The target of rapamycin (TOR) pathway, which is known to regulate cell growth in eukaryotes, is shown to affect cell wall structure in plants. ROL5 (REPRESSOR of LRX1) is identified as a possible mitochondrial component of the TOR pathway.
Abstract
Plant cell growth is limited by the extension of cell walls, which requires both the synthesis and rearrangement of cell wall components in a controlled fashion. The target of rapamycin (TOR) pathway is a major regulator of cell growth in eukaryotes, and inhibition of this pathway by rapamycin reduces cell growth. Here, we show that in plants, the TOR pathway affects cell wall structures. LRR-extensin1 (LRX1) of Arabidopsis thaliana is an extracellular protein involved in cell wall formation in root hairs, and lrx1 mutants develop aberrant root hairs. rol5 (for repressor of lrx1) was identified as a suppressor of lrx1. The functionally similar ROL5 homolog in yeast, Ncs6p (needs Cla4 to survive 6), was previously found to affect TOR signaling. Inhibition of TOR signaling by rapamycin led to suppression of the lrx1 mutant phenotype and caused specific changes to galactan/rhamnogalacturonan-I and arabinogalactan protein components of cell walls that were similar to those observed in the rol5 mutant. The ROL5 protein accumulates in mitochondria, a target of the TOR pathway and major source of reactive oxygen species (ROS), and rol5 mutants show an altered response to ROS. This suggests that ROL5 might function as a mitochondrial component of the TOR pathway that influences the plant's response to ROS.
INTRODUCTION
Plant cell growth is tightly linked to the expansion of the cell wall. Cell walls are complex structures that resist internal turgor pressure and, for cell enlargement to take place, have to incorporate new material and rearrange internal linkages between the different components (Martin et al., 2001). In dicotyledonous plants, the primary cell wall is composed of cellulose microfibrils that are interconnected by hemicelluloses, mainly xyloglucan. This is considered to be the load-bearing structure and is embedded in a matrix of pectic polysaccharides (Carpita and Gibeaut, 1993). The pectic matrix has three major components: homogalacturonan, rhamnogalacturonan-I (RGI), which contains side chains of galactan and arabinan, and rhamnogalacturonan-II. Pectins influence cell wall rigidity and strength as well as cell–cell adhesion. In addition, RGI regulates wall porosity, which in turn influences the mobility of cell wall–modifying proteins and, thus, cell wall expansion (Baron-Epel et al., 1988; Ridley et al., 2001; Willats et al., 2001; McCartney et al., 2003). Structural cell wall proteins such as hydroxyproline-rich glycoproteins (HRGPs) influence the mechanical properties of cell walls but can also be involved in cell elongation and signaling as exemplified by arabinogalactan proteins (AGPs). These are GPI-anchored proteins of the HRGP family that are extensively glycosylated with arabinose and galactose (Ding and Zhu, 1997; Majewska-Sawka and Nothnagel, 2000; van Hengel and Roberts, 2002).
The structure of cell walls, which influences the cell walls' properties, is in a constant flow of remodeling as it adapts to the prevailing functional requirements. Therefore, plants have evolved a sensing system to monitor cell wall composition and to regulate cell wall modification and restructuring. These activities are likely to involve transmembrane or membrane-anchored proteins. Receptor-like kinases, such as THESEUS and wall-associated kinases, have been shown to sense and modify cell elongation (Kohorn et al., 2006; Hematy et al., 2007), as have lectins and GPI-anchored proteins, such as AGPs (Humphrey et al., 2007; Hematy and Höfte, 2008). LRR-extensins (LRXs) are extracellular proteins consisting of an N-terminal leucine-rich repeat domain and a C-terminal extensin domain typical of HRGPs (Baumberger et al., 2003a). This particular structure suggests that LRX proteins might have a signaling or regulatory function during cell wall development (Ringli, 2005). Indeed, Arabidopsis thaliana LRX1 is predominantly expressed in root hairs, and lrx1 mutants develop defective cell walls resulting in aberrant root hair formation (Baumberger et al., 2001, 2003b).
The TOR (for target of rapamycin) pathway is a major growth regulator in eukaryotes that senses nutrient availability and growth stimulators, regulates the translational machinery, and modulates cell growth. The Ser/Thr kinase TOR is central to the TOR pathway and is inhibited by the specific inhibitor rapamycin, resulting in reduced cell growth. Rapamycin inhibits the TOR kinase by forming a ternary complex with the immunophilin protein FKBP12 (FK506 binding protein 12) and TOR (Huang et al., 2003; Wullschleger et al., 2006). An important function of the TOR pathway is the regulation of mitochondrial activity and, hence, the production of reactive oxygen species (ROS), which affect life span (Schieke and Finkel, 2006; Cunningham et al., 2007) and, in plants, have an impact on oxidative stress, cell wall extension, and cell growth (for review, see Gapper and Dolan, 2006; Rhoads et al., 2006).
Recent analyses in Candida albicans have provided evidence for the participation of the TOR pathway in cell wall integrity sensing in yeast (Tsao et al., 2009). Numerous components of the TOR pathway were identified in yeast based on rapamycin hypersensitivity of the corresponding mutants (Chan et al., 2000). Mutations in NCS6 (needs Cla4 to survive 6) of yeast induce rapamycin hypersensitivity and influence cell growth under nutrient-limited conditions (Chan et al., 2000; Goehring et al., 2003a). Recently, Ncs6p has been shown to be important for the modification of cytoplasmic tRNAs. tRNAs are frequently modified, mostly at the wobble position (position 34) or next to and 3′ of the anticodon (position 37). tRNAs specific for Glu, Glc, and Lys have a 2-thiouridine derivative as wobble nucleoside, which helps to effectively read the corresponding codons on the mRNAs (Björk et al., 2007). Ncs6p and homologous proteins in other organisms are involved in the thiolation of U34, and mutations in the corresponding genes lead to the absence of thiolation (Björk et al., 2007; Schlieker et al., 2008; Leidel et al., 2009). Even though mutating ncs6 only affects cytoplasmic tRNAs (Noma et al., 2009), Ncs6p is also found in mitochondria (Huh et al., 2003). Dual localization of proteins in different compartments frequently has been observed (Krause and Krupinska, 2009). Thus, it remains to be shown whether the effect of the ncs6 mutant on TOR signaling is an indirect effect induced by the lack of tRNA modification or a second activity of the protein, reflected by its presence in mitochondria.
The TOR pathway has also been identified in plants, and some of the proteins involved in this process have been characterized (Anderson et al., 2005; Deprost et al., 2005; Ingram and Waites, 2006; Mahfouz et al., 2006). While a tor knockout mutant in Arabidopsis is embryo-lethal, modified TOR expression strongly influences plant growth, emphasizing the importance of TOR during plant development (Menand et al., 2002; Deprost et al., 2007). Arabidopsis is not sensitive to the specific TOR inhibitor rapamycin as rapamycin cannot form the ternary complex with FKBP12 and TOR. However, expression of yeast FKBP12 induces rapamycin sensitivity in Arabidopsis (Mahfouz et al., 2006; Sormani et al., 2007).
