In this study, the characterization of OrrDs, a dominant transposon-tagged tomato mutant deficient in the NDH-M subunit, provided evidence that the plastid NAD(P)H dehydrogenase (NDH) complex is essential for the fruit ripening process and related metabolism.
Abstract
In higher plants, the plastidial NADH dehydrogenase (Ndh) complex supports nonphotochemical electron fluxes from stromal electron donors to plastoquinones. Ndh functions in chloroplasts are not clearly established; however, its activity was linked to the prevention of the overreduction of stroma, especially under stress conditions. Here, we show by the characterization of OrrDs, a dominant transposon-tagged tomato (Solanum lycopersicum) mutant deficient in the NDH-M subunit, that this complex is also essential for the fruit ripening process. Alteration to the NDH complex in fruit changed the climacteric, ripening-associated metabolites and transcripts as well as fruit shelf life. Metabolic processes in chromoplasts of ripening tomato fruit were affected in OrrDs, as mutant fruit were yellow-orange and accumulated substantially less total carotenoids, mainly β-carotene and lutein. The changes in carotenoids were largely influenced by environmental conditions and accompanied by modifications in levels of other fruit antioxidants, namely, flavonoids and tocopherols. In contrast with the pigmentation phenotype in mature mutant fruit, OrrDs leaves and green fruits did not display a visible phenotype but exhibited reduced Ndh complex quantity and activity. This study therefore paves the way for further studies on the role of electron transport and redox reactions in the regulation of fruit ripening and its associated metabolism.
INTRODUCTION
The shift to ripening in fleshy fruit involves multiple metabolic changes, including the accumulation and degradation of pigments such as carotenoids and chlorophylls in tomato (Solanum lycopersicum) (Adams-Phillips et al., 2004). The pathway of carotenoid biosynthesis has been intensively studied, particularly in tomato fruit (Fraser et al., 2009). While most structural genes of the pathway have been characterized (Hirschberg, 2001), very little is known regarding the regulatory network that controls carotenogenesis. The control of carotenoid biosynthesis via the redox status of components of the photosynthetic electron transport has been established during etioplast-to-chloroplast transition (Woitsch and Romer, 2003). In earlier studies, the involvement of quinones in carotenoid desaturation was demonstrated biochemically (Mayer et al., 1990) and genetically (Norris et al., 1995). It was further discovered that phytoene desaturase activity responded to the redox state of the plastoquinone (PQ) pool (Nievelstein et al., 1995). In chloroplasts, PQ oxidoreduction status can be modulated by linear electron transfer and also by one of the two alternative electron pathways operating around photosystem I (PSI; Bukhov and Carpentier, 2004; Johnson, 2005). Moreover, it was proposed that chlororespiration, a respiratory electron transfer chain in the thylakoids, whose functioning has been clearly established in Chlamydomonas reinhardtii (Nixon, 2000; Peltier and Cournac, 2002) could also modulate PQ pool redox status in chloroplasts of higher plants. Although it has been demonstrated that the oxidative reaction of chlororespiration involving a plastid-located terminal oxidase (PTOX; Wu et al., 1999; Carol and Kuntz, 2001) deeply affected carotenogenesis in leaves (Wu et al., 1999; Josse et al., 2000), the participation of the other reactions in modulating the redox state of PQ has not been investigated.
For the last 10 years, many efforts have been pursued to identify the molecular entities underlying the cyclic electron transfer reactions around PSI (for reviews, see Shikanaï, 2007a, 2007b; Suorsa et al., 2009). Several Arabidopsis mutants affected in these pathways have been studied (Munekage et al., 2002; Rumeau et al., 2005; Ishihara et al., 2007; Shikanaï, 2007a; DalCorso et al., 2008; Peng et al., 2009; Sirpiö et al., 2009; Takabayashi et al., 2009) and showed no change in their carotenoid content. However, participation of PQ redox state in carotenogenesis in other tissues than leaves cannot be excluded. Indeed, plant genetic modification has shown that in some cases carotenogenesis was differentially affected in fruit and in leaves, suggesting that the metabolic pathways and/or their regulation are different in both organs (Bramley et al., 1992).
The NADH dehydrogenase (Ndh) complex, likely originating from the respiratory electron transfer chain of a cyanobacterial ancestor, catalyzes PQ reduction using soluble electron donors (Burrows et al., 1998; Endo et al., 2008). It participates in one of the two electron pathways operating around PSI. The other pathway likely involves a still uncharacterized ferredoxin-plastoquinone reductase and the two interacting proteins PGR5 and PGRL1 (Munekage et al., 2002; DalCorso et al., 2008). Because Ndh and PTOX colocalize in the stroma lamellae (Lennon et al., 2003), they have been implicated in the chlororespiratory pathway, although direct evidence of electron transfer between Ndh and PTOX is still lacking.
Accumulation of the Ndh complex within etiolated leaf tissue (Lennon et al., 2003) led to the hypothesis that the corresponding electron pathway may participate in the etioplast-to-chloroplast transition process by energizing the plastid membrane and favoring synthesis and/or insertion of the photosynthetic complexes. However, the absence of any obvious phenotype related to greening in Ndh-deficient plants (Ishikawa et al., 2008) appears to contradict this hypothesis. By contrast, PTOX is essential during greening and carotenoid biosynthesis (Wu et al., 1999; Carol and Kuntz, 2001). It was suggested to be a component of a redox chain responsible for the desaturation of phytoene and perhaps ζ-carotene (Wu et al., 1999; Carol and Kuntz, 2001). In this chain, electrons would be transferred from phytoene (and ζ-carotene) to the PQ pool via phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS) and from the PQ pool to molecular oxygen via PTOX. The Ndh complex subunits were also detected in nonphotosynthetic tissues, such as maize (Zea mays) etioplasts and tomato fruit (Guera and Sabater, 2002). In tomato fruit, the same authors found that the Ndh complex is mainly present in the outer pericarp tissue and that its amount and activity in the plastids decreased during the transition from chloroplasts to chromoplasts. These findings imply that the Ndh complex may be important for several processes in plant development, including fruit maturation.
As part of our efforts to understand the regulation of metabolic pathways in tomato fruit, we identified a novel transposon-tagged mutant. It was named Orange Ripening (OrrDs) due to its typical pigmentation of the fruit. The transposon insertion created a dominant mutation that altered carotenoid biosynthesis as well as other metabolic pathways in the fruit, such as those producing flavonoids and tocopherols. The ORR protein showed sequence similarity to the Arabidopsis thaliana NDH-M subunit of the Ndh complex. Detailed characterization of the OrrDs mutant and the corresponding gene implies the association of the Ndh complex in the process of fruit ripening and related metabolism.
RESULTS
Isolation of the OrrDs Tomato Fruit Mutant
To identify mutants altered in metabolic pathways associated with tomato fruit ripening, we screened an Ac/Ds transposon-based mutant population for changes in fruit appearance (Meissner et al., 1997). The fruit of one mutant line (termed OrrDs) exhibited orange and yellow sectors, suggesting an insertion in a gene involved in fruit carotenogenesis (Figure 1A). A transposase-free plant recovered from the mutant offspring lines displayed a yellow-orange color in the entire fruit (Figure 1B), and its selfing segregated as expected from a codominant trait, χ2 = 0.9 for 1 (red fruit; wild type; 9/40): 2 (yellow fruit; heterozygote; 22/40): 1 (orange fruit, homozygote; 9/40). In addition, crossing orange fruit plants with the wild type resulted in a population that contained only plants with yellow fruit, whereas crossing plants producing yellow fruit with wild-type plants gave two groups of F1 progeny (total 43 plants): one displaying red fruit (26 plants) and the other with yellow fruit (17 plants). Thus, we concluded that the mutation OrrDs exhibits an overdominant mode of inheritance.
Figure 1.
Phenotypes of the OrrDs Mutation and Complementation of the Phenotype by the ex-35S-Orr Allele and ORR Gene Overexpression.
(A) Somatic activity (sectors) in fruit of a plant (OrrDs) harboring both the Ds and Ac elements.
(B) Fruit phenotypes in progeny of an OrrDs/ORR plant. Orange-colored fruit (homozygous, left), yellow-colored fruit (heterozygous, middle), and red (wild type, right).
(C) The phenotype of peel isolated from mature fruit of the plants shown in (B).
(D) Whole ripe fruit and isolated peel derived from the ex-35S-Orr plant compared with the homozygous mutant fruit (OrrDs/OrrDs).
(E) Whole ripe fruit and isolated peel derived from a plant in which the full-length ORR cDNA (starting from AUG-I) was overexpressed in the OrrDs/OrrDs plant.
(F) Whole ripe fruit and isolated peel derived from an F1 line obtained by a cross between a plant in which the ORR cDNA (starting from AUG-II) was overexpressed in the OrrDs/OrrDs plant.
(G) Whole ripe fruit and isolated peel derived from a plant in which the ORR cDNA (starting from AUG-II) was overexpressed in the OrrDs/OrrDs plant. In (E) to (G), the plus and minus symbols represent the presence or absence of the transgene conferring ORR gene overexpression, respectively.
The OrrDs mutants (particularly the heterozygotes) contained reduced levels of the yellow flavonoid pigment that normally accumulates in the peel (Figure 1C). The color of the OrrDs ripe fruit was strongly dependant on the environmental conditions (radiation, shading, and temperature; see Supplemental Figure 1 online). Moreover, compared with the wild type, the levels of total soluble solids (TSSs) in the fruit were reduced by 23 and 15% in OrrDs/Orr and OrrDs/OrrDs, respectively (see Supplemental Figure 2 online). Detached ripe OrrDs/Orr fruit exhibited a strong delay in shrinking when left to dry at room temperature (see Supplemental Figure 2 online) and were often smaller in size.
ORR Corresponds to the Subunit M of the Tomato Ndh Complex
Inverse PCR analysis identified a Ds insertion in the 5′ end of a putative tomato homolog (SGN-U579052) of the Arabidopsis gene for subunit M of the Ndh complex (Figure 2). The ORR gene was mapped in the center of tomato chromosome 4 and was found to contain a single intron. Based on the sequence of the entire tomato genome, ORR is a unique locus (see Supplemental Figure 3 online). It putatively encodes a 211–amino acid protein that is 62% identical (75% similar) to the characterized Arabidopsis NDH-M protein (At4g37925; Rumeau et al., 2005) and shows the highest homology (72% identity and 85% similarity) to an olive (Olea europaea) protein (see Supplemental Figure 5 online).
Figure 2.
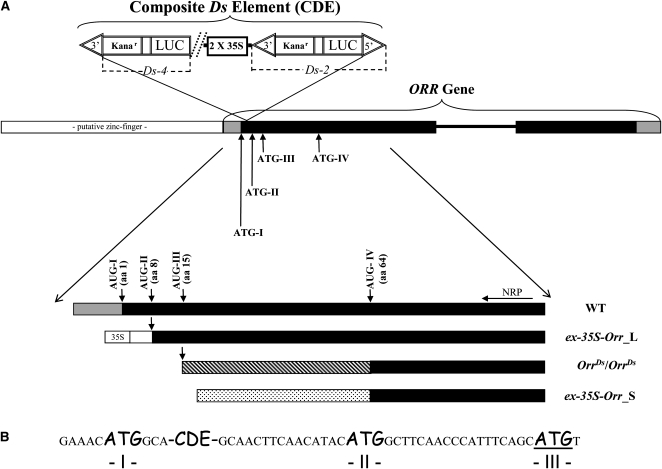
Structure of the OrrDs Allele and the 5′ Regions Obtained by RACE Analysis in the Different OrrDs Genotypes and the Wild Type.
