al., 1999) was transformed with a cDNA expression library generated from arsenate-grown gametophytes (Ellis et al., 2006). A transformed yeast colony that was able to grow in the presence of 10 μM arsenite, a concentration of arsenic that is lethal to MG102 yeast, was selected and characterized further. The transforming plasmid partially suppresses the arsenite-sensitive phenotype of yeast strain MG100, which is deleted for only ACR3 (Ghosh et al., 1999) when grown in varying concentrations of arsenite in liquid culture (Figure 1A). The same plasmid also partially suppresses the arsenic hyperaccumulation phenotype of Δacr3 yeast over short time periods (Figure 1B) and in a dose-dependent manner (Figure 1C). Together, these results indicate that the protein encoded by this cDNA has a similar function as the yeast ACR3 protein.
Figure 1.
ACR3 Partially Rescues the Arsenic-Sensitive and Hyperaccumulation Phenotypes of Δacr3 Yeast.
(A) Growth curves of the wild type (diamonds), Δacr3 (squares), and Δacr3 expressing ACR3t (triangles) yeast grown in liquid cultures containing varying concentrations of arsenite.
(B) Arsenic accumulation in the wild type (diamonds), Δacr3 (squares), and Δacr3 expressing ACR3t (triangles) yeast cultured in 10 μM arsenite over time.
(C) Arsenic accumulation in the wild type (diamonds), Δacr3 (squares), and Δacr3 expressing ACR3t (triangles) in varying concentrations of arsenite.
All assays were done in triplicate. Data represent means, and error bars represent sd.
The P. vittata cDNA encodes a protein similar in size and sequence to yeast ACR3 but appeared to lack 5′ DNA sequences representing the N-terminal end of ACR3. Additional 5′ sequences were sought and obtained by 5′ rapid amplification of cDNA ends. A second P. vittata gene also was identified from a small collection of P. vittata ESTs based on sequence similarity to the P. vittata ACR3 gene. The full-length P. vittata gene is referred to here as ACR3, the original truncated form as ACR3t, and the second P. vittata ACR3 gene as ACR3;1. ACR3 and ACR3t were both able to suppress the arsenite-sensitive phenotype of Δacr3 yeast (see Supplemental Figure 1 online). We were unable to test whether Pv ACR3;1 could suppress the arsenite-sensitive phenotype of Δacr3 yeast since we were unable to select MG100 yeast transformed with the purified Pv ACR3;1 plasmid after several attempts.
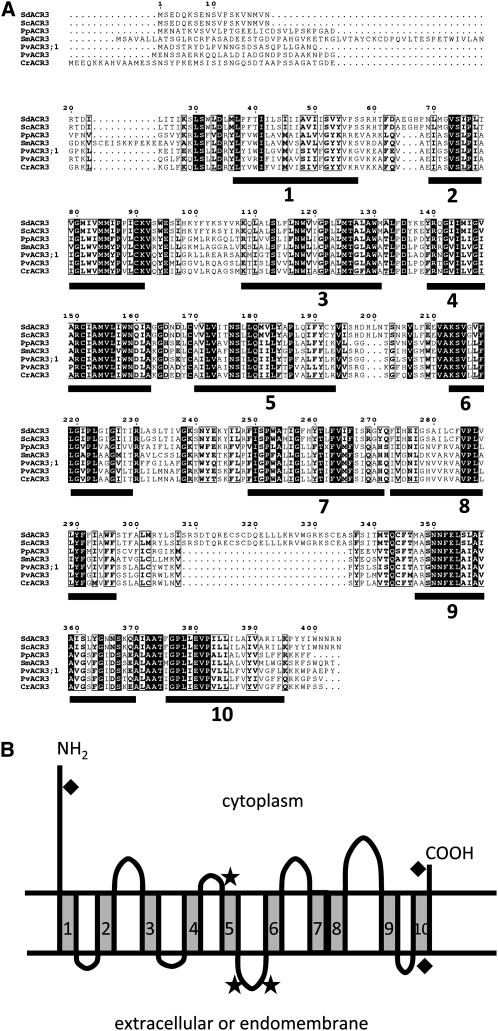
The Structure of the Fern and Yeast ACR3 Proteins
The ACR3 genes encode proteins that are similar to other ACR3 proteins both in sequence and in size (Figure 2A). Like the yeast ACR3 protein, the Pv ACR3 proteins have 10 predicted transmembrane domains (Figure 2A), as determined by the membrane topology prediction software SOUSI (Hirokawa et al., 1998; Mitaku and Hirokawa, 1999; Mitaku et al., 2002), HMMTOP (Tusnady and Simon, 1998, 2001) and TMHMM (Sonnhammer et al., 1998). Based on predictions by HMMTOP and TMHMM, the N and C termini of both Pv ACR3 proteins are localized to the cytoplasm, similar to the yeast ACR3 protein (Wysocki et al., 1997). WoLF PSORT (Horton et al., 2007), a protein localization predictor, predicted that the P. vittata ACR3 proteins are equally likely to be localized to the vacuolar and plasma membranes.
Figure 2.
ACR3 Proteins Are Conserved Membrane Proteins.
(A) Alignment of ACR3 proteins from S. cerevisiae (Sc), S. douglasii (Sd), P. patens (Pp), S. moellendorffii (Sm), P. vittata (Pv), and C. richardii (Cr). White letters indicate residues identical in all six proteins, and bold letters represent amino acids with similar chemical properties. The 10 putative transmembrane domains are indicated by black bars.
(B) The predicted topology of Pv ACR3. Putative phosphorylation site (stars) and O-glycoslyation sites (diamonds) are indicated.
The greatest differences among the ACR3 proteins reside within their N-terminal cytoplasmic domains, which vary in length from 27 to 81 amino acids. The putative transmembrane domains of all ACR3 proteins are similar in size, as are all but one of the loops, the exception being the loop between transmembrane domains 8 and 9. This loop in yeast is more than twice the size of the P. vittata ACR3 loops at the same position (Figure 2A). Two of the eight putative loops of the P. vittata ACR3 proteins have three predicted phosphorylation sites (Figure 2B) that are also predicted in the yeast ACR3 protein. Three predicted O-glycosylation sites are present in the Pv ACR3 proteins; two of them are also predicted in the yeast ACR3 protein (Hirokawa et al., 1998) (Figure 2B). While the Pv ACR3 and ACR3;1 proteins are 84% similar at the amino acid level, their transmembrane domains are highly conserved (85% similarity), whereas their N-terminal ends are not (22% similarity).