Here, we provide evidence for a role of the plant TOR pathway in modulating cell wall structures. A suppressor screen on the root hair cell wall formation mutant lrx1 resulted in the identification of the rol5 (for repressor of lrx1) mutant. The rol5 mutation induced changes in cell wall structure that might be the basis of suppression of lrx1. ROL5 is functionally similar to Ncs6p, which influences TOR signaling in yeast and is required for the modification of tRNAs in Arabidopsis. Interfering with TOR signaling by the addition of rapamycin in yeast FKBP12-expressing lrx1 mutant plants relieved the lrx1 root hair phenotype and induced specific changes in cell wall structure similar to rol5. Together, these data indicate that interfering with TOR signaling induces changes in cell walls and provide evidence for a role of the TOR pathway in the regulation of cell wall structure and properties.
RESULTS
Identification of rol5, a Suppressor of the lrx1 Root Hair Phenotype
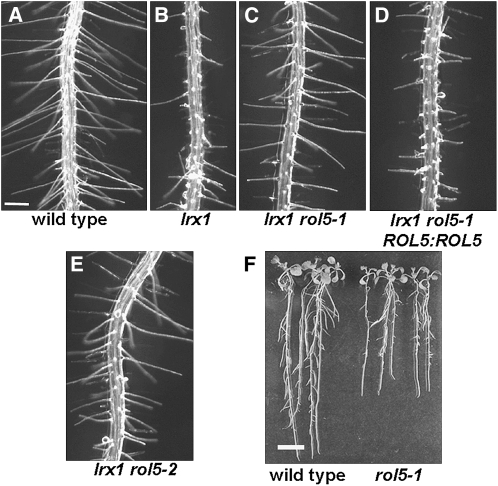
As a result of the defective cell wall structure, lrx1 mutants form root hairs that are short and deformed and frequently burst (Figures 1A and 1B). To identify new loci that are involved in regulating cell wall formation and structure, a suppressor screen was performed on the lrx1 mutant. As described previously (Diet et al., 2006), an lrx1 missense allele was used for ethyl methanesulfonate (EMS) mutagenesis, and M2 seedlings displaying a suppressed lrx1 phenotype were isolated. The rol5-1 mutant was identified in this screen as it suppressed the lrx1 phenotype. lrx1 rol5-1 double mutants developed root hairs that were comparable to those of wild-type seedlings (Figure 1C). The rol5-1 mutation was found to be recessive, since the F1 generation of a backcross with lrx1 developed an lrx1 phenotype and seedlings of the F2 generation segregated 3:1 for lrx1:wild-type-like root hairs. To characterize the effect of the rol5-1 mutation in more detail, a rol5-1 single mutant was identified after backcrossing with wild-type Columbia (see Methods).
Figure 1.
Suppression of the lrx1 Root Hair Phenotype by Mutations in rol5.
Seedlings were grown for 5 d ([A] to [E]) and 10 d (F) in a vertical orientation. The wild type (A) developed regular root hairs, whereas root hairs of the lrx1 mutant (B) were severely deformed. The EMS missense allele rol5-1 (C) was complemented with a ROL5 genomic clone, inducing an lrx1-like phenotype (D). The rol5-2 T-DNA knockout mutant (E) also suppressed the lrx1 mutant phenotype. The rol5 mutation (F) leads to shorter roots as shown for rol5-1. Bars = 0.5 mm in (A) to (E) and 10 mm in (F).
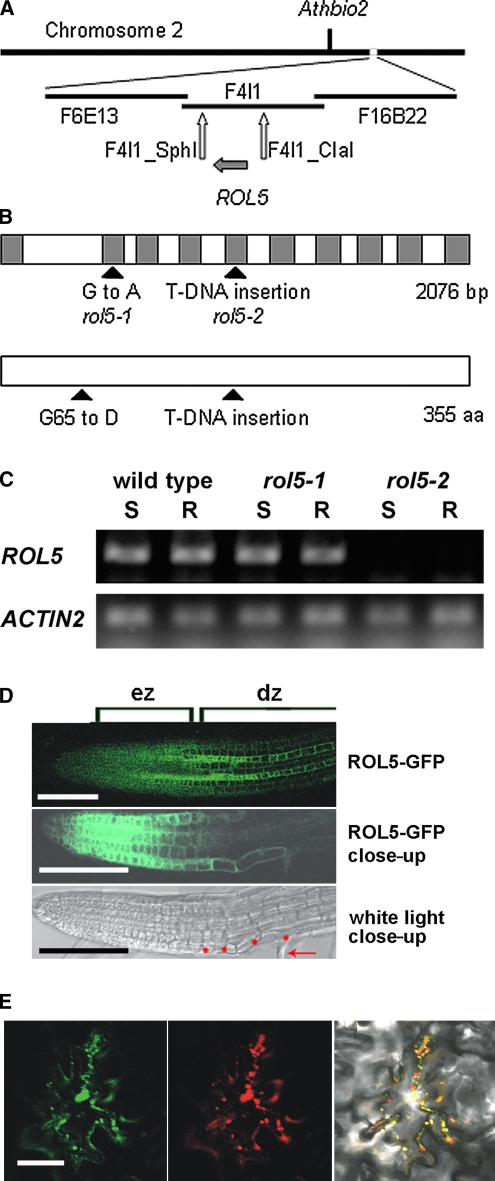
Map-Based Cloning of rol5
The rol5 gene was identified by map-based cloning and initially localized to a region on chromosome 2, south of AthBio2. Further mapping revealed two flanking markers on the BAC F4I1, F4I1-SphI, and F4I1-ClaI at positions 16,700 and 43,500, respectively (Figure 2A). This interval was sequenced and a single point mutation was identified in the gene At2g44270 encoding a protein of 355 amino acids (Figure 2B). Transformation of the lrx1 rol5-1 mutant with a wild-type genomic copy of this gene led to the development of an lrx1 root hair phenotype, confirming that the identified gene represents the ROL5 locus (Figure 1D). The mutation in rol5-1 results in an amino acid change from Gly-65 to Asp. A rol5 T-DNA mutant with the insertion site 3′ adjacent to the Glu-170 codon was identified and named rol5-2 (Figure 2B). RT-PCR on RNA isolated from wild-type and mutant seedlings revealed the presence of ROL5 RNA in wild-type and rol5-1 mutant seedling root and shoot tissue but not in rol5-2 seedlings (Figure 2C). Together with the position of the T-DNA in an exon, this suggests that rol5-2 is a null allele. The lrx1 phenotype was also suppressed by rol5-2 (Figure 1E), revealing that suppression is not dependent on the particular missense mutation present in the rol5-1 allele.
Figure 2.
Identification of the ROL5 Locus.
(A) The rol5 locus was identified by map-based cloning on the long arm of chromosome 2, south of Athbio2. BAC clones in the region of ROL5 are indicated. For mapping, cleaved-amplified polymorphic sequence and simple sequence length polymorphism markers were established, of which F4I1-Sph and F4I1-Cla were the closest flanking markers identified.
(B) The ROL5 gene consists of 10 exons encoding a protein of 355 amino acids. The G-to-A mutation in rol5-1 is located in the second exon and changes Gly-65 to Asp. rol5-2 represents a T-DNA insertion line that interrupts the reading frame at the amino acid codon Glu-170. Gray boxes, exons.
(C) RT-PCR experiments on RNA isolated from shoots (S) and roots (R) of 1-week-old seedlings demonstrated that the ROL5 gene is expressed in the wild type and the rol5-1 mutant but not to detectable levels in rol5-2. RT-PCR on the ACTIN2 gene was performed to confirm the use of similar amounts of RNA in the different samples. One of two biological replicates is shown.
