This study describes a newly isolated altered auxin response mutant. It finds that the mutant exhibits a reduction in polar auxin transport caused by a decrease in the levels of members of the PIN family of auxin efflux proteins.
Abstract
Auxin is an essential phytohormone that regulates many aspects of plant development. To identify new genes that function in auxin signaling, we performed a genetic screen for Arabidopsis thaliana mutants with an alteration in the expression of the auxin-responsive reporter DR5rev:GFP (for green fluorescent protein). One of the mutants recovered in this screen, called weak auxin response1 (wxr1), has a defect in auxin response and exhibits a variety of auxin-related growth defects in the root. Polar auxin transport is reduced in wxr1 seedlings, resulting in auxin accumulation in the hypocotyl and cotyledons and a reduction in auxin levels in the root apex. In addition, the levels of the PIN auxin transport proteins are reduced in the wxr1 root. We also show that WXR1 is ROOT UV-B SENSITIVE2 (RUS2), a member of the broadly conserved DUF647 domain protein family found in diverse eukaryotic organisms. Our data indicate that RUS2/WXR1 is required for auxin transport and to maintain the normal levels of PIN proteins in the root.
INTRODUCTION
The plant hormone auxin regulates essential aspects of plant growth and development, including embryo patterning, root and shoot elongation, tropic response, and vascular differentiation (Davies, 1995). Recent studies indicate that auxin controls development through a complex regulatory network involving auxin biosynthesis, transport, and perception (Benjamins and Scheres, 2008). In both the root and shoot, these processes contribute to the formation of an auxin concentration gradient required for patterning of developing tissues. Formation and maintenance of auxin gradients is dependent on polar cell-to-cell auxin transport mediated by special transporter proteins, including the auxin influx carriers AUX/LAX, the efflux facilitators PIN FORMED (PIN), and several ABCB proteins (Gälweiler et al., 1998; Marchant et al., 1999; Noh et al., 2001, 2003; Geisler et al., 2005; Bandyopadhyay et al., 2007; Petrasek and Friml, 2009; Robert and Friml, 2009). Moreover, modeling studies suggest that polar auxin transport is necessary to generate an auxin maximum and concentration gradient in the root tip (Grieneisen et al., 2007; Robert and Friml, 2009).
The patterns of expression and cellular localization of the AUX1 and PIN proteins are key to their function. For example, in Arabidopsis thaliana roots, AUX1 is expressed in stele, columella, epidermis, and lateral root cap cells and is polarly localized on the apical side of the protophloem cells (Swarup et al., 2001). The cellular localizations of PIN proteins vary depending on the protein and cell type. In general, however, the localization of the auxin efflux carriers correlates with and determines the direction of auxin transport (Gälweiler et al., 1998; Friml et al., 2002a, 2002b;Wisniewska et al., 2006).
Localization of the PIN proteins is a dynamic process that responds rapidly to physiological and environmental changes (Paciorek et al., 2005; Kleine-Vehn et al., 2008; Laxmi et al., 2008; Pan et al., 2009; Petrasek and Friml, 2009; Robert and Friml, 2009). PIN1 and related proteins are constitutively internalized by clathrin-dependent endocytosis and relocalized to the plasma membrane by ARF-GEF–dependent (guanine-nucleotide exchange factors for ADP-ribosylation factor GTPase) recycling (Geldner et al., 2001; Dhonukshe et al., 2007). Studies using yellow fluorescent protein (YFP)-tagged PIN1 and inducible PIN1 proteins indicate that PIN polar localization involves a two-step mechanism: the newly synthesized PIN proteins are localized to the plasma membrane nonpolarly, and their polar localization is mediated by the Rab5 GTPases (named ARA7 and RHA1 in Arabidopsis) endosome pathway (Dhonukshe et al., 2008). ARA7 and the putative retromer complex components SORTING NEXIN1 and VACUOLAR PROTEIN SORTING29, which are localized in late endosomes in the plant (Jaillais et al., 2006, 2007), are thought to be necessary for retrieval of PIN2 from late endosomes and thus prevent it from being internalized into lytic vacuoles (Abas et al., 2006; Kleine-Vehn et al., 2008). Moreover, this vacuolar internalization pathway mediates aspects of the response to gravity stimulation. A gravity stimulus increases the amount of PIN2 internalized into vacuoles on the upper side of epidermal cells after reorientation of root growth (Kleine-Vehn et al., 2008; Pan et al., 2009).
Once auxin is transported to a target tissue, auxin interacts with the TIR1/AFB auxin receptors and promotes degradation of the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins to allow ARF-dependent regulation of transcription (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Genetic studies of the TIR1/AFB, Aux/IAA, and ARF genes indicate that auxin is required for establishment of the root meristem and postembryonic root growth. Because previous genetic screens for auxin-resistant mutants involved examination of auxin response in the seedling root, it is possible that mutants with severe defects in auxin response were not recovered. In an attempt to circumvent this problem, we used the well-characterized auxin reporter DR5rev:GFP (for green fluorescent protein) to screen for mutants with altered auxin response. We isolated several previously uncharacterized mutants with short primary roots and auxin response defects. Here, we report the isolation and characterization of weak auxin response1 (wxr1), an allele of the ROOT UV-B SENSITIVE2 (RUS2) gene (Leasure et al., 2009). RUS2/WXR1 encodes a protein belonging to the DUF647 family (for Domain of Unknown Function 647). In this study, we show that the wxr1 mutant has reduced levels of the auxin efflux proteins PIN1, PIN2, and PIN7. This defect results in a reduction in polar auxin transport and, as a consequence, altered auxin responses.
RESULTS
The wxr1 Mutant Has Short Roots and an Auxin Response Defect
To discover new genes involved in auxin signaling, we mutagenized seeds carrying the auxin-responsive reporter DR5rev:GFP with ethyl methanesulfonate and screened M2 plants on medium containing 75 nM 2,4-D. Five-day-old seedlings with reduced GFP signal in the root were identified and transferred to medium without auxin for further analysis. Eight single gene recessive mutants were ultimately recovered, called wxr mutants.
The wxr1 mutant displays severe defects in root development and slight defects in leaf and inflorescence development (Figure 1). To characterize the wxr1 phenotype in more detail, M3 plants were backcrossed to Columbia-0 (Col-0) (DR5rev:GFP) plants three times. When 5-d-old seedlings were placed on 75 nM 2,4-D medium for 12 h, wild-type plants carrying DR5rev:GFP display an increase in GFP signal in the root tip, whereas wxr1 plants do not exhibit an obvious change (Figures 1B and 1D). A similar result was obtained when wxr1 plants were tested on medium containing IAA or 1-napthalene acetic acid (see Supplemental Figure 1 online).
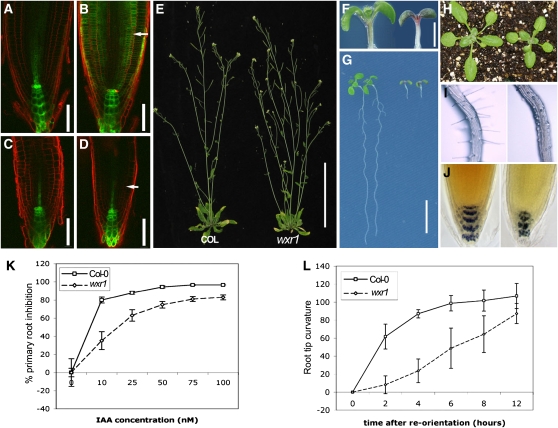
Figure 1.