The ACR3 Proteins of Fungi and Plants
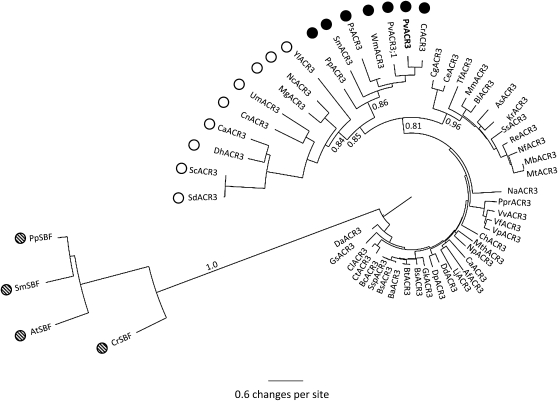
Genes similar to ACR3 were sought from GenBank using BLAST tools (Altschul et al., 1990). Single ACR3 genes were found in bacteria, fungi, in the whole-genome sequences of the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii, and in EST collections from the fern Ceratopteris richardii and the gymnosperms Welwitschia milabilis and Picea sitchensis. Interestingly, ACR3 genes were not found in angiosperms, including Arabidopsis, rice, black cottonwood (Populus trichocarpa), maize (Zea mays), grape (Vitis vinifera), and Sorghum bicolor (sorghum for which whole-genome sequences exist. In Arabidopsis, the protein most similar protein to ACR3 is a bile acid:sodium symporter family protein (AT1G78560), with an E value of 0.59 in a BLAST search. Apparent orthologs of this bile acid:sodium symporter protein were found in P. patens and S. moellendorffii as well as other angiosperms. A phylogenetic tree of ACR3 and bile acid:sodium symporter proteins from many organisms was generated (Figure 3; see Supplemental Figure 2 and Supplemental Data Set 1 online). The fungal and nonangiosperm plant ACR3 proteins form a clade that is split into fungal and nonangiosperm plant subclades. This clade is also distinct from the land plant bile acid:sodium symporter clade proteins, which form a strongly supported clade. The absence of ACR3 genes in monocots and dicots and their presence in P. sitchensis and W. mirabilis in the nonangiosperm ACR3 subclade suggests that ACR3 was lost in the angiosperms shortly after diverging from the gymnosperms. Within the nonangiosperm ACR3 subclade, the Pv ACR3 protein is more closely related to ACR3 from C. richardii, which is also a member of the Pteridaceae, than it is to ACR3;1 (Figure 3). To date, P. vittata is the only known plant with two ACR3 genes; only single ACR3 genes were found in all other nonangiosperm species. This suggests that a duplication of the ACR3 gene is likely to have occurred within the fern lineage.
Figure 3.
Neighbor-Joining Phylogenetic Tree of ACR3 Proteins and Sodium Bile Ttransporter Proteins.
Plant ACR3 proteins are indicated by black circles, fungal ACR3 proteins by open circles, bacterial ACR3 proteins with no circles, and plant sodium bile proteins by hatched circles. Bootstrap values are based on 10,000 bootstrap replications. Key bootstrap values are displayed on the branches. Supplemental Figure 2 online contains the alignments used to generate this phylogenetic tree.
The Expression of ACR3 and ACR3;1 Genes in P. vittata Gametophytes and Sporophytes
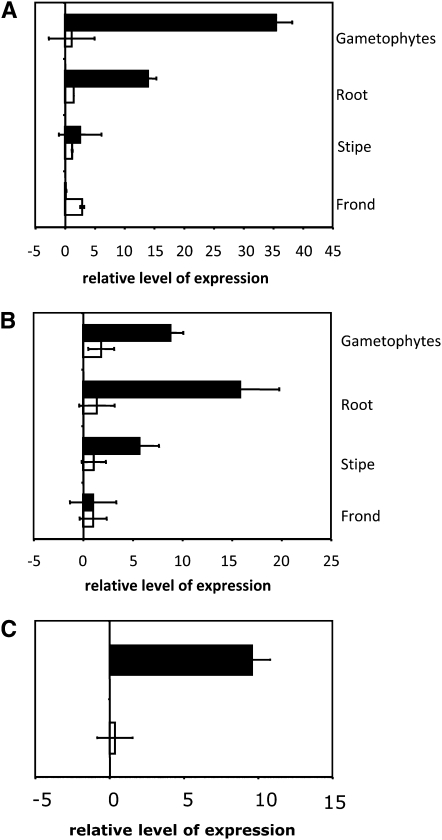
The effects of arsenic on the expression of the ACR3 and ACR3;1 genes in P. vittata gametophytes were assessed by quantitative RT-PCR (qRT-PCR). Gametophytes grown in media containing 10 mM arsenate accumulated ∼35 times more ACR3 transcripts than gametophytes cultured in the absence of arsenate (Figure 4A). The accumulation of ACR3;1 transcripts was comparable in gametophytes grown in either the absence or presence of arsenate (Figure 4A), demonstrating that the steady state levels of ACR3 transcripts, but not those of ACR3;1, are upregulated in the gametophyte by arsenate. Arsenite also upregulates ACR3, but not ACR3;1, expression in the gametophyte, with arsenite-grown gametophytes accumulating ∼10 times more ACR3 transcripts than nonarsenite-grown gametophytes (Figure 4C).
Figure 4.
Expression of ACR3 and ACR3;1 in P. vittata Gametophytes and Sporophyte Tissues as Determined by qRT-PCR.
(A) Each bar indicates the fold change in the expression of ACR3 (black bars) or ACR3;1 (white bars) in various tissues of plants grown in 10 mM arsenate relative to plants grown in the absence of arsenic.
(B) The difference in expression of ACR3 (black bars) or ACR3;1 (white bars) in various tissues of plants grown in the presence of 10 mM arsenate relative to their expression in the fronds of arsenate-grown plants.
(C) The fold change in the expression of ACR3 (black bars) or ACR3;1 (white bars) in gametophytes grown in 0.5 mM arsenite relative to gametophytes grown in the absence of arsenite.
For (A) to (C), three technical replicates of three biological replicates per treatment were performed. Error bars represent the range around the mean calculated as a composite sd from the P. vittata histone gene and ACR3.
The change in expression of ACR3 in P. vittata sporophytes in response to arsenate was also examined by qRT-PCR. Roots, stipes, and fronds from plants grown hydroponically for 7 d in the presence or absence of 10 mM arsenate were sampled. Roots of plants exposed to arsenate accumulated ∼12 times more ACR3 transcripts than roots of plants grown in the absence of arsenate (Figure 4A). The accumulation of ACR3 transcripts in the stipes and fronds of the same plants was much less affected or unaffected by arsenic (Figure 4A). Levels of ACR3;1 transcripts, by contrast, showed only small increases in response to arsenate in all sporophyte tissues examined (Figure 4A).
To compare the relative levels of ACR3 and ACR3;1 transcripts in different tissues of arsenate-exposed plants, the ΔCT value obtained from each tissue was divided by the value obtained from the tissue having the lowest ΔCT values (i.e., frond tissue). Arsenate-grown gametophytes and sporophyte roots accumulated 9- and 15-fold more ACR3 transcripts than frond tissue, respectively (Figure 4B). While arsenate has only a small effect on ACR3 transcript abundance in the stipe (Figure 4A), the levels of this transcript were ∼5 times higher in the stipe than the fronds of arsenate exposed plants (Figure 4B). The levels of ACR3;1 transcripts, on the other hand, were similar in all tissues (Figure 4). These results reveal distinct tissue-specific and arsenate-responsive differences in the accumulation of ACR3 transcripts, whereas the abundance of ACR3;1 transcripts varied little (less than threefold).