(A) The diagram displays the structure of the OrrDs allele. The ORR gene has two exons (black closed boxes) flanking one intron (black line between the exons). The position of composite Ds element (CDE) in the ORR 5′ end is marked. The CDE is composed of the CaMV 35S promoter flanked (downstream) by a complete Ds element and another element (possibly partial) upstream. ATG-I to ATG-IV are marked. At the bottom part, results of 5′ RACE analysis of the various OrrDs genotypes and the wild type (WT) are depicted. Long and short 5′ RACE products were obtained in the ex-35S-Orr genotype (ex-35S-Orr_L and ex-35S-Orr_S, respectively). The location of the corresponding Met along the amino acid (aa) sequence is indicated in parentheses next to each AUG codon. NRP, nested RACE primer. Black areas mark the putative protein formed in each transcript.
(B) The exact position of the CDE in the ORR gene sequence between ATG-I and ATG-II. The CDE and the three ATGs (I, II, and III) are marked by a bigger and different font.
As shown in Figure 2, we discovered a complex insertion with one intact element (Ds-2) flanked by the double 35S cauliflower mosaic virus (CaMV) promoter (2X 35S) and another truncated element (Ds-4). Such composite element structures have been previously described (Huang and Dooner, 2008); the most likely explanation for its formation is described in Supplemental Figure 3 online.
A single line that was recovered following a transposon excision event in the OrrDs mutant exhibited red pigmented fruit (a revertant phenotype; ex-35S-Orr; Figure 1D), in which the Ds-2 element had excised and the ORR was consequently driven by the 35S CaMV promoter (see Supplemental Figure 4 and Supplemental Table 1 online). This excision event generated a typical excision footprint, leaving most of the host duplication intact with a small deletion (see Supplemental Figure 6 online).
We further demonstrated that the insertion cosegregated with the OrrDs mutation and was only present in plants that exhibited the mutant phenotype of orange or yellow fruit (see Supplemental Figure 6 online). This validation was performed by examining plants segregating from the selfing of OrrDs/Orr fruit (total 40 plants; see above). Selected plants (20 in total) were chosen randomly for evaluation of cosegregation between the insertion and fruit pigmentation using DNA gel blots and PCR analyses. These experiments as well as others performed in the course of this study involving genotyping and cosegregation assays of hundreds of plants for the different OrrDs genotypes showed complete linkage between the genotype and the phenotype.
Examination of the ORR full-length transcript revealed that it contained three in-frame AUG codons at its 5′ end (AUG- I, II, and III, amino acids 1, 8, and 15 of the putatively encoded protein, respectively, counting from the 5′ end). The transposon insertion in the OrrDs mutants was located in between AUG-I and AUG-II (Figure 2). To examine which ORR transcripts are formed in the case of each genotype (wild type; OrrDs/OrrDs and ex-35S-Orr), rapid amplification of cDNA ends–PCR (RACE-PCR) experiments were performed in which the ORR gene 5′ upstream region was isolated (Figure 2; see Supplemental Figure 5 online). The wild-type ORR transcript untranslated region contained AUG- I, II, and III as described above. The homozygous genotype produced a shorter transcript that initiated (i.e., the mRNA) exactly at AUG-III, suggesting that either the transcript will be degraded or a protein starting from the next, downstream AUG, will be generated (termed AUG-IV; amino acid position 64; Figure 2). In the case of the ex-35S-Orr genotype, we identified a long transcript (ex-35S-Orr_L) that contained a small part of the 35S promoter region and a region of the wild-type untranslated region directly upstream of AUG-II (AUG-I was not present; Figure 2; see Supplemental Figure 4 online). A second transcript detected in the ex-35S-Orr genotype was shorter (ex-35S-Orr_S) and started downstream of AUG-III, suggesting that if a protein is produced in this case, its translation would start from AUG-IV (similar to the homozygous transcript; see above).
The capability to genetically complement the OrrDs mutation was tested by generating transgenic plants, which overexpress two different ORR transcripts, starting either from AUG-I or AUG-II, that were driven by the 35S CaMV promoter (35S:AUGI-ORR and 35S:AUGII-ORR, respectively). Transformation of 35S:AUGI-ORR in the OrrDs/OrrDs background resulted in plants with red fruit that accumulated the typical orange color of the peel (Figure 1E). Complementation of the phenotype was also observed in F1 fruit obtained through a cross between 35S:AUGII-ORR transgenic lines (#1919) and a OrrDs/OrrDs plant (Figure 1F) and a T1 plant obtained after transforming OrrDs/OrrDs with the 35S:AUGII-ORR construct (Figure 1G).
To further exclude the possibility that a gene flanking the insertion site was affected in the mutant, we examined the expression of a putative zinc-finger protein (the closest gene to ORR on its 5′ end; see Figure 2) in the mutant and the wild-type fruit using quantitative RT-PCR (qRT-PCR) and detected no difference. In addition, we annotated a large region flanking ORR on chromosome 4 and found a unigene (SGN-U576377) located 12.2 kb downstream from ORR in the same orientation. It encodes a protein that is 56% identical and 73% similar to the peroxisome biogenesis protein PEX1 from Arabidopsis. Another unigene located 9.6 kb upstream to ORR in the opposite orientation is SGN-U580948, which encodes a putative RPS15A (ribosomal protein S15A). While we cannot completely exclude effects of the Ds insertion on expression of the latter two genes, this possibility is unlikely because of the significant distance between them and the Ds element. Moreover, the nature of the products of these genes does not conform to the unique phenotypes seen in heterozygous and homozygous plants.
This set of results provided strong evidence that the insertion in the putative tomato NDH-M-like ORR gene is responsible for the OrrDs phenotype.
ORR Gene Expression and Subcellular Localization of Its Corresponding Protein Variants
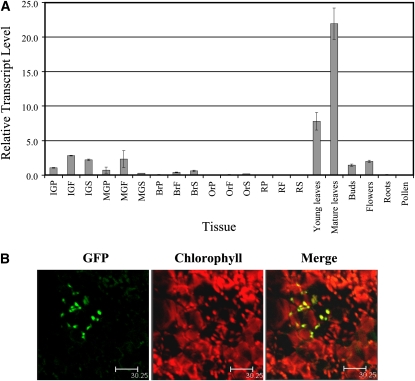
qRT-PCR analysis showed that ORR is preferentially expressed in green, chlorophyll-containing tissues, with highest levels detected in mature leaves (Figure 3A). ORR expression was detected in young leaves, immature and mature green fruit (either peel, flesh, or seeds), stems (data not shown), and in flower buds and open flowers (including sepals), while no or traces levels of transcripts were evident in mature fruit tissues (i.e., peel, flesh, and seeds), roots, and pollen. Higher ORR expression in the early green stages of fruit development and subsequent decline in the maturation phase (mature green and breaker stages) was further supported by microarray data (Mintz-Oron et al., 2008; see Supplemental Figure 7 online).
Figure 3.
ORR Gene Expression and Protein Subcellular Localization.
(A) Expression of ORR in various plant tissues (cv MicroTom) detected by qRT-PCR. Peel (P), flesh (F), and seeds (S) were dissected from IG (immature green), MG (mature green), Br (breaker), Or (orange), and R (red) fruit. Expression data were normalized to the expression of the ASR1 gene, and values are means ± se (n = 3).
(B) The subcellular localization of ORR (starting from AUG-I) was investigated using confocal microscopy after transient expression of ORR-GFP fusions in tobacco leaves. The natural chlorophyll red fluorescence signal was used to visualize chloroplasts and was merged with the GFP signal. The pattern of ORR-GFP starting from AUG-I coincided with that of chlorophyll confirming its chloroplast localization. Bars = 30.25 μm.
In silico prediction tools (i.e., iPSORT and TargetP) suggested that the putative ORR protein (starting from AUG-I) is targeted to the plastids. To experimentally determine the subcellular localization of the ORR protein, a construct was generated in which the entire ORR open reading frame, starting from the AUG-I codon, was fused to the 5′ end of a green fluorescent protein (GFP) reporter. Subcellular localization was investigated using confocal microscopy after transient expression of the ORR (AUG-I)-GFP fusion in tobacco (Nicotiana tabacum) leaves (Figure 3B). The natural chlorophyll red fluorescence signal was used to visualize chloroplasts and was overlaid with the GFP signal. The pattern of ORR (AUG-I)-GFP coincided with that of chlorophyll, confirming its chloroplast localization.
Since the Ds transposon insertion was located in between the ATG-I and ATG-II codons, we examined if the putative proteins starting at either AUG-II or AUG-III are still directed to the plastids as detected for the entire ORR protein (starting from AUG-I). In silico analysis predicted low likelihood of targeting to plastids for a protein starting at either AUG-II or from AUG-III. The subcellular localization of the putative ORR proteins starting from AUG-II and AUG-III was further investigated by transient assays (see Supplemental Figure 8 online). The results showed that ORR proteins initiating at either AUG-II or AUG-III are directed to the nucleus and cytoplasm.
The OrrDs Mutation Results in Reduced ORR Gene Expression and Ndh Complex Quantity and Activity
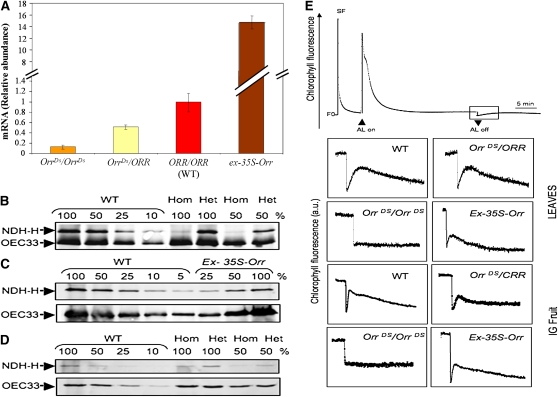
We further examined ORR gene expression in green fruit derived from plants of the OrrDs mutants compared with the wild type (Figure 4A). The results showed that ORR expression is reduced to 13 and 50% in the OrrDs/OrrDs and OrrDs/ORR genotypes, respectively (compared with the wild type). By contrast, ORR expression was strongly induced (15-fold compared with expression in the wild type) in green fruit derived from the ex-35S-Orr genotype (Figure 4A).
Figure 4.
ORR Gene Expression and Activity and Levels of the Ndh Complex in the OrrDs Mutants.
(A) Expression of ORR in immature green fruit (30 d after anthesis) of the wild type (WT) and OrrDs mutants. qRT-PCR expression data were normalized to the expression of the ASR1 gene and presented as percentage of the wild type, which was set at 100% (values are means ± se; n = 3). Asterisk indicates values that are significantly different from the wild type (P < 0.01). The amplified part is located in the coding region downstream of the insertion.