Phenotype of the wxr1 Mutant.
(A) to (D) DR5rev:GFP expression in 5-d-old wxr1 and wild-type seedlings. Before auxin treatment, the GFP signal (green) is similar in wild-type (A) and in wxr1 (C) root tips. Treatment with 75 nM 2,4-D for 12 h results in a strong increase in GFP signal in the wild type (B) but not in the mutant (D). Roots were stained with propidium iodide before observation. Arrows indicate cortical cells. Bars = 50 μm; n ≥ 30.
(E) Five-week-old wxr1 plants (right) display decreased apical dominance compared with wild-type plants (left). Bar =10 cm.
(F) 7-day-old wxr1 (right) seedlings accumulate more pigment in cotyledons and hypocotyls than does the wild type (left). Bar = 1 mm.
(G) The growth of wxr1 roots (right, 7 d old) is dramatically reduced. Bar = 1 cm.
(H) The rosette leaves and petioles of 3-week-old wxr1 plants (right) are smaller than the wild type (left).
(I) wxr1 root hairs (right) exhibit reduced root hair elongation compared with the wild type (left).
(J) Wild-type (left) and wxr1 roots (7 d after germination) stained with Lugol's solution.
(K) Effect of auxin on root elongation in wild-type and wxr1 seedlings grown in yellow light. Bars represent se, n = 14
(L) The roots of wxr1 seedlings exhibit a reduced response to gravity after 90° reorientation on half-strength MS medium in the dark. Error bars represent se, n = 25
When grown using our typical growth conditions (continuous white light, 80 to ∼90 μE m−2 s−1, 22°C), the young wxr1 seedlings accumulate anthocyanin in the cotyledons and hypocotyls (Figure 1F) and primary root elongation is dramatically reduced. The roots of wild-type seedlings were 5.5 ± 0.5 cm (mean ± se, n = 16) after 7 d on ATS medium, whereas wxr1 roots were only 0.5 ± 0.1 cm (mean ± se, n = 15) (Figure 1G). Furthermore, root hairs on mutant seedlings initiate but do not elongate (Figure 1I). Lugol staining revealed that 7-d-old mutant roots had fewer columella cells than did wild-type roots and that those cells were disorganized (Figure 1J). The cell pattern was irregular in the wxr1 root tip, and the meristem region was much smaller (Figures 2A and 2B). Five-day-old wild-type plants had 49 ± 4.5 (mean ± se, n = 10) meristem cortex cells, whereas wxr1 plants had 22 ± 2 meristem cortex cells (mean ± se, n = 10, t test, P < 0.005). The rosette leaves of 3-week-old wxr1 mutants were smaller than those of wild-type plants (Figure 1H), and at 5 weeks, mutant plants displayed a slight decrease in apical dominance. Shoot number, including primary and axillary shoots, was 3.88 ± 0.60 in wxr1 and 1.63 ± 0.99 in wild-type plants (mean ± se, n = 50, t test, P < 0.005). These data indicate that the wxr1 mutation affects mainly root growth and has a smaller effect on development of the rosette and inflorescence.
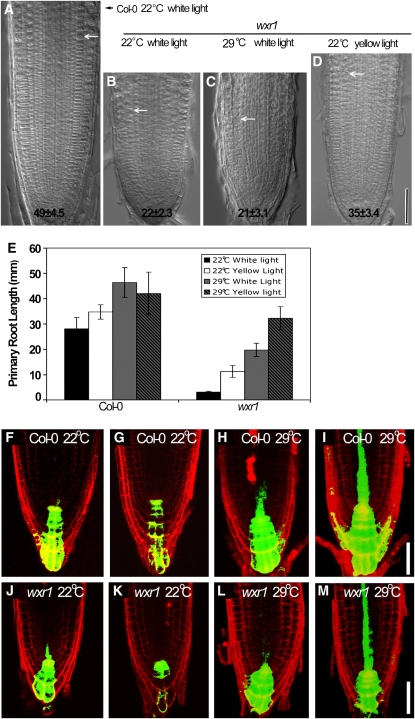
Figure 2.
The wxr1 Mutation Causes Short Primary Roots and Root Meristem Cell Proliferation Defects.
(A) to (D) Five-day-old wild-type and wxr1 seedlings grown under indicated light and temperature conditions. Arrows show the boundary between the meristem zone and elongation zone. Numbers indicate cortical cell number in the meristem zone (mean ± se). Bar = 50 μm.
(E) Elongation of 7-d-old wxr1 roots was rescued by low intensity light or high temperature. Error bars represent se, n = 30.
(F) to (M) DR5rev:GFP expression in 5 d after germination wxr1 and wild-type seedlings grown at indicated temperature. Seedlings in (F), (H), (J), and (L) were grown in 80 to 90 μE m−2 s−1, while those in (G), (I), (K), and (M) were grown at 30 μE m−2 s−1. Bars = 50 μm.
During the course of our studies, we found that wxr1 plants are hypersensitive to both light and temperature. When grown at the elevated temperature of 29°C in white light, the meristem cortex cell number of wxr1 plants did not change (21 ± 3.1, mean ± se, n = 10), but primary root elongation increased dramatically (Figures 2C and 2E). When wxr1 plants were grown at lower light levels (∼50 μE m−2 s−1, 24 h/day, 22°C, yellow filters), both primary root length and meristem cell number were increased relative to the wild type (Figures 2D and 2E). To determine if this effect was specific for yellow light, we also grew seedlings in low levels of far-red, red, and blue light, as well as in dark conditions. In each case, the effect of wxr1 on root growth was reduced relative to that in high levels of white light, indicating that wxr1 roots were responding to reduced light intensity rather than to a specific wavelength (see Supplemental Figure 2 online). In yellow light and high temperature (∼50 μE m−2 s−1, 24 h/day, 29°C), wxr1 root elongation was further stimulated (Figure 2E). High temperature has been shown to cause an increase in auxin levels in seedlings, and light affects both local auxin levels as well as auxin transport (Gray et al., 1998; Gil et al., 2001; Tao et al., 2008). Thus, it is possible that increased temperature and reduced light result in increased levels of endogenous auxin and that this change partially restores root growth. To investigate this possibility, we used the DR5rev:GFP reporter. Surprisingly, we found that GFP levels were decreased in both wild-type and wxr1 roots grown at low light (∼30 μE m−2 s−1, 22°C) (Figures 2F, 2G, 2J, and 2K). However, GFP levels were increased at elevated temperature in both genotypes (Figures 2H and 2L). Furthermore, this increase was even more dramatic in roots grown in low light and high temperature, suggesting a synergism between these two environmental conditions (Figures 2I and 2M).
