This study demonstrates that the HopF2 effector of Pseudomonas syringae can interact with MAP KINASE KINASE5 to weaken pathogen-associated molecular pattern–triggered immunity in Arabidopsis and identifies key residues required for this interaction.
Abstract
The successful recognition of pathogen-associated molecular patterns (PAMPs) as a danger signal is crucial for plants to fend off numerous potential pathogenic microbes. The signal is relayed through mitogen-activated protein kinase (MPK) cascades to activate defenses. Here, we show that the Pseudomonas syringae type III effector HopF2 can interact with Arabidopsis thaliana MAP KINASE KINASE5 (MKK5) and likely other MKKs to inhibit MPKs and PAMP-triggered immunity. Inhibition of PAMP-induced MPK phosphorylation was observed when HopF2 was delivered naturally by the bacterial type III secretion system. In addition, HopF2 Arg-71 and Asp-175 residues that are required for the interaction with MKK5 are also necessary for blocking MAP kinase activation, PAMP-triggered defenses, and virulence function in plants. HopF2 can inactivate MKK5 and ADP-ribosylate the C terminus of MKK5 in vitro. Arg-313 of MKK5 is required for ADP-ribosylation by HopF2 and MKK5 function in the plant cell. Together, these results indicate that MKKs are important targets of HopF2.
INTRODUCTION
Many phytopathogenic microbes deliver a suite of effector proteins into the host cell to modulate host cellular processes in favor of pathogen infection or proliferation (Hogenhout et al., 2009). These effector proteins collectively enable a parasitic lifestyle for the pathogens. For instance, Gram-negative bacterial pathogens use the type III secretion system to inject dozens of effectors into host cells. Oomycete pathogens potentially deliver hundreds of effectors into host cells through an unknown mechanism (Hogenhout et al., 2009). Understanding the biochemical function and host targets of these pathogen effectors has been a major focus of the field.
Plants possess highly sensitive surveillance systems that monitor threats from potential pathogenic microbes and activate effective defense responses. The surface-localized immune receptors detect a variety of pathogen-associated molecular patterns (PAMPs), such as bacterial flagellar protein and elongation factor-Tu (EF-Tu), lipopolysaccharides, and fungal cell wall fragments. PAMP-triggered immunity (PTI) is critically important for plant resistance to numerous potential pathogens (Chisholm et al., 2006; Boller and Felix, 2009). Pseudomonas syringae carries ∼30 effectors, many of which are now known to inhibit PTI (Zhou and Chai, 2008; Boller and He, 2009). The ability of various pathogen effector proteins to suppress PTI within the plant cell has driven the evolution of plant disease resistance (R) proteins, which serve as cytoplasmic immune receptors that either directly or indirectly recognize effector proteins to trigger a second layer of host immunity termed effector-triggered immunity (ETI). Increasing evidence suggests that a number of P. syringae effectors are also capable of inhibiting ETI (Guo et al., 2009). To date, the biochemical functions of only a handful of P. syringae effectors have been experimentally elucidated, and a smaller number of these effectors have been shown to target defined host proteins to enhance virulence.
Several effectors have been shown to target key components of the PTI signaling pathway, which consists of receptor kinases such as FLAGELLIN SENSING2 (FLS2), EF-Tu receptor (EFR), and Chitin Elicitor Receptor Kinase 1 (CERK1), coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1, and MITOGEN-ACTIVATED PROTEIN KINASES (MPKs). FLS2 and EFR are receptors that recognize the bacterial flagellar and EF-Tu peptides flg22 and elf18, respectively (Chinchilla et al., 2006; Zipfel et al., 2006), whereas CERK1 is the receptor for the fungal cell wall component chitin (Miya et al., 2007; Wan et al., 2008; Iizasa et al., 2010). AvrPto acts as a kinase inhibitor of target FLS2, EFR, and other receptor kinases, thereby blocking PAMP-triggered immunity (Xiang et al., 2008). Likewise, AvrPtoB uses an E3 ubiquitin ligase activity to target FLS2 and/or CERK1 for degradation (Gohre et al., 2008; Gimenez-Ibanez et al., 2009). In addition, HopAI1 dephosphorylates mitogen-activated protein (MAP) kinases to inhibit PTI signaling (Zhang et al., 2007). Furthermore, the analyses of HopU1 and HopM1 led to the identification of novel components important for plant immunity. HopU1 is an ADP-ribosyltransferase that modifies the RNA binding protein GRP7, which is required for PTI defenses (Fu et al., 2007). HopM1 targets and leads to the degradation of MIN7, an ADP-ribosylation factor guanine exchange factor protein that likely regulates vesicle trafficking (Nomura et al., 2006). Host targets for the majority of P. syringae effectors, however, remain unknown.
The P. syringae pv tomato DC3000 type III effector HopF2 enhances virulence in tomato (Solanum lycopersicum) plants (Robert-Seilaniantz et al., 2006). When expressed in Arabidopsis thaliana protoplasts, HopF2 strongly inhibits flg22-induced expression of the basal resistance marker gene, NONHOST RESISTANCE1 (NHO1), suggesting a role in PTI inhibition (Li et al., 2005). Delivery of HopF2 by the bacterial type III secretion system or transgenic expression in Arabidopsis inhibited PAMP-induced callose deposition (Guo et al., 2009). In tobacco (Nicotiana tabacum) plants, HopF2 induces the hypersensitive response (HR), which is indicative of ETI activation (Robert-Seilaniantz et al., 2006). Similarly, the homologous effector HopF1 (AvrPphF) carried by P. syringae pv phaseolicola triggers R1-specified ETI in bean (Phaseolus vulgaris) plants but promotes virulence in cultivars lacking the R1 gene (Tsiamis et al., 2000). Crystal structure analysis of HopF1 revealed marginal structure similarity to the catalytic domain of diphtheria toxin, an ADP-ribosyltransferase (Singer et al., 2004). However, the enzymatic activity of HopF1 has not been detected. The biochemical function of HopF2 and HopF1 remains unknown.
Here, we show that HopF2 is capable of targeting multiple MAP kinase kinases (MKKs). Consistent with a role in inhibiting MKKs, HopF2 delivered from Pseudomonas bacteria strongly inhibits the activation of MAP kinases in plants. We also demonstrate that HopF2 ADP-ribosylates MKK5 in vitro to block its kinase activity in a manner dependent on HopF2 Arg-71 and Asp-175, two residues required for the interaction with MKK5. These residues are also necessary for inhibiting PTI, virulence function in tomato plants, and HR elicitation in tobacco plants, suggesting the importance of HopF2 enzymatic activity for virulence and ETI induction.