(B) to (D) Protein gel blot analysis. Thylakoids were extracted from wild-type, homozygous (Hom; OrrDs/OrrDs), heterozygous (Het; OrrDs/ORR), and the ex-35S-Orr mutants leaves and immature green fruit. Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membrane, and analyzed using anti-NDH-H antibodies. The lanes were loaded with samples corresponding to 15 μg of chlorophyll (100%) and a series of dilutions as indicated. Immunodetection of OEC33 was used as a loading control.
(E) In vivo detection of Ndh complex activity by chlorophyll fluorescence measurements. Leaf and immature green fruits chlorophyll fluorescence of wild-type and mutant tomato plants was monitored with a PAM fluorometer. The top curve is a typical trace of chlorophyll fluorescence in the wild type. After a 5-min actinic illumination (AL; 250 μmol photons m−2 s−1), Fs levels reached similar levels in wild-type and mutant leaves. After switching the light off (AL off), transient increases in chlorophyll fluorescence were recorded under low nonactinic light. Insets are magnified traces from the boxed area. SF, saturating flash of white light.
[See online article for color version of this figure.]
Since it has been well established that the presence of all NDH subunits is required for the assembly and functioning of the complex (Rumeau et al., 2005; Ishihara et al., 2007; Shikanaï, 2007a, 2007b; Sirpiö et al., 2009; Takabayashi et al., 2009), the presence of the Ndh complex in the different genotypes was investigated using an anti-NDH-H (a different subunit of the Ndh complex) antiserum (Figure 4). Compared with the levels of the wild-type protein, the homozygous line retained <10 and 50% of the protein (NDH-H) in leaves and the immature green fruit, respectively (Figures 4B and 4D). The fact that levels of the NDH-H subunit were reduced signifies that the integrity of the entire complex was disrupted in the homozygous line. The levels of the NDH-H protein was slightly reduced in the heterozygous line compared with the wild type in the leaves but was not significantly different in the immature green fruit (Figures 4B and 4D).
Subsequently, the Ndh complex activity was probed in vivo using chlorophyll fluorescence measurements (Figure 4E). The transient increase in the basal level of chlorophyll fluorescence after a light-to-dark transition has been specifically attributed to PQ and QA reduction via the Ndh complex activity. Accordingly, the postillumination fluorescence rise could not be detected in the homozygous mutant line leaves and green fruit, demonstrating the absence of a functional Ndh complex (Figure 4E). The heterozygous mutant line showed very slight reduction in the levels of the NDH-H in leaves and no difference in the green fruit, and it was not altered in chlorophyll fluorescence in either tissue (Figure 4E). Thus, the chlorophyll fluorescence measurement corroborated the results obtained by the examination of the NDH-H subunit (see above).
In the case of the ex-35S-Orr genotype, the analyses of both NDH-H subunit levels (in leaves) and chlorophyll fluorescence (in leaves and immature green fruit) showed that these two parameters were similar to the ones observed in wild-type plants (Figures 4C and 4E). Thus, the excision event resulted in the induction of ORR transcript levels and restoration of the wild-type phenotype.
Analysis of the Redox Status in the Different OrrDs Genotypes
Levels of the reduced and oxidized forms of pyridine nucleotides (NAD+, NADH, NADP+, and NADPH) were measured in leaves of wild-type, OrrDs/ORR, and OrrDs/OrrDs mutant genotypes (Table 1). A significant decrease in the reduced forms was detected in both the homozygous and heterozygous OrrDs mutants.
Table 1.
Analysis of Pyridine Nucleotides in Leaves of the OrrDs Mutant Genotypes
| Genotype | NAD+ (nmol g−1 FW) | NADH (nmol g−1 FW) | NADP+ (nmol g−1 FW) | NADPH (nmol g−1 FW) |
| Wild type | 16.50 ± 1.80 | 1.40 ± 0.11 | 11.90 ± 3.70 | 2.06 ± 0.25 |
| OrrDs/OrrDs | 16.95 ± 2.30 | 0.92 ± 0.05 | 10.34 ± 1.70 | 1.89 ± 0.10 |
| OrrDs/ORR | 18.08 ± 0.73 | 1.08 ± 0.17 | 11.80 ± 2.50 | 1.70 ± 0.30 |
The values are the mean ± sd of three replicates. Values of the mutant lines that differ significantly (P < 0.05) from the wild type are depicted in bold. FW, fresh weight.
Having established that the mutant genotypes exhibited altered cellular levels of the pyridine nucleotides, we next determined the activation states of the reactions catalyzed by the plastidial malate dehydrogenase and ADP glucose pyrophosphorylase (AGPase). Both reactions have been suggested previously as markers for shifts in the plastidial redox status (Scheible and Stitt, 1988; Tiessen et al., 2002). Thus, we extracted leaves and assayed both total and initial enzyme activities and subsequently deduced their activation states (Table 2). Compared with the wild type, neither the heterozygote nor the complemented ex-35S-Orr line displayed any changes in activities or activation state. By contrast, the homozygote OrrDs/OrrDs line was characterized by an elevated total enzyme activity and a decreased activation state of AGPase. This was coupled to increased total and initial activities and decreased activation state of the plastidial malate dehydrogenase. Taken together, these changes are strongly indicative of an altered redox status of the plastids, suggesting that the plastids of the OrrDs/OrrDs mutant constitute a more reductive environment.
Table 2.
Enzyme Activities in Wild-Type and OrrDs Mutant Genotypes in Leaves
| Activities (nmol min−1 g−1 FW) | ORR/ORR (Wild Type) | OrrDs/ORR | OrrDs/OrrDs | ex-35S-Orr |
| AGPase initial | 85.96 ± 2.800 | 85.96 ± 5.45 | 71.15 ± 6.36 | 73.32 ± 7.29 |
| AGPase total | 139.19 ± 11.19 | 147.28 ± 22.11 | 375.09 ± 44.22 | 248.24 ± 23.32 |
| AGPase % of activation | 69.40 ± 9.46 | 77.30 ± 12.17 | 21.10 ± 5.20 | 30.20 ± 6.86 |
| NADP-MDH initial | 45.83 ± 4.12 | 43.82 ± 2.36 | 72.30 ± 1.52 | 67.89 ± 7.16 |
| NADP-MDH total | 106.73 ± 19.66 | 101.96 ± 8.32 | 205.61 ± 15.74 | 179.24 ± 11.51 |
| NADP-MDH % of activation | 53.40 ± 6.09 | 46.00 ± 4.35 | 35.80 ± 2.17 | 38.00 ± 3.47 |
Data presented are means ± se of measurements from four independent plants per genotype; values set in bold were determined by the t test to be significantly different (P < 0.05) from the wild type. MDH, malate dehydrogenase; FW, fresh weight.
Primary Metabolism Is Only Moderately Altered in Fruits of the OrrDs Mutants
To assess the broad-range consequences of a mutation in the ORR gene on metabolism, we measured the levels of primary metabolites in fruits of the OrrDs genotypes using an established gas chromatography–mass spectrometry (GC-MS) method (Fernie et al., 2004). The results revealed that primary (or central) metabolism was essentially unaltered in the OrrDs genotypes (Table 3; see Supplemental Table 5 online). Although levels of several of the metabolites were significantly different from those observed in the wild type (of 46 metabolites measured, 16 were different in the heterozygote mutant, 9 in the homozygote, and 9 in the ex-35S-Orr line), most of these changes were relatively small (Table 3). An exception was tyramine, whose levels in the heterozygote and homozygote were threefold and twofold, respectively, higher than in the wild type. Tyramine was reduced to 0.6 of the wild-type level in the complemented line. Intriguingly, significant decrease was observed in the levels of Ala, Lys, and Pro (in both OrrDs/ORR and OrrDs/OrrDs); in Asp, Trp, succinate, and glucose (in OrrDs/ORR); and in glutamate and maltose (in OrrDs/OrrDs). Given that these metabolites normally increase during ripening (Roessner-Tunali et al., 2003; Carrari et al., 2006), this result suggests an alteration in the nature of ripening of the OrrDs fruit.
Table 3.
The Effect of the OrrDs Mutation on Levels of Primary Metabolites
| ORR/ORR (Wild Type) | se | OrrDs/ORR | se | OrrDs/OrrDs | se | ex-35S-Orr | se | |
| Amino acids | ||||||||
| Ala | 1.00 | 0.08 | 0.68 | 0.11 | 0.62 | 0.05 | 0.95 | 0.04 |
| Asp | 1.00 | 0.10 | 0.61 | 0.10 | 0.71 | 0.24 | 0.72 | 0.10 |
| Glu | 1.00 | 0.06 | 0.86 | 0.12 | 0.71 | 0.11 | 0.82 | 0.07 |
| Ile | 1.00 | 0.11 | 0.51 | 0.19 | 0.57 | 0.21 | 0.71 | 0.14 |
| Lys | 1.00 | 0.07 | 0.49 | 0.09 | 0.66 | 0.11 | 0.86 | 0.11 |
| Pro | 1.00 | 0.11 | 0.59 | 0.06 | 0.62 | 0.10 | 0.72 | 0.09 |
| Ser | 1.00 | 0.08 | 0.68 | 0.05 | 0.94 | 0.09 | 0.86 | 0.06 |
| Trp | 1.00 | 0.10 | 0.56 | 0.09 | 1.02 | 0.08 | 0.80 | 0.09 |
| Tyr | 1.00 | 0.02 | 3.31 | 0.17 | 2.36 | 0.17 | 0.60 | 0.11 |
| Organic acids | ||||||||
| Citric acid | 1.00 | 0.09 | 0.80 | 0.14 | 0.88 | 0.12 | 0.65 | 0.10 |
| 2-oxo-glutaric acid | 1.00 | 0.06 | 1.03 | 0.08 | 0.97 | 0.04 | 0.61 | 0.13 |
| Dehydroascorbic acid | 1.00 | 0.03 | 1.41 | 0.10 | 1.02 | 0.11 | 0.95 | 0.05 |
| Phosphoric acid | 1.00 | 0.07 | 0.74 | 0.12 | 0.86 | 0.07 | 0.62 | 0.10 |
| Succinic acid | 1.00 | 0.09 | 0.55 | 0.12 | 0.67 | 0.18 | 0.68 | 0.12 |
| Sugars and sugar alcohols | ||||||||
| Fruc | 1.00 | 0.10 | 0.71 | 0.09 | 0.91 | 0.11 | 0.70 | 0.08 |
| Fuc | 1.00 | 0.04 | 0.97 | 0.05 | 0.83 | 0.06 | 0.92 | 0.03 |
| Glc | 1.00 | 0.07 | 0.69 | 0.12 | 0.86 | 0.10 | 0.65 | 0.08 |
| Glc-6-P | 1.00 | 0.06 | 0.66 | 0.07 | 0.94 | 0.04 | 1.03 | 0.05 |
| Maltose | 1.00 | 0.07 | 0.91 | 0.05 | 0.76 | 0.05 | 0.75 | 0.10 |
| Xyl | 1.00 | 0.04 | 1.36 | 0.11 | 1.72 | 0.08 | 0.94 | 0.06 |
| Miscellaneous | ||||||||
| Calystegine A3 | 1.00 | 0.09 | 0.61 | 0.11 | 0.95 | 0.06 | 0.94 | 0.16 |
| Guanidine | 1.00 | 0.12 | 0.48 | 0.08 | 0.72 | 0.06 | 0.87 | 0.16 |
| Inositol, myo | 1.00 | 0.04 | 0.96 | 0.11 | 1.02 | 0.03 | 0.85 | 0.05 |
Metabolite levels were determined in immature green fruit. Data are normalized to the mean response calculated for the wild type. Values presented are mean ± se of four replicates. Values in bold denote significant differences that were determined by t test analysis (P < 0.05). For details on the entire set of metabolites detected in this experiment, see Supplemental Table 5 online.