Localization of the ACR3 Protein in Gametophytes
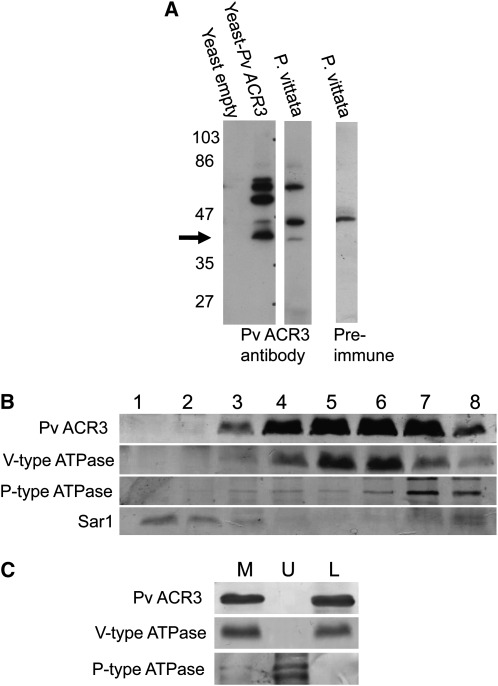
To determine the subcellular localization of ACR3, a peptide antibody corresponding to a cytoplasmic loop between amino acid residues of 200 and 213 (LKVVSRGKGFHVSS) was generated. The specificity of the antibody to ACR3 was determined by probing total microsomal proteins from yeast expressing ACR3 and yeast transformed with empty vector. As shown in Figure 5A, the antibody detected several proteins in Pv ACR3–expressing yeast, including a 37-kD protein (the expected size of Pv ACR3), and none in the empty vector–transformed yeast, indicating that the antibody has reactivity against Pv ACR3 (Figure 5A). The same antibody detected several proteins in P. vittata gametophytes, including a 37-kD protein, a 47-kD protein, and a 75-kD protein (Figure 5A). Preimmune serum also detected a 47-kD protein (Figure 5A), indicating that the 47-kD protein is not ACR3. As the size of the 75-kD protein is approximately twice the size of the ACR3 proteins, this band could represent a ACR3 protein dimer. Given the similarity in amino acid sequence and size, we also cannot rule out the possibility that the ACR3 antibody also cross-reacts with ACR3;1.
Figure 5.
Immunodetection and Localization of ACR3 in P. vittata Gametophytes.
(A) Protein immunoblot of total microsomal extracts from yeast and P. vittata gametophytes were probed either with the Pv ACR3 peptide antibody or with preimmune serum from the rabbit used to generate the Pv ACR3 antibody. All lines were loaded with equal amounts of protein. Arrow represents the expected molecular weight of Pv ACR3.
(B) Protein immunoblot following sucrose step gradient fractionation of microsomal membranes isolated from P. vittata gametophytes. Probes used were as follows: the ACR3 antibody (37 kD); tonoplast marker, V-type ATPase (27 kD); plasma membrane marker, P-type ATPase (100 kD); and endoplasmic reticulum marker, Sar1 (21 kD). Fractions from 1 to 8 correspond to 13/22%, 22%, 22/32%, 32%, 32/38%, 38%, 38/45%, and 45% sucrose, respectively. All lanes were loaded with equal amounts of protein.
(C) Protein immunoblot of aqueous two-phase partitioning of P. vittata gametophyte microsomal membranes into upper (U) and lower (L) phases. Probes used were as follows: the Pv ACR3 antibody (37 kD); tonoplast marker, V-type ATPase (27 kD); and plasma membrane marker, P-type ATPase (100 kD). M represents total microsomal membranes and all lanes were loaded with equal amounts of protein.
To determine where ACR3 localizes in the cell, total microsomal membranes from gametophytes were fractionated on a step sucrose gradient (22, 32, 38, and 45%) and fractions examined by protein immunoblotting for the presence of ACR3 and specific membrane marker proteins. Blots were probed with the ACR3 antibody and with antibodies specific to proteins in three subcellular compartments: the plasma membrane marker H+ATPase (95 kD), the endoplasmic reticulum marker Sar1 (21 kD), and the vacuolar membrane marker V-ATPase (27 kD). The results of these blots, shown in Figure 5B, demonstrate that membranes containing ACR3 copurify with vacuolar membranes containing the V-ATPase.
To further establish that ACR3 is not localized at the plasma membrane, we generated a highly purified preparation of plasma membranes from P. vittata gametophytes using an aqueous two-phase system as described in Methods. In this purification system, plasma membranes are recovered in the upper phase and all other membranes are recovered in the lower phase. We examined by protein immunoblot the upper and lower phases and the total microsomal membrane preparation used for the two-phase separation for the presence of ACR3, the P-type ATPase, and the V-type ATPase (Figure 5C). The results of this immunoblot confirmed that ACR3 does not occur in the plasma membrane. We conclude from these results that the ACR3 protein, as detected by the ACR3 antibody, localizes to the vacuoles of the P. vittata gametophyte.
Effects of Knocking Down ACR3 and ACR3;1 Transcript Levels in P. vittata Gametophytes
While ACR3 is able to complement the ACR3 deficiency in yeast, and its expression is upregulated in response to arsenate in sporophyte roots and gametophytes, its exact function in the fern is unknown. To determine whether the ACR3 or ACR3;1 genes are necessary for arsenic tolerance in P. vittata gametophytes, a method for systemically silencing gene expression developed in C. richardii gametophytes (Rutherford et al., 2004) was adapted for P. vittata gametophytes. The efficiency of this method was first tested by silencing the expression of the P. vittata protophorphrin magnesium chelatase IX (MgChl) gene. Because this gene is required for chlorophyll biosynthesis, systemic silencing of the endogenous MgChl gene should result in the development of a colorless P. vittata gametophyte. P. vittata gametophytes were cobombarded with both the reporter construct 35S:DsRed2 and a construct designed to drive the expression of a P. vittata MgChl antisense mRNA. In two independent experiments, gametophytes with a red fluorescent cell were selected from the rest of the population 1 d after bombardment. Of these, 85% (n = 446) and 94% (n = 239) were colorless 6 d after bombardment (Figure 6A). Furthermore, the accumulation of MgChl RNA in colorless gametophytes was found to be threefold less than in green gametophytes bombarded with 35S-DsRed alone, as determined by qRT-PCR (Figure 6A). These experiments demonstrate three important points about this experimental setup: that DsRed can be used as a marker for selecting transformed gametophytes, that the efficiency of cobombarding the reporter and gene silencing plasmids is very high, and that the expression of an antisense gene in a single cell of the gametophyte is sufficient to systemically spread and knock down the expression of the corresponding endogenous gene in almost all cells of the gametophytes.
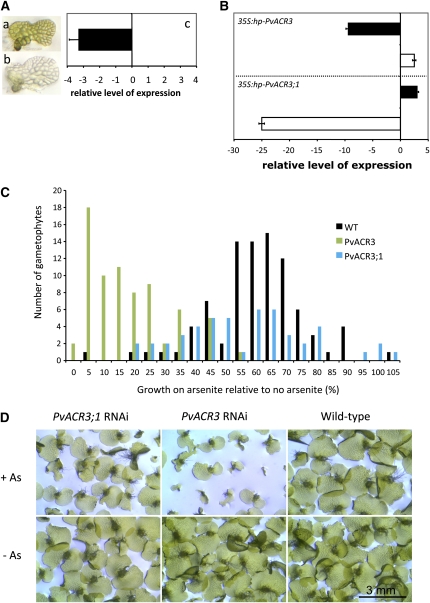
Figure 6.
Development and Use of RNAi to Determine the Effects of Knocking Down the Expression of ACR3 and ACR3;1 in P. vittata Gametophytes.
(A) Knocking down the expression of the P. vittata MgChI gene in gametophytes by RNAi results in a colorless phenotype. (a) Wild-type P. vittata gametophyte. (b) Gametophyte bombarded with 35S:antisense-PvChlI 6 d after bombardment. (c) Relative level of MgChI expression in pools of colorless gametophytes relative to expression in green wild-type gametophytes (n = 3) as determined by qRT-PCR.