RESULTS
HopF2 Inhibits PTI Responses
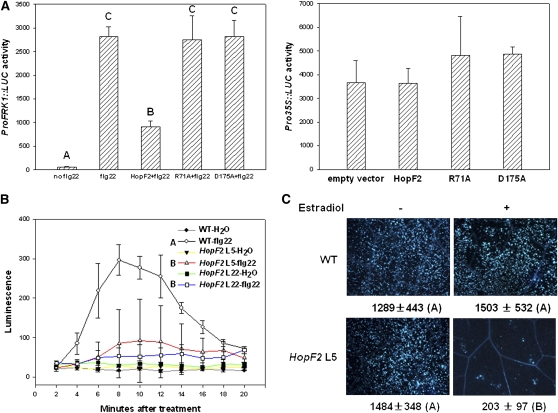
We previously showed that transient expression of HopF2 in Arabidopsis protoplasts can suppress flg22-induced transcription of a basal resistance marker gene, NHO1 (Li et al., 2005). To further investigate the role of HopF2 in PTI inhibition, we tested the effect of HopF2 on the expression of another PAMP-induced reporter gene construct, FRK1 promoter-luciferase (ProFRK1-LUC; Asai et al., 2002). Figure 1A (left panel) shows that transient expression of HopF2 blocked flg22-induced ProFRK1-LUC expression. HopF2 does not affect the expression of luciferase driven by the cauliflower mosaic virus 35S promoter (Figure 1A, right panel), indicating that the reduced expression of ProFRK1-LUC was not caused by a general reduction of gene transcription. Two surface-exposed residues in pocket A of HopF1, Arg-72 and Asp-174, are invariant residues in HopF alleles and are required for both virulence and ETI function of HopF1 (Singer et al., 2004). We therefore tested if corresponding residues in HopF2 (Arg-71 and Asp-175) are required for the inhibition of flg22-induced ProFRK1-LUC expression by substituting these residues with Ala. The HopF2R71A and HopF2D175A mutants were completely unable to inhibit ProFRK1-LUC expression, indicating that these residues are essential for inhibiting flg22-induced signaling.
Figure 1.
HopF2 Suppresses flg22-Induced Resistance Responses.
(A) HopF2 inhibits flg22-induced FRK1 expression. Left: Protoplasts were transfected with ProFRK1:LUC reporter construct along with HopF2-FLAG, HopF2R71A-FLAG, or HopF2D175A-FLAG and induced with 100 nM flg22 for 2 h before ProFRK1-LUC activity was measured. Right: Pro35S:LUC reporter control. Each data point represents the mean of three replicates.
(B) HopF2 inhibits the flg22-induced oxidative burst. Four-week-old wild-type (WT), HopF2 transgenic line 5, and line 22 T3 plants were sprayed with 50 μM β-estradiol 48 h before leaves were excised. Leaf strips were treated with water or 1 μM flg22, and the production of H2O2 was measured at the indicated time points and expressed as relative luminescence units. Each time point represents the data from eight replicates. Error bars indicate sd. Different letters indicate significant difference at a P value <0.01 (Student's t test). The experiments were repeated twice with similar results.
(C) HopF2 inhibits flg22-induced callose deposition. Six-week-old wild-type and HopF2 transgenic line 5 T3 plants were sprayed with water (−) or 50 μM β-estradiol (+) 48 h before infiltration with 1 μM flg22. Leaves were stained for callose 6 h after flg22 treatment. The number below each microscopy photograph indicates the average number of callose deposits/0.1 mm2 leaf from eight independent leaves.
We next generated transgenic plants expressing HopF2 and examined PAMP-induced immune responses in addition to ProFRK1-LUC expression. Oxidative burst is an early response occurring within 5 min of flg22 treatment (Felix et al., 1999). The estradiol-induced expression of HopF2 in two independent transgenic lines suppressed flg22-induced H2O2 accumulation to ∼20 to 30% that of the wild type (Figure 1B). Consistent with a previous report (Guo et al., 2009), the estradiol-induced expression of HopF2 also suppressed flg22-induced callose deposition, a late response to PAMPs (Figure 1C). Furthermore, the HopF2-expressing plants showed enhanced bacterial growth when inoculated with a nonpathogenic strain of DC3000, hrpL− (Zwiesler-Vollick et al., 2002; see Supplemental Figure 1 online), a result consistent with previous reports that HopF2-expressing plants support DC3000 hrcC− mutant bacterial growth (Guo et al., 2009; Wilton et al., 2010). Together, these results indicate that HopF2 may target PTI signaling components.
HopF2 Inhibits MAP Kinase Activity in Plants
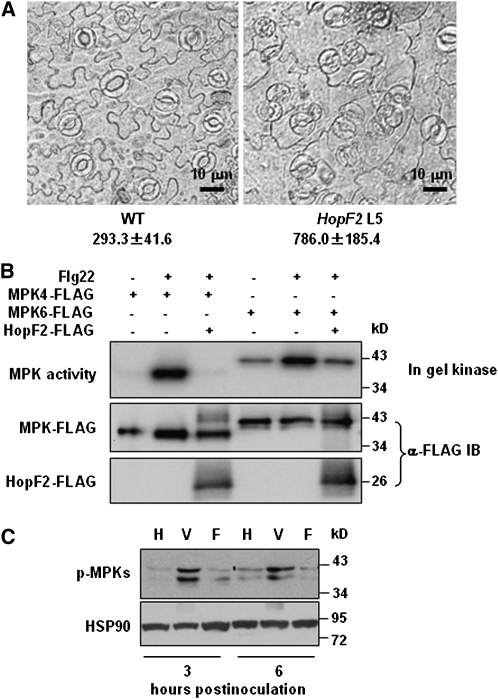
When germinated in the presence of estradiol, the HopF2 transgenic seedlings developed curly leaves and exhibited dwarfism (see Supplemental Figure 2 online). An examination of the abaxial epidermis showed that plants expressing HopF2 formed clustered stomata that defied the one-cell spacing rule (Figure 2A), a phenotype reminiscent of mutants defective in a MAP kinase cascade composed of a MAP kinase kinase kinase YODA, MAP kinase kinases MKK4 and MKK5, and MAP kinases MPK3 and MPK6 (Bergmann et al., 2004; Wang et al., 2007). These results raised the possibility that expression of HopF2 interferes with MAP kinase cascade components.
Figure 2.
HopF2 Function Is Linked to the Inhibition of MAP Kinase Cascades.
(A) HopF2 transgenic plants display clustered stomatal cells. The micrographs of the abaxial epidermis were taken 2 weeks after germination on plates containing estradiol. Numbers below the photographs are stomatal density/mm2 in HopF2 and wild-type (WT) plants, which are significantly different at a P value <0.01 (Student's t test). The results shown are representative of four independent experiments.