(D) In roots, ROL5 is predominantly expressed in the elongation zone (ez) and in a striped pattern in the differentiation zone (dz) (top panel). A close-up of the root (GFP fluorescence in the middle panel; bright field in the bottom panel) revealed overlapping GFP fluorescence and root hair formation. Red dots, root hair–forming trichoblasts; arrow, root hair structure. Bar = 0.3 mm.
(E) When transiently expressed in Arabidopsis epidermal cells, ROL5-GFP (left panel) and a mitochondrial marker protein (for details, see Methods) fused to red fluorescent protein (middle panel) display overlapping fluorescence patterns (right panel). Bar = 50 μm.
The RT-PCR data indicated that ROL5 is expressed in various tissues. For a more detailed analysis, a ROL5:ROL5-GFP (green fluorescent protein) fusion construct was transformed into wild-type Arabidopsis and roots of transgenic seedlings were analyzed. Fluorescence was found to be predominant in the elongation zone and to expand in a striped pattern into the differentiation zone. These stripes overlapped with the arrangement of root hair cells (Figure 2D), which initiate root hair elongation in the differentiation zone (Dolan et al., 1994). Thus, ROL5 is predominantly expressed in elongating cells, suggesting an important function during cell expansion. Indeed, compared with wild-type seedlings, rol5-1 mutants had shorter roots, root epidermal cells, and root hairs (Figure 1F, Table 1).
Table 1.
Length of Roots, Trichoblasts, and Root Hairs of the rol5-1 Mutant
| Genotype | Root Length (mm) | Epidermal Cell Length (Trichoblasts) (μm) | Root Hair Length (μm) |
| Wild Type | 15 ± 0.2 | 147 ± 29 | 700 ± 80 |
| rol5-1 | 10 ± 0.2 | 126 ± 24 | 480 ± 120 |
Seedlings were grown for 5 d in a vertical orientation. Values represent means ± sd. Differences are significant (t test, P < 0.05).
ROL5 Is Structurally and Functionally Similar to the Yeast Ncs6p
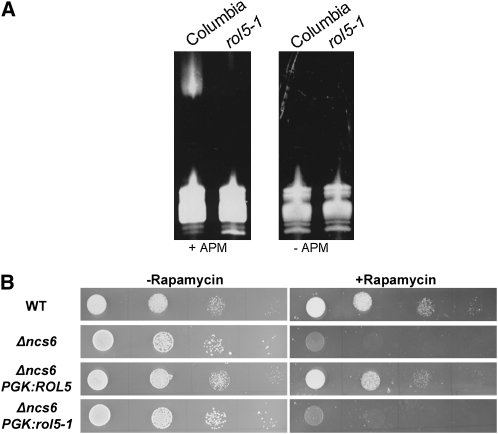
ROL5 shows 54% identity and 70% similarity to Ncs6p/Tuc1p of yeast (Saccharomyces cerevisiae), subsequently referred to as Ncs6p (Figure 3). The Ncs6p-like proteins of different organisms share conserved motifs, including a PP-loop domain with ATP pyrophosphatase activity (Bork and Koonin, 1994; Björk et al., 2007), which are also conserved in ROL5. The Gly-65 to Asp mutation in rol5-1 is adjacent to the PP-loop motif SGGxDS (Figure 3). Ncs6p-like proteins have been found to be involved in the thiolation of the uridine residue 34 of a subset of cytoplasmic tRNAs (Björk et al., 2007; Schlieker et al., 2008; Leidel et al., 2009). To investigate whether ROL5 is involved in tRNA modification in Arabidopsis, the tRNA fraction of wild-type and rol5-1 mutant seedlings was isolated. tRNAs containing this modification can be detected in an acrylamide gel containing N-acryloylamino phenyl mercuric chloride (APM), a compound that interacts with 2-thiouridine and retards migration in the gel. While a band shift can be observed with wild-type tRNAs in gels containing APM, it is absent from rol5-1 tRNA extracts. In the absence of APM, as expected, no shift is detectable in either of the extracts (Figure 4A). Hence, ROL5 is involved in this tRNA modification process in Arabidopsis.
Figure 3.
ROL5 Is Homologous to Ncs6p of Yeast.
The alignment of ROL5 with Ncs6p of S. cerevisiae reveals 54% identity and 70% similarity between the two proteins. The Ncs6p-like proteins of different organisms share common motifs that are indicated below the sequences [(CxxC)2 – SGGxDS – CxxC – GH – PL – C – (CxxC)2], all of which are conserved between the two proteins. The motif PL is not fully conserved in Ncs6p and ROL5. Sequences important for protein activity (Björk et al., 2007) are boxed. The Gly-65 to Asp mutation in rol5-1 (star) is adjacent to the PP-loop motif SGGxDS, which is important for ATP binding.
Figure 4.
Ncs6p and ROL5 Have Similar Functions.
(A) tRNA was extracted from 7-d-old wild-type and rol5-1 seedlings and separated on an acrylamide gel with (left panel) or without (right panel) APM, a compound that interacts with the 2-thiouridine and retards migration in the gel.
(B) Wild-type (WT) and Δncs6 mutant yeast was grown in the absence or presence of rapamycin. The Δncs6 mutant grew normally on control medium but was hypersensitive to rapamycin compared with the wild type. Expression of ROL5 but not rol5-1 under the control of the yeast PHOSPHOGLYCERATE KINASE promoter in the Δncs6 mutant suppressed this rapamycin hypersensitivity phenotype. Spots on a line represent serial dilutions (10-fold).
Since Ncs6p was also identified as a component of the TOR pathway, we assessed whether Ncs6p and ROL5 have similar functions with respect to TOR signaling. The Δncs6 mutant yeast strain, which is hypersensitive to rapamycin (Chan et al., 2000; Goehring et al., 2003a), was complemented with ROL5 under the control of a constitutive yeast promoter. On standard growth medium, the wild type, the Δncs6 mutant, and the complemented Δncs6 mutant showed comparable growth properties, while in the presence of rapamycin, growth of the Δncs6 mutant was considerably retarded. This effect was compensated for by expressing ROL5 in the Δncs6 mutant, but not by expressing the rol5-1 missense allele (Figure 4B).
Ncs6p appears to accumulate in mitochondria, which prompted us to investigate the subcellular localization of ROL5. A ROL5-GFP fusion construct was transiently expressed in Arabidopsis epidermal cells, and colocalization with well-established organellar marker proteins was investigated. A clear overlap was found for ROL5-GFP and a mitochondrial protein (for details, see Methods) fused to red fluorescent protein (Figure 2E). This suggests that ROL5, similar to Ncs6p in yeast, translocates to mitochondria. Together, these data demonstrate that Ncs6p and ROL5 have very similar functions in their respective organisms.
Interfering with TOR Signaling Leads to Suppression of lrx1
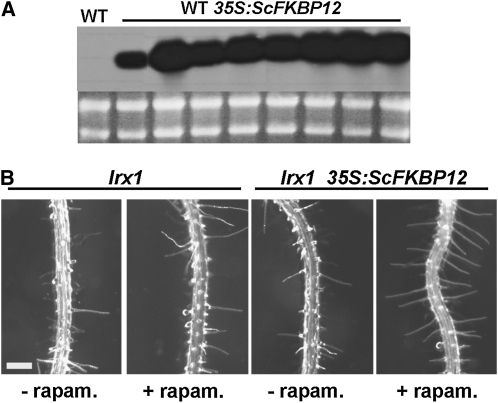
The functional similarity between ROL5 and Ncs6p suggested that the rol5 mutant might be impaired in a TOR-related process. This led us to further investigate whether the rol5 mutations suppress the lrx1 root hair phenotype by influencing TOR signaling. To this end, the TOR-specific inhibitor rapamycin was used to interfere with TOR signaling in Arabidopsis. This required transformation of Arabidopsis with the yeast FKBP12 under the control of the ubiquitously active 35S promoter. Wild-type Columbia plants expressing FKBP12 were produced (Figure 5A), and the lrx1 mutation was crossed into two independent transgenic lines. While lrx1 mutants expressing FKBP12 developed typical lrx1 root hairs under normal growth conditions, the presence of rapamycin led to a clear suppression of the lrx1 phenotype in both transgenic lines. In nontransgenic lrx1 mutants, rapamycin had no effect on the root hair phenotype (Figure 5B). This shows that interfering with TOR signaling suppresses the lrx1 phenotype.