(B) Relative levels of ACR3 and ACR3;1 expression in wild-type and RNAi gametophytes as determined by qRT-PCR. Relative change in ACR3 (solid bar) and ACR3;1 (white bar) expression in ACR3-RNAi gametophytes (top panel) and ACR3;1 RNAi gametophytes (bottom panel) relative to similarly treated wild-type gametophytes (relative change = 2ΔΔCT; ΔΔCT = ΔCT RNAi gametophytes − ΔCT wild-type gametophytes). All gametophytes were grown in the presence of arsenite.
(C) The tolerance of gametophytes to arsenite was measured as the change in growth of the prothallus. Wild-type (WT) gametophytes are indicated by black bars (n = 88 individual gametophytes with As and n = 96 without As), ACR3 RNAi gametophytes by green bars (n = 73 individual gametophytes with As and n = 72 without As), and ACR3;1 RNAi gametophytes by blue bars (n = 50 individual gametophytes with As and n = 71 without As).
(D) Phenotypes of gametophytes 12 d after transfer to no or 0.5 mM arsenite. The top panels show the gametophytes on 0.5mM arsenite, and the bottom panels show the gametophytes on no arsenite. The genotypes of the gametophytes are labeled above the panels.
Error bars in (A) and (B) represent the range around the mean calculated as a composite sd from the P. vittata histone gene and ACR3.
The effects of reducing the expression of ACR3 and ACR3;1 on arsenic tolerance in the gametophyte were determined by cobombarding gametophytes with the reporter 35S:DsRed2 plasmid plus constructs that drive the expression of hairpin-forming ACR3 or ACR3;1 mRNA. We previously established that hairpin-forming constructs are more efficient in systemic gene silencing than antisense constructs in fern gametophytes (Rutherford et al., 2004). One day after bombardment, gametophytes with a red fluorescent cell were transferred to plates and cultured for 2 d. Gametophytes were then transferred to fresh media with or without 0.5 mM arsenite to establish the effects of reduced levels of ACR3 or ACR3;1 transcripts on the tolerance of gametophytes to arsenite. Arsenite, rather than arsenate, was used in these experiments for several reasons. After examining the effects of different concentrations of arsenate and arsenite on gametophyte growth, we observed that gametophytes were uniformly affected by arsenite but not arsenate, making the screen for RNA interference (RNAi) phenotypes on arsenite more robust. Furthermore, as arsenate is rapidly reduced to arsenite in the gametophyte, with >95% of arsenic stored as arsenite in arsenate grown gametophytes (Gumaelius et al., 2004), the use of arsenite allowed us to directly assay the proposed function of ACR3 as an arsenite transporter required for arsenic tolerance.
The P. vittata gametophyte prothallus develops as a flat, single layer of cells during the first 2 weeks following germination. We therefore determined the growth rate of each gametophyte by measuring the perimeter of each gametophyte after 4 and 6 d of growth on media with or without arsenite. The growth rate of each gametophyte was calculated as the change in gametophyte perimeter per day, and the difference in growth rate calculated as a percentage of difference in the growth rates of arsenite-grown gametophytes relative to nonarsenite-grown gametophytes. A histogram illustrating the frequency distribution of the growth rates of individual gametophytes among different genotypic populations is shown in Figure 6C. The growth rate of ACR3-silenced gametophytes was reduced on average by 85% due to arsenite exposure. This represents a significant increase (t test, P < 0.001) in growth inhibition in response to arsenite when compared with wild-type gametophytes, which showed on average a 42% reduction in growth.
While arsenite inhibits the growth rate of ACR3-silenced gametophytes, the effects of arsenic on the growth rate of ACR3;1-silenced gametophytes were not significantly different from that of wild-type (t test, P = 0.5) gametophytes, showing on average a 44% reduction in growth. After prolonged growth (22 d) on arsenite-containing media, at least 60% of the ACR3 silenced gametophytes remained small compared with ACR3-silenced gametophytes grown in the absence of arsenite (Figure 6D). Reactivation of silenced endogenous ACR3 transcript likely occurs over time after bombardment (Rutherford et al., 2004), and such reactivated gametophytes are expected to regain their tolerance to arsenic. We note that ACR3 and ACR3;1 silenced gametophytes both showed small but significant reductions in growth (13 to 32%) compared with wild-type gametophytes between 4 and 6 d after bombardment, most likely due to the bombardment process. However, this growth difference between ACR3 and ACR3;1 silenced gametophytes compared with the wild type in the absence of arsenite was not observed after 22 d (Figure 6D). Only ACR3-silenced gametophytes showed a consistent reduction in growth on arsenite-containing media (Figures 6C and 6D).
To confirm that the transcript levels of ACR3 or ACR3;1 were reduced in silenced gametophytes, the relative levels of ACR3 and ACR3;1 transcripts were determined by qRT-PCR 5 d after transfer to media with or without arsenite. In wild-type gametophytes, arsenite increased levels of ACR3 transcripts ∼10-fold but had little effect on ACR3;1 transcript levels (Figure 4C). In ACR3-silenced gametophytes grown in the presence of arsenite, levels of ACR3 were reduced ∼10-fold compared with arsenite-grown wild-type gametophytes (Figure 6B). In ACR3;1-silenced gametophytes grown in the presence of arsenite, levels of ACR3;1 transcripts were reduced ∼25-fold (Figure 6B). These results demonstrate that the method of gene silencing used effectively reduces the accumulation of the target transcripts in the gametophyte. Neither levels of ACR3;1 transcripts in ACR3-silenced gametophytes, or ACR3 transcripts in ACR3;1-silenced gametophytes were dramatically altered (Figure 6B), indicating that silencing of targeted genes is relatively gene specific. Upregulation of ACR3 by arsenite was observed in DsRed2-expressing gametophytes bombarded with 35S:DsRed2 alone (see Supplemental Figure 3 online), demonstrating that the observed gene silencing is not attributed to the act of bombardment alone.
DISCUSSION
We have shown that ACR3 is necessary for arsenic tolerance in P. vittata. ACR3 likely functions as an arsenite effluxer based upon its ability to suppress the arsenite sensitivity and arsenic hyperaccumulation phenotypes of Δacr3 yeast, which lack an endogenous arsenite effluxer. Furthermore, the abundance of the ACR3 transcripts in P. vittata increases in response to arsenic in gametophytes and the roots of sporophytes, organs, and structures that naturally come in direct contact with arsenic in the soil. Finally, reduction in ACR3 transcript levels in gametophytes by RNAi results in an arsenite sensitive phenotype, clearly establishing the role of this protein in arsenic tolerance in P. vittata.
ACR3 is a membrane protein and is similar in amino acid sequence to the yeast arsenite effluxer ACR3. In yeast, arsenate taken up from the media is rapidly reduced to arsenite by the arsenate reductase ACR2, whereupon ACR3 functions to efflux the arsenite from the cell. In P. vittata, arsenate is also rapidly reduced to arsenite, possibly by the previously identified arsenate reductase ACR2 (Ellis et al., 2006). Based upon the phenotype of ACR3 knockdown gametophytes and the localization of ACR3 to the vacuolar membrane, we predict that ACR3 functions to transport arsenite rapidly from the cytoplasm to the vacuole for long-term storage. This is consistent with our observation that arsenite accumulates at high levels in the gametophyte, most likely in the vacuole (Pickering et al., 2006). The loss of ACR3 (in ACR3 knockdown gametophytes) may prevent the transport of arsenite from the cytoplasm to the vacuole, leading to increased sensitivity to arsenite. Determination of the subcellular distribution of arsenic in ACR3 knockdown gametophytes, by subcellular fractionation or direct imaging using high resolution x-ray spectroscopy (e.g., Pickering et al., 2006), would help to further establish this model of the function of ACR3.