(B) HopF2 inhibits flg22-induced activation of MPK4 and MPK6. Protoplasts were transfected with the MPK4-FLAG or MPK6-FLAG construct in the presence (+) or absence (−) of the HopF2-FLAG construct, treated with water (−) or 1 μM flg22 (+) for 10 min, and the total protein extract was subjected to anti-FLAG immunoprecipitation. The purified MPK4-FLAG and MPK6-FLAG protein was then subjected to an in-gel kinase assay using myelin basic protein as a substrate and anti-FLAG immunoblot analysis.
(C) Bacterial PAMP-induced MAPK activation is suppressed by TTSS-delivered HopF2. Leaves of 6-week-old Col-0 plants were infiltrated with the indicated bacteria at 1 × 107 cells/mL. Leaf total protein was extracted at the indicated time points and subjected to immunoblot analysis with antiphospho-ERK antibodies. H, water control; V, P. fluorescens strain (pLN1965) bacteria lacking effector genes but containing an empty vector; F, P. fluorescens strain (pLN1965) carrying the ShcF-HopF2 construct. Equal loading was indicated by immunoblot with anti-HSP90 antibodies.
To test this, we coexpressed MPK6-FLAG along with HopF2-FLAG in protoplasts and treated the protoplasts with flg22. An in-gel kinase assay using myelin basic protein as a substrate indicated that MPK6 activity was strongly activated by flg22 in control protoplasts. The coexpression of HopF2 significantly reduced MPK6 activity (Figure 2B). Interestingly, HopF2 also abolished MPK4 activity in this assay, indicating that HopF2 may inhibit multiple MPKs.
The amount of P. syringae effector proteins delivered into the host cell by the type three secretion system (TTSS) is likely much lower than that in the transgenic plants. We therefore determined if HopF2 delivered by the bacterial TTSS is capable of inhibiting the activation of MPKs. Because it was previously shown that AvrPto, AvrPtoB, and HopAI1 can all block MPK activation, we used an engineered Pseudomonas fluorescens strain containing functional TTSS-coding genes but lacking any effector genes (Guo et al., 2009). This strain allowed us to determine whether HopF2 alone secreted from the bacterium is capable of inhibiting MPK activation. As expected, the P. fluorescens strain lacking hopF2 induced phosphorylation of MPKs by 3 and 6 h postinoculation, presumably as a result of recognition of PAMPs carried by P. fluorescens bacteria. By contrast, the strain containing hopF2 inhibited MPK phosphorylation to near background levels (Figure 2C). These results demonstrated that HopF2 indeed inhibits the activation of MPKs.
HopF2 Directly Targets MKKs
MPK3 and MPK6 can be activated by the expression of MKK5DD, a constitutively active form of MKK5 (Liu and Zhang, 2004). To further dissect the mechanism by which HopF2 inhibits PTI signaling, we tested whether HopF2 can inhibit MKK5DD-induced MPK activation and defenses.
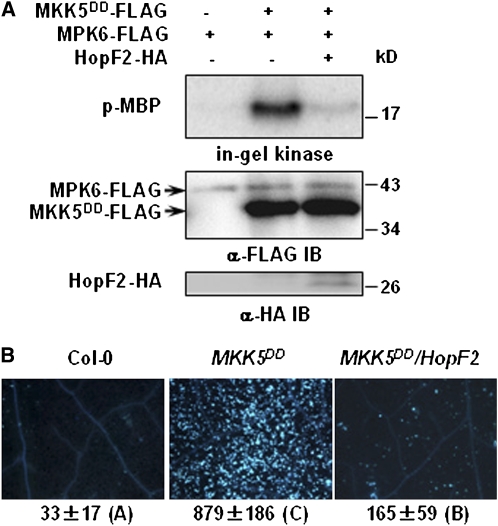
To determine if HopF2 is capable of inhibiting MKK5DD activity, we separately expressed FLAG-tagged MKK5DD and MPK6 in protoplasts. The MKK5DD and MPK6 proteins were isolated by immunoprecipitation and incubated in an in vitro kinase assay. MKK5DD from protoplasts lacking HopF2 constitutively activated MPK6 (Figure 3A), whereas MKK5DD coexpressed with HopF2 did not. To determine if HopF2 impacts defense responses activated by MKK5DD, the HopF2 transgene was introduced into an MKK5DD transgenic line by crossing. While dexamethasone-inducible expression of the MKK5DD transgene strongly induced callose deposition (at 24 h) and cell death (at 36 h) in the absence of HopF2 (Zhang et al., 2007; Figures 3B; see Supplemental Figure 3 online), the estradiol-inducible expression of the HopF2 transgene severely inhibited the callose deposition. Together, these results demonstrated that HopF2 acts on or downstream of MKK5 to block immune responses.
Figure 3.
HopF2 Blocks MKK5 Function.
(A) HopF2 inactivates MKK5 in protoplasts. Protoplasts were transfected with MKK5DD-FLAG alone or in combination with HopF2-HA, and MKK5DD-FLAG protein was isolated by immunoprecipitation. MPK6-FLAG was expressed alone in protoplasts and isolated by immunoprecipitation. The isolated MPK6 and MKK5DD were incubated in a kinase reaction system before an in-gel kinase assay was performed. The amount of MPK6 and MKK5DD in the in vitro kinase reaction assay was indicated by immunoblot (IB). An aliquot of total protein extract was subjected to immunoblot analysis with anti-HA antibody to determine the expression of HopF2-HA.
(B) HopF2 inhibits the callose deposition triggered by the constitutively active form of MKK5. Six-week-old Col-0, MKK5DD, and MKK5DD/HopF2 transgenic plants were induced with both 30 μM dexamethasone and 50 μM β-estradiol for 24 h before callose deposition was examined. The number below each microscopy photograph indicates the average number of callose deposits/0.1 mm2 leaf from eight independent leaves. Error bars indicate sd. Different letters indicate significant difference at a P value <0.01 (Student's t test). The results shown are representative of two independent experiments.
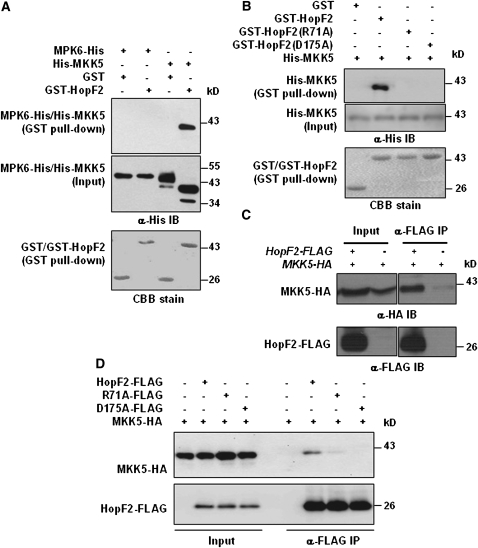
A glutathione S-transferase (GST) pull-down assay was used to test whether HopF2 can interact with MKK5. His-MKK5 was coexpressed with GST or GST-HopF2 in Escherichia coli, and His-MKK5 copurified with GST-HopF2 but not GST in glutathione agarose (Figure 4A). By contrast, MPK6-His was not pulled down by GST-HopF2. The His-MKK5 protein that was coexpressed with GST migrated slower than that coexpressed with GST-HopF2. A treatment with shrimp alkaline phosphatase eliminated the difference (see Supplemental Figure 4 online), indicating that His-MKK5 is phosphorylated when coexpressed with GST, likely because of autophosphorylation. The results also indicated that the His-MKK5 autophosphorylation was abolished in the presence of GST-HopF2. Arabidopsis contains 10 MKKs divided into seven subfamilies (Hamel et al., 2006). We therefore tested whether HopF2 interacts with additional MKKs. A GST pull-down assay indicated that HopF2 interacts with MKK3, MKK4, MKK6, and MKK10, but not MKK9 (see Supplemental Figure 5A online). Among the MKKs tested, HopF2 interacted with MKK4 and MKK5 most reliably, indicating that MKK4 and MKK5 may be the preferred targets of HopF2.
