Transposable elements (TEs) have surprised and intrigued biologists since their discovery, and our understanding of the roles of transposons in genome dynamics has continued to evolve in the elapsed years. Continued research on transposons and insights from genome sequencing have banished the “junk” designation and produced a more nuanced view of the interaction between transposons of different types and their host genomes. Specifically, it was found that the retrotransposons (REs), which move through an RNA intermediate, and the DNA TEs can produce strikingly different effects on genome dynamics. Although transposons can create adaptive variation, they also adversely affect the host genome by causing mutations and genome expansion. To mitigate these effects, plants typically use epigenetic control to limit element replication by silencing the elements. In this issue, two papers examine different aspects of transposon biology and its role in genome evolution: one by examining the locations of thousands of new insertions of the maize (Zea mays) Dissociation (Ds) element, and the other by profiling the distribution of transposons and genes in megabase tracts of sequence from the tremendously expanded and repeat-rich wheat (Triticum aestivum) genome.
The DNA TEs tend to associate with gene space, and these elements can produce changes in gene regulation and coding by multiple mechanisms (reviewed in Dooner and Weil, 2007; Feschotte and Pritham, 2007). TE insertion causes mutations, ranging from loss of function to alterations in gene regulation. The excision of a cut-and-paste TE also causes double-stranded DNA breaks, which may be imprecise or exact and can be repaired with mismatched templates. Some classes of DNA TE capture and move genomic fragments, thereby producing rearrangements and gene duplications. Transposons may even contribute protein domains, as in the cases of transposases coopted as DNA binding proteins. Moreover, complex transpositions involving two or more intact or fragmented elements can cause large-scale genomic rearrangements. All of these transposon-induced changes can create adaptive variation by shuffling both coding sequences and regulatory elements, thereby producing new genes with different regulatory controls.
One way to examine the effects of DNA transposons on genome dynamics is to determine where the transposons land when they move. The Ds element is a DNA transposon that replicates by a cut-and-paste mechanism and requires exogenous transposase from another source, such as an autonomous Activator (Ac) element. One of the first transposons identified, Ds is a useful tool for mutagenesis, as insertions can be stabilized by removal of the transposase source. Vollbrecht et al. (pages 1667–1685) generated thousands of Ds insertion lines and characterized the insertion site of the TE in each line, thereby producing both a valuable resource for mutant studies and a comprehensive picture of the insertion site preferences of the Ds element. Because the Ds element favors local hopping, the authors also designed a genetic scheme to isolate Ds insertions at sites unlinked to the donor site. Starting with more than 18,000 lines in which the Ds had excised from the donor site, the authors identified 1785 lines with a Ds insertion, most at an unlinked site.
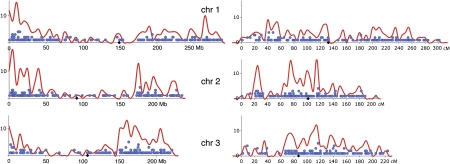
Analysis of sequences flanking the insertion sites of unlinked events showed that the Ds element preferred to transpose into gene-rich regions, as only ∼10% of the adjacent sequences were repetitive, although repetitive sequences make up ∼80% of the maize genome. By contrast, sequences adjacent to insertions of TEs of the Mutator (Mu) family (derived from publicly available sequences) showed a much higher repeat content. Also, Ds inserted nonrandomly across the chromosomes (Figure 1), preferring the regions toward the telomeres, likely following the distribution of maize genes. The Ds insertions also showed preferences for some “hot spots” across the chromosomes. Comparison of Ds insertions with maize gene models confirmed that the uneven distribution of Ds insertions follows the distribution of genes and revealed a preference for Ds insertions in exons and introns and against insertions into 3′-untranslated regions. By contrast, Mu showed a strong preference for insertions into promoters and 5′-untranslated regions. Finally, analysis of the genomic sequences flanking the insertion sites confirmed prior findings that Ac/Ds have no consensus target sequence but found preferred structural parameters of the target site, including fluctuations in protein-DNA twist, which measures DNA deformability.
Figure 1.
Distribution of Ds Insertions on Maize Chromosomes 1 to 3.
Three maize chromosomes are shown, with physical distance in megabases (Mb) on the horizontal axis (left) and genetic distance in centimorgans (cM) at right. The number of Ds insertions per 5-Mb or 5-cM bin is shown as a smoothed curve, in red; the number and positions of BACs hit by a Ds insertion are indicated by the vertical and horizontal positions of the blue dots. The centromere is marked by a black diamond. [Reprinted from Vollbrecht et al. (2010).]
While DNA TEs mostly inhabit and affect the “gene space” of the genome, REs in plants are mainly found in heterochromatic regions, centromeres, and the large tracts of repetitive DNA between so-called gene islands (Du et al., 2010). Although this dichotomy is not exclusive, REs can contribute to genome dynamics in a different way; specifically, RE proliferation and elimination can lead to vast differences in genome sizes, magnifying distances between genes or groups of genes by waves of nested RE insertions. Some of the most affected, and thereby most interesting, genomes are so large and repeat-rich as to pose a substantial barrier to genome sequencing. For example, bread wheat has an enormously expanded repeat-rich genome (17 Gb) that has not been fully sequenced, leaving wheat genomics lagging behind the other important cereal crops.