The function and localization of ACR3 in the sporophyte is likely to be more complex, as the movement of arsenic in the sporophyte requires transporting arsenic from the root into the xylem for translocation to the shoot and to the cells of the lamina where it is likely sequestered in the vacuole. In addition to a possible role in arsenite efflux into the vacuole, ACR3 may also function in the root to load arsenite into the xylem in a similar manner to Lsi2 in rice (Ma et al., 2008). While ACR3 is expressed in all organs of the sporophyte, its expression is strongly upregulated by arsenite in the root, where it accumulates at high levels compared with other organs (fronds and stipes). Determining the tissue and cellular localization of the ACR3 proteins in both phases of the life cycle of P. vittata is critical to test this model of ACR3 function and should provide insights into the mechanism of arsenic hyperaccumulation in P. vittata.
Phylogenetically, the P. vittata ACR3 proteins cluster with the ACR3 family of the BART (bile/arsenite/riboflavin/transporter) protein superfamily (Mansour et al., 2007). The BART superfamily of proteins includes five other families (BASS, RFT, TMS, UNK, and SHK) that have plant orthologs (Mansour et al., 2007). Until now, the ACR3 family was thought to be present only in archaea, bacteria, and yeast. These organisms have only one ACR3 gene with the exception of some bacteria, which have two ACR3 genes. Searches of whole-genome and EST databases revealed that ACR3 genes are present in a variety of plants, including the moss P. patens, the lycophyte S. moellendorffii, and the gymnosperms W. mirabilis and P. sitchensis, yet are absent in angiosperm genomes. The absence of ACR3 genes in both monocots and eudicots may preclude their ability to hyperaccumulate arsenic, which is consistent with the observation that no arsenic-hyperaccumulating angiosperm has ever been identified. Why the highly conserved ACR3 gene was lost in the angiosperm lineage may never be known, but its loss coincides with their reliance on insects and other animals for pollination and fruit dispersal. The selection of arsenic hyperaccumulation in P. vittata, on the other hand, may have evolved as a deterrent and in response to herbivores, as it has recently been shown that insects avoid eating P. vittata grown in the presence of arsenic, preferring those not grown in arsenic (Rathinasabapathi et al., 2007). The adaptive significance of arsenic hyperaccumulation in natural populations of P. vittata, which is native to China, remains to be determined, and the possibility that this trait evolved as an adaptation to arsenic-contaminated soils cannot be ruled out.
With the exception of P. vittata, only a single ACR3 gene has been identified in plant species. P. vittata is exceptional in having at least two ACR3 genes, only one of which (ACR3) is necessary for arsenic tolerance in the gametophyte. As all other plants have only one ACR3 gene and arsenic hyperaccumulation has only been observed in P. vittata and its relatives, we speculate that the duplication of ACR3 gene, followed by neofunctionalization of one of the duplicates, may have played a role in its unique ability to hyperaccumulate arsenic. Since species in the Pteridaceae vary in their ability to hyperaccumulate arsenic, we are testing this hypothesis by identifying ACR3 genes from hyperaccumulating and nonaccumulating species in the Pteridaceae and testing the function of these genes in gametophytes using RNAi.
METHODS
Bacterial Strains, Yeast Strains, and Plasmids
The genotypes of strains and plasmids used in this study and their sources are listed in Supplemental Table 1 online.
Cloning of ACR3 Genes in Pteris vittata
A yeast (Saccharomyces cerevisiae) expression library was constructed from RNA purified from P. vittata gametophytes grown in liquid culture containing arsenate (Ellis et al., 2006) and was used to transform the yeast strain MG102. Arsenate-tolerant transformants were selected on yeast minimal media lacking Trp as described by Ellis et al. (2006). The plasmids were then isolated, transformed into Escherichia coli, isolated from recombinant E. coli colonies, and transformed into the yeast strain MG100. Plasmids from recombinant arsenic-tolerant MG100 colonies were isolated and their inserts sequenced. The SMART 5′ RACE kit (Clontech) was used to clone the 5′ end of the ACR3 cDNA. The ACR3-like cDNA from a P. vittata EST library was identified based on its sequence similarity to ACR3.
Yeast Growth Experiments
Yeast growth experiments followed the protocol of Mukhopadhyay et al. (2000) with the following modifications. Yeast were transformed with a recombinant plasmid vector (pCM184; see Supplemental Table 1 online) that drives the expression of the yeast and P. vittata ACR3 genes then cultured in the presence of Na2HAsO4 (25 μM in plates and 10 μM in liquid media). Primers used to generate these plasmid constructs are listed in Supplemental Table 3 online. For plate cultures, 7 μL of several fivefold serial dilutions of each yeast culture was applied to agar plates containing minimal media without Trp and with arsenate. The plates were incubated at 30°C for 4 to 5 d. For liquid culture growth assays, yeast strains were cultured for 24 h in 5 mL of liquid minimal media without Trp and with 10 μM Na2HAsO4. Once the absorbance (OD600) of each culture reached 0.05, it was measured every 1.5 h for 12 h to generate a growth curve. All strains were cultured in triplicate. Arsenic content in yeast was determined by inductively coupled plasma–mass spectroscopy, as previously described (Ellis et al., 2006).
Phylogenetic Analysis
Phylogenetic analyses were conducted using MEGA version 4.0 (Tamura et al., 2007). Protein sequences were aligned using MAFFT and MUSCLE (Edgar, 2004b, 2004a) as shown in Supplemental Figure 2 online. The tree was constructed using distance methods of neighbor joining (Saitou and Nei, 1987) performed on a Kimura-2 parameter (pairwise distances; Kimura, 1980). Bootstraps were performed with 10,000 replications.
Growth of Plant Tissues
P. vittata spores were harvested, stored, and sterilized and gametophytes grown in medium as previously described (Gumaelius et al., 2004) without or with various concentrations KH2AsO4 (arsenate) or NaAsO2 (arsenite). Greenhouse grown P. vittata sporophytes were removed from pots, their roots extensively washed, and then hydroponically grown in liquid half-strength Murashige and Skoog medium, pH 6.5, without or with 10 mM KH2AsO4.
qRT-PCR
For qRT-PCR on gametophytes used in the RNAi experiments, total RNA was extracted from 100 pooled RNAi gametophytes per treatment. For all other qRT-PCR experiments total RNA was extracted from ∼100 mg of gametophyte or sporophyte tissues. Total RNA was extracted using the RNeasy plant mini kit (Qiagen) and treated with DNase. First-strand cDNAs were synthesized using the SuperScript III kit (Invitrogen). The ACR3-specific primers used for qRT-PCR are listed in Supplemental Table 3 online. A P. vittata histone gene was used as an internal control in all qRT-PCR experiments because its expression is not altered by arsenic in P. vittata and it fulfills the requirements for an internal standard as defined by the ABI Prism 7000 Manual (Applied Biosystems). Each primer pair was optimized for the amplification of a single PCR product; the efficiencies of all PCR reactions were between 95 and 105%. qRT-PCRs were prepared with a SYBR-green master mix (Applied Biosystems) and performed using the Prism 7000 (Applied Biosystems). The Prism 7000 manual baseline was set to 9 and 25. Three technical replicates of three biological replicates per treatment were performed. The ΔCT values were averaged, and the ΔΔCT value calculated as the difference between the ΔCT values for two different treatments (ΔCTtreatment1 − ΔCTtreatment2). The relative fold change of transcript abundance between two treatments was calculated as 2ΔΔCT.