Figure 4.
HopF2 Targets MKKs.
(A) HopF2 interacts with MKK5 but not MPK6 in E. coli. GST or GST-HopF2 was coexpressed with His-MKK5 or MPK6-His in E. coli, purified with glutathione agarose, and subjected to immunoblot analysis using anti-His antibody to detect His-MKK5 or MPK6-His. Coomassie blue (CBB) staining indicates equal amounts of bait proteins.
(B) HopF2 Arg-71 and Asp-175 are required for the interaction with MKK5 in E. coli. His-MKK5 was coexpressed with GST, GST-HopF2, GST-HopF2R71A, or GST-HopF2D175A in E. coli, and protein was subjected to a GST pull-down assay.
(C) HopF2 interacts with MKK5 in plants. Transgenic plants carrying the HopF2-FLAG and MKK5-HA transgenes were analyzed by a co-IP assay.
(D) HopF2 Arg-71 and Asp-175 are required for the interaction with MKK5 in protoplasts. Protoplasts were transfected with MKK5-HA alone or in combination with HopF2-FLAG, HopF2R71A-FLAG, or HopF2D175A-FLAG, and protein–protein interaction was analyzed by co-IP.
We further tested if Arg-71 and Asp-175 are required for the HopF2-MKK5 interaction. Figure 4B shows that the HopF2R71A and HopF2D175A mutants were completely unable to interact with MKK5 in GST pull-down assays. To determine if HopF2 can interact with MKKs in plant cells, FLAG-tagged HopF2 and HA-tagged MKK1, MKK4, MKK5, and MKK5DD were coexpressed in Arabidopsis protoplasts. Coimmunoprecipitation (co-IP) assay showed that HopF2 can interact with all four constructs tested, indicating that HopF2 is capable of interacting with MKKs both before and after activation (see Supplemental Figure 5B online). To determine if HopF2 interacts with MKK5 in plants, we constructed an HA-tagged MKK5 transgenic line under the control of the native MKK5 promoter and crossed this line with a HopF2 transgenic line. A co-IP assay detected HopF2-MKK5 interaction in plants (Figure 4C). We further used the protoplast system to determine if HopF2 Arg-71 and Asp-175 are required for the interaction with MKK5 in plant cells. Figure 4D shows that the HopF2R71A mutant interacted less with MKK5, whereas the HopF2D175A mutant failed to interact with MKK5 in protoplasts, indicating that the interaction detected is highly specific and required the surface-exposed residues Arg-71 and Asp-175 in HopF2. Together, these results indicated that HopF2 can target MKKs to inhibit MPK activation.
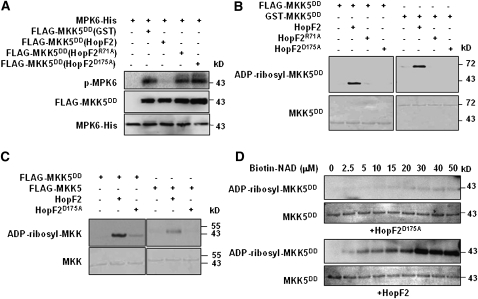
HopF2 Possesses ADP-Ribosyltransferase Activity and Blocks MKK5 Kinase Activity
To explore the biochemical function of HopF2, we coexpressed a FLAG-tagged recombinant MKK5DD with GST or GST-HopF2 in E. coli and examined MKK5DD kinase activity following affinity purification. The FLAG-MKK5DD coexpressed with GST strongly phosphorylated the recombinant MPK6-His in an in vitro kinase assay (Figure 5A). By contrast, FLAG-MKK5DD coexpressed with GST-HopF2 was completely inactive in MPK6 phosphorylation. Because the HopF2 protein was largely removed by the thorough wash during the isolation of FLAG-MKK5DD, the results suggested that the inhibitory effect of HopF2 on MKK5DD was not a result of physical interference. Thus HopF2 may possess enzymatic activity that covalently modifies MKK5. Although HopF2 can inhibit MKK5 autophosphorylation in E. coli (see Supplemental Figure 4 online), it is unlikely that HopF2 acted merely by preventing MKK5 activation because the constitutive active MKK5DD mutant was similarly inactivated.
Figure 5.
HopF2 ADP-Ribosylates MKK5 in Vitro.
(A) HopF2 inactivates MKK5 kinase activity in vitro. FLAG-MKK5DD was coexpressed with GST or GST-tagged wild-type HopF2, HopF2R71A, or HopF2D175A in E. coli and purified with anti-FLAG M2-agarose. The activity of FLAG-MKK5DD was measured by its ability to phosphorylate MPK6-His in vitro, as indicated by autoradiography. Amounts of protein were detected by immunoblot analysis with anti-FLAG and anti-MPK6 antibodies.
(B) HopF2 ADP-ribosylates MKK5 in vitro. Purified HopF2, HopF2R71A, or HopF2D175A was incubated with purified FLAG-MKK5DD or GST-MKK5DD at a substrate-to-enzyme ratio of 10:1 in the presence of biotinylated NAD. ADP-ribosylation was detected by immunoblot using horseradish peroxidase–conjugated streptavidin. Amounts of MKK5DD protein are indicated by CBB staining.
(C) MKK5DD is a better substrate for HopF2 than is WT MKK5. FLAG-tagged MKK5DD or WT MKK5 was incubated with HopF2 or HopF2D175A in the presence of biotinylated NAD, and ADP-ribosylation was detected as in (B). Amounts of MKK5 protein are indicated by Coomassie blue staining.
(D) HopF2 ADP-ribosylates MKK5 in a NAD concentration–dependent manner. Purified HopF2 or HopF2D175A was incubated with FLAG-MKK5DD in the ADP-RT reaction system containing the indicated concentrations of biotinylated NAD.