Since primary root elongation is reduced so dramatically under our standard light conditions, it was difficult to measure changes in primary root length in wxr1. As an alternative, we analyzed root growth under yellow light. The results in Figures 1K and 1L show that wxr1 seedlings have reduced responses to IAA and gravity, consistent with a defect in auxin response. To characterize further the gravitropic defect, we also constructed double mutants with axr2-1, aux1-7, and abcb1. Each of these mutants is affected in gravitropism due to a defect in auxin signaling or transport (Pickett et al., 1990; Wilson et al., 1990; Noh et al., 2001). The double mutants were all more severely affected than were the corresponding single mutants (see Supplemental Figure 3 online).
The wxr1 Mutant Exhibits a Synergistic Interaction with Mutations in Auxin Receptor Genes
To examine the function of WXR1 in auxin signaling, we introduced wxr1 into tir1-1, tir1-1 afb3-1, and tir1-1 afb2-1 afb3-1 mutants defective in auxin perception (Dharmasiri et al., 2005b). The seedling roots of tir1-1 and tir1-1 afb3-1 were similar to those of wild-type plants except that they had fewer lateral roots (Dharmasiri et al., 2005b). By contrast, 24% of wxr1 tir1-1 afb3-1 seedlings (n = 147) did not develop a root and died on the plates. Similarly, introduction of wxr1 into tir1-1 afb2-1 afb3-1 had a dramatic effect on seedling phenotype. Greater than 50% of tir1-1 afb2-1 afb3-1 survived beyond the seedling stage, whereas <12% of wxr1 tir1-1 afb2-1 afb3-1 seedlings survived (n = 176) and these had much more severely affected primary roots than those of wxr1. The rosettes of 3-week-old wxr1 tir1-1 afb2-1 afb3-1 plants had extremely small rosette leaves compared with those of tir1-1 afb2-1 afb3-1 and wxr1 plants and exhibited an early yellowing phenotype (Figures 3A to 3F). The inflorescences of wxr1 tir1-1 afb2-1 afb3-1 were also very small, whereas those of wxr1 tir1-1 afb3-1 were similar to those of tir1-1 afb2-1 afb3-1 plants after flowering (Figure 3G). These results confirm that wxr1 is deficient in auxin-related processes.
Figure 3.
The wxr1 Mutation Displays a Synergistic Interaction with the tir1-1 afb2-1 afb3-1 Mutant.
(A) to (F) Three-week-old wxr1 tir1-1 afb2-1 afb3-1 plants have extremely small curled-down rosette leaves. The plants from (A) to (F) are Col-0, tir1-1 afb2-1 afb3-1, wxr1, wxr1 tir1-1, wxr1 tir1-1 afb3-1, and wxr1 tir1-1 afb2-1 afb3-1, respectively. Bars = 1 cm.
(G) Five-week-old wxr1 tir1-1 afb2-1 afb3-1 plants produce extremely short inflorescences. Plants from left to right are Col-0, wxr1, tir1-1 afb2-1 afb3-1, wxr1 tir1-1 afb3-1, and wxr1 tir1-1 afb2-1 afb3-1, respectively. Bar = 10 cm.
Aux/IAA Degradation in the wxr1 Mutant
To investigate further the effects of WXR1 on auxin response, the HS:AXR3NT-GUS (for β-glucuronidase) transgene was introduced into wxr1 mutant plants. AXR3NT-GUS is a substrate for SCFTIR1/AFBs and provides a useful tool to assess SCFTIR1/AFB function in the plant (Gray et al., 2001; Dharmasiri et al., 2005b). Seven-day-old wild-type and wxr1 seedlings carrying the HS:AXR3NT-GUS construct were heat shocked for 2 h at 37°C in the dark. Seedlings were transferred to plates with or without IAA at 22°C and stained for GUS at time intervals. The roots of wxr1 seedlings accumulated much more AXR3NT-GUS than did those of wild-type plants following heat shock (Figure 4A). However, upon auxin treatment, GUS staining rapidly decreased in both wild-type and mutant roots. Measurement of GUS activity by fluorescent β-galactosidase methylumbelliferylglucuronide (MUG) assay showed that AXR3NT-GUS was degraded at a similar rate in both wild-type and wxr1 plants (Figure 4H). These results indicate that the auxin-resistant aspects of the wxr1 phenotype cannot be explained by a reduction in SCFTIR1/AFB-dependent degradation of Aux/IAA proteins. However, AXR3NT-GUS accumulation was reduced in the cotyledons and hypocotyls of wxr1 plants compared with the wild type, suggesting that degradation of the protein was accelerated in the aerial parts of the wxr1 mutant (Figures 4B and 4C).
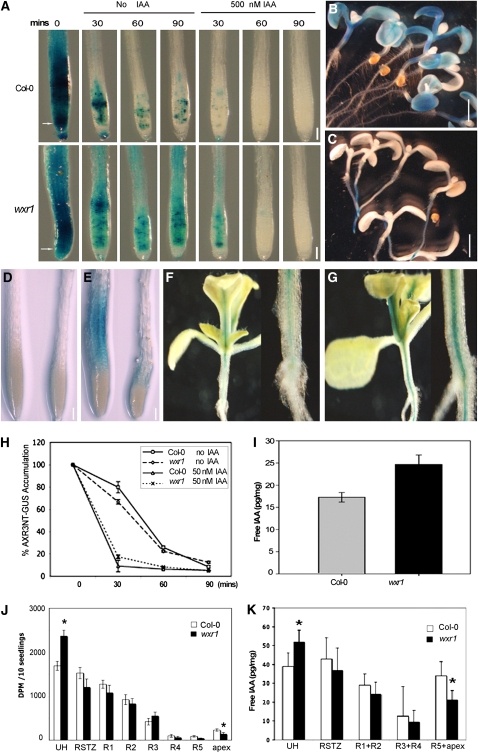
Figure 4.
Auxin Distribution Is Altered in the wxr1 Mutant.
(A) GUS staining of HS:AXR3NT-GUS lines after heat shock in the absence and presence of 500 nM IAA. AXRNT-GUS levels were higher in the untreated mutant than in the wild type but decreased upon auxin treatment.
(B) and (C) AXR3NT-GUS staining in wild-type (B) and wxr1 cotyledons (C) after a 2-h heat shock and 20-min recovery at room temperature.
(D) and (E) Expression of BA3:GUS in Col-0 and wxr1 roots after 24 h treatment with buffer (D) or 100 nM IAA (E). In each panel, Col-0 root is on the left and the wxr1 root on the right.
(F) and (G) Expression of BA3:GUS in shoots of Col-0 (F) and wxr1 plants (G). The image on the right of each panel is a close up of the hypocotyl.
(H) GUS activity in the root tips of seedlings from (A) measured by MUG assay. Error bars represent se, n = 30.
(I) The levels of IAA are increased in the shoots of wxr1 plants compared with the wild type. Error bars represent sd, n = 10
(J) Transport of 3H-IAA was reduced in wxr1 seedlings. UH, upper hypocotyl; RSTZ, root shoot transition zone; apex, root apex. The asterisks indicate significant differences between wild-type and mutant values based on Student's t test, P < 0.05. Error bars represent se, n = 10.
(K) IAA levels in segments as described for (J). Error bars represent se, n = 3
Bars = 100 μm in (A), (D), and (E) and 1 mm in (B) and (C).