RNAi in P. vittata Gametophytes
RNAi constructs were delivered to 7-d-old P. vittata gametophytes grown on FM plates by biolistic bombardment as previously described (Rutherford et al., 2004). Hairpin constructs targeting ACR3 and ACR3-like were made using the pKannibal plasmid (Wesley et al. 2001) and are referred to as 35S:hpACR3 and 35S:hpACR3;1. The primers used in constructing these plasmids are listed in Supplemental Table 2 online. The hairpin construct was made by sequential digestions and ligations; the antisense fragment was digested with ClaI and XbaI and then ligated into ClaI-XbaI sites of pKannibal. This plasmid was then digested with XhoI and KpnI and the ACR3 sense XhoI-EcoRI–digested fragment inserted. A similar strategy was used to generate 35S:hpACR3;1. The primers used to generate the antisense P. vittata CHLI1 plasmid, named 35S:antiMgChl, are listed in Supplemental Table 3 online. After digestion with SpeI and SalI, the PCR fragment was cloned into the same sites of the 35S:irint plasmid (Rutherford et al., 2004). All RNAi plasmids were verified by sequencing.
Measuring the Growth of Gametophytes
One day after bombardment with 35S:DsRed2 (Tzfira et al., 2005), or after cobombardment with 35S:DsRed2 plus 35S:hpACR3 or 35S:hpACR3;1, gametophytes with a red fluorescing cell were selected using a Leica fluorescence binocular microscope equipped with a Texas Red filter, then transferred to plates containing 0.5 mM arsenite or no arsenite. Images of individual gametophytes 4 and 6 d after transfer to fresh media were captured using a SPOT RT camera (Diagnostic Instruments) attached to the microscope. The perimeter of each gametophyte prothallus was measured from these images using ImageJ software (W.S. Rasband, National Institutes of Health; Abramoff et al., 2004). The change in growth per day of each gametophyte was calculated as the change in gametophyte perimeter divided by two. At least 57 gametophytes were assayed per treatment, and all gametophyte growth experiments were replicated two times.
Protein Analysis
Membrane proteins from yeast were isolated according to Kim et al. (2004). To isolate P. vittata membrane proteins, 5 g of 4-week-old gametophytes grown in the presence of 2 mM arsenate were washed twice with deionized water, filtered, and homogenized at 4°C in a prechilled glass homogenizer with grinding buffer (25 mM HEPES-KOH, pH 8.5, 0.29 M sucrose, 3 mM DTT, 4 mM EDTA, 0.5% polyvinylpolypyrrolidone, 0.1 mM phenylmethylsulfonyl fluoride, and Plant Protease Inhibitor Cocktail [Sigma-Aldrich P9599]). Homogenized tissue was passed through a Miracloth filter and centrifuged twice at 8000g for 10 min. Supernatants were combined and centrifuged at 100,000g for 1 h to collect the microsomal fraction. The microsomal pellet was resuspended in microsomal buffer (10 mM BTP-MES, pH 7.8, 13% [w/v] sucrose, 5 mM EDTA, 3 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, and Plant Protease Inhibitor Cocktail).
For sucrose gradient analysis, the microsomal membranes were separated using a sucrose step gradient (22, 32, 38 and 45% [w/v] sucrose with 10 mM BTP-MES, pH 7.8, and 5 mM EDTA) (Gustin et al., 2009). Gradients were spun at 100,000g for 4 h at 4°C. Eight fractions representing the entire gradient were collected and sucrose removed according to Gustin et al. (2009). Each fraction was then centrifuged at 100,000g for 1 h at 4°C and the pellets resuspended in microsomal buffer lacking DTT and sucrose, and the protein concentration determined using the BCA protein assay kit according to the manufacturer's instructions (Pierce). Triton X-100 (0.1%), 100 μM NaCl, 3 mM DTT, and Plant Protease Inhibitor Cocktail were added to the samples and proteins were dissociated from the membranes by gentle shaking at 4°C for 1 hour. Proteins where then stored at −80°C.
For aqueous two-phase separation, 0.9 g of microsomal membranes in microsomal buffer was added to a 2.7 g phase system (6.6% [w/v] dextran [Mr = 500,000], 6.6% [w/v] polyethylene glycol (Mr = 3350), 0.4 M sucrose, 5 mM K-phosphate, pH 7.8, and 3 mM KCl) to create 3.6 g experimental system (Larsson et al., 1987). The phase system was thoroughly mixed by inverting 100 times without introducing air bubbles, and phase separation was hastened by centrifugation at 2500g for 6 min. The upper phase (U1) was transferred to a clean tube and extracted three times (U2, U3, and U4) with fresh lower phase. The lower phase (L1) was also transferred to a separate clean tube and extracted three times (L2, L3, and L4) with fresh upper phase. Fresh upper and lower phases were obtained from a bulk phase system of identical composition prepared separately. The final upper and lower phases were diluted with 10 times the volume of microsomal buffer without sucrose and centrifuged at 100,000g for 1 h at 4°C. Pellets were resuspended in microsomal buffer lacking DTT and sucrose, and the protein concentration determined use the BCA Protein Assay Kit. Quantified proteins were stored at −80°C until analyzed.
To generate an antibody specific to ACR3, the peptide LKVVSRGKGFHVSS, which corresponds to residues 200 to 213 of ACR3, was synthesized, conjugated to keyhole limpet hemocyanin, and used to generate two rabbit polyclonal antibodies (GenScript). The antibody was affinity purified using a protein A column with binding and elution buffers according to the manufacturer's instructions (Sigma-Aldrich). Immunoblot analyses were performed according to Sambrook and Russel (2002). The plant H+ATPase, Sar1, and V-ATPase antibodies were obtained from Agrisera (Sweden). All antibodies were detected using horseradish peroxidase–conjugated secondary antibody with the VisiGlo developing kit (Amresco).
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: P. vittata ACR3, GI:224814383; P. vittata ACR3;1, GI:224814385; S. cerevisiae ACR3, Gi:2498103; P. vittata magnesium chelatase, HM179992. The GenBank accession numbers used in the phylogeny are as follows: Marchantia polymorpha (BJ846622), Physcomitrella patens (BJ965142 and XM001752302), Selaginella moellendorffii (DN837757 and FJ751633), Ceratopteris richardii (CV736091 and CV735945), Welwitschia milabilis (CK760764), Pinus sitchensis (FD735511), Oriza sativa (NP_001043701), Arabidopsis (At1g78560), Aspergillus clavatus (XM_001269582), Gibberella zeae (XP_386755), Magnaporthe grisea (XP_360856), Cryptococcus neoformans (XP_772635), Neurospora crassa (XP_963886), and S. cerevisiae (NP_015527). All other ACR3 sequences used in Figure 3 are listed in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Yeast Growth Curve.
Supplemental Figure 2. Sequence Alignment of ACR3 and Sodium Bile Proteins.
Supplemental Figure 3. Expression of ACR3 and ACR3;1 in Gametophytes Transformed with the DsRed2 Plasmid as Determined by qRT-PCR.
Supplemental Table 1. Plasmids and Strains Used in This Study.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Proteins and GI Numbers for the Phylogenetic Analysis.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 3 and Supplemental Figure 2 Online.