The similarity of HopF1 structure to diphtheria toxin prompted us to test if HopF2 possesses ADP-ribosyltransferase (ADP-RT) activity. We developed an in vitro ADP-RT reaction assay using the biotin-labeled NAD as the donor of ADP-ribose. Figure 5B shows that both FLAG- and GST-tagged MKK5DD proteins were ADP-ribosylated by recombinant HopF2. Interestingly, the wild-type MKK5 protein appeared to be ADP-ribosylated to a lesser extent (Figure 5C), suggesting that the activated form of MKK5 is a preferred substrate of HopF2. Optimum enzyme activity toward MKK5DD was achieved in the presence of ∼30 μM or greater NAD (Figure 5D). The HopF2R71A and HopF2D175A mutants, which do not interact with MKKs, did not ADP-ribosylate MKK5DD (Figure 5B).
A recent report showed that HopF2 can interact with the RPM1-interacting protein RIN4 to inhibit ETI (Wilton et al., 2010), suggesting that HopF2 may target different classes of host proteins. An ADP-RT assay showed that, like MKK5DD, RIN4 was ADP-ribosylated by HopF2 (see Supplemental Figure 6 online). These results indicate that HopF2 possesses the ADP-RT activity targeted to both MKK5 and RIN4.
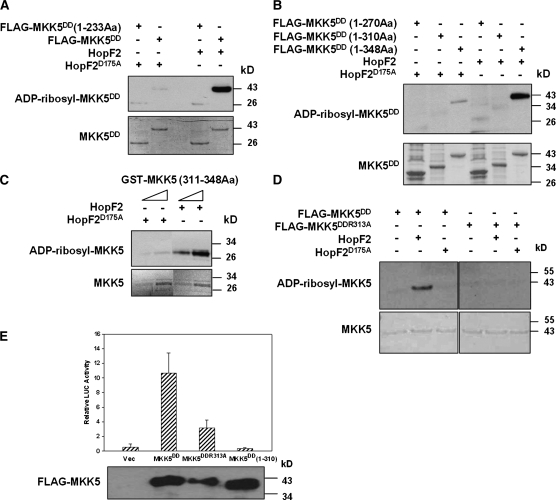
MKK5 Arg313 Is Required for ADP-Ribosylation
We next sought to determine putative ADP-ribosylation site(s) in MKK5. Deletion analyses indicated that the C-terminal 38 amino acids of MKK5DD are required for in vitro ADP-ribosylation by HopF2 (Figures 6A and 6B). A GST fusion protein containing the last 38 amino acids was ADP-ribosylated by wild-type HopF2 but not HopF2D175A (Figure 6C), indicating that the C terminus is sufficient for the modification. An examination of the MKK5 C-terminal sequence revealed an invariant Arg (Arg-313) and two nonconserved Arg residues (Arg-335 and Arg-342; see Supplemental Figure 7A online). Because Arg is a preferred residue for ADP-ribosyltransferases, we introduced Arg to Lys substitutions into MKK5DD and performed ADP-RT assays. The R313K substitution diminished ADP-ribosylation (see Supplemental Figure 7B online), whereas the R335K and R342K substitutions did not affect the modification (see Supplemental Figure 7C online), indicating that only the invariant Arg-313 is required for the modification. Similarly, the MKK5DD protein containing an R313A substitution was not ADP-ribosylated by HopF2 (Figure 6D). Together, these results suggest that Arg-313 may be the ADP-ribosylation site. To test if the C terminus and Arg-313 are required for MKK5DD to induce FRK1 expression, we transfected protoplasts with MKK5DD mutants lacking the C-terminal 38 amino acids or containing the R313A substitution and measured their ability to stimulate ProFRK1-LUC expression. Figure 6E shows that the R313A mutation diminished MKK5DD-triggered FRK1 expression, whereas the C-terminal truncation completely abolished FRK1 expression. These results indicate that Arg-313 and the C terminus play an important role in MKK5 function.
Figure 6.
Arg-313 of MKK5 Is Required for ADP-Ribosylation and Function.
(A) and (B) The C terminus of MKK5DD is required for ADP-ribosylation by HopF2. FLAG-tagged MKK5DD full-length (1-348Aa) or deletion constructs (1-270Aa and 1-310Aa) were incubated with wild-type HopF2 or HopF2D175A in an ADP-RT reaction. Aa, amino acids. ADP-ribosylated MKK5 and total MKK5 protein were detected as in Figure 5.
(C) The C-terminal 38 amino acids of MKK5 are sufficient for ADP-ribosylation. The GST-MKK5(311-348Aa) fusion protein was incubated with the wild-type HopF2 or HopF2D175A in an ADP-RT reaction.
(D) Arg-313 of MKK5DD is required for ADP-ribosylation by HopF2. The MKK5DD mutant protein containing the R313A substitution was incubated with wild-type HopF2 or HopF2D175A in an ADP-RT reaction.
(E) The C-terminal 38 amino acids of MKK5DD are required for the activation of FRK1 expression. FRK1-LUC and the indicated constructs were cotransfected into Arabidopsis protoplasts, and LUC activity was measured 18 h later. An empty vector (Vec) was included as a negative control. Each data point represents the average of four replicates. Error bars indicate sd.
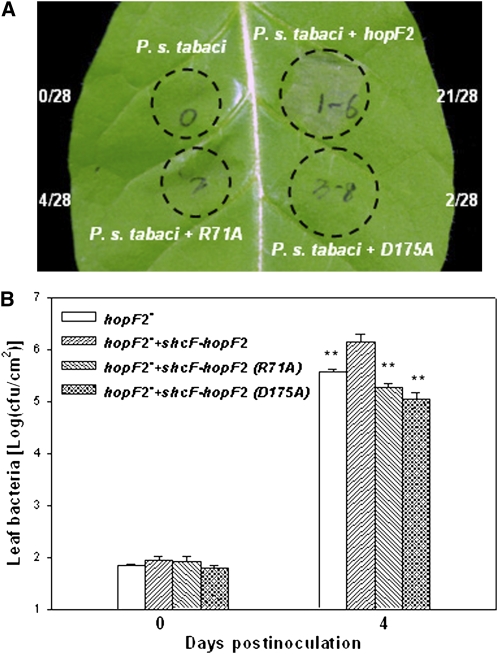
HopF2 R71 and D175 Are Required for Both Virulence and ETI Functions in Plants
HopF1 Arg-72 and Asp-174 are required for virulence on susceptible plants and ETI on resistant plants (Singer et al., 2004). Our results described above indicated that HopF2 can directly target MKK5 for ADP-ribosylation. Arg-71 and Asp-175 are required for the interaction with and/or ADP-ribosylation of MKK5. We therefore verified if HopF2 Arg-71 and Asp-175 are similarly required for nonhost HR in tobacco plants and virulence function in tomato plants (Robert-Seilaniantz et al., 2006).