Auxin Accumulates in the Shoots of wxr1 Plants
To explore how auxin distribution is affected in the wxr1 mutants, BA3:GUS, a sensitive auxin-responsive reporter, was introduced into wxr1 plants (Oono et al., 1998). As for the DR5rev:GFP reporter, GUS staining was weaker in auxin-treated mutant roots compared with the wild type (Figures 4D and 4E). By contrast, GUS staining was stronger in the leaves and hypocotyls of wxr1 plants than in those of the wild type (Figures 4F and 4G).
One interpretation of these results is that wxr1 affects transport of IAA from sites of production in the shoot to the root. To assess this possibility, we directly measured free IAA levels in aerial tissues of wild-type and mutant seedlings. The results show that IAA levels are increased in the mutant compared with the wild type (Figure 4I). To test our hypothesis that the wxr1 mutant is deficient in auxin transport, we directly measured transport in seedlings using 3H-IAA. The results in Figure 4J show that transport of applied IAA out of the hypocotyl into the root is reduced in wxr1 plants. In addition, we found a reduction in accumulation of IAA in the root tip (Figure 4K). Together, these results indicate that wxr1 interferes with IAA distribution in the Arabidopsis seedling by reducing auxin transport.
The wxr1 Mutant Has Reduced Levels of PIN and AUX1 Protein in the Plasma Membrane
Auxin efflux carriers of the PIN family and the auxin influx carrier AUX1 both contribute to polar auxin transport. Because wxr1 plants are deficient in auxin transport, we next examined the levels and distribution of several PIN proteins and AUX1 in the mutant. Localization of AUX1 using a AUX1:AUX1-YFP line revealed slightly reduced AUX1 levels in the wxr1 background (Figures 5G and 5H) (Swarup et al., 2004). By contrast, in situ immunodetection using anti-PIN1 and anti-PIN2 antisera revealed that the levels of these two proteins were dramatically decreased in 3- to 4-d-old wxr1 roots compared with the wild type (Figures 5A to 5D). To examine PIN7 levels, we introduced PIN7:PIN7-GFP into wxr1 plants. As for the other PIN proteins, the level of PIN7-GFP was clearly reduced in 5-d-old wxr1 root tips (Figures 5E and 5F). To determine if these differences reflect changes in transcription, the transcript levels of AUX1, PIN1, PIN2, and PIN7 were examined in 5-d-old wxr1 seedlings by quantitative RT-PCR. The results indicate that transcript levels are similar in Col-0 and wxr1 plants, suggesting that the effect on protein level is posttranscriptional (Figure 5I).
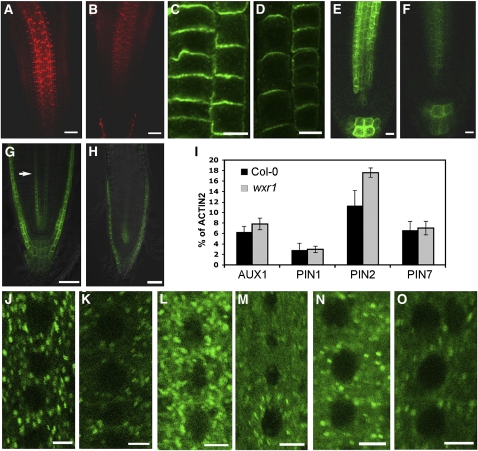
Figure 5.
The Levels of Auxin Transport Carriers Are Reduced in wxr1 Plants.
(A) to (H) The levels of PIN and AUX1 proteins are reduced in wxr1 plants. Each experiment was repeated at least three times with at least 30 roots observed for each treatment.
(A) and (B) PIN1 visualized with anti-PIN1 antibody in the wild type (A) and wxr1 (B). Bars = 50 μm.
(C) and (D) PIN2 visualized with anti-PIN2 antibody in the wild type (C) and wxr1 (D); epidermal cells on the left and cortical cells on the right of each panel. Bars = 10 μm.
(E) and (F) PIN7:PIN7-GFP in wild-type (E) and wxr1 (F) roots. Bars = 10 μm.
(G) and (H) AUX1:AUX1-YFP in wild-type (G) and wxr1 (H) seedlings (arrow indicates protophloem cells). Bars = 50 μm.
(I) AUX1, PIN1, PIN2, and PIN7 transcript levels were assessed by real-time RT-PCR. ACT2 was used as a control.
(J) to (O) Comparison of VHA-α1-GFP ([J] and [K]), GN-GFP ([L] and [M]), and ARA-7-GFP ([N] and [O]) levels in root epidermal cells of wild-type ([J], [L], and [N]) and wxr1 ([K], [M], and [O]) plants. Bars = 5 μm.
To determine if wxr1 has a specific effect on proteins involved in auxin transport, we crossed transgenes expressing the marker proteins VHA-a1-GFP (trans-Golgi network), GNOM-GFP (recycling endosomes), or ARA7-GFP (prevacuolar compartment) into mutant plants (Geldner et al., 2003; Lee et al., 2004; Dettmer et al., 2006). Surprisingly, the levels of these proteins were also reduced in wxr1, indicating that the effects of the mutation are not specific to auxin transport proteins (Figures 5J to 5O). By contrast, the levels of the tonoplast marker vac-yb was increased in wxr1 roots (see Supplemental Figure 4 online).
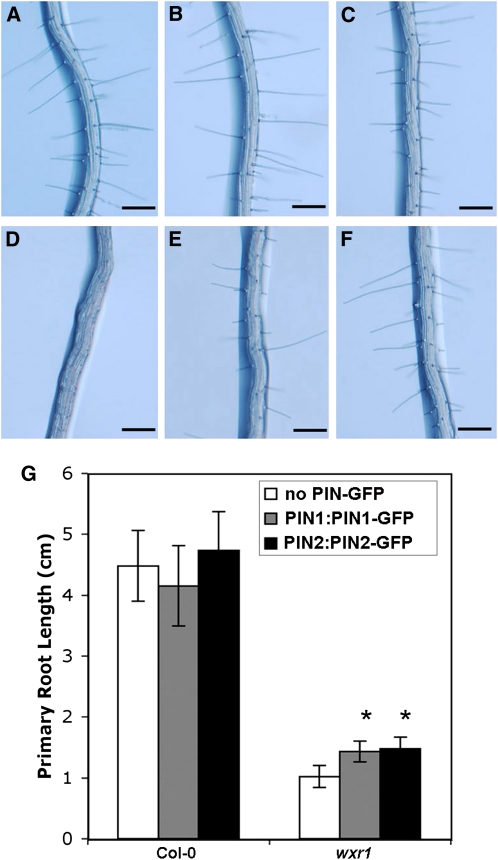
If the wxr1 phenotype is related to reduced PIN levels, it is possible that more PIN protein may partially suppress the phenotype. To test this possibility, we crossed PIN1:PIN1-GFP and PIN2:PIN2-GFP into wxr1 plants. Plants homozygous for these transgenes, carrying four copies of the PIN1 and PIN2 genes, respectively, displayed increased root elongation compared with wxr1 (n ≥ 14, t test, P < 0.001), indicating partial rescue of the mutant phenotype (Figure 6G). In addition, the root hairs of wxr1 PIN1:PIN1-GFP and wxr1 PIN2:PIN2-GFP were nearly the same as those of the wild type (Figures 6A to 6F).