Figure 5.
Rapamycin Treatment Suppresses the lrx1 Root Hair Phenotype.
(A) RNA gel blot of wild-type (WT) Arabidopsis transformed with a 35S promoter:FKBP12 construct.
(B) lrx1 mutants expressing FKBP12 were sensitive to rapamycin (rapam.) and showed a suppressed lrx1 root hair phenotype (right). In nontransgenic lrx1 mutants, the root hair phenotype was not affected by rapamycin (left). Bar = 0.5 mm.
The treatment with rapamycin had additional effects on root development that were similar to those observed in the rol5-1 mutant. In the presence of rapamycin, FKBP12-expressing wild-type plants developed shorter roots and shorter epidermal cells (Table 2) as previously observed by Sormani et al. (2007), confirming the involvement of TOR signaling in plant cell elongation.
Table 2.
Length of Roots and Trichoblasts Due to Rapamycin Treatment
| 35S:ScFKBP12 | Root Length (mm) | Epidermal Cell Length (Trichoblasts) (μm) |
| − Rapamycin | 15 ± 0.2 | 140 ± 12 |
| + Rapamycin | 11 ± 0.2 | 90 ± 9 |
Seedlings were grown for 5 d in a vertical orientation. Values represent means ± sd. Differences are significant (t test, P < 0.05).
rol5-1 and Rapamycin Treatment Lead to Changes in Cell Wall Components
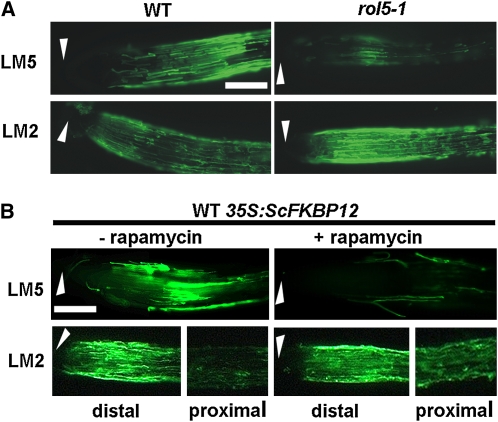
A possible mechanism of suppression of the lrx1 root hair phenotype might be through compensation of the cell wall defects in lrx1 mutants by the introduction of additional changes to cell walls. To identify potential alterations in cell wall structures, root surfaces were analyzed in the wild type and rol5-1 mutant using a series of monoclonal antibodies targeted to different cell wall polysaccharide components. These were antipectic homogalacturonan JIM5 and JIM7 (Knox et al., 1990), anti-(1→4)-β-d-galactan LM5 (Jones et al., 1997), anti-(1→5)-α-l-arabinan LM6 (Willats et al., 1998), antixyloglucan LM15 (Marcus et al., 2008), and anti-AGP LM2 (Yates et al., 1996). Four of these epitopes were detected at equivalent levels on wild-type and rol5-1 mutant root surfaces (see Supplemental Figure 1 online), whereas the LM5 galactan and the LM2 AGP epitopes displayed differential modulation in response to the mutation. Detection of the LM5 galactan epitope decreased and that of the LM2 AGP epitope increased at the root surface of the rol5-1 mutant (Figure 6A). The same monoclonal antibodies were used to analyze root surfaces upon interfering with TOR signaling by rapamycin. Treatment of seedlings expressing FKBP12 with rapamycin resulted in alterations in immunolabeling that were similar to those observed in the rol5-1 mutant. The LM5 galactan epitope showed a marked decrease in occurrence, whereas the LM2 AGP epitope was increased in proximal parts of the root (Figure 6B) compared with nontreated seedlings expressing yeast FKBP12. The other antibodies showed equal labeling for both conditions (see Supplemental Figure 2A online). Immunodetection with LM5 and LM2 was identical between nontransgenic wild-type seedlings grown with or without rapamycin and FKBP12-expressing seedlings grown without rapamycin (see Supplemental Figure 2B online). Thus, the observed modulations of these two cell wall epitopes were specifically induced by rapamycin and only in those seedlings that were expected to be rapamycin sensitive, suggesting that they were the result of impaired TOR signaling.
Figure 6.
Immunolabeling of Cell Walls of rol5-1 and Rapamycin-Treated Wild-Type Seedlings.
Immunolabeling of 4-d-old roots with monoclonal antibodies (1→4)-β-d-galactan side chains of RG I (LM5) and glucuronic acid–containing side chains of arabinogalactan proteins (LM2).
(A) Compared with the wild type (WT), the rol5-1 mutant root surface revealed reduced detection of the LM5 epitope and stronger detection of the LM2 epitope.
(B) Roots of FKBP12-expressing wild-type seedlings grown in the absence (left) and presence (right) of rapamycin. The presence of rapamycin led to a reduced labeling with LM5 and a stronger labeling in proximal parts with LM2.
Arrowheads, root apex. Bars = 0.3 mm.
rol5-1 Mutants Are Affected in Their Response to ROS and ROS Scavengers
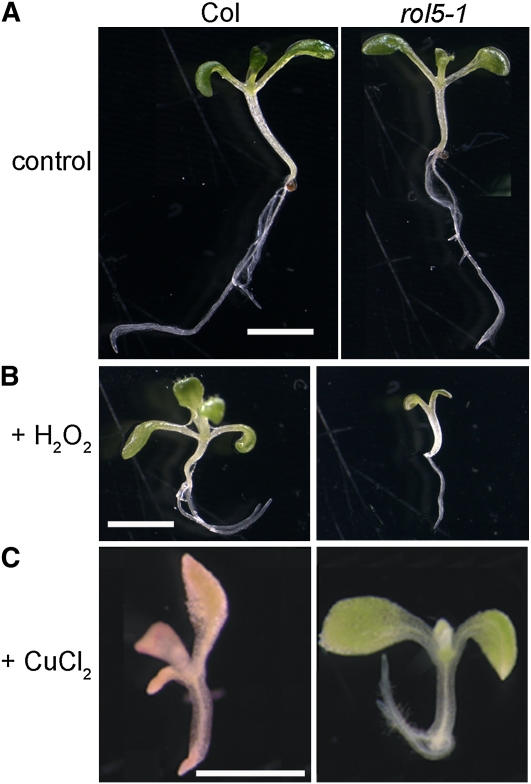
One possible crossing point of TOR signaling and ROL5 is the mitochondrial localization of ROL5, since the TOR pathway is a regulator of mitochondrial activity and hence the production of ROS. To investigate this possibility, ROS levels in roots of wild-type and rol5-1 mutant seedlings were analyzed using different ROS-sensitive staining substrates. Under the growth conditions used, none of these stainings revealed a clear, reproducible change in ROS levels in rol5-1 seedlings (see Supplemental Figure 3 online). Next, the effect of ROS on seedling growth was tested in liquid culture. This experiment revealed an increased susceptibility of rol5-1 seedlings to hydrogen peroxide. While wild-type seedlings showed a similar development with or without 8 mM H2O2, the development of rol5-1 seedlings was retarded in the presence of H2O2. By contrast, rol5-1 seedlings were revealed to be more tolerant to ROS scavengers. While wild-type seedlings barely grew and failed to accumulate chlorophyll in the presence of 100 μm CuCl2, rol5-1 seedlings turned green and grew considerably better (Figure 7). This indicates that ROL5 is important for the sensing of, and the response to, ROS.