The OrrDs Mutants Are Altered in the Levels of Isoprenoids and Flavonoids
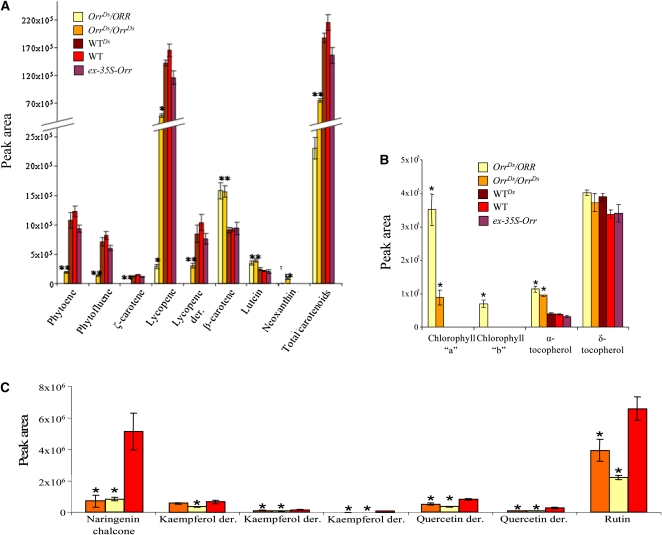
The levels of isoprenoids, including carotenoids, that typically accumulate in tomato fruit were measured in green and ripe fruit of the wild type and OrrDs mutants (Figure 5; see Supplemental Figure 9 online). Compared with wild-type plants, a reduction of 90 and 60% in total carotenoid level was observed in ripe fruit of OrrDs/ORR and OrrDs/OrrDs, respectively. This severe phenotype was mainly due to a decrease in phytoene, phytofluene, ζ-carotene, a lycopene derivative, and lycopene that was reduced dramatically (to 2 and 33% of its wild-type levels in OrrDs/ORR and OrrDs/OrrDs, respectively). By contrast, an increase in both β-carotene (40%) and lutein (30%) was detected in the pericarp of the OrrDs/ORR and OrrDs/OrrDs, respectively (Figure 5; see Supplemental Figure 9 online). In most of the cases, the effect in fruit of the heterozygote plants (OrrDs/ORR) was more severe compared with fruit of the homozygote plants (OrrDs/OrrDs). Carotenoid levels in genotypes producing red fruit, including the wild type, wild-typeDs (siblings of the OrrDs/ORR mutant that contain the wild-type ORR alleles), and ex-35S-Orr plants were not altered. No difference in carotenoid levels was observed between either one of the OrrDs mutants in the green stage of development (data not shown). Apart from the various carotenoids, we also detected altered levels of other isoprenoid pathway compounds, including chlorophylls and tocopherols (Figure 5). Compared with red fruit of the wild type, in which chlorophylls could not be detected, chlorophyll-a accumulated in both the homozygote and heterozygote mutants, while chlorophyll-b accumulated only in the OrrDs/ORR genotype. Levels of α-tocopherol were doubled in the mutant fruit compared with fruit of the wild type.
Figure 5.
The Effect of the OrrDs Mutation on Metabolite Levels in the Isoprenoid and Flavonoid Pathways.
(A) HPLC analysis of various flavonoids in extracts (ripe fruit, 14 d after breaker stage) of the wild type (WT; red bars) and OrrDs mutants (OrrDs/ORR, yellow bars; OrrDs/OrrDs, orange bars). Numbers correspond to the peak area representing each metabolite; data are shown as means ± se (n = 4). Asterisk indicates values that are significantly different from the wild type (P < 0.01).
(B) Composition of carotenoids in mature fruit derived from the different OrrDs genotypes and wild-type plants detected by HPLC. Total carotenoids are the sum of all the peaks area of each genotype. Data are shown as means ± se (n = 4). Asterisk indicates values that are significantly different from the wild type (P < 0.01).
(C) Chlorophylls and tocopherols levels in mature fruit derived from the various OrrDs genotypes and wild-type plants detected by HPLC. Data are shown as means ± se (n = 4). Asterisk indicates values that are significantly different from the wild type (P < 0.01).
[See online article for color version of this figure.]
Fruit peels of the OrrDs mutants plants showed a strong reduction in the content of flavonoids compared with the wild type, and levels of the yellow pigment naringenin chalcone dropped by 85% (Figure 5). Levels of the flavonol rutin were reduced to 33 and 60% in the OrrDs/ORR and OrrDs/OrrDs peels, respectively. The same trend was also observed in the case of several other flavonoids, including various kaempferol derivatives, quercetin trisaccharides, and quercetin derivatives. The more severe phenotype of the heterozygote genotype compared with that of the homozygote in terms of carotenoid and flavonoid levels raises the possibility of posttranslational regulation. However, this is not surprising when taking into consideration that ORR represents one of the numerous Ndh complex subunits and thus changes in its quantity alters protein–protein interactions. A possible explanation for this phenomenon is detailed below (see Discussion).
Gene Expression Changes in the OrrDs Mutant Fruit
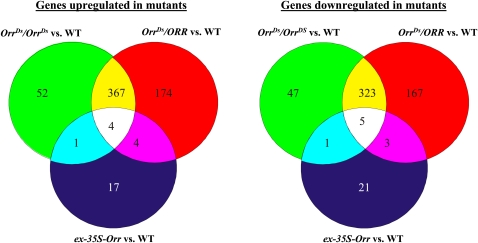
Microarray gene expression analysis was performed to evaluate the transcriptional changes in the OrrDs mutants (breaker stage fruit). Compared with the wild type, 1048 and 800 genes were altered in the OrrDs heterozygous and homozygous mutants, respectively. A total of 549 and 424 genes were upregulated, while 499 and 376 genes were downregulated in the OrrDs heterozygous and homozygous lines, respectively (Figure 6; see Supplemental Table 2 online). Out of the 602 and 546 genes that were up- or downregulated in the two mutant genotypes, respectively, 371 genes were upregulated in both genotypes, whereas 328 genes were downregulated in both. Conversely, when comparing the ex-35S-Orr line to the wild-type, only 26 genes were upregulated and 30 downregulated when compared with the wild type. Furthermore, most of these changes were restricted to the ex-35S-Orr line and were not shared with either of the OrrDs insertion lines, indicating that the ex-35S-Orr is more similar to the wild type. This was also supported by principal component analysis (see Supplemental Figure 10 online).
Figure 6.
Gene Expression Changes in the OrrDs Mutants.
Venn diagrams of genes changed in expression in fruit (breaker stage) of the three OrrDs genotypes compared with the wild type (WT). The Venn diagrams show significantly up- and downregulated genes in fruit of the OrrDs/ORR (red), OrrDs/OrrDs (green) and the ex-35S-Orr (blue) lines when compared with the wild type. Genes being significantly up- or downregulated in several of the genotypes are shown in the respective overlap regions.
[See online article for color version of this figure.]
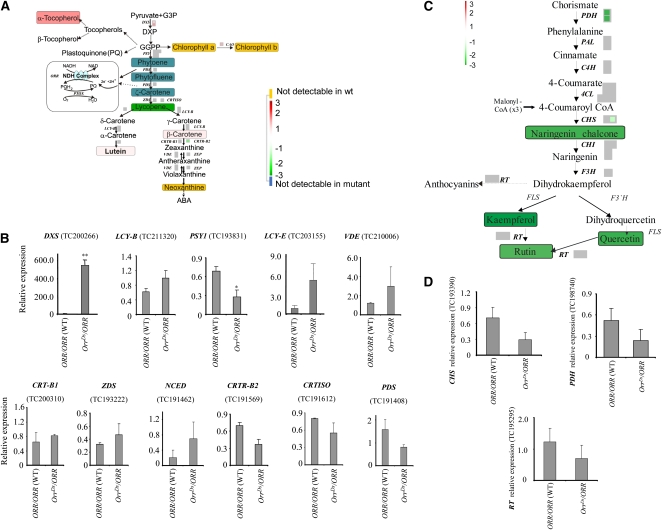
The changes in expression of genes involved in carotenoid and flavonoid biosynthesis was of particular interest since these two pathways had a major contribution to the OrrDs mutant phenotype (Figure 1). Out of 13 carotenoid biosynthesis genes evaluated by the array analysis, the expression of 1-DEOXY-D-XYLULOSE 5-PHOSPHATE SYNTHASE (DXS) and of β-CAROTENE HYDROXYLASE (CRTR-B2) was statistically significantly different between the mutants and the widl type (DXS increased and CRTR-B2 decreased; Figure 7A). The results showed that the effect on gene expression in the carotenoid pathway was mild and possibly posttranscriptional. qRT-PCR was subsequently performed to further evaluate the expression of eleven carotenoid-related genes (Figure 7B; see Supplemental Figure 11 online). As detected in the array analysis, expression of DXS was strongly upregulated, while CRTR-B2 expression showed a trend of decreasing in the OrrDs heterozygous mutant fruit. One major difference was in PSY1 expression, which was downregulated in the qRT-PCR analysis but not in the array data.
Figure 7.
The Effect of the OrrDs Mutation on Gene Expression in the Isoprenoid and Phenylpropanoid/Flavonoid Biosynthesis Pathways.
(A) Representation of gene expression and metabolite levels changes in fruit derived from the OrrDs/ORR genotype compared with that of wild-type plants. Microarray probe sets reporting the gene expression of likely pathway genes are depicted by color-coded boxes indicating the log2 fold change of the respective transcripts according to a false-color scale reproduced in the figure, where white represents no change, red an upregulation, and green a downregulation. Each individual box represents a unique probe set present on the chip hybridizing to isoforms of the respective gene indicated next to it. Nonsignificantly changed probe sets are depicted in gray. Metabolites where quantitative data were available are represented as black text on colored boxes, where the box color indicates log2 fold changes following the same color scheme as transcripts. Metabolites not detectable in the mutant are painted on a blue box, whereas the ones not detectable in the wild type are painted on a yellow box. The electron transport pathway involving the Ndh complex and PTOX (adapted from Carol and Kuntz, 2001) is represented in a box on the left. GGPP, geranylgeranylpyrophosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; DXS, 1-deoxy-d-xylulose 5-phosphate synthase; PSY, phytoene synthase; CrtISO, carotene isomerase; CRTL-E, LCY-E, lycopene e-cyclase; LCY-B, lycopene β-cyclase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; CAO, chlorophyll a oxidase.