Figure 6.
Increased Expression of PIN1 and PIN2 Partially Rescues Aspects of the wxr1 Phenotype.
(A) to (F) Root hairs of Col-0 (A), PIN1:PIN1-GFP (B), PIN2:PIN2-GFP (C), wxr1 (D), wxr1 PIN1:PIN1-GFP (E), and wxr1 PIN2:PIN2-GFP (F); n = 25. Bars = 200 μm.
(G) Root length is increased in 8-d-old PIN1:PIN1-GFP wxr1 and PIN2:PIN2-GFP wxr1 compared with wxr1. Bars represent se; n ≥ 14, t test, P < 0.001.
[See online article for color version of this figure.]
WXR1 Is a Member of the DUF647 Family and Is Expressed in All Plant Organs
To map the wxr1 mutation, genomic DNA was isolated from 438 F2 plants generated in a cross of wxr1 (Col-0) with Landsberg erecta. Homozygous wxr1 plants were identified based on the short-root phenotype. The wxr1 mutation was first mapped between simple sequence length polymorphism markers nga168 and nga1126. Additional fine mapping placed the mutation in a 90-kb region containing 42 genes. The coding regions of all 42 genes were sequenced. A single mutation was found in At2g31190 that results in the substitution of Ser-105 with Leu. At2g31190 is identical to the previously identified RUS2 gene, implicated in response to UV light (Leasure et al., 2009). Ser-105 is located in a highly conserved motif, TQXLL, present in most members of this family (see Supplemental Figure 5A online).
DUF647 protein family members are encoded by most of the eukaryotic genomes sequenced to date (see Supplemental Data Set 1 online). Phylogenetic analysis indicates that several subfamilies diverged very early in eukaryotic evolution, but different eukaryotic lineages exhibit different propensities to retain members of the different DUF647 clades (see Supplemental Figure 6 online). Whereas plants and red algae have retained multiple lineages of DUF647 proteins, most animals and fungi have only retained a single DUF647 protein, and several species, including yeasts and nematodes, lack DUF647 proteins. Thus, DUF647 proteins are probably involved in a process common to most, but not all, eukaryotes.
The At2g31190 cDNA was isolated by RT-PCR and used to generate C-terminal GFP and GUS fusions under control of the cauliflower mosaic virus 35S promoter or the WXR1 promoter and introduced into wxr1 plants. Two WXR1:WXR1-GFP, four 35S:WXR1-GFP, and eight 35S:WXR1-GUS transgenic lines were analyzed (see Supplemental Figure 5B online). Root growth was partially restored in each line. However, the WXR1-GFP proteins were much more effective in rescuing the mutant phenotype than the WXR1-GUS line, perhaps because of the large size of the GUS protein. Overall, these results confirm that At2g31190 is WXR1.
To analyze expression of the WXR1 gene, we generated a WXR1:WXR1-GUS construct in which the same WXR1 genomic DNA sequence of WXR1:WXR1-GFP was fused in frame with the GUS reporter gene and introduced into wxr1 plants. GUS staining revealed that the transgene is active in most plant organs, including cotyledons, hypocotyls, roots, stems, flowers, and leaves (see Supplemental Figures 7A to 7D online). In particular, root tips, vascular tissues of all organs, and lateral root primordia showed strong GUS staining (see Supplemental Figures 7A to 7E online). Strong staining was also observed in root pericycle cells and lateral root primorda, suggesting that WXR1 may play a role in lateral root initiation, perhaps through its effects on auxin transport.
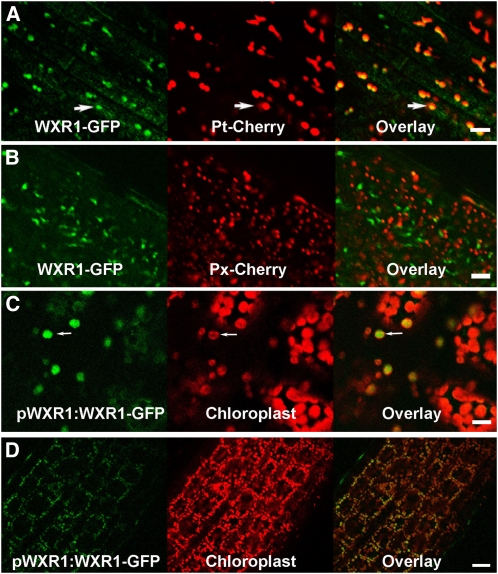
WXR1 Is Present in Plastids
Several proteomic studies suggest that WXR1 is localized to the chloroplast envelope (Ferro et al., 2003; Dunkley et al., 2006). To investigate this possibility further, we introduced WXR1:WXR1-GFP into wxr1 as well as into Col-0 plants expressing pt-rb and px-rb proteins, mCherry-based markers for the plastid and peroxisome, respectively (Nelson et al., 2007). The data in Figures 7A and 7B show that WXR1-GFP colocalizes with pt-rb but not px-rb, indicating that WXR1 is localized to the plastid. The distribution of WXR1-GFP was also examined in hypocotyls and young rosette leaves. In this case, chloroplasts were visualized by autoflorescence. In both tissues, WXR1-GFP is associated with the chloroplast, confirming that the protein is plastid localized (Figures 7C and 7D).
Figure 7.
WXR1 Localizes to Plastids.
(A) and (B) WXR1-GFP colocalizes with plastid marker Pt-Cherry in epidermal cells of wild-type roots (A) but not with the peroxisome marker PX-Cherry (B).
(C) and (D) WXR1-GFP colocalizes with autofluorescence of the chloroplast in wxr1 WXR1:WXR1-GFP leaves (C) and hypocotyls (D). Bars = 10 μm in (A) to (C) and 20 μm in (D).
Removal of UV-B Light Does Not Restore the wxr1 Phenotype
Previous studies of the rus2/wxr1 and rus1 mutants suggested that the mutant phenotype is primarily the result of hypersensitivity to very-low-fluence-rate UV-B (Tong et al., 2008; Leasure et al., 2009). To explore this possibility further with the wxr1 mutant, we examined the response of seedlings to various growth conditions. To test the effects of UV light on root growth, we compared seedlings grown in dark conditions, white light, or white light passing through a UV filter that removes essentially all UV-B. Our results show that dark conditions increase root growth in wxr1 seedlings but do not restore it to wild-type levels (see Supplemental Figure 8 online). By contrast, the removal of UV-B light resulted in only a modest stimulation of root growth in both wild-type and wxr1 seedlings. These results suggest that factors other than UV light contribute to the wxr1 phenotype.
DISCUSSION
Auxin has an essential role in many plant growth processes, including establishment of the root meristem during embryogenesis subsequent patterning of the meristem, and other aspects of root development (Sabatini et al., 1999). In fact, many genes involved in auxin signaling have been identified in screens for mutants that display auxin-resistant root growth (Mockaitis and Estelle, 2008). However, mutants with severe defects in auxin perception or response, such as tir1 afb2 afb3 or bdl, either lack a root completely or produce a very short root and thus would not be recovered in a simple screen for auxin resistance (Hamann et al., 1999; Dharmasiri et al., 2005b). To identify new genes in auxin signaling, we used the DR5rev:GFP reporter and concentrated on mutants with short roots. In this study, we focus on the role of RUS2/WXR1 in auxin-regulated root development.