Acknowledgments
This work was supported by a grant to J.A.B. and D.E.S. from the U.S. National Science Foundation (IOS 0844413) and a grant to D.E.S. from the U.S. National Institutes of Health (P42ES007373). We thank Brett Lahner at the Purdue Ionomics Center for inductively coupled plasma–mass spectroscopy analysis.
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with Image. J. Biophotonics International 11: 36–42 [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bleeker P.M., Hakvoort H.W.J., Bliek M., Souer E., Schat H. (2006). Enhanced arsenate reduction by a CDC25-like tyrosine phosphatase explains increased phytochelatin accumulation in arsenate-tolerant Holcus lanatus. Plant J. 45: 917–929 [DOI] [PubMed] [Google Scholar]
- Catarecha P., Segura M.D., Franco-Zorrilla J.M., Garcia-Ponce B., Lanza M., Solano R., Paz-Ares J., Leyva A. (2007). A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Huang Z., Huang Y., Lei M. (2005). Distributions of arsenic and essential elements in pinna of arsenic hyperaccumulator Pteris vittata L. Sci. China C Life Sci. 48: 18–24 [DOI] [PubMed] [Google Scholar]
- Cullen W.R., Reimer K.J. (1989). Arsenic speciation in the environment. Chem. Rev. 89: 713–764 [Google Scholar]
- Dhankher O.P., Li Y., Rosen B.P., Shi J., Salt D., Senecoff J.F., Sashti N.A., Meagher R.B. (2002). Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and g-glutamylcysteine synthetase expression. Nat. Biotechnol. 20: 1140–1145 [DOI] [PubMed] [Google Scholar]
- Dhankher O.P., Rosen B.P., McKinney E.C., Meagher R.B. (2006). Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2). Proc. Natl. Acad. Sci. USA 103: 5413–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G.L., Zhou Y., Tong Y.P., Mukhopadhyay R., Rosen B.P., Zhu Y.G. (2007). A CDC25 homologue from rice functions as an arsenate reductase. New Phytol. 174: 311–321 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004a). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004b). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.R., Gumaelius L., Indriolo E., Salt D., Banks J.A. (2006). A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol. 141: 1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi K., Visoottiviseth P., Sridokchan W., Goessler W. (2002). Arsenic species in an arsenic hyperaccumulating fern, Pityrogramma calomelanos: A potential phytoremediator of arsenic-contaminated soils. Sci. Total Environ. 284: 27–35 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Shen J., Rosen B.P. (1999). Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96: 5001–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga M.I., Santos J.A., Ma L.Q. (2008). Phytoextraction by arsenic hyperaccumulator Pteris vittata L. from six arsenic-contaminated soils: Repeated harvests and arsenic redistribution. Environ. Pollut. 154: 212–218 [DOI] [PubMed] [Google Scholar]
- Gumaelius L., Lahner B., Salt D.E., Banks J.A. (2004). Arsenic hyperaccumulation in gametophytes of Pteris vittata. A new model system for analysis of arsenic hyperaccumulation. Plant Physiol. 136: 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin J.L., Loureiro M.E., Kim D., Na G., Tikhonova M., Salt D.E. (2009). MTP1-dependent Zn sequestration into shoot vacuoles suggests dual roles in Zn tolerance and accumulation in Zn-hyperaccumulating plants. Plant J. 57: 1116–1127 [DOI] [PubMed] [Google Scholar]
- Ha S.-K., Smith A., Howden R., Dietrich W.M., Bugg S., O'Connell M.J., Goldsbrough P.B., Cobbett C.S. (1999). Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermine O., et al. (2004). Phase II trial of arsenic trioxide and alpha interferon in patients with relapsed/refractory adult T-cell leukemia/lymphoma. Hematol. J. 5: 130–134 [DOI] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. (1998). SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35: W585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T., Tanaka M., Mitani N., Ma J.F., Maeshima M., Fujiwara T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284: 2114–2120 [DOI] [PubMed] [Google Scholar]
- Kertulis-Tartar G.M., Ma L.Q., Tu C., Chirenje T. (2006). Phytoremediation of an arsenic-contaminated site using Pteris vittata L.: A two-year study. Int. J. Phytoremediation 8: 311–322 [DOI] [PubMed] [Google Scholar]
- Kim D., Gustin J.L., Lahner B., Persans M.W., Baek D., Yun D.J., Salt D.E. (2004). The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 39: 237–251 [DOI] [PubMed] [Google Scholar]
- Kimura M.A. (1980). Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de The H. (2008). Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10: 547–555 [DOI] [PubMed] [Google Scholar]
- Landrieu I., da Costa M., De Veylder L., Dewitte F., Vandepoele K., Hassan S., Wieruszeski J.M., Corellou F., Faure J.D., Van Montagu M., Inzé D., Lippens G. (2004b). A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieu I., Hassan S., Sauty M., Dewitte F., Wieruszeski J.M., Inzé D., De Veylder L., Lippens G. (2004a). Characterization of the Arabidopsis thaliana Arath; CDC25 dual-specificity tyrosine phosphatase. Biochem. Biophys. Res. Commun. 322: 734–739 [DOI] [PubMed] [Google Scholar]
- Larsson C., Widell S., Kjellbom P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol. 148: 558–568 [Google Scholar]
- Li W., Chen T., Chen Y., Lei M. (2005). Role of trichome of Pteris vittata L. in arsenic hyperaccumulation. Sci. China C Life Sci. 48: 148–154 [DOI] [PubMed] [Google Scholar]
- Lombi E., Zhao F.-J., Fuhrman M., Ma L., McGrath S.P. (2002). Arsenic distribution and speciation in the fronds of the hyperaccumulator Pteris vittata. New Phytol. 156: 195–203 [DOI] [PubMed] [Google Scholar]
- Ma J.F., Yamaji N., Mitani N., Xu X.Y., Su Y.H., McGrath S.P., Zhao F.J. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. USA 105: 9931–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.Q., Komar K.M., Tu C., Zhang W., Cai Y., Kennelley E.D. (2001). A fern that hyperaccumulates arsenic. Nature 409: 579. [DOI] [PubMed] [Google Scholar]
- Mansour N.M., Sawhney M., Tamang D.G., Vogl C., Saier M.H., Jr. (2007). The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 274: 612–629 [DOI] [PubMed] [Google Scholar]
- Marin A.R., Masscheleyn P.H., Patrick W.H. (2003). Soil redox-pH stability of arsenic species and its influence on arsenic uptake by rice. Plant Soil 152: 245–253 [Google Scholar]
- Meharg A.A., Hartley-Whitaker J. (2002). Tansley review no. 133: Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154: 29–43 [Google Scholar]
- Meharg A.A., Macnair M.R. (1992). Suppression of the high-affinity phosphate-uptake system - A mechanism of arsenate tolerance in Holcus lanatus L. J. Exp. Bot. 43: 519–524 [Google Scholar]
- Mitaku S., Hirokawa T. (1999). Physicochemical factors for discriminating between soluble and membrane proteins: hydrophobicity of helical segments and protein length. Protein Eng. 12: 953–957 [DOI] [PubMed] [Google Scholar]