(B) Expression of genes in the isoprenoid pathway (including carotenoid-related) in the hetrozygous OrrDs mutant and wild-type breaker stage fruit as detected by qRT-PCR. Full names of genes are given in (C). Expression data were normalized to the expression of the CLATHRIN ADAPTOR COMPLEXES SUBUNIT (CAC) gene; values are means ± se (n = 3). Asterisks indicate values that are significantly different from the wild type at *P < 0.05 and **P < 0.01.
(C) Representation of gene expression and metabolite levels changes in fruit derived from the OrrDs/ORR genotype compared with wild-type plants. The array and metabolite analysis data are presented in the same way as in Figure 7A. PAL, Phe-ammonia lyase; 4CL, 4-coumaroyl-CoA synthase 2; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; 3GT, anthocyanidin 3-O-glucosyltransferase; RT, rhamnosyltransferase; CAD, cinnamyl-alcohol dehydrogenase; F3′5'H, flavanone 3′, 5′hydroxylase.
(D) Expression of genes related with phenylpropanoids/flavonoids pathway in the hetrozygous OrrDs mutant and wild-type breaker stage fruit as detected by qRT-PCR. Full names of genes are given in (B). Expression data were normalized to the expression of the CAC gene; values are means ± se (n = 3).
The flavonoid pathway was also affected at the transcriptional level as expression of CHALCONE SYNTHASE2 (CHS2) and PREPHENATE DEHYDROGENASE (PDH) upstream of Phe was downregulated compared with the wild type (Figure 7C). It is important to note that although most of the flavonoid pathway genes displayed a reduction in expression, this was relatively mild and for most genes not statistically significant. qRT-PCR analysis confirmed that expression of CHS and PDH was reduced in the OrrDs heterozygous mutant fruit (Figure 7D).
Apart from the targeted analysis of transcripts involved in the carotenoid and flavonoid pathways described above, an unbiased investigation of the differences in the two mutant genotypes and the wild type was performed. To this aim, a pathway enrichment analysis of differentially expressed genes and a Wilcoxon test on the magnitude of changes within a pathway were performed to identify grossly changed categories (Usadel et al., 2006). Both analyses identified that the light reactions of photosynthetic processes as well as tetrapyrroll biosynthesis were upregulated, whereas transcripts encoding for enzymes involved in the calvin cycle reactions were unchanged in both mutants, which is in line with the apparent upregulation of chlorophyll in the mutant (Figure 5). Interestingly, both analyses consistently highlighted changes in cell wall metabolism. Here, a general downregulation of cell wall related processes, such as nucleotide sugar metabolism, providing precursors for cell wall metabolism, as well as many pectin methyl and acetyl esterases, was observed. However, the potentially pectin derived rhamnose that was measured by means of GC-MS was unchanged (see Supplemental Table 2 online), and galacturonic acid, which is a major constituent of pectin, was not detected. Nevertheless, several enzymes acting on xyloglucan were changed; indeed, three of these were amongst the genes most strongly downregulated in the heterozygous mutant (Table 4). This might be in line with the observed changes of free xylose that was identified by means of GC-MS.
Table 4.
The Most Altered Genes in the Heterozygous OrrDs Mutant Fruit
| Probe Set Name | OrrDs/OrrDs | OrrDsORRr | ex-35S-Orr | MapMan Bin | Annotation |
| Les.3652.1.S1_at | 5.8 | 6.0 | 0.1 | β-1,3 glucan hydrolases | β- 1,3-glucanase (SGN-U577505) |
| Les.5934.1.S1_at | 5.5 | 5.4 | −0.8 | Omega 6 desaturase | Omega 6 fatty acid desaturase (SGN-U574777) |
| Les.4829.1.S1_at | 3.4 | 5.1 | −1.4 | Ethylene synthesis degradation | E8 homolog (SGN-U579236) |
| LesAffx.56637.1.S1_at | 3.6 | 4.7 | −0.1 | Heat stress | DNAJ heat shock N-terminal domain-containing protein (SGN-U573193) |
| Les.4489.1.S1__at | 5.6 | 4.4 | 0.5 | Unknown | SGN-U578947, SGN-U590412, SGN-U579487 |
| Les.1175.2.S1_at | 5.5 | 4.2 | −0.1 | Signaling in sugar and nutrient physiology | PAR-1c (SGN-U569163) |
| Les.435.1.S1_at | 3.6 | 4.2 | -2.0 | Stress, biotic | Chitinase-endo (SGN-U566861) |
| LesAffx.70768.1.S1_at | 3.4 | 4.1 | −0.2 | AP2/EREBP, APETALA2/Ethylene-responsive family | DNA-binding protein (SGN-U573405) |
| Les.931.1.A1_at | −0.2 | 3.9 | 0.0 | WRKY transcription factor | WRKY (SGN-U589067, SGN-U573117) |
| LesAffx.10279.1.A1_at | 3.1 | 3.8 | 0.0 | Transport nucleotides | Purine permease (PUP1) (SGN-U568852) |
| LesAffx.18067.1.S1_at | −2.9 | −4.0 | −0.7 | Unknown | SGN-U566840 |
| LesAffx.30832.1.S1_at | −2.7 | −4.0 | −1.2 | Unknown | SGN-U576446 |
| Les.4530.1.S1_at | −3.4 | −4.0 | 0.0 | Cell wall modification | Xyloglucan endotransglucosylase-hydrolase (XTH9) (SGN-U577928, SGN-U581575) |
| Les.5038.1.S1_at | −3.5 | −4.2 | 0.2 | Unknown | SGN-U571966 |
| Les.4353.1.S1_at | −3.6 | −4.3 | 0.0 | Cell wall modification | Xyloglucan endotransglycosylase (XET1) (SGN-U579684, SGN-U579445) |
| LesAffx.11542.1.S1_at | −3.7 | −4.4 | −0.1 | Calcium signaling | Calcium-binding EF hand family protein (SGN-U566130) |
| Les.429.1.S1_at | −3.9 | −4.5 | 0.4 | Cell wall modification | Xyloglucan endo-1,4-β-d-glucanase (SGN-U581358) |
| Les.2038.1.A1_at | −4.8 | −4.5 | 0.2 | GDSL-motif lipase | Lipase/hydrolase, GDSL-motif (SGN-U568434) |
| LesAffx.63587.1.S1_at | −4.1 | −4.9 | −2.0 | AP2/EREBP, APETALA2/ethylene-responsive family | ERF (ethylene response factor) subfamily B-4 (SGN-U5864)39 |
| LesAffx.58308.1.S1_at | −4.5 | −5.8 | −1.0 | AP2/EREBP, APETALA2/ethylene-responsive family | CBF4-like, DREB subfamily A-1 of ERF/AP2 factors (SGN-U563215) |
The 10 most strongly up- and downregulated Affymetrix probes sets that could be assigned to an SGN unigene in the heterozygous OrrDs/ORR mutant compared to the wild type (breaker stage). The log2 fold changes of these probe sets in the homozygous OrrDs/OrrDs, the heterozygous OrrDs/ORR, and the ex-35S-Orr line in comparison to the wild type and their inferred MapMan classes and annotations are reproduced. Values in bold denote significant changes (P < 0.01 after false discovery rate control). The annotations were retrieved from SGN, where available manual annotation was reproduced; otherwise, the best BLAST hit against GenBank or Arabidopsis was imported from the SGN website.
Another interesting observation revealed by the global pathway-based tests was changes in protein synthesis and degradation. In the case of protein synthesis, this was largely attributable to a strong (more than fourfold) upregulation of Elongation factor 1-α (Les.48761.1.s1_at, SGN-U580191). However, in the case of protein degradation, a strong coordinated upregulation of many transcripts, which encode either proteasomal subunits or regulatory particles, could be observed (data not shown).
Further inspection of changed metabolites revealed that consistent changes in both mutant lines were largely confined to amino acid metabolism (Table 3; see Supplemental Table 5 online). These were primarily restricted to a downregulation in levels of the branched chain amino acids Ile and Lys. However, inspection of biosynthesis and degradation pathways for these amino acids did not reveal marked, significant changes in the respective genes. One potential exception was the probe set LesAffx.9260.1.S1_at (SGN-U578917) being similar to dihydrodipicolinate reductase. Interestingly, this unigene was most similar the Arabidopsis gene chlororespiration reduction 1, which has been shown not to be involved in Lys biosynthesis but rather plays a role in accumulation of the chloroplast Ndh complex (Shimizu and Shikanai, 2007) consistent with the changes in the ORR gene.
Further investigation of the genes that were most strongly changed in the heterozygous mutant (Table 4) revealed that apart from genes involved in cell wall modification (mainly ones encoding for enzymes acting on xyloglucan), a relatively large number of genes putatively encoding transcription factors were changed, many of them belong to the ethylene responsive factor gene family.
The Effect on Ripening-Related Gene Expression and Ethylene Levels in the OrrDs Mutant Fruit
To examine the effect on the ripening process in the OrrDs mutant fruit, genes known or previously suggested to be involved in ripening were checked for differential expression (see Supplemental Data Set 1 online). Here, Les.4450.1.S1_at, which potentially queries a MADS box gene similar to the RIPENING INHIBITOR (RIN) and Les.132.1.S1_at potentially reporting the expression of a gene similar to tomato ACO1, both playing a role in fruit ripening (Nakatsuka et al., 1998; Vrebalov et al., 2002), were more than fivefold upregulated (data not shown). However, comparing the individual oligonucleotide sequences present in these probe sets with the deposited nucleotide sequences for RIN and ACO1 revealed that these were unlikely to reflect their expression. This is in accordance with the fact that other probe sets interrogating these genes seemed unchanged (see Supplemental Table 4 online). Furthermore, the probe sets reporting the expression of ERT1b and ERT10 also failed to detect any changes (Picton et al., 1993). However, three probe sets reporting the expression of the E8 ripening-related gene (Deikman and Fischer, 1998) were consistently upregulated in the OrrDs mutants and downregulated in the ex-35S-Orr line, albeit these findings were not significant. Genes short-listed by Alba et al. (2005) as good candidates for genes involved in ripening regulation were also tested for differential expression (see Supplemental Table 3 online, bottom part). However, none of these genes were significantly changed, and with the exception of MBF1 and bZIP1, no trend was visible either (see Supplemental Table 3 online).
To obtain a more conclusive picture regarding the effect on ripening-associated gene expression, genes shown by Alba et al. (2005) to change during ripening or fruit developmental were also examined. Interestingly, of the 336 probe sets that could be mapped to the original TOM1-based list of genes changing during ripening or development, 54 and 47 were flagged as differentially expressed in the OrrDs/ORR and OrrDs/OrrDs mutants, respectively, indicating a mild enrichment (P < 0.05) of potentially ripening-related genes (see Supplemental Data Set 1 online).