Figure 7.
Altered Response of the rol5-1 Mutant to ROS and ROS Scavenger.
Seedlings were grown in liquid culture for 10 d. Under control conditions (A), growth of the wild type and rol5-1 is comparable. rol5-1 seedlings are hypersensitive to H2O2 (8 mM), revealed by the reduced growth (B), and hyposensitive to the ROS scavenger CuCl2 (100 μm), indicated by better growth and the development of green cotyledons (C). col, wild-type Columbia.
DISCUSSION
The work presented here suggests that the TOR pathway is a process that can lead to the specific modification of cell wall components. The rol5 locus was identified in a suppressor screen on the lrx1 mutant, which is affected in cell wall formation in root hairs (Baumberger et al., 2001, 2003b). This screen was performed with the aim of identifying novel loci involved in cell wall formation, as suppressors can reveal a functional relationship between genetic loci (Huang and Sternberg, 1995). After the previous identification of rol1, which encodes RHAMNOSE SYNTHASE1 (Diet et al., 2006), rol5 is the second identified suppressor of lrx1 and also affects cell wall structure.
The TOR Pathway Is a Regulator of Cell Wall Development
ROL5 is homologous to the yeast Ncs6p, and these proteins have similar functions in their respective organisms. These functions include the modification of tRNAs and the effect on TOR signaling. It is likely that suppression of lrx1 is induced via a modification of TOR signaling, as the lrx1 mutant root hair phenotype can be suppressed by the TOR kinase inhibitor rapamycin. Rapamycin is a macrocyclic lactone originally identified in the bacterium Streptomyces hygroscopicus and one of the most specific kinase inhibitors known (Heitman et al., 1991; Huang et al., 2003), making additional effects of rapamycin very unlikely. The TOR pathway is a central regulator of eukaryotic growth processes (Wullschleger et al., 2006), and previous work in Arabidopsis has revealed the importance of this signaling pathway, including the TOR kinase itself, for plant development (Menand et al., 2002; Bögre et al., 2003; Mahfouz et al., 2006). Our analysis shows that the TOR pathway is able to modify cell wall components, suggesting that it is part of the regulatory process that coordinates cell wall structure with cell growth and development. In fact, recent work in C. albicans suggests that the TOR pathway is involved in sensing cell wall integrity. The rhb1 mutant, affected in a small G-protein of the RAS superfamily, was found to be hypersensitive to drugs interfering with cell wall formation and showed a modified cell wall integrity signaling previously identified in this organism. The rhb1 mutation induces rapamycin hypersensitivity in C. albicans, indicating a function of RHB1 in TOR signaling (Tsao et al., 2009). Plants clearly have mechanisms to monitor, sense, and modify cell wall composition and structures. Proteins involved in this process have been found to be transmembrane or membrane associated, such as wall-associated kinases, the receptor kinase THESEUS, GPI-anchored proteins, or lectins (Kohorn et al., 2006; Hematy et al., 2007; Humphrey et al., 2007; Hematy and Höfte, 2008). These proteins are probably directly involved in the regulatory or sensing process, since they localize to cell surfaces. Considering the importance in regulating cell growth, the TOR pathway might function as a relay system that integrates the signals of sensing mechanisms into cellular responses and developmental processes.
The core component of the TOR pathway is the Ser/Thr kinase protein TOR. Only yeast encodes two separate but functionally similar TOR proteins. In yeast and mammals, TOR forms two distinct multiprotein complexes, TORC1 and TORC2, of which only TORC1 is rapamycin sensitive. While TORC1 is involved in regulating translation, nutrient import, or stress responses, TORC2 influences the actin cytoskeleton (Loewith et al., 2002; Wullschleger et al., 2006). Also in Arabidopsis, the diverse functions of this pathway are likely to require the establishment of distinct multiprotein complexes with the TOR protein. The TOR-interacting proteins RICTOR and RAPTOR were identified in TORC1 and TORC2 (Wullschleger et al., 2006), and RAPTOR has been shown to undergo interactions with the Arabidopsis TOR protein. Mutations in RAPTOR affect plant development, corroborating the importance of this protein for the TOR pathway (Deprost et al., 2005; Mahfouz et al., 2006). It is likely that TOR-interacting proteins establish further protein–protein interactions as shown for other organisms (Wullschleger et al., 2006), leading to diverse signaling outputs. A better understanding of the signaling network is required to identify the mechanism by which the TOR pathway senses and influences cell wall structures.
Interfering with TOR Modifies Cell Walls and Plant Growth
Plant cell walls are complex, structurally variable organelles that underpin many aspects of cell and organ growth. Cell wall extension is the limiting factor in cell enlargement (Carpita and Gibeaut, 1993; Martin et al., 2001). Monoclonal antibodies are a useful tool to analyze cell walls as they can detect changes in the composition or the accessibility of cell wall structures. The LM5 galactan epitope that is a component of RGI has been specifically detected in the elongation zone at the Arabidopsis root surface and implicated in the onset of the acceleration of cell elongation (McCartney et al., 2003). The occurrence of the LM5 epitope has also been observed to correlate with modified mechanical properties of cell walls and to be reduced in different mutants that show reduced root epidermal cell growth (McCartney et al., 2000, 2003; Diet et al., 2006). Thus, the short root phenotype observed in the rol5-1 mutant and in rapamycin-treated seedlings correlates well with reduced LM5 labeling and the known involvement of this cell wall component with growth. Pectin regulates the porosity of cell walls and hence influences the mobility of cell wall modifying enzymes necessary for cell wall expansion (Baron-Epel et al., 1988). RGI is thought to modify this porosity, which would serve as a possible explanation of why changes in the RGI structure influence cell growth (Ridley et al., 2001; Willats et al., 2001). AGPs are a second cell wall component found to be altered due to modified TOR signaling. The LM2 antibody, which binds to a glucuronic acid–containing epitope of AGPs (Yates et al., 1996), showed increased immunolabeling of rol5-1 mutants and rapamycin-treated seedlings. The modified distribution and/or abundance of AGPs has been shown to correlate with aberrant cell growth in roots, root hairs, and pollen tubes as demonstrated by the analysis of agp mutants as well as the use of Yariv reagent that precipitates AGPs and blocks their action (Willats and Knox, 1996; Ding and Zhu, 1997; van Hengel and Roberts, 2002; McCartney et al., 2003; Levitin et al., 2008). Reducing root epidermal cell expansion with Yariv reagent also modifies the occurrence of the LM5 epitope, indicating some linkage between RGI and AGPs (McCartney et al., 2003) that has also now been shown in the rol5-1 mutant and rapamycin-treated seedlings. The reduced cell elongation phenotypes observed in the rol5 mutants and upon rapamycin treatment therefore correlate with changes to specific cell wall components. Thus, the TOR pathway might be a regulatory mechanism to modulate these two factors in cell walls. It is possible that the observed changes in root surface detection of the LM5 galactan and LM2 AGP epitopes are mechanistically involved in the suppression of the lrx1 root hair phenotype. As lrx1 mutants develop aberrant cell walls, it can be hypothesized that secondary modifications overcome the defects induced by the lack of LRX1.