The wxr1 mutant was isolated based on reduced expression of DR5rev:GFP in the root. In addition, we show that WXR1 is required for a number of auxin-regulated processes, such as root hair and lateral root production, root gravitropism, and root meristem maintenance. In addition, we observe a synergistic interaction between wxr1 and mutations in the TIR1/AFB family of auxin receptors, consistent with a role for WXR1 in some aspect of auxin biology.
Although it is clear that expression of both DR5rev:GFP and BA3:GUS is reduced in the root tips of wxr1 plants, these reductions, as well many aspects of the pleiotropic growth phenotype, are better explained by a reduction in auxin transport rather than response. In young seedlings, the cotyledons and developing leaves are the primary sites of auxin production. This auxin is transported through the hypocotyl into the root system where it supports root elongation and the formation of lateral roots. Transport assays indicate that IAA transport from the apex of the seedling is reduced in wxr1 seedlings. Indeed, free IAA levels are increased in the aerial portion of mutant seedlings compared with the wild type but decreased in the root apex. However, it is important to note that WXR1 is unlikely to have a specific role in auxin transport, and some aspects of the phenotype are probably not related to auxin (see below).
Auxin transport is mediated by the AUX1/LAX influx carriers as well as the PIN and ABCB efflux carriers (Bandyopadhyay et al., 2007; Robert and Friml, 2009). The cellular localizations of both AUX1 and the PINs are dynamically regulated in a cell-type-specific manner. In the case of the PINs, localization occurs through a GNOM-dependent recycling pathway and is regulated by the PID kinase (Friml et al., 2004; Michniewicz et al., 2007; Kleine-Vehn et al., 2009; Sukumar et al., 2009; Zhang et al., 2010). AUX1 trafficking is not dependent on GNOM but does require an endoplasmic reticulum–localized protein called AXR4 (Dharmasiri et al., 2006; Kleine-Vehn et al., 2008). In our analysis, we find that WXR1 is required for normal accumulation of both PINs and AUX1 on the plasma membrane, providing a likely explanation for the observed decrease in auxin transport. In addition, we find that the levels of several other proteins involved in the endosomal pathway are reduced, including VHA-α1-GFP, GNOM-GFP, and ARA7-GFP, indicating that the effect of wxr1 is not restricted to auxin transport proteins.
WXR1 is a member of a family of six DUF647 proteins in Arabidopsis and identical to the previously identified RUS2 protein. Mutations in RUS2 and another member of the family called RUS1 result in hypersensitivity to UV light (Tong et al., 2008; Leasure et al., 2009). For a variety of reasons, it is unlikely that the primary function of RUS2/WXR1 is to mediate response to UV light. First, we were unable to demonstrate UV-hypersensitive root growth in wxr1. In fact, even in complete darkness, wxr1 roots are shorter than wild-type roots. It is possible that this difference is related to the specific allele tested or an unknown difference in growth conditions. Second, we find that conditions that increase auxin movement into the root, such as elevated temperature or overexpression of PIN1, restore root growth in mutant roots. Third, the wxr1 mutation enhances the strong embryonic phenotype of the receptor tir1 afb2 afb3 mutants, indicating that WXR1 contributes to embryogenesis. It seems likely that both the defect in auxin transport/distribution and the UV hypersensitivity are consequences of a defect in cellular regulation, perhaps in protein trafficking. Until we obtain information on the biochemical function of the DUF647, this issue will remain unresolved.
Proteomic studies suggest that WXR1 is localized to the chloroplast envelope (Ferro et al., 2003; Dunkley et al., 2006). We confirmed this using a WXR1-GFP line. Because the biochemical function of RUS2/WXR1 and other members of the DUF647 family is unknown, there is no clear model for how the loss of a chloroplast-localized protein impacts the levels of proteins in other membranes. DUF647-containing proteins are present in other plants, as well as most animals and fungi, and in the future it will be interesting to determine if these proteins also function in endocytosis and protein trafficking.
METHODS
Plant Materials and Conditions
All Arabidopsis thaliana lines used in this article were generated in the Col-0 ecotype with the exception of afb2-1 and afb3-1 lines, which are T-DNA insertion lines in Wassilewskija ecotype. The transgenic plants GN:GN-GFP, VHA-α1-GFP, and ARA-7-GFP (Geldner et al., 2003; Lee et al., 2004; Dettmer et al., 2006) were kindly provided by Gerd Jurgens. The PIN1:PIN1-GFP, PIN2:PIN2-GFP, PIN7:PIN7-GFP, DR5rev:GFP, and AUX1:AUX1-YFP have been described (Swarup et al., 2001; Benkova et al., 2003; Blilou et al., 2005). The HS:AXR3NT-GUS wxr1 and BA3:GUS wxr1 lines were made by crossing war plants to HS:AXR3NT-GUS and BA3:GUS plants (Oono et al., 1998; Gray et al., 2001). The axr2-1, abcb1/pgp1, and aux1-7 mutants have been described (Pickett et al., 1990; Wilson et al., 1990; Geisler et al., 2005). Seedlings were grown in a growth chamber (80 to ∼90 μE/m2s measured by LI-185B Quantum/Radiometer/photometer [LI-COR], 24 h/day, 22°C) on ATS medium plates except where indicated (Lincoln et al., 1990). Yellow light (∼50 μE/m2s measured by LI-185B Quantum/Radiometer/photometer; LI-COR) was produced using a medium yellow filter, 480 (Gamproducts). 1-napthalene acetic acid, 2,4-D, and IAA treatment were performed by adding different auxins to half-strength Murashige and Skoog (MS) (1% sucrose and 0.8% agar) plates.
35S:WXR1-GFP and 35S:WXR1-GUS constructs were generated by introducing full length of WXR1 from pENTR-D (Invitrogen) into pMDC83 and pMDC140 using the Gateway LR reaction kit (Invitrogen). WXR1:WXR1-GFP and WXR1:WXR1-GUS constructs were made by introducing WXR1 gnomic DNA sequence plus 2.0 kb upstream genomic DNA sequence into pMDC107 and pMDC163. The primers (WXR1CDS, sense, 5′-CACCATGCAGTTTCTTCAGGAGAAGG-3′, antisense, 5′-AGCAAATCTTGTCCCCGTGC-3′; WXR1 genomic DNA with promoter, sense, 5′-CACCGGTCATGCAATCTCGCTTTTG-3′, antisense, 5′-AGCAAATCTTGTCCCCGTGC-3′) were synthesized by Invitrogen. pt-rb, px-rb, and vac-yb were constructed as described (Nelson et al., 2007).
To measure the gravitropic response, seedlings were grown under yellow light for 5 d. After rotating 90° (Figure 1L) or 135° (see Supplemental Figure 3 online) clockwise, the root tip position was recorded with a digital camera once every 2 h for 12 h (Figure 1L) or after 12 h (Supplemental Figure 3 online) and measured using ImageJ. Plants in soil were grown with continuous light in a growth room at 22°C.