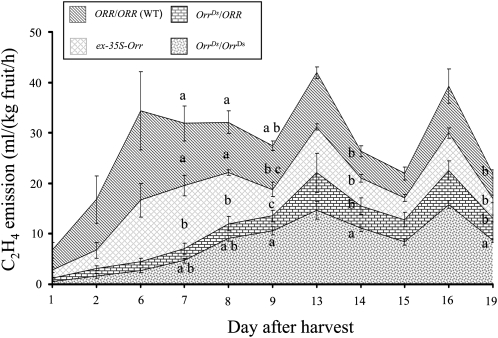
We also performed measurements of ethylene, which marks the typical climacteric during fruit ripening in tomato (Figure 8). Ethylene emission levels were measured using GC coupled to a flame ionization detector 11 times starting from day 1 after the harvest of green fruit. The results showed that the peak in ethylene emission was delayed and reduced in fruit of both OrrDs/ORR and OrrDs/OrrDs (stronger in the former), although not diminished, compared with its peak in wild-type fruit 6 d after harvest in the wild type compared with 13 to 16 d after harvest of the mutant fruit. The ethylene emission profile of ex-35S-Orr fruit was very similar to the one observed in wild-type fruit.
Figure 8.
Ethylene Emission from the OrrDs Mutants and Wild-Type Fruit.
The chart shows ethylene emission from fruit of the OrrDs genotypes and the wild type (WT) measured 11 times during 19 d after the mature green fruit stage. Emission of each line is represented by the patterned strip between two graphs or between the graph and x axis (in the case of the OrrDs/OrrDs genotype). The letters a to c represent significance groups of ethylene emission per day (n = 3; P < 0.05; Student's t test).
Examining the correspondence in fruit development (from flowering to ripening and from the breaker to ripening) between the wild-type and the OrrDs genotypes showed that the mutants were slightly delayed in their development, although this was not statistically significant.
DISCUSSION
Altered NDH Activity in Plastids of Mature Green Fruit Impinges on Subsequent Metabolism in Chromoplasts
Characterization of the OrrDs mutant and its corresponding gene encoding the NDH-M subunit described in this study indicate that Ndh complex and redox control play essential roles during tomato fruit maturation in ripening-related metabolism, most notably carotenoid biosynthesis. Interestingly, although other Ndh subunit mutations have been isolated, unlike OrrDs, these mutants do not exhibit a clear visual phenotype (Burrows et al., 1998; Sazanov et al., 1998a; Shikanaï et al., 1998; Horváth et al., 2000). The specific expression of ORR in chlorophyll-containing tissues (i.e., stems, leaves, young flowers, and immature green fruits) is consistent with the involvement of the Ndh complex in alternative electron pathways around PSI (Nixon, 2000; Peltier and Cournac, 2002; Bukhov and Carpentier, 2004; Johnson, 2005). At the onset of fruit maturation and degreening, Ndh-M expression is downregulated, in agreement with the data of Guera and Sabater (2002) that demonstrated reduction in the Ndh complex quantity and activity during fruit maturation. Interestingly, publicly available gene expression data (Tomato Functional Genomics Database; http://ted.bti.cornell.edu/) indicate that expression of Ndh-N and Ndh-O does not change during fruit development and maturation; see Supplemental Table 7 online).
The visual and biochemical phenotypes of the OrrDs mutant imply that in addition to its importance during the early green stages of fruit development (Guera and Sabater, 2002), the Ndh complex also influences the mature fruit once the transition of chloroplasts to chromoplasts takes place. These observations in OrrDs resemble the effects seen in fruits of the tomato ghost (gh) mutant when averted from light (Shahbazi et al., 2007). This mutant is impaired in the plastid terminal oxidase gene (PTOX). When green stage gh fruit are covered and mature in the dark, they remain green and do not exhibit the typical white gh phenotype. Similar to OrrDs, covered fruit of the gh mutant are severely affected in their carotenoid composition (Barr et al., 2004). Authors of this article suggested that the gh mutation irreversibly changes the plastid ultrastructure in green fruits exposed to light. Consequently, carotenogenesis, which is performed by membrane-associated enzymes, is severely affected in mature fruits. Although the green-stage fruits of the OrrDs mutant visually showed a wild-type phenotype, we cannot rule out a possible damage to plastids that slows the conversion of chloroplasts to chromoplasts.
Lack of a significant difference between OrrDs and the wild type in carotenoid content and composition in green fruits indicates that the Ndh complex affects the carotenoid biosynthesis pathway in fruit chromoplasts and not in chloroplasts. Nevertheless, no alterations in carotenoids levels and content were observed in OrrDs flowers that also contain chromoplasts. In an ndh-h-defective mutant of tobacco (Zapata et al., 2005), the carotenoid profile was not changed in leaves nor when we examined its chromoplast-containing nectary tissue. Thus, it appears that the effect of reduced Ndh complex activity on carotenoid pigment composition is restricted to the mature tomato fruit tissue. The reduction in levels of the red carotenoid lycopene, combined with the increase in the yellow-orange carotenoids β-carotene and lutein, and the large reduction in the levels of several flavonoids, particularly of the yellow flavonoid pigment naringenin chalcone, explain the pale-yellow/orange appearance of the OrrDs mature fruit. The fruit phenotype is similar to the one observed when the tomato y, colorless peel, and the r mutations are combined (Rick and Butler, 1956).
OrrDs Is a Unique Gain-of-Function Mutation
The OrrDs mutant is not a null mutation, as we detected reduced expression in the OrrDs/OrrDs genotype compared with the wild type. The residual ORR expression is most probably due to the fact that the promoter region of ORR is located in the 5′ region of the Ds element. In addition, transposon-induced transcription of the neighboring region (i.e., readout transcription) has been reported previously in transgenic tomato plants (Rudenko et al., 1994).
This mutation results in an overdominant phenotype that is reflected by the enhancement of several but not all of the parameters examined in this study. Most traits that exhibited a more severe phenotype in the OrrDs/ORR heterozygous fruit than in the homozygous ones were observed in fruit that contained chromoplasts. These traits included carotenoid and flavonoid composition, global gene expression changes, levels of chlorophylls, α-tocopherol, and TSS, and fruit texture (postharvest shrinking). On the other hand, traits that were more affected in the OrrDs/ OrrDs fruit than in the heterozygous are those associated with chloroplast-containing tissues (leaves and green fruit), such as chlorophyll fluorescence, Ndh complex levels, and Ndh activity. A possible explanation for the overdominance phenomenon may be that ORR is only one out of 14 subunits of the Ndh complex, and changes in ORR quantity likely alter the complex activity and levels. The ORR proteins that are likely to be incorporated into the Ndh complex in either the homozygous or heterozygous OrrDs lines are different in their quantity. In the homozygous lines, either no proteins or alternatively small amounts of much shorter ones in the case of plastidic localization (starting at AUG-IV, from amino acid residue 64) will be incorporated into the complex. However, in the heterozygous line, it is expected that a half of the proper protein dose will be incorporated, while the shorter protein (from AUG-IV) might also be inserted to the complex (if localized to the plastids). It is also likely that the short protein (from AUG-IV) will be localized to a different subcellular compartment (e.g., the cytosol). Another possible explanation is the activity of a compensatory process that reduces the effect on the Ndh complex function in the green tissues (in the case of the heterozygous) and in the mature fruit (in the case of the homozygous). Such a mechanism could be related to other alternative electron pathways, such as those involving PGR5/PGRL1 (Munekage et al., 2002; DalCorso et al., 2008), NDH-2 [a type II NAD(P)H dehydrogenase; Corneille et al., 1998; Desplats et al., 2009], and/or ferredoxin NADPH reductase (Zhang et al., 2001).
Since the OrrDs mutation exhibits an overdominant phenotype, it could not be as a result of a loss of function (i.e., haplo-insufficient), but rather a consequence of either a gain-of-function mutation (whose gene product become constitutively active due to a gain of a novel function normally not found in the wild-type protein phenotype) or dominant-negative (whose gene product adversely affects the normal wild-type gene product) mutations. If OrrDs was a dominant-negative mutation, we should have observed the OrrDs phenotype by overexpression of the ORR gene either in the wild type or in the mutant backgrounds. However, plants overexpressing the ORR gene in the wild-type background and the ex-35S-Orr line displayed a wild-type phenotype. In addition, the OrrDs phenotype was complemented by overexpression of ORR in the mutant background. Thus, OrrDs is not a consequence of a dominant-negative mutation but more likely a gain-of-function mutation.
The Mutation in ORR Affects the Redox State of Fruit Plastids
The role of the Ndh complex in chlororespiration and cyclic electron transport around PSI suggested that alterations in this complex could influence the redox state of its cellular environment. The NDH-M subunit encoded by ORR in tomato is most likely directed to the plastids, as indicated by localization assays of transiently expressed full-length transcript (starting from AUG-I) (Figure 3). The consequences of the lack of Ndh activity on the redox status of chloroplasts has not been reported probably since the electron donors in the stroma have not been identified and also because the alternative dark electron flux proceeds at very low rate. The levels of NADH and NADPH were reduced in the OrrDs mutant lines. Consistent with these changes in the plastidial redox status was the alteration in the activities and activation state of the AGPase and plastidial malate dehydrogenase (Scheible and Stitt, 1988; Tiessen et al., 2002). Together, these results indicate that in tomato, the lack of the Ndh complex alters the plastid redox status.
Involvement of ORR in the Carotenoid Biosynthetic Pathway
The changes observed in the carotenoid pathway do not seem to result from altered transcript levels, as expression of most genes of the pathways were not altered in the OrrDs mutant. More probably, the effect on carotenoid composition is instigated by obstructing enzymes in the pathway that require redox electron chain for their activity. Desaturation of phytoene to lycopene catalyzed by PDS and ZDS necessitates acceptors for eight hydrogen atoms. It has been demonstrated that PQ serves as an intermediate in transferring electrons between phytoene desaturase and molecular oxygen (Mayer et al., 1990; Norris et al., 1995), a process mediated by a bound flavin (Hugueney et al., 1992). In chloroplasts, oxidized PQ required for PDS activity is regenerated from plastoquinol by electron transfer to PSI. In the dark and in chromoplasts, where photosynthetic electron transport is inactive, oxidation of plastoquinol is performed by PTOX.
The interaction of PDS with the electron transfer chain is unspecific as evidenced by the fact that when expressed in Escherichia coli cells both plant enzymes are able to mechanistically link to the bacterial respiratory redox chains (Mayer et al., 1990; Nievelstein et al., 1995). An analogous situation exists in cyanobacteria where plastoquinol can be oxidized in darkness because the electron transport reactions of photosynthesis and respiration are linked in the same membranes (Sandmann and Malkin, 1984). Similar to PDS and ZDS, other enzymes involved in carotenoid biosynthesis also contain a conserved FAD binding motif in their N termini, for example, CRTISO, LCY-E, LCY-B, and ZEP, suggesting the involvement of redox balance in these reactions even though they do not involve net electron transfer. This is the case in lycopene β-cyclase, which was active in vitro only in the presence of NADH or NADPH (Schnurr et al., 1996), and carotenoid isomerase, which was dependent on redox driving force (Isaacson et al., 2004). Involvement of FAD in ZEP activity has also been demonstrated (Buch et al., 1996). It has been recently demonstrated that the bacterial lycopene cyclase (CRTY) requires a reduced FAD as a catalytic cofactor probably for stabilization of a transition state in an acid-base reaction (Yu et al., 2010). A similar phenomenon has also been discovered in the case of the carotene isomerase (CRTISO; Q. Yu, P. Beyer, and J. Hirschberg, unpublished results). The role of the redox state in the case of these enzymes is unknown. However, the above results imply that it is indirect and not associated with net electron transfer.