ROL5 Might Have Dual Functions
It remains unclear exactly how ROL5 affects TOR signaling. The rol5-1 mutant, similar to the yeast Δncs6 mutant, fails to properly modify tRNAs. The uridine residue 34 of several tRNAs is modified to 5-methoxycarbonylmethyl-2-thiouridine to improve translational efficiency. Ncs6p, together with other proteins, has been shown to transfer the sulfur group during 2-thiouridine formation (Björk et al., 2007; Schlieker et al., 2008; Leidel et al., 2009; Noma et al., 2009). The TOR pathway is involved in regulating the translational machinery in different organisms, including plants (Mahfouz et al., 2006; Wullschleger et al., 2006; Dinkova et al., 2007), and the lack of tRNA modifications might trigger signals that feed back into TOR signaling. This indirect effect on the TOR pathway is a possible explanation for the rapamycin hypersensitivity of the Δncs6 mutant (Chan et al., 2000; Goehring et al., 2003a). Alternatively, Ncs6p and ROL5 might have an additional, so far unidentified function that links protein activity to the TOR signaling network. Indicative for this hypothesis is the localization of these proteins to mitochondria (Huh et al., 2003; this work). Previous work has shown that Ncs6p is dispensable for the thiolation of mitochondrial tRNA (Noma et al., 2009), suggesting an additional function of the protein in this organelle. The dual localization in different compartments has been shown for a number of proteins (Krause and Krupinska, 2009). Goehring et al. (2003a) reported on the influence of Ncs6p on protein conjugation by Urm1p, a ubiquitin-related modifier protein. Yeast Urm1p has recently been shown to be involved in the same sulfur carrier process as Ncs6p (Leidel et al., 2009). In addition, however, Urm1p is also conjugated to Ahp1p, which is not involved in tRNA modification but is likely to have a function in TOR signaling (Goehring et al., 2003b). Our analysis points to an additional effect of ROL5 in a ROS-related process as the rol5-1 mutant showed an increased sensitivity to ROS and an increased tolerance to ROS scavengers compared with the wild type. The mitochondrial localization of ROL5 is consistent with this additional function since mitochondria are a major source of ROS (Rhoads et al., 2006). ROS are not just byproducts of the respiratory chain but revealed to serve as signaling molecules that can affect cell elongation and cell wall development (Liszkay et al., 2004; Takeda et al., 2008). ROL5 might regulate the response of the cell to ROS signaling, which is in agreement with the reduced cell growth observed in rol5-1 mutants. A major function of the TOR pathway is the regulation of mitochondrial activity (Schieke et al., 2006; Cunningham et al., 2007), and ROL5 might be part of this regulatory mechanism.
The TOR pathway is a central regulator of eukaryotic cell growth. The analyses presented here suggest that the TOR pathway has the ability to modify cell wall structure and specifically components implicated in cell elongation. The TOR pathway appears to be one mechanism of connecting plant cell growth processes with specific changes to cell wall structure. Further analyses are necessary to identify the proteins that establish the link between the TOR signaling network and the extracellular proteins that sense and survey cell wall developmental processes. Moreover, the possible multiple activities of ROL5-like proteins need to be elucidated in greater detail to identify their precise roles during cell growth.
METHODS
Plant Growth, EMS Mutagenesis, and Mapping
The lrx1 missense allele and the EMS mutagenesis procedure are described by Diet et al. (2004). The lrx1 mutant and all other Arabidopsis thaliana lines used are in the Columbia genetic background, except for the line used for mapping, which is Landsberg erecta (Ler). The rol5-2 allele (line 709D04) was obtained from the GABI collection (Rosso et al., 2003). Phenotypic analysis was performed on lrx1 rol5-1 and rol5-1 mutant plants backcrossed twice with the lrx1 mutant and wild-type Columbia, respectively. lrx1 rol5-1 and lrx1 rol5-2 double mutants and rol5-1 single mutants were identified with molecular markers for the mutations (see below). For growth of plants in sterile conditions, seeds were surface sterilized with 1% sodium hypochlorite and 0.03% Triton X-100, stratified for 3 d at 4°C, and grown for 5 d on half-strength Murashige and Skoog (MS) medium containing 0.6% Phytagel (Sigma-Aldrich), 2% sucrose, and 100 mg/L myo-inositol, or, for liquid culture, in half-strength MS medium, 1% sucrose, and 100 mg/L myo-inositol, with a 16-h-light/8-h-dark cycle at 22°C. For crosses and propagation of the plants, seedlings were transferred to soil and grown in growth chambers with a 16-h-light/8-h-dark cycle at 22°C. Plant transformation was performed as described by Diet et al. (2006).
For mapping, the lrx1 rol5-1 mutant was crossed with Ler and propagated to the F2 generation. Five hundred F2 seedlings displaying a wild-type root hair phenotype were selected and screened for homozygous lrx1 mutant plants with a PCR-based marker (Diet et al., 2004). These plants were assumed to be homozygous mutants for rol5-1 and were thus used for initial mapping. Once the approximate map position of rol5 was identified, F2 plants displaying an lrx1 mutant phenotype (i.e., being homozygous mutant lrx1) were selected, and those heterozygous Columbia/Ler in the region containing the rol5 locus were propagated to the F3 generation. As expected, seedlings of the F3 population segregated 3:1 for lrx1 versus wild-type root hairs. One thousand wild-type-like F3 seedlings were selected for detailed mapping of rol5. Mapping was performed using standard simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers developed based on the Columbia/Ler polymorphism databank (Jander et al., 2002).
Molecular Markers for Genotyping
The marker for lrx1 was previously described (Diet et al., 2004). The rol5-1 mutation was detected by PCR with the primers rol5BanI_F (5′-ACAATCTTAAGAGGCAAACC-3′) and rol5BanI_R (5′-CATATTAAGCAGAAGCTTGG-3′), followed by digestion with the enzyme BanI, which only cuts wild-type ROL5 but not the rol5-1 DNA. The T-DNA insertion in rol5-2 is 3′ adjacent to the sequence 5′-GTTATTGAAAGTAGAGA-3′. Homozygous rol5-2 mutants were identified by DNA gel blotting using genomic DNA digested with BglII and a fragment of the ROL5 gene 3′ adjacent of the T-DNA insertion site as a specific probe for hybridization.