As previously reported (Robert-Seilaniantz et al., 2006), the P. syringae pv tabaci strain was unable to induce the nonhost HR in tobacco W38 plants (Figure 7A). Introduction of wild-type hopF2, but not of the hopF2R71A and hopF2D175A mutants, into this strain induced a strong HR, indicating an essential role of Arg-71 and Asp-175. We also introduced the hopF2R71A and hopF2D175A mutants into a DC3000 derivative lacking hopF2 (hopF2−) and inoculated the resulting bacteria on tomato plants. While the wild-type hopF2 enhanced bacterial growth by approximately sevenfold, the two hopF2 mutants were completely unable to enhance bacterial growth (Figure 7B), indicating an essential role of Arg-71 and Asp-175 in the virulence function of hopF2. These results indicated that the ability of HopF2 to interact with and ADP-ribosylate MKK5 is correlated with both the HR elicitation and virulence function in plants.
Figure 7.
HopF2 Arg-71 and Asp-175 Are Required for Virulence and ETI Functions.
(A) HopF2 Arg-71 and Asp-175 are required for its ability to induce nonhost HR in tobacco W38 plants. Tobacco leaves were infiltrated with the P. syringae pv tabaci strain 1152 containing the indicated constructs at 2 × 108 colony-forming units/mL. Photographs were taken at 14 h after inoculation. The numbers on the side indicate areas showing HR/total injected areas. Infiltrated leaf areas are indicated by circles. The data shown are representative of two independent experiments.
(B) HopF2 Arg-71 and Asp-175 are required for virulence function in tomato plants. Tomato plants were infiltrated with the mutant strain lacking hopF2 (hopF2−), the hopF2− strain complemented with the wild-type schF-hopF2 locus, or the hopF2− strain complemented with the two mutant forms of schF-hopF2 at 5 × 104 colony-forming units/mL. Leaf bacterial numbers were determined at the indicated time points (four replicates/data point). Error bars indicate sd. ** Significant difference compared with the hopF2− strain carrying wild-type hopF2 at a P value <0.01 (Student's t test). The experiment was repeated three times with similar results.
DISCUSSION
In this study, we showed that the P. syringae effector HopF2 can target Arabidopsis MKK5 and likely other MKKs to inhibit PAMP-induced MPK activation. These findings are relevant to the natural infection process because HopF2 delivered by Pseudomonas TTSS is sufficient to block MPK activation. HopF2 possesses ADP-RT activity, and its ability to interact with and ADP-ribosylate MKK5 is correlated with the inhibition of PAMP-induced defenses and the virulence function in susceptible plants, indicating that MKKs are important host targets for HopF2.
Arabidopsis contains 10 MKKs that fall into four clades. Genetic and biochemical analyses have established that the closely related MKK1 and MKK2 proteins play an essential role in the activation of MPK4, which negatively regulates defenses against P. syringae (Petersen et al., 2000; Ichimura et al., 2006; Ichimura et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008). Gain-of-function studies suggested that MKK4, MKK5, and MKK9 can all activate MPK3 and MPK6, two MPKs thought to contribute positively to resistance to P. syringae (Asai et al., 2002; Xu et al., 2008). Our experiments suggested that HopF2 potentially targets multiple MKKs, although we failed to detect a HopF2-MKK9 interaction in GST pull-down assays. This is consistent with our finding that transient expression of HopF2 leads to the inhibition of both MPK4 and MPK6 in protoplasts.
A recent report showed that HopF2 is capable of inhibiting AvrRpt2-triggered ETI responses by interacting with RIN4 (Wilton et al., 2010). On rps2/rpm1/rin4 mutant plants, hopF2 no longer enhances virulence to DC3000 bacteria (Wilton et al., 2010), indicating that RIN4 is required for HopF2 virulence function in Arabidopsis. Because DC3000 bacteria can trigger weak but significant ETI (Zhang et al., 2010), the HopF2 virulence function reported by Wilton et al. (2010) may be indicative of an RIN4-dependent inhibition of ETI by this effector. Our results provide strong evidence that HopF2 targets MKKs to inhibit PTI. Thus, it is possible that HopF2 can target at least two classes of proteins, thereby inhibiting both PTI and ETI. It cannot be ruled out, however, that the HopF2–RIN4 interaction also contributes to the inhibition of PTI. We recently found that RIN4 can be phosphorylated by MPK4 in vitro, raising the possibility that one or more MKKs could act upstream of RIN4 to regulate plant immunity.
In vitro ADP-RT assays and in vivo labeling using radiolabeled NAD failed to detect an ADP-RT activity of HopF2 on RIN4 or other host proteins (Wilton et al., 2010). Using biotin-labeled NAD, we showed that HopF2 is also capable of ADP-ribosylating both MKK5 and RIN4 in vitro. One possibility is that recombinant HopF2 possesses relatively weak ADP-RT activity that was not detected by radiolabeling. Biotin labeling coupled with immunoblot analysis may provide greater sensitivity in the assay.
An important function of HopF1 is its ability to activate ETI in plants carrying a cognate R gene. HopF2 is also likely to trigger ETI in tobacco plants, as indicated by the nonhost HR. The conserved Arg-72 and Asp-174 in HopF1 and Arg-71 and Asp-175 in HopF2 are required for ETI. These results raise a tantalizing possibility that the cognate R protein may detect HopF2 and HopF1 by associating with an MKK.
HopF1 assumes a mushroom-like structure containing head and stalk subdomains, with the head subdomain displaying marginal similarity to diphtheria toxin ADP-RT (Singer et al., 2004). Our in vitro assay demonstrated that HopF2 indeed possesses ADP-RT activity. Results from Singer et al. (2004) and this study demonstrate the importance of Arg-72 and Asp-174 in HopF1 and Arg-71 and Asp-175 in HopF2 in ETI and virulence. The alignment of these residues to the diphtheria toxin catalytic sites His-21 and Glu-148 led to the speculation that Arg-72 and Asp-174 function as ADP-RT catalytic sites in HopF1. HopF2 Arg-71 and Asp-175 are required for ADT-ribosylation of MKK5. However, Arg-71 and Asp-175 are also required for binding to MKK5, suggesting that these residues are not the catalytic sites and that a specific interaction with MKKs is required for ADP-ribosylation.
It is interesting to note that hopF2 is located in an operon containing hopU1, which encodes another ADP-RT unrelated to HopF2 in amino acid sequence. HopU1 inhibits PTI by targeting RNA binding proteins such as GRP7 (Fu et al., 2007). Thus, the entire hopF2-hopU1 operon encodes two ADP-RTs that target distinct host proteins to inhibit PTI.