The changes in the carotenoid pathway intermediates we observed in the OrrDs mutants cannot be explained solely by altered activities of PDS, ZDS, and CRTISO. If this was the case, we should have detected an accumulation of the PDS substrate phytoene, as seen in the gh leaves, flowers, and fruit (Zapata et al., 2005) and reduced levels of metabolites downstream to ζ-carotene. In addition to the expected reduced levels of phytofluene, ζ-carotene, and lycopene, we also measured reduced concentration of phytoene. Interestingly, levels of β-carotene, neoxanthin, and lutein, all derived from lycopene, were increased. It is also important to note that expression of ORR and genes encoding PDS, ZDS, and CRITSO are not correlated during development; while expression of ORR is reduced after the green stage, PDS, ZDS, and CRITSO show an increase in their transcript levels (Josse et al., 2000).
It was earlier reported that alteration in PQ redox induces intracellular signaling leading to changes in chloroplast and nuclear genes expression (Rodermel, 2001; Pfannschmidt, 2003). Thus, an alternative explanation to the involvement of ORR in carotenoid biosynthesis might be that Ndh-catalyzed PQ reduction in plastids of fruit triggers the expression of genes encoding enzymes involved in carotenoid biosynthesis. Steinbrenner and Linden (2003) demonstrated that in Haemotococcus pluvialis, an astaxanthin-accumulating green alga, the photosynthetic PQ pool functions as a redox sensor for the upregulation of carotenoid biosynthesis genes. In the OrrDs mutant, changes in the expression of carotenoid biosynthetic genes were detected in fruit at breaker stage, although not many genes of the pathway were altered in expression.
The changes in the OrrDs carotenoid composition together with the previous reports with respect to the chlororespiratory function of the Ndh complex suggest that the PTOX encoded by the tomato GH gene and ORR are part of the same pathway. Publicly available microarray data indicated that expression of GHOST gradually increases from early fruit development until ripening before it declines sharply (Tomato Functional Genomics Database; see Supplemental Table 7 online). Thus, expression of GH and ORR only partially overlaps in the early stages of fruit development. The interaction between ORR and GH should therefore be examined in subsequent work through the generation of double OrrDs/gh mutant lines.
A Proposed Link between NDH Complex Activity, Antioxidant Systems, and the Process of Fruit Ripening
Apart from changes in carotenoid composition, OrrDs displayed alteration in additional metabolite classes in fruit, including chlorophylls, tocopherols, and flavonoids. Changes in specialized metabolism that increased total antioxidant content have been demonstrated in fruits of high-pigment (hp) tomato mutants (Bino et al., 2005). Downregulation of the DE-ETIOLATED1 gene that corresponds to the hp2 mutation through an RNA interference approach resulted in similar metabolic changes, including an increase in levels of carotenoids, flavonoids, and chlorogenic acid (Davuluri et al., 2005; Enfissi et al., 2010). An increase in chloroplast size and number in fruits of the high-pigment mutants hp1 and hp3 was suggested to trigger the changes in metabolism of antioxidants (Cookson et al., 2003; Galpaz et al., 2008). This combined effect on pathways generating antioxidant or photoprotective activities might represent a compensatory mechanism between the different classes of protecting compounds that is perhaps due to a change in plastid structure or number at early fruit development. In the case of metabolites that showed elevated levels in mature OrrDs fruits (e.g., chlorophylls), we cannot exclude the possibility that they were derived from the progenitor chloroplast.
The reduced postharvest shrinking and TSS levels in the OrrDs fruit suggested that the process of ripening is also affected by ORR activity. This was further corroborated by the relatively low and delayed emission of the climacteric ethylene and changes to ripening-associated genes detected by array analysis. The nature of such a link between the perturbation of the Ndh complex, carotenoid, ethylene, and additional metabolic pathways in fruit and the ripening process is difficult to explain at this stage. In one scenario, the altered ORR gene activity will directly affect ethylene biosynthesis or signaling that mediate climacteric ripening. Even so, the effect on ethylene is most probably through its biosynthesis and not signaling, as OrrDs mutants seedlings were not altered in the triple response (see Supplemental Figure 2 online). In a second scenario, as suggested above, if the reduced activity of the Ndh complex results in damage to chloroplasts, it could affect ripening-associated processes in chromoplasts later in fruit development (e.g., carotenoid formation and Met metabolism for ethylene production). A third possibility might be that these processes are affected simultaneously through the requirement for a particular redox environment by multiple enzymes of the different pathways.
In conclusion, our study on tomato plants deficient in the Ndh complex through a mutation in its subunit M provided evidence that specialized (secondary) metabolism in tomato fruit, including carotenoid biosynthesis, depends on the complex activity that modulates the redox state of PQ. While the Ndh complex is highly active in chloroplasts of the early green stages of tomato fruit development, its inactivation also affects the accumulation of the typical carotenoid pigments in fruit chromoplasts during ripening. This work therefore opens the way for the future investigation of the interaction between the two redox chain components, namely, the Ndh complex and PTOX, in the regulation of fruit carotenogenesis.
METHODS
Plant Material
The OrrDs mutant (Solanum lycopersicum cv Micro-Tom) was identified among the transposon insertion population generated by Meissner et al. (1997). For metabolic analysis, peel and flesh were dissected manually as described by Mintz-Oron et al. (2008). Fruit at a particular developmental stage up to breaker were harvested according to their appearance; from the breaker stage and further days were counted following breaker.
Isolation of the OrrDs Insertion and Genotyping
DNA was extracted from leaves as described by Eshed and Zamir (1995). Isolation of the transposon flanking regions was performed by long-range inverse PCR as described previously (Meissner et al., 2000). The wild-type ORR locus and the insertion allele were genotyped by oligonucleotides 1 and 2 (1149-bp amplicon) and 1 and 3 (808 bp), respectively (see Supplemental Table 4 online). DNA gel blot analysis was according to Sambrook et al. (1989), using the kanamycin gene as a probe.
Isoprenoid and Flavonoid Extraction and Analyses
Isoprenoid extraction was performed as described by Bino et al. (2005). Analysis was performed by the use of an HPLC-PDA detector and a YMC C30 column as described by Fraser et al. (2000). Peak areas of the carotenoid compounds were determined at the wavelength providing maximum absorbance (see Supplemental Methods online) using Waters Millennium32 software. Flavonoids were extracted in methanol and profiled with UPLC-PDA as described previously by Mintz-Oron et al. (2008) for the profiling of semipolar compounds. Detailed description of sample preparation and injection and compound quantitation can be found in the supplemental figures and Supplemental Methods online.
RACE and Gene Expression Analyses
The 5′ RACE-PCR was performed with the SMARTer RACE cDNA amplification kit according to the manufacturer's instructions (Clontech; see Supplemental Table 4 online for oligonucleotides). Fruit RNA was extracted as described by Verwoerd et al. (1989) while from other tissues using the Tri-reagent method (Molecular Research Center). qRT-PCR for the ORR gene (oligonucleotides 7 and 8; see Supplemental Table 3 online) and ASR1 the endogenous control (oligonucleotides 11and 12) was performed as described by Mintz-Oron et al. (2008). Global gene expression was analyzed in OrrDs homozygote and heterozygote mutants, ex-35S-Orr, and the wild type (10 pooled breaker stage fruit collected from five plants in three replicates for each genotype). Sample preparation and hybridization to the Affymetrix Tomato GeneChip was as described by Mintz-Oron et al. (2008). Normalization of the raw expression intensity values was performed using RMA analysis (Irizarry et al., 2003) using the BioConductor software suite (Gentleman et al., 2004). Subsequently, arrays were quality controlled using standard procedures, such as RNA degradation plots, relative log expression, and others (see Supplemental Figure 7 online). Even though two arrays showed somewhat lower quality in the normalized unscaled errors of the means plots (see Supplemental Figure 7D online), and one of these also showed a somewhat stronger RNA degradation (see Supplemental Figure 7C online), they were included for final analysis. Subsequently, differentially expressed genes were extracted as follows: first, a linear model was fitted to the data using the limma package (Smyth, 2004), which included the single factor genotype. P values for differentially expressed genes were then extracted using the empirical Bayes procedure of the limma package, for the contrasts OrrDs/OrrDs versus the wild type, OrrDs/ORR versus the wild type, and ex-35S-Orr versus the wild type. The resulting P values were globally corrected using false discovery rate control at a level of 1% (Benjamini and Yekutieli, 2001).
Pathway Analyses
To investigate changes in processes and to identify co-coordinated changes in pathways and processes, we used PageMan software (Usadel et al., 2006) and the according tomato ontology (Urbanczyk-Wochniak et al., 2006) to perform a Wilcoxon test on the log-fold change values and a Fisher's exact test for an enrichment of categories, harboring significantly up- and downregulated genes.
Comparison with Ripening Genes
To test for ripening genes, SGN unigenes potentially involved in ripening were extracted from Supplemental Table 1 of Alba et al. (2005). These SGN release #1 unigenes were converted to SGN release #4 unigene versions using the SGN resource (Mueller et al., 2005) and finally matched to Affymetrix probe set identifiers (see Supplemental Data Set 1 online). A Fisher's exact test was performed to test for overrepresentation of ripening related genes differentially expressed in the OrrDs heterozyogous mutant.
Chlorophyll Fluorescence
Chlorophyll fluorescence was measured at room temperature on attached leaves using a PAM-2000 modulated fluorometer (Walz Effeltrich) as previously described by Rumeau et al. (2005). Postillumination chlorophyll fluorescence rises were recorded under low nonactinic light following a 5-min actinic illumination (250 μmol photons m−2 s−1).
Preparation of Chloroplasts and Thylakoid Membranes and Immunoblotting
Chloroplasts were extracted from tomato leaves using the protocol described for tobacco (Nicotiana tabacum) by Sazanov et al. (1998b). Briefly, tomato leaves were ground in 50 mM Tricine buffer at pH 7.8 containing 0.33 M sorbitol, 2 mM EDTA, 1 mM MgCl2, 2 mM ascorbate, and 5 mM DTT using a Waring blender. Chloroplasts were collected by three successive low-speed centrifugations to avoid mitochondria contamination. Chloroplasts were osmotically lysed in 10 mM Tricine buffer at pH 7.8 containing 10 mM NaCl and 10 mM MgCl2. Thylakoid membranes were collected by centrifugation. Chloroplasts were extracted from green tomatoes according to Bathgate et al. (1985). Tomatoes were sliced and the gelatinous material and seeds were discarded. The tomato pericarp was homogenized in buffer containing 0.6 M sucrose and 0.1 M Na-phosphate, pH 7.5 (2 mL/g fresh material), using a Waring blender. The homogenate was filtered twice through three layers of Miracloth and centrifuged 3 min at 1500g. The pellet was resuspended in buffer containing 0.6 M sucrose and 0.01 M Na-phosphate, pH 7.5, and centrifuged as before. Washed chloroplasts were resuspended in a small volume of resuspension buffer and loaded onto a discontinuous sucrose gradient prepared using 0.01 M Na-phosphate, pH 7.5, and consisting of three layers of 30, 45, and 55% sucrose, respectively. Following centrifugation for 15 min at 2500g, bands formed at the 30/45% sucrose and at the 45/55% sucrose interfaces were collected and pooled. Finally, chloroplasts were recovered by centrifugation for 5 min at 1500g. Proteins were subjected to SDS-PAGE (15% polyacrylamide) and then electrotransferred onto nitrocellulose membranes (0.45 μm; Biotrace NT). Bound proteins were subsequently probed with the antiserum and visualized with 1:5000 diluted anti-rabbit IgG-IRDye800 antibody (Invitrogen). Detection was enabled by the Odyssey infrared imaging system (LI-COR Biosciences).