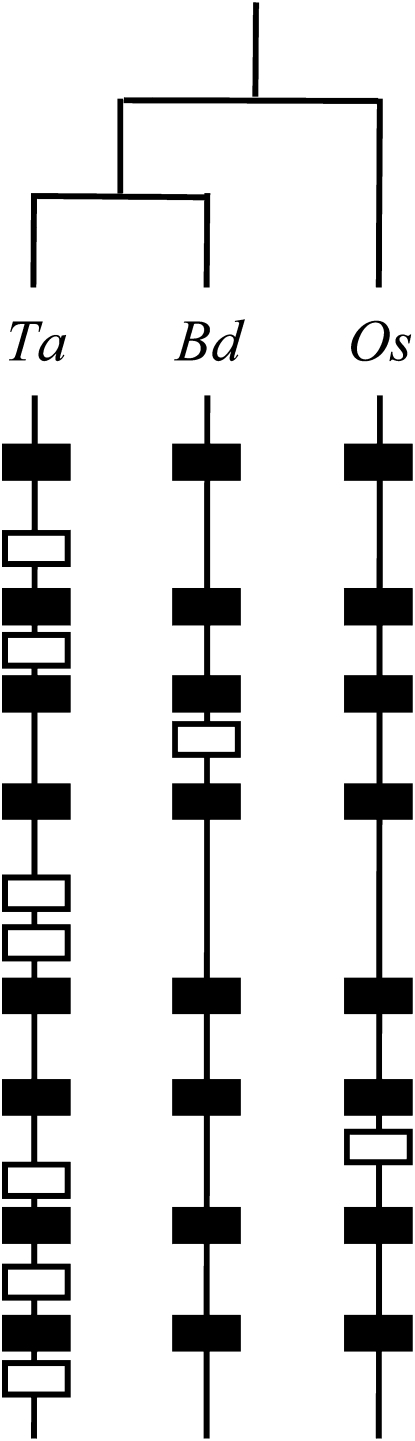
Although this lag frustrates genomics efforts in wheat, it also suggests interesting questions on genome dynamics. How did this expansion occur, and what roles did transposons play? Are expanded areas evenly distributed, or are genes clustered into “islands”? Does wheat have more genes than other grasses? To examine these questions, Choulet et al. (pages 1686–1701) produced megabase tracts of DNA sequence from chromosome 3B of wheat. They selected 13 BAC contigs from chromosome 3B, sampling areas along both arms and in the centromere, and supplementing this with Illumina/Solexa sequencing of flow-sorted and amplified 3B DNA. Within this sequence, they identified 175 putative genes, at one gene per 104 kb, with genes closer in the distal contigs (one gene per 86 kb) and farther apart in the centromere-proximal contigs (one gene per 184 kb). Genes also clustered into small islands of 2 to 10, with few isolated genes. Examination of the transposon content surrounding the gene islands showed that the wheat genome has had multiple waves of transposon activity followed by silencing. Moreover, by comparison with orthologous regions from rice, the authors found that expansion of the wheat genome was nonuniform, where some regions expanded dramatically, thereby contributing to the formation of gene islands. Interestingly, comparison of wheat with rice and Brachypodium revealed that wheat has a large number of noncollinear genes interspersed among the ancestral conserved genes (Figure 2), indicating that wheat has had a large number of gene duplication events, likely mediated by transposon activity. Thus, this good look at a part of the challenging wheat genome has yielded an abundance of information on the dynamics that lead to genome expansion.
Figure 2.
Syntenic and Nonsyntenic Loci in Wheat, Rice, and Brachypodium.
Orthologous chromosomes from wheat (Ta), Brachypodium distachyon (Bd), and rice (Os), with syntenic genes displayed as black boxes and nonsyntenic genes in white. [Reprinted from Choulet et al. (2010).]
The habits of transposons produce seemingly paradoxical effects: for example, genome expansion is coupled with gene rearrangements and duplications that may provide adaptive variation. Given recent advances in synthetic biology, one is left to wonder about a plant genome without transposons. Aside from the consideration of what sort of centromere one might design without transposons, would a transposon-free plant be a parasite-free wonder with a streamlined, efficient genome? Or would it be an evolutionary dead end, frozen without the ability to generate adaptive variation such as gene duplications and rearrangements? We may never know, as after millions of years of coevolution there is no separating the parasite from the host; there is only the evolution of our understanding.
References
- Choulet F., Wicker T., Rustenholz C., Paux E., Salse J., Leroy P., Schlub S., Le Paslier M.-C., Magdelenat G., Gonthier C., Couloux A., Budak H., et al. (2010). Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Weil C.F. (2007). Give-and-take: Interactions between DNA transposons and their host plant genomes. Curr. Opin. Genet. Dev. 17: 486–492 [DOI] [PubMed] [Google Scholar]
- Du J., Tian Z., Hans C.S., Laten H.M., Jackson S.A., Cannon S.B., Shoemaker R.C., Ma J. (2010). Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: Insights from genome-wide analysis and multi-specific comparison. Plant J. 10.1111/j.1365–313X.2010.04263.x [DOI] [PubMed] [Google Scholar]
- Feschotte C., Pritham E.J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41: 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Duvick J., Schares J.P., Ahern K.R., Deewatthanawong P., Xu L., Conrad L.J., Kikuchi K., Kubinec T.A., Hall B.D., Weeks R., Unger-Wallace E., et al. (2010). Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22: 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]