DNA Constructs
For complementation of the rol5-1 mutant, a ROL5 genomic clone including 1.8 kb of the promoter region and 400 bp of terminator sequence was amplified by PCR using the primer pair Rol5R1Not (5′-ATTGCGGCCGCTGGGCTGGTGATGAAAGTTG-3′), Rol5F1NotI (5′-ATTGCGGCCGCCAGAGTGTCTTGATTGGTTCG-3′). The PCR product was digested by NotI and cloned into the pART27 plant transformation vector (Gleave, 1992) cut with the same enzyme. For the ROL5-GFP fusion constructs, the genomic clone of ROL5 that was used for complementation was subjected to site-directed mutagenesis (QuikChange; Stratagene) to introduce one BamHI site using the primer pair mutBamHI-midF (5′-GAATCTCCTCCTCGGATCCAAAAACCTCATAAAAGC-3′) and mutBamHI-midR (5′-GCTTTTATGAGGTTTTTGGATCCGAGGAGGAGATTC-3′). The GFP gene was amplified from the vector pMDC83 (Curtis and Grossniklaus, 2003) using the primer pair GFP-F (5′-TATGGATCCATGAGTAAAGGAGAAGAACTTTTC-3′), GFP-R (5′-AATGGATCCGTGGTGGTGGTGGTGGTGTTTG-3′) and cloned into the BamHI site of the ROL5 gene. The resulting ROL5-GFP construct was cloned into the binary vector pART27 with the restriction enzyme NotI. For transient expression in Arabidopsis epidermal cells, the ROL5-GFP construct was ligated into the overexpression cassette of pART7 (Gleave, 1992) and used for particle bombardment. CoxIV-DsRed with a yeast COXIV presequence tag for mitochondrial localization (Mollier et al., 2002) was used as the mitochondrial marker protein. The overexpression construct for Sc FKBP12 was obtained by PCR amplification of FKBP12 from yeast with the primer pair ScFKBP12_F (5′-GAATTCATGTCTGAAGTAATTGAAGGTAAC-3′), ScFKBP12_R (5′-TCTAGATTAGTTGACCTTCAACAATTCGAC-3′) and cloning of the PCR product into pART7 containing a 35S cauliflower mosaic virus promoter:ocs terminator cassette (Gleave, 1992) by digestion with EcoRI and XbaI. A correct pART7-ScFKBP12 clone was digested with NotI, and the excised 35S:ScFKBP12:ocs cassette was inserted into the binary vector pART27 digested with NotI. For expression in yeast, the coding sequence of a ROL5 cDNA was amplified with the primers ROL5-pFL61_F (5′-GCGGCCGCATGGAGGCCAAGAACAAGAAAGC-3′) and ROL5-pFL61_R (5′-GCGGCCGCTTAGAAATCCAGAGATCCACATTG-3′) and cloned into the expression vector pFL61 (Minet et al., 1992) by digestion with NotI.
Yeast Strain and Growth Conditions
Yeast strains used in this study were obtained from EUROSCARF, Frankfurt, Germany. The wild-type strain is BY4741 with the relevant genotype MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0, and the ncs6Δ strain has the relevant genotype MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGL211w::kanMX4. Yeast strains were grown using standard methods. Synthetic yeast media was prepared with 2.4 nM rapamycin where indicated. Yeast strains were grown at 30°C until log phase and drops of an OD600 of 0.8 and three subsequent 10-fold dilutions were spotted onto synthetic solid medium and grown for 3 d at 30°C.
Transient Gene Expression in Arabidopsis Epidermal Cells
For transient gene expression, Arabidopsis leaf epidermal cells were transformed by particle bombardment as described (Escobar-Restrepo et al., 2007). Bombarded tissue was incubated for 2 d at room temperature and the fluorescence analyzed using confocal microscopy.
ROS Staining
For ROS staining, 0.1 mg/mL NBT was directly dissolved in 0.1 M K-phosphate buffer, pH 7, stirred for 60 min at room temperature, and filtered through a 0.2-μm pore size filter. Seedlings were incubated at room temperature for 60 min. The reaction was stopped by washing twice with 100% ethanol.
Microscopy
GFP fluorescence was analyzed by confocal microscopy (DMIRE2; Leica) and analysis of immunolabeling on a LM510 (Zeiss). Phenotypic observations were performed with a Leica LZ M125 stereomicroscope. For cell and root hair length measurements, pictures were taken by differential interference contrast microscopy using an Axioplan microscope (Zeiss). Over 30 data points from ≥5 seedlings were collected. Root length was manually determined, using ≥20 seedlings per data point. The t test was used for statistical analysis, and the values are given with ± sd, P = 0.05. Confocal microscopy was performed on a DMIRE2 (Leica).
Immunolabeling
Immunolabeling of surfaces of intact Arabidopsis seedling roots was performed using six rat monoclonal antibodies directed to cell wall components. Arabidopsis seedlings were prepared for immunofluorescence microscopy as described (McCartney et al., 2003). Seedlings were vertically grown for 4 d prior to immunolabeling. An FITC-linked anti-rat antibody (Sigma-Aldrich) was used as secondary antibody. Seedlings were mounted in a glycerol antifade solution (Citifluor AF1; Agar Scientific) for microscopy observation.
RNA Extraction and RT-PCR
Seedlings were grown vertically on half-strength MS plates for 2 weeks. Approximately 150 seedlings of each plant line were cut at the hypocotyl to separate shoot and root tissue, and the tissues were used for extraction of total RNA using the TRIzol method (Gibco BRL). The reverse transcription was performed using the SuperScriptTM II RNase reverse transcriptase kit (Invitrogen). The resulting single-stranded DNA was used for PCR with 30 cycles. ACTIN2 was amplified as a control using the primer pair Actin2F (5′-AATGAGCTTCGTATTGCTCC-3′) and Actin2R (5′-GCACAGTGTGAGACACACC-3′). Levels for the ROL5 transcript were checked using the primer pair Rol5NorthF3 (5′-CCAAGATGTAAACCTTTCAAG-3′) and RolNorth2R (5′-GCTTCTTTGTTTCCTTATTATG-3′).
tRNA Extraction and Analysis
Whole seedlings grown for 14 d in half-strength liquid medium containing 1% sucrose and 100 mg/L myo-inositol were collected for tRNA extraction. Plant material was frozen in liquid nitrogen, ground, and stored at −80°C. Total RNA was extracted two times with acidic phenol and chloroform and one time with chloroform. tRNA was purified using Nucleobond AX 100 columns (Macherey-Nagel) and precipitated overnight at −20°C. One microgram of tRNA per sample was separated on an 8% polyacrylamide gel containing 7 M urea and 1 μg/μL APM chloride where indicated. For tRNA visualization, the gel was stained with SYBR Gold (Invitrogen).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ROL5, At2G44270; LRX1, At1G12040; Sc FKBP12, YNL023C; and Sc NCS6, YGL211W.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cell Wall Epitopes Not Affected by the rol5-1 Mutation.
Supplemental Figure 2. Effect of Rapamycin on Cell Wall Epitopes.
Supplemental Figure 3. ROS Staining in Wild-Type and rol5-1 Mutant Roots.
Acknowledgments
This work was supported by the Swiss National Science Foundation (R.-M.L., F.J., and C.R.) and Marie Curie WallNet MRTN-CT-2004512265 (Y.V.). We thank Sebastian Leidel for providing APM, Geneviève Ephritikhine for the vector encoding the mitochondrial marker protein, Markus Klein for the pFL61 yeast expression vector, and Robert Dudler for helpful discussions and critical reading of the manuscript. The service provided by The Arabidopsis Information Resource (www.arabidopsis.org) was crucial for this work.