Extraction, Derivatization, and Analysis of Polar Metabolites Using GC-MS
Metabolite analysis by GC-MS was performed essentially as described by Lisec et al. (2006) with the exception that the method was optimized for tomato fruit tissue (Schauer et al., 2006). The mass spectra were cross-referenced with those in the Golm Metabolome Database (Kopka et al., 2005).
Analysis of Enzyme Activities
Enzyme extracts were prepared as described previously by Gibon et al. (2004), except that Triton X-100 was used at a concentration of 1% and glycerol at 20%. AGPase was determined as described by Gibon et al. (2004). Malate dehydrogenase (NADP) was assayed as described by Scheible and Stitt (1988).
Measurements of Pyridine Nucleotides and Ethylene
Pyridine nucleotides were extracted as described by Gibon and Larher (1997). Briefly, five leaf disks (9-mm diameter) were cut from fully expanded leaves and ground in a mortar with pestle in 1 mL of 0.2 M HCl for NAD+ and NADP+ determination or in 1 mL of 0.2 M NaOH for NADH and NADPH determination. Following heat treatment (4 min at 100°C), homogenates were centrifuged for 10 min at 12,000g at 4°C. The oxidized and reduced forms of the nucleotide were determined by enzymatic cycling according to the method described by Matsumara and Miyachi (1980). The enzyme reaction was stopped by addition of highly concentrated NaCl (6 M). In these conditions, formazan that precipitates is sedimented by centrifugation (15 min at 14,000g) and solubilized in ethanol as described by Gibon and Larher (1997). Ethylene emission from breaker stage fruit was measured as described by Itkin et al. (2009).
Protein Localization Assays
The ORR cDNA fragment, starting from either one of the 3 ATGs in its 5′ upstream region, was amplified from tomato young leaf cDNA (for primer sequences, see Supplemental Table 4 online), digested with BglI and SmaI, and cloned between the 5′ of GFP and 3′ of the CaMV 35S promoter in the pUC19 vector (BamHI and SmaI digested). The pUC19 construct was introduced into tobacco leaf cells via particle bombardment. Leaves were left in the dark for 36 h before observation. Images of green and chlorophyll red autofluorescence were monitored using a confocal laser scanning microscope (LCTCS-SP2; Leica) with a 488-nm argon laser excitation. The fluorescence signals were collected from 500 to 535 nm for GFP and from 680 to 720 nm for chlorophyll.
Gene Expression Analysis by Real-Time PCR
For two-step, real-time qRT-PCR, a known amount of DNase-treated total tomato fruit RNA was reverse transcribed using Invitrogen's Superscript II RTase kit. Platinum SYBR SuperMix (Invitrogen) RT-PCR reactions were tracked on an ABI 7300 instrument (Applied Biosystems). Each sample was PCR amplified using the same amount of cDNA template in triplicate reactions. Gene-specific quantitative PCR primer pairs for the 19 selected genes encoding peel-associated proteins were designed by the Primer Express software (Applied Biosystems). Tomato ASR1 and CAC were used as endogenous controls. Following an initial step in the thermal cycler for 15 min at 95°C, PCR amplification proceeded for 40 cycles of 15 s at 95°C and 30 s at 60°C and was completed by a melting curve analysis to confirm specificity of PCR products. The baseline and threshold values were adjusted according to the manufacturer's instructions. Similar results were obtained from relative quantification of transcript abundance determined independently by the standard curve method described in Applied Biosystems User Bulletin 2 (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf). Oligonucleotide sequences designed for the amplification of the transcripts are listed in Supplemental Table 4 online.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL or Sol Genomics Network databases under the following accession numbers: Arabidopsis NDH-M protein, NP_001031804, and gene, NM_001036727; tomato ORR, SGN-U579052; putative tomato Elongation factor 1-α, SGN-U580191; putative tomato dihydrodipicolinate reductase, SGN-U578917; 1-deoxy-d-xylulose 5-phosphate synthase (DXS), SGN-U567647; phytoene synthase (PSY), SGN-U580527; PDS, SGN-U565586; carotene isomerase (CrtISO), SGN-U584987; ZDS, SGN-U568537; Lycopene e-cyclase (CRTL-E and LCY-E), SGN-U567885; Lycopene β-cyclase (LCY-B), SGN-U563340; CRTR-B1, SGN-U568606; CRTR-B2, SGN-U568611; zeaxanthin epoxidase (ZEP), SGN-U569421; violaxanthin deepoxidase (VDE), SGN-U577684; PDH, SGN-U573964; Phe-ammonia lyase (PAL), SGN-U577677; Cinnamic acid 4-hydroxylase (C4H), SGN-U585432; 4-coumaroyl-CoA synthase 2 (4CL), SGN-U578380; CHS1, SGN-U579222; CHS2, SGN-U580856; chalcone isomerase (CHI), SGN-U573550; flavanone 3-hydroxylase (F3H), SGN-U563669; flavonol synthase (FLS), SGN-U563669; anthocyanidin 3-O-glucosyltransferase (3GT), SGN-U563149; and rhamnosyltransferase (RT), SGN-U580217. Microarray data have been deposited in the Gene Expression Omnibus under accession code Gse22110.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Effect of Season and Light on the OrrDs Phenotype.
Supplemental Figure 2. Examination of Shelf Life, TSS, and Triple Response of the OrrDs Mutants.
Supplemental Figure 3. The Proposed Mechanism That Led to the Unique OrrDs Mutation and the Excision Event (ex-35S-Orr).
Supplemental Figure 4. ORR Gene Sequences and Transcripts.
Supplemental Figure 5. Homology between ORR and Other Plant NDH-M Proteins.
Supplemental Figure 6. Segregation of Genotypes and Phenotypes in an Offspring Derived from the OrrDs Mutant.
Supplemental Figure 7. ORR Gene Expression in the Peel and Flesh Tissues during Five Stages of Fruit Development.
Supplemental Figure 8. Evaluation of the Subcellular Localization of ORR (Starting from Either AUG-I, AUG-II, or AUG-III).
Supplemental Figure 9. Analysis of Carotenoids in Extracts of Wild-Type and OrrDs Mutants (Presented as a Percentage of Total Carotenoids).
Supplemental Figure 10. Routine Quality Controls Performed on the Array Data.
Supplemental Figure 11. Quantitative Real-Time PCR Results of Selected Genes.
Supplemental Table 1. Genotype and Phenotypes of Plants Belonging to the Progeny of an ex-35S-Orr Line.
Supplemental Table 2. The Effect of the OrrDs Mutation on Primary Metabolite Levels.
Supplemental Table 3. Fold Changes in the OrrDs Mutant Lines Compared with the Wild Type for Probe Sets That Report Expression of Genes That Are Potentially Implicated or Change in Ripening.
Supplemental Table 4. List of Oligonucleotides Used in This Study.
Supplemental Table 5. Expression of Nuclear Genes Putatively Encoding Ndh Complex Subunits and PTOX during Tomato Fruit Development.
Supplemental Data Set 1. Comparison of Genes That Were Altered in Alba et al. (2005) to Their Alterations in This Experiment.
Supplemental Data Set 2. Detailed Results of the Tomato Array Experiments.
Supplemental Methods.
Acknowledgments
We thank Tali Mandel, Michal Mintz, and Lior Tal for their valuable help. We are grateful to members of the Aharoni, Levy, and Wolf laboratories for useful discussions, Ester Feldmesser for assistance with array analysis, Merav Yativ for the carotenoid analysis, and Dani Zamir and Tzili Pleban for their assistance with mapping the ORR locus. A.A. is an incumbent of the Adolfo and Evelyn Blum Career Development Chair. A.A.L. is an incumbent of the Gilbert de Botton chair in plant sciences. Research in the A.A. laboratory was supported by a grant from the Israel Ministry of Science (IMOS Project 3-2552) and the Y. Leon Benoziyo Institute for Molecular Medicine. Research in the A.A.L. laboratory was supported by the German Federal Ministry of Education and Research within the framework of German-Israeli Project Cooperation (DIP Grant E.3.1). Research in the J.H. laboratory was supported by Israel Science Foundation Grant 548/05.
References
- Adams-Phillips L., Barry C., Giovannoni J. (2004). Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G.B., Tanksley S.D., Giovannoni J.J. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J., White W.S., Chen L., Bae H., Rodermel S. (2004). The GHOST terminal oxidase regulates developmental programming in tomato fruit. Plant Cell Environ. 27: 840–852 [Google Scholar]
- Bathgate B., Purton M.E., Grierson D., Goodenough P.W. (1985). Plastid changes during the conversion of chloroplasts to chromoplasts in ripening tomatoes. Planta 165: 197–204 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188 [Google Scholar]
- Bino R.J., Ric de Vos C.H., Lieberman M., Hall R.D., Bovy A., Jonker H.H., Tikunov Y., Lommen A., Moco S., Levin I. (2005). The light-hyperresponsive high pigment-2dg mutation of tomato: Alterations in the fruit metabolome. New Phytol. 166: 427–438 [DOI] [PubMed] [Google Scholar]
- Burrows P.A., Sazanov L.A., Svab Z., Maliga P., Nixon P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley P.M., Teulières C., Blain I., Bird C., Schuch W. (1992). Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through expression of antisense RNA to pTOM5. Plant J. 2: 343–349 [Google Scholar]
- Buch K., Stransky H., Hager A. (1996). FAD is a further essential cofactor of the NAD(P)H and O2-dependent zeaxanthin-epoxidase. FEBS Lett. 27: 45–48 [DOI] [PubMed] [Google Scholar]
- Bukhov N.G., Carpentier R. (2004). Alternative photosystem I-driven electron transport routes: Mechanisms and functions. Photosynth. Res. 82: 17–33 [DOI] [PubMed] [Google Scholar]
- Carol P., Kuntz M. (2001). A plastid terminal oxidase comes to light: Implications for carotenoid biosynthesis and chlororespiration. Trends Plant Sci. 6: 31–36 [DOI] [PubMed] [Google Scholar]
- Carrari F., Baxter C., Usadel B., Urbanczyk-Wochniak E., Zanor M.I., Nunes-Nesi A., Nikiforova V., Centero D., Ratzka A., Pauly M., Sweetlove L.J., Fernie A.R. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson P.J., Kiano J.W., Shipton C.A., Fraser P.D., Romer S., Schuch W., Bramley P.M., Pyke K