MAP kinases play an important role in both plant and animal innate immunity. Not surprisingly, components of the MAP kinase cascade are frequently targeted by pathogen effectors. We previously showed that the P. syringae effector HopAI1 directly targets MPK3 and MPK6 to inhibit PTI (Zhang et al., 2007). HopAI1 family members from animal bacterial pathogens also target MPKs to inhibit immune responses (Li et al., 2008). We recently found that another P. syringae effector, AvrB, directly interacts with and activates MPK4 to alter cellular responses in favor of bacterial proliferation (Cui et al., 2010). In addition, the Yersinia effector YopJ is known to inhibit MKKs. The results reported here indicate that HopF2 is a novel P. syringae effector that targets components in MAP kinase cascades. However, YopJ and HopF2 use distinct mechanisms to inhibit MKKs. YopJ acetylates phosphorylation sites of MKKs, thereby preventing the activation of MKKs (Mukherjee et al., 2006). Our results showed that HopF2 can interact with both the wild-type and the constitutively active form of MKK5. The constitutively active form of MKK5 can be readily ADP-ribosylated by HopF2, indicating that HopF2 can block MKK5 even after it has been activated by upstream MKKKs. Our analyses indicate that the ADP-ribosylation occurs to the last 38 amino acids of MKK5 and that Arg-313 is required for ADP-ribosylation. Importantly, Arg-313 and the C terminus are required for MKK5DD to activate PAMP response gene expression. The results thus uncover a regulatory role of the C terminus of MKK5 and suggest that ADP-ribosylation impedes this function.
METHODS
Plant Materials and Bacterial Strains
Arabidopsis thaliana plants used in this study include wild-type Columbia-0 (Col-0) and a dexamethasone-inducible MKK5DD transgenic line (Liu and Zhang, 2004). To generate HopF2-FLAG transgenic plants, the HopF2-FLAG fragment was excised from the pUC19-35S-HopF2-FLAG-RBS plasmid (Li et al., 2005) with XhoI and SpeI and inserted into pER8 (Zuo et al., 2000). The resulting construct was transformed into Col-0 plants by Agrobacterium tumefaciens–mediated transformation. Tobacco (Nicotiana tabacum) cultivar W38 was used for HR assay and tomato (Solanum lycopersicum) cultivar Money Maker was used for bacterial growth assay.
Pseudomonas syringae strains used in this study include P. s. pv tabaci R1152, P. s. pv tomato DC3000, DC3000 HopF2− mutant, DC3000+pLK-hrp-shcF-hopF2ATG-HA (Robert-Seilaniantz et al., 2006), Pseuodmonas fluorescens (pLN1965+pML123), and P. fluorescens (pLN1965+pLN1420) (Guo et al., 2009). To generate P. syringae hopF2R71A and hopF2D175A mutant strains, the hopF2 mutations were introduced into the pMOD-hrp-shcF-hopF2ATG construct (Shan et al., 2004) by site-directed mutagenesis, and the mutant fragments were excised with EcoRI and BamHI. The hrp-shcF-hopF2ATG fragment in the pLK-hrp-shcF-hopF2ATG-HA construct was replaced with the hrp-shcF-hopF2R71A or hrp-shcF-hopF2D175A fragment. The resulting pLK constructs were then transformed into the P. s. pv tabaci R1152 or P. s. pv tomato DC3000 hopF2− mutant strain.
Oxidative Burst
Four-week-old wild-type and hopF2 transgenic line 5 and line 22 T3 plants were sprayed with 50 μM β-estradiol. Leaves were collected 48 h later, sliced into 1-mm strips, and kept in water in a 96-well plate overnight before being treated with 100 μL solution containing 10 μg/mL peroxidase (Sigma-Aldrich) and 20 nM luminol in the presence of 1 μM flg22. Luminescence was measured and calculated using a GLOMAX 96 microplate luminometer (Promega).
Callose Staining
Six-week-old wild-type or transgenic plants were presprayed with corresponding inducer (50 μM β-estradiol, 30 μM dexamethasone, or both) to induce protein expression for 12 to 24 h. Leaves were then infiltrated with 1 μM flg22 for 6 h, stained with aniline blue, and visualized with a fluorescence microscope as described (Hauck et al., 2003). Callose deposits were calculated using Image J software (http://www.uhnresearch.ca/wcif).
MAPK Assay
Protoplasts were isolated from 6-week-old Col-0 plants and transfected with MPK4-FLAG or MPK6-FLAG along with HopF2-FLAG as described (Li et al., 2005). The transfected protoplasts were treated with water or 1 μM flg22 for 10 min before the FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2-agarose (Sigma-Aldrich) and eluted with 3×FLAG peptide (Sigma-Aldrich). The purified MPK4-FLAG or MPK6-FLAG proteins were subjected to an in-gel kinase assay as described (Zhang and Klessig, 1997).
A HopF2-HA construct was made by PCR amplifying the HopF2 coding sequence from the pUC19-35S-HopF2-FLAG-RBS construct, digesting it with KpnI and XhoI, and inserting it into the pUC19-35S-HA-RBS plasmid between KpnI and SalI. Protoplasts were separately transfected with MKK5DD-FLAG (Zhang et al., 2007) alone or together with the HopF2-HA construct. A separate set of protoplasts was transfected with MPK6-FLAG. The FLAG-tagged proteins were isolated by anti-FLAG IP. Purified MKK5DD and MPK6 were incubated in a kinase reaction buffer containing 50 mM HEPES, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 10 μM ATP at 30°C for 30 min. The reaction was terminated by adding SDS sample buffer, and the products were subjected to an in-gel kinase assay.
GST Pull-Down Assay
HopF2, HopF2R71A, and HopF2D175A were PCR amplified from pUC19-35S-FLAG constructs containing HopF2, HopF2R71A, and HopF2D175A and inserted between EcoRI and XhoI sites of pGEX-6p-1 (Pharmacia) to generate GST-HopF2, GST-HopF2R71A, and GAT-HopF2D175A, respectively. MKK5 was PCR amplified from Arabidopsis cDNA and inserted between NdeI and XhoI sites of pET28b (Novagen) to generate His-MKK5. Primer sequences are presented in Supplemental Table 1 online.
For GST pull-down, MPK6-His (Zhang et al., 2007) was coexpressed with GST or GST-HopF2, and His-MKK5 was coexpressed with GST, GST-HopF2, GST-HopF2R71A, and GST-HopF2D175A, respectively, in Escherichia coli BL21 strain. The GST-tagged proteins were purified with glutathione agarose (Sigma-Aldrich) from the bacterial lysate, washed five times with a buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, and 0.1% (v/v) Triton X-100, and eluted by 10 mM GSH (Sigma-Aldrich). Alternatively, GST- and His-tagged proteins were separately purified and incubated in the buffer described above. Glutathione agarose was then added for GST pull down. Immunoblot analysis was used to detect the presence of MPK6-His or His-MKK5 bound to the GST-tagged proteins with anti-His antibody (Sigma-Aldrich).