Ethyl Methanesulfonate Mutagenesis and Mutant Screen
Approximately 10,000 DR5rev:GFP seeds were treated with 0.3% ethyl methanesulfonate for 10 h. After extensive washing, the mutagenized seeds were planted in soil. Approximately 40,000 M2 seeds were germinated on ATS medium. Seedlings with a primary root less than half the length of the control root were transferred to ATS medium supplemented with 75 nM 2,4-D.
After a 12-h treatment, seedlings with reduced levels of GFP signal compared with the control were transferred to soil. Each candidate mutant was crossed to Col-0 DR5rev:GFP and HS:AXR3NT-GUS lines (Gray et al., 2001). The genetic behavior of each mutant was assessed in the F2 generation. Homozygous F2 plants were backcrossed to Col-0 DR5rev:GFP three times prior to analysis.
GUS Histochemical Staining
Six-day-old wild-type and wxr1 plants homozygous for the HS:AXR3NT-GUS were heat shocked for 2 h at 37°C in the dark to induce AXR3NT-GUS expression. Treated seedlings were transferred to control ATS medium ±50 nM IAA. At time intervals (0, 20, 40, and 60 min), whole plants were collected and stained overnight at 37°C with staining solution [100 mM Na2PO4, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, and 1.0 mM K3Fe(CN)6] containing 2 mM X-Gluc to visualize GUS activity. GUS activity was measured in extracts prepared from the terminal 0.5 cm of the root. Dissected tips from ∼100 roots were homogenized in 100 μL extraction buffer (50 mM Na2PO4, pH 7.0, 10 mM DTT, 1 mM EDTA, 0.1% sodium lauryl sarcosine, and 0.1%Triton X-100). Twenty microliters of extract were added into 180 μL MUG assay buffer (1 mM MUG in extraction buffer). After incubation for 4 h at 37°C in the dark, an equal amount of 0.2 M Na2CO3 was added to stop the reactions. Enzyme activity in each sample was measured using a spectrofluorimeter. This experiment was repeated three times with the same result.
To examine BA3:GUS activity, 6-d-old wild-type and wxr1 plants carrying the transgene were treated for 12 h on ATS plates with or without 100 nM IAA. Whole plants were stained overnight with X-Gluc staining buffer containing 2 mM X-Gluc. Over 50 roots from wxr1 and wild-type plants were observed.
Auxin Transport Assay
Auxin transport was assayed using 3H-IAA radiotracer assays as described (Blakeslee et al., 2007). Seedlings used for transport assays were grown for 5 d at 120 μE/m2s white light, 0.5% sucrose, quarter-strength MS, pH 5.5. Data are from three independent experiments. A 2-mm section centered on the root-shoot transition zone was first excised, followed by an upper hypocotyl section excised immediately below the cotyledon petioles. The root apex was then excised (0.5 mm) followed by serial 2-mm sections up the root. Sampled sections from 10 seedlings were pooled for each measurement and counted in a low-range scintillation counter (Tri-Carb; Perkin-Elmer). Values presented are means and standard deviations from three independent experiments and are presented in order downward from upper hypocotyls to root apex.
IAA Determinations
To measure free auxin levels in aerial tissues (Figure 4I), seedlings were grown on half-strength MS plates (Caisson Laboratories) with 1% sucrose and 0.5% phytagel (Sigma-Aldrich). The Petri dishes were incubated vertically at 22°C for 7 d under continuous light (70 μmol m−2 s−1) provided by cool-white florescent tubes. Fresh whole seedlings (25 to 50 mg) were harvested, weighed, and frozen in liquid nitrogen. After adding 3 ng of internal standard, [13C6] IAA, each sample was homogenized in a Mixer-mill (Qiagen) extracted, purified by two sequential solid phase extraction steps, methylated, and then analyzed using selected ion monitoring gas chromatography–mass spectrometry on an Agilent 5973A instrument, exactly as described by Barkawi et al. (2008). At least 10 biological replicates were analyzed for each treatment.
Free IAA determinations in hypocotyl and root segments (Figure 4K) were performed as described (Kim et al., 2007). Seedlings were grown as described for transport assays. Data are from three independent experiments. Results were analyzed for statistical significance in pairwise Student's t tests followed by Newman Keuls posthoc analysis.
Microscopy and Drug Treatment
Confocal imaging was performed on an inverted SP/2 confocal microscope (Leica). At least 30 roots were observed for each sample, and each experiment was repeated at least three times. Propidium iodide staining was performed by incubating roots samples in 10 μg/mL propidium iodide (Sigma-Aldrich) and half-strength MS (1% sucrose) solution for 15 min. For Lugol staining, seedlings were stained in Lugol solution (Sigma-Aldrich; L-6146) for 1 h at room temperature. Immunofluorescence analysis of Arabidopsis roots was performed using anti-PIN1 (1:400) and anti-PIN2 (1:800) as described by Dhonukshe et al. (2007). Excitation wavelengths were 488 nm (argon laser); emission was detected at 520 to 560 nm for GFP. Pinhole and gain settings were identical among treatments.
Real-Time RT-PCR Assay
AUX1, PIN1, PIN2, and PIN7 levels were determined in root tips of 7-d-old Col-0 and wxr1 seedlings grown under yellow light. The terminal 0.5 to 1.0 cm of roots was collected by dissection using a razor blade. Total RNA was isolated from ∼50 mg of root tips using the RNAeasy plant mini kit (Qiagen). The cDNA was obtained from ∼2 ug total RNA using a Superscript Double cDNA synthesis kit (Invitrogene). Real-time RT-PCR reactions were performed on a Mastercycler realplex4 (Eppendorf) with 40 cycles of 95°C 15 s, 55°C 10 s, 72°C 20 s. ACT2 (sense, 5′-ATTCAGATGCCCAGAAGTCTTGTTC-3′; antisense, 5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′), PIN1 (sense, 5′-TGGTCCCTCATTTCCTTCAA-3′; antisense, 5′-GGCAAAGCTGCCTGGATAAT-3′), PIN2 (sense, 5′-CAATAATAGTGTTCCGTCGTACCC-3′; antisense, 5′-GCATCGCTTTAGTAGCGAGGTT-3′), PIN7 (sense, 5′-TTTGGGCTCTTGTTGCTTTCA-3′; antisense, 5′-CAGCTTGAACAATGGCCACA-3′) and AUX1 (sense, 5′-GCCTCCGCTCGTCAGAAT-3′; antisense, 5′-ACGGTGGTGTAAAGCGGAGA-3′) primers were synthesized by Sangong Bio-Technologies. SYBR Premix Ex Taq II was ordered from TaKaRa. The real-time RT-PCR experiment was repeated three times using independently isolated RNA samples with similar results.
Light Experiments
Yellow light and UV-deficient conditions were established using Medium Yellow filter 480 (GamProducts) and UV shield filter 1510 (GamProducts) over white fluorescence tube F17T8/TL741 (Philips). Light intensity was measured by a LI-185B Quantum/Radiometer/photometer (LI-COR). The dark treatment involved wrapping plates in aluminum foil. Over 30 roots were measured for each treatment.