- Mitaku S., Hirokawa T., Tsuji T. (2002). Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18: 608–616 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R., Shi J., Rosen B.P. (2000). Purification and characterization of Acr2p, the Saccharomyces cerevisiae arsenate reductase. J. Biol. Chem. 275: 21149–21157 [DOI] [PubMed] [Google Scholar]
- Nordstrom D. (2002). Worldwide occurences of arsenic in ground water. Science 296: 2143–2144 [DOI] [PubMed] [Google Scholar]
- Pickering I.J., Gumaelius L., Harris H.H., Prince R.C., Hirsch G., Banks J.A., Salt D.E., George G.N. (2006). Localizing the biochemical transformations of arsenate in a hyperaccumulating fern. Environ. Sci. Technol. 40: 5010–5014 [DOI] [PubMed] [Google Scholar]
- Pickering I.J., Prince R.C., George M.J., Smith R.D., George G.N., Salt D.E. (2000). Reduction and coordination of arsenic in Indian mustard. Plant Physiol. 122: 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaghebeur M., Rengel Z. (2004). Arsenic uptake, translocation and speciation in pho1 and pho2 mutants of Arabidopsis thaliana. Physiol. Plant. 120: 280–286 [DOI] [PubMed] [Google Scholar]
- Raab A., Feldmann J., Meharg A.A. (2004). The nature of arsenic-phytochelatin complexes in Holcus lanatus and Pteris cretica. Plant Physiol. 134: 1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab A., Schat H., Meharg A.A., Feldmann J. (2005). Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): Formation of arsenic-phytochelatin complexes during exposure to high arsenic concentrations. New Phytol. 168: 551–558 [DOI] [PubMed] [Google Scholar]
- Rahman M.M., Ng J.C., Naidu R. (2009). Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ. Geochem. Health. 31 (suppl.): 189–200 [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B., Rangasamy M., Froeba J., Cherry R.H., McAuslane H.J., Capinera J.L., Srivastava M., Ma L.Q. (2007). Arsenic hyperaccumulation in the Chinese brake fern (Pteris vittata) deters grasshopper (Schistocerca americana) herbivory. New Phytol. 175: 363–369 [DOI] [PubMed] [Google Scholar]
- Rutherford G., Tanurdzic M., Hasebe M., Banks J.A. (2004). A systemic gene silencing method suitable for high throughput, reverse genetic analyses of gene function in fern gametophytes. BMC Plant Biol. 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Salt D.E., Rauser W.E. (1995). MgATP-dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiol. 107: 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. (2002). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Shelmerdine P.A., Black C.R., McGrath S.P., Young S.D. (2009). Modelling phytoremediation by the hyperaccumulating fern, Pteris vittata, of soils historically contaminated with arsenic. Environ. Pollut. 157: 1589–1596 [DOI] [PubMed] [Google Scholar]
- Singh N., Ma L.Q. (2006). Arsenic speciation, and arsenic and phosphate distribution in arsenic hyperaccumulator Pteris vittata L. and non-hyperaccumulator Pteris ensiformis L. Environ. Pollut. 141: 238–246 [DOI] [PubMed] [Google Scholar]
- Smith A.H., Hira-Smith M.M. (2004). Arsenic drinking water regulations in developing countries with extensive exposure. Toxicology 198: 39–44 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175–182 [PubMed] [Google Scholar]
- Sorrell D.A., Chrimes D., Dickinson J.R., Rogers H.J., Francis D. (2004). The Arabidopsis CDC25 induces a short cell length when overexpressed in fission yeast: Evidence for cell cycle function. New Phytol. 165: 425–428 [DOI] [PubMed] [Google Scholar]
- Srivastava M., Ma L.Q., Santos J.A. (2006). Three new arsenic hyperaccumulating ferns. Sci. Total Environ. 364: 24–31 [DOI] [PubMed] [Google Scholar]
- Su Y.H., McGrath S.P., Zhu Y.G., Zhao F.J. (2008). Highly efficient xylem transport of arsenite in the arsenic hyperaccumulator Pteris vittata. New Phytol. 180: 434–441 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Geoffroy M.C., Shen L., Plechanovova A., Hattersley N., Jaffray E.G., Palvimo J.J., Hay R.T. (2008). RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Tu C., Ma L.Q. (2005). Effects of arsenic on concentration and distribution of nutrients in the fronds of the arsenic hyperaccumulator Pteris vittata L. Environ. Pollut. 135: 333–340 [DOI] [PubMed] [Google Scholar]
- Tusnady G.E., Simon I. (1998). Principles governing amino acid composition of integral membrane proteins: Application to topology prediction. J. Mol. Biol. 283: 489–506 [DOI] [PubMed] [Google Scholar]
- Tusnady G.E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Visoottiviseth P., Francesconi K., Sridokchan W. (2002). The potential of Thai indigenous plant species for the phytoremediation of arsenic contaminated land. Environ. Pollut. 118: 453–461 [DOI] [PubMed] [Google Scholar]
- Wang H.B., Wong M.H., Lan C.Y., Baker A.J., Qin Y.R., Shu W.S., Chen G.Z., Ye Z.H. (2007). Uptake and accumulation of arsenic by 11 Pteris taxa from southern China. Environ. Pollut. 145: 225–233 [DOI] [PubMed] [Google Scholar]
- Wang H.B., Ye Z.H., Shu W.S., Li W.C., Wong M.H., Lan C.Y. (2006). Arsenic uptake and accumulation in fern species growing at arsenic-contaminated sites of southern China: Field surveys. Int. J. Phytoremediation 8: 1–11 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao F.J., Meharg A.A., Raab A., Feldmann J., McGrath S.P. (2002). Mechanisms of arsenic hyperaccumulation in Pteris vittata. Uptake kinetics, interactions with phosphate, and arsenic speciation. Plant Physiol. 130: 1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S.M., Gaillard J.F., Ma L., Tu C. (2003). XAS speciation of arsenic in a hyperaccumulating fern. Environ. Sci. Technol. 37: 754–760 [DOI] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Williams P.N., Villada A., Deacon C., Raab A., Figuerola J., Green A.J., Feldmann J., Meharg A.A. (2007). Greatly enhanced arsenic shoot assimilation in rice leads to elevated grain levels compared to wheat and barley. Environ. Sci. Technol. 41: 6854–6859 [DOI] [PubMed] [Google Scholar]
- Wysocki R., Bobrowicz P., Ulaszewski S. (1997). The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 272: 30061–30066 [DOI] [PubMed] [Google Scholar]
- Xu X.Y., McGrath S.P., Meharg A.A., Zhao F.J. (2008). Growing rice aerobically markedly decreases arsenic accumulation. Environ. Sci. Technol. 42: 5574–5579 [DOI] [PubMed] [Google Scholar]
- Zhang W., Cai Y., Tu C., Ma L.Q. (2002). Arsenic speciation and distribution in an arsenic hyperaccumulating plant. Sci. Total Environ. 300: 167–177 [DOI] [PubMed] [Google Scholar]
- Zhao F.-J., Dunham S., McGrath S.P. (2002). Arsenic hyperaccumulation by different fern species. New Phytol. 156: 27–31 [Google Scholar]
- Zhao F.J., Ma J.F., Meharg A.A., McGrath S.P. (2008). Arsenic uptake and metabolism in plants. New Phytol. 181: 777–794 [DOI] [PubMed] [Google Scholar]
- Zhao F.J., Wang J.R., Barker J.H.A., Schat H., Bleeker P.M., McGrath S.P. (2003). The role of phytochelatins in arsenic tolerance in the hyperaccumulator Pteris vittata. New Phytol. 159: 403–410 [DOI] [PubMed] [Google Scholar]
- Zhu Y.G., Williams P.N., Meharg A.A. (2008). Exposure to inorganic arsenic from rice: A global health issue? Environ. Pollut. 154: 169–171 [DOI] [PubMed] [Google Scholar]