Co-IP
MKK5 was PCR amplified from Arabidopsis cDNA and inserted between the KpnI and SalI sites of the pUC19-35S-HA-RBS plasmid to generate MKK5-HA. Protoplasts were transfected with MKK5-HA alone or in combination with HopF2-FLAG, HopF2R71A-FLAG, or HopF2D175A-FLAG. Total protein was immunoprecipitated with anti-FLAG antibody, and the presence of FLAG-tagged proteins and MKK5-HA in the same immunocomplex was detected by immunoblot with a monoclonal anti-FLAG antibody (Sigma-Aldrich) and anti-HA antibody (Tiangen), respectively. Approximately 1% of input and one-third of eluted protein were analyzed by immunoblot.
In Vitro Kinase Assay
MKK5DD was fused to the C-terminal of the FLAG tag of pET28a (Xu et al., 2008), between NdeI and SalI digestion sites, to generate the FLAG-MKK5DD construct. FLAG-MKK5DD was coexpressed with GST, GST-HopF2, GST-HopF2R71A, and GST-HopF2D175A, respectively, in E. coli BL21 strain and purified with anti-FLAG M2-agarose. One microgram of purified MPK6-His was incubated with 100 ng FLAG-MKK5DD in a kinase reaction buffer containing 40 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM DTT, 50 μM ATP, and 0.1 μCi/μL [γ-32P]ATP at 30°C for 30 min. The reaction was terminated by adding SDS sample buffer and separated by SDS-PAGE. MPK6 phosphorylation by MKK5DD was detected by autoradiography.
ADP-Ribosyltransferase Activity Assay
MKK5DD was PCR amplified from the FLAG-MKK5DD construct and inserted between the EcoRI and XhoI sites of pGEX-6p-1 (Pharmacia) to generate GST-MKK5DD. GST-RIN4 was purified as described (Cui et al., 2010), and GST was removed by PreScission Protease (GE Healthcare) cleavage, following the manufacturer's instructions. One microgram of purified FLAG-MKK5DD, GST-MKK5DD, or RIN4 recombinant protein was incubated with 100 ng HopF2, HopF2D175A, or HopF2R71A in a 25-μL ADP-RT reaction buffer containing 40 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM DTT, 60 μM ATP, 3 mM ADP-ribose (Sigma-Aldrich), and 30 mM (or the indicated concentrations) of 6-biotin-17-NAD (Trevigen) at room temperature for 40 min. The reaction was terminated by boiling in SDS sample buffer and subjected to SDS-PAGE. The ADP-ribosylated MKK5DD proteins were analyzed by immunoblot with horseradish peroxidase–conjugated streptavidin (Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HopF2, AAO54046; MKK1, AtG26070; MKK2, At4G29810; MKK3, At5G40440; MKK4, At1G51660; MKK5, At3G21220; MKK6, At5G56580; MKK7, At1G18350; MKK8, At3G06230; MKK9, At1G73500; and MKK10, At1G32320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transgenic Expression of HopF2 Compromises Plant Resistance to a Nonpathogenic P. syringae Mutant Strain.
Supplemental Figure 2. Transgenic Expression of HopF2 Results in Dwarfism.
Supplemental Figure 3. HopF2 Blocks MKK5DD-Induced Cell Death.
Supplemental Figure 4. HopF2 Prevents MKK5 Autophosphorylation in E. coli.
Supplemental Figure 5. HopF2 Interacts with MKKs in Vitro and in Protoplasts.
Supplemental Figure 6. HopF2 ADP-Ribosylates RIN4 in Vitro.
Supplemental Figure 7. Arg-313 of MKK5DD Is Required for ADP-Ribosylation.
Supplemental Table 1. Primers Used.
Acknowledgments
We thank James Alfano for helpful comments and bacterial strains. D.R. was supported by grants from National Natural Science Foundation of China (30870220 and 30721062). J.-M.Z. was supported by a grant from the Chinese Ministry of Science and Technology (2003-AA210080).
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boller T., He S.Y. (2009). Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.-M. (2010). A Pseudomonas syringae protein perturbs Arabidopsis hormone signaling by activating MAP KINASE 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fu Z.Q., Guo M., Jeong B.R., Tian F., Elthon T.E., Cerny R.L., Staiger D., Alfano J.R. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288 [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Gohre V., Spallek T., Haweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Guo M., Tian F., Wamboldt F., Alfano J.R. (2009). The majority of the Type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microbe Interact. 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L.P., et al. (2006). Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Hauck P., Thilmony R., He S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout S.A., Van der Hoorn R.A.L., Terauchi R., Kamoun S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Iizasa E., Mitsutomi M., Nagano Y. (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xu H., Zhou Y., Zhang J., Long C., Li S., Chen S., Zhou J.M., Shao F. (2008). The phosphothreonine lyase activity of a bacterial type III effector family. Science 315: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Li X., Lin H., Zhang W., Zou Y., Zhang J., Tang X., Zhou J.M. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. (2006). Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312: 1211–1214 [DOI] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., MacKinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Shan L., Zhou J.-M., Tang X. (2006). The Pseudomonas syringae pv. tomato DC3000 Type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19: 130–138 [DOI] [PubMed] [Google Scholar]
- Shan L., Oh H.-S., Chen J., Guo M., Zhou J.-M., Alfano J.R., Collmer A., Jia X., Tang X. (2004). The HopPtoF locus of Pseudomonas syringae pv tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant Microbe Interact. 7: 447–455 [DOI] [PubMed] [Google Scholar]
- Singer A.U., Desveaux D., Betts L., Chang J.H., Nimchuk Z., Grant S.R., Dangl J.L., Sondek J. (2004). Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12: 1669–1681 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiamis G., Mansfield J.W., Hockenhull R., Jackson R.W., Sesma A., Athanassopoulos E., Bennett M.A., Stevens C., Vivian A., Taylor J.D., Murillo J. (2000). Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv phaseolicola, the cause of bean halo-blight disease. EMBO J. 19: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X., Neece D., Ramonrll K.M., Clough S., Kim S., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Subramanian R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J.M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Zhang J., Lu H., Li X., Li Y., Cui H., Wen C.K., Tang X., Su Z., Zhou J.M. (2010). Effector-triggered and pathogen-associated molecular pattern-triggered immunity differentially contribute to basal resistance to Pseudomonas syringae. Mol. Plant Microbe Interact. 23: 940–948 [DOI] [PubMed] [Google Scholar]
- Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., Zou Y., Long C., Lan L., Chai J., Zhou J.M. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.M., Chai J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11: 179–185 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.-W., Chau N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
- Zwiesler-Vollick J., Plovanich-Jones A.E., Nomura K., Bandyopadhyay S., Joardar V., Kunkel B.N., He S.Y. (2002). Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv tomato DC3000 genome. Mol. Microbiol. 45: 1207–1218 [DOI] [PubMed] [Google Scholar]