Phylogenetic Analysis
An alignment was generated using T-Coffee (Notredame et al., 2000) with the default parameters and adjusted manually in MacClade (Maddison and Maddison, 1989) to remove poorly conserved regions. The phylogeny was inferred with a parallelized version of the MrBayes program (version 3.1.2) using the following parameters: nruns=2 nchains=4 ngen=300000 nst=6 rates=invgamma, samplefreq=100 with burnin=1700. The number of generations was selected using the MCMC convergence diagnostic program AWTY (Nylander et al., 2008). The support values indicate the posterior probabilities based upon the 2600 post-burnin trees in the two runs. The tree was drawn with midpoint rooting using TreeView (Page, 1996) and formatted using OmniGraffle (http://www.omnigroup.com).
Accession Numbers
The GenBank accession numbers for the amino acid sequences used in the phylogenetic analysis are as follows: AAO42280, BAD44143, AAD20664, AAP40410, ACI88737, and AAL24114 (Arabidopsis); BAD61065, BAD82127, EER92646, CAE02373, BAF15233, and AAV25645 (Oryza sativa); EDQ79928, EDQ75723, EDQ71156, EDQ69089, EDQ72935, and EDQ70190 (Physcomitrella patens); EEC48020, EEC44574, EEC43483, EEC43518 (Phaeodactylum tricornutum); EAL68775 and EAL68330 (Dictyostelium discoideum); EAW52119 (Homo sapiens); AAF53692 (Drosophila melanogaster); and EDQ87821 and EDQ93134 (Monosiga brevicollis). The Phanerochaete chrysosporium sequence (Phchr1|4271) and the Volvox carteri sequences (Volca1|80645, Volca1|90632, Volca1|90737, Volca1|119273, and Volca1|121395) were downloaded from the Department of Energy Joint Genome Institute's websites http://genome.jgi-psf.org/Phchr1 and http://genome.jgi-psf.org/Volca1. The Cyanidioschyzon merolae sequences (CMI082C, CMI124C, CMM174C, CMN030C, CMP326C, CMQ317C, and CMR069C) were downloaded from http://merolae.biol.s.u-tokyo.ac.jp/. The Arabidopsis Genome Initiative locus number for the major genes discussed in this article are as follows: RUS2/WXR1 (at2g31190), TIR1 (at3g62980), AFB1 (at4g03190), AFB2 (at3g26810), AFB3 (at1g12820), AXR2/IAA7 (at3g23050), ABCB1/PGP1 (at2g36910), PIN1 (at1g73590), PIN2 (at5g57090), PIN7 (at1g23080), AUX1 (at2g38120), VHA-A1 (at2g28520), ARA7 (at4g19640), and GNOM (at1g13980).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. NAA and IAA Response in DR5rev:GFP and DR5rev:GFPwxr1 Plants.
Supplemental Figure 2. Effect of Light Wavelength on wxr1Root Growth.
Supplemental Figure 3. The wxr1 Mutation Enhances the Gravitropic Defect Observed in the aux1, pgp1, and axr2 Mutants.
Supplemental Figure 4. Levels of the Tonoplast Marker vac-yb Is Slightly Increased in wxr1 Plants.
Supplemental Figure 5. WXR1 Is a Member of the DUF647 Family.
Supplemental Figure 6. The DUF647 Protein Family Diverged Early in Eukaryote Evolution.
Supplemental Figure 7. WXR1 Is Expressed in All Organs in Arabidopsis.
Supplemental Figure 8. Growth of wxr1 Seedlings in UV-Deficient Light.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 6.
Acknowledgments
Work in the authors' labs was supported by grants from the National Institutes of Health (GM43644 to M.E.), the National Science Foundation (MCB0725149 and DBI-PGRP-0606666 to J.D.C.), and the USDA (National Research Initiative, 2005-35318-16197, to J.D.C.).
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wisniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., et al. (2007). Interactions of PIN and PGP auxin transport mechanisms. Biochem. Soc. Trans. 35: 137–141 [DOI] [PubMed] [Google Scholar]
- Barkawi L.S., Tam Y.Y., Tillman J.A., Pederson B., Calio J., Al-Amier H., Emerick M., Normanly J., Cohen J.D. (2008). A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal. Biochem. 372: 177–188 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Davies P.J. (1995). The plant hormones: Their nature, occurrence and functions. Plant Hormones Physiology, Biochemistry and Molecular Biology, Davies P.J., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 1–12 [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jurgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S., Swarup R., Mockaitis K., Dharmasiri N., Singh S.K., Kowalchyk M., Marchant A., Mills S., Sandberg G., Bennett M.J., Estelle M. (2006). AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunkley T.P., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Brugiere S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Friml J., Benkova E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jurgens G., Palme K. (2002a). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J., Wisniewska J., Benkova E., Mendgen K., Palme K. (2002b). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geisler M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jurgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jurgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Gil P., Dewey E., Friml J., Zhao Y., Snowden K.C., Putterill J., Palme K., Estelle M., Chory J. (2001). BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15: 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Maree A.F., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jurgens G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miege C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Santambrogio M., Rozier F., Fobis-Loisy I., Miege C., Gaude T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130: 1057–1070 [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kim J.I., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure C.D., Tong H., Yuen G., Hou X., Sun X., He Z.H. (2009). ROOT UV-B sensitive2 acts with root UV-B sensitive1 in a root ultraviolet B-sensing pathway. Plant Physiol. 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.J., Sohn E.J., Lee M.H., Hwang I. (2004). The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W.P., Maddison D.R. (1989). Interactive analysis of phylogeny and character evolution using he computer program MacClade. Folia Primatol. (Basel) 53: 190–202 [DOI] [PubMed] [Google Scholar]
- Marchant A., Kargul J., May S.T., Muller P., Delbarre A., Perrot-Rechenmann C., Bennett M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenfuhr A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Noh B., Bandyopadhyay A., Peer W.A., Spalding E.P., Murphy A.S. (2003). Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002 [DOI] [PubMed] [Google Scholar]
- Noh B., Murphy A.S., Spalding E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Nylander J.A., Wilgenbusch J.C., Warren D.L., Swofford D.L. (2008). AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583 [DOI] [PubMed] [Google Scholar]
- Oono Y., Chen Q.G., Overvoorde P.J., Kohler C., Theologis A. (1998). age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Zazimalova E., Ruthardt N., Petrasek J., Stierhof Y.D., Kleine-Vehn J., Morris D.A., Emans N., Jurgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Page R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pan J., Fujioka S., Peng J., Chen J., Li G., Chen R. (2009). The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pickett F.B., Wilson A.K., Estelle M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94: 1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sukumar P., Edwards K.S., Rahman A., Delong A., Muday G.K. (2009). PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 150: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Leasure C.D., Hou X., Yuen G., Briggs W., He Z.H. (2008). Role of root UV-B sensing in Arabidopsis early seedling development. Proc. Natl. Acad. Sci. USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.K., Pickett F.B., Turner J.C., Estelle M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertova D., Brewer P.B., Ruzicka K., Blilou I., Rouquie D., Benkova E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencik A., Rolcik J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]