References
- Anderson G.H., Veit B., Hanson M.R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Epel O., Gharyal P.K., Schindler M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta 175: 389–395 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Doesseger B., Guyot R., Diet A., Parsons R.L., Clark M.A., Simmons M.P., Bedinger P., Goff S.A., Ringli C., Keller B. (2003a). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice: A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Ringli C., Keller B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Steiner M., Ryser U., Keller B., Ringli C. (2003b). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35: 71–81 [DOI] [PubMed] [Google Scholar]
- Björk G.R., Huang B., Persson O.P., Bystrom A.S. (2007). A conserved modified wobble nucleoside (mcm(5)s(2)U) in lysyl-tRNA is required for viability in yeast. RNA 13: 1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L., Okresz L., Henriques R., Anthony R.G. (2003). Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 8: 424–431 [DOI] [PubMed] [Google Scholar]
- Bork P., Koonin E.V. (1994). A p-loop-like motif in a widespread ATP pyrophosphatase domain: Implications for the evolution of sequence motifs and enzyme-activity. Proteins 20: 347–355 [DOI] [PubMed] [Google Scholar]
- Carpita N.C., Gibeaut D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Chan T.F., Carvalho J., Riles L., Zheng X.F.S. (2000). A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. USA 97: 13227–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1 alpha transcriptional complex. Nature 450: 736–740 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D., Truong H.N., Robaglia C., Meyer C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolai M., Bedu M., Robaglia C., Meyer C. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A., Brunner S., Ringli C. (2004). The enl mutants enhance the lrx1 root hair mutant phenotype of Arabidopsis thaliana. Plant Cell Physiol. 45: 734–741 [DOI] [PubMed] [Google Scholar]
- Diet A., Link B., Seifert G.J., Schellenberg B., Wagner U., Pauly M., Reiter W.D., Ringli C. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Zhu J.K. (1997). A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203: 289–294 [DOI] [PubMed] [Google Scholar]
- Dinkova T.D., de la Cruz H.R., Garcia-Flores C., Aguilar R., Jimenez-Garcia L.F., de Jimenez E.S. (2007). Dissecting the TOR-S6K signal transduction pathway in maize seedlings: Relevance on cell growth regulation. Physiol. Plant. 130: 1–10 [Google Scholar]
- Dolan L., Duckett C.M., Grierson C., Linstead P., Schneider K., Lawson E., Dean C., Poethig S., Roberts K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Gapper C., Dolan L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goehring A.S., Rivers D.M., Sprague G.F. (2003a). Urmylation: A ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell 14: 4329–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring A.S., Rivers D.M., Sprague G.F. (2003b). Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot. Cell 2: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Movva N.R., Hall M.N. (1991). Targets for cell-cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hematy K., Höfte H. (2008). Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 11: 321–328 [DOI] [PubMed] [Google Scholar]
- Hematy K., Sado P.E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J.P., Höfte H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17: 922–931 [DOI] [PubMed] [Google Scholar]
- Huang L.S., Sternberg P.W. (1995). Genetic dissection of developmental pathways. Methods in Cell Biology, Epstein H.F., Shaker C.C., (San Diego, CA: Academic Press; ), pp. 97–122 [DOI] [PubMed] [Google Scholar]
- Huang S., Bjornsti M.A., Houghton P.J. (2003). Rapamycins. Cancer Biol. Ther. 2: 222–232 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. (2003). Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Humphrey T.V., Bonetta D.T., Goring D.R. (2007). Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 176: 7–21 [DOI] [PubMed] [Google Scholar]
- Ingram G.C., Waites R. (2006). Keeping it together: Co-ordinating plant growth. Curr. Opin. Plant Biol. 9: 12–20 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Seymour G.B., Knox J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1->4)-beta-D-galactan. Plant Physiol. 113: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.P., Linstead P.J., King J., Cooper C., Roberts K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Kohorn B.D., Kobayashi M., Johansen S., Riese J., Huang L.F., Koch K., Fu S., Dotson A., Byers N. (2006). An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 46: 307–316 [DOI] [PubMed] [Google Scholar]
- Krause K., Krupinska K. (2009). Nuclear regulators with a second home in organelles. Trends Plant Sci. 14: 194–199 [DOI] [PubMed] [Google Scholar]
- Leidel S., Pedrioli P.G.A., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. (2009). Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232 [DOI] [PubMed] [Google Scholar]
- Levitin B., Richter D., Markovich I., Zik M. (2008). Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J. 56: 351–363 [DOI] [PubMed] [Google Scholar]
- Liszkay A., van der Zalm E., Schopfer P. (2004). Production of reactive oxygen intermediates (O2·-, H2O2, and ·OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 136: 3114–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., Hall M.N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Mahfouz M.M., Kim S., Delauney A.J., Verma D.P.S. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A., Nothnagel E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.E., Verhertbruggen Y., Herve C., Ordaz-Ortiz J.J., Farkas V., Pedersen H.L., Willats W.G.T., Knox J.P. (2008). Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Bhatt K., Baumann K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4: 540–549 [DOI] [PubMed] [Google Scholar]
- McCartney L., Ormerod A.P., Gidley M.J., Knox J.P. (2000). Temporal and spatial regulation of pectic (1->4)-beta-D-galactan in cell walls of developing pea cotyledons: Implications for mechanical properties. Plant J. 22: 105–113 [DOI] [PubMed] [Google Scholar]
- McCartney L., Steele-King C.G., Jordan E., Knox J.P. (2003). Cell wall pectic (1 -> 4)-β-D-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33: 447–454 [DOI] [PubMed] [Google Scholar]
- Menand B., Desnos T., Nussaume L., Berger F., Bouchez D., Meyer C., Robaglia C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M., Dufour M.E., Lacroute F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Mollier P., Hoffmann B., Debast C., Small I. (2002). The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 40: 405–409 [DOI] [PubMed] [Google Scholar]
- Noma A., Sakaguchi Y., Suzuki T. (2009). Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 37: 1335–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.M., Umbach A.L., Subbaiah C.C., Siedow J.N. (2006). Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley B.L., O'Neill M.A., Mohnen D.A. (2001). Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57: 929–967 [DOI] [PubMed] [Google Scholar]
- Ringli C. (2005). The role of extracellular LRR-extensin (LRX) proteins in cell wall formation. Plant Biosyst. 139: 32–35 [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schieke S.M., Finkel T. (2006). Mitochondrial signaling, TOR, and life span. Biol. Chem. 387: 1357–1361 [DOI] [PubMed] [Google Scholar]
- Schieke S.M., Phillips D., McCoy J.P., Aponte A.M., Shen R.F., Balaban R.S., Finkel T. (2006). The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281: 27643–27652 [DOI] [PubMed] [Google Scholar]
- Schlieker C.D., Van der Veen A.G., Damon J.R., Spooner E., Ploegh H.L. (2008). A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. USA 105: 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani R., Yao L., Menand B., Ennar N., Lecampion C., Meyer C., Robaglia C. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Tsao C.C., Chen Y.T., Lan C.Y. (2009). A small G protein Rhb1 and a GTPase-activating protein Tsc2 involved in nitrogen starvation-induced morphogenesis and cell wall integrity of Candida albicans. Fungal Genet. Biol. 46: 126–136 [DOI] [PubMed] [Google Scholar]
- van Hengel A.J., Roberts K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 32: 105–113 [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., Knox J.P. (1996). A role for arabinogalactan-proteins in plant cell expansion: Evidence from studies on the interaction of beta-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9: 919–925 [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., Marcus S.E., Knox J.P. (1998). Generation of a monoclonal antibody specific to (1->5)-alpha-L-arabinan. Carbohydr. Res. 308: 149–152 [DOI] [PubMed] [Google Scholar]
- Willats W.G.T., McCartney L., Mackie W., Knox J.P. (2001). Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 47: 9–27 [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. (2006). TOR signaling in growth and metabolism. Cell 127: 5–19 [DOI] [PubMed] [Google Scholar]
- Yates E.A., Valdor J.F., Haslam S.M., Morris H.R., Dell A., Mackie W., Knox J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6: 131–139 [DOI] [PubMed] [Google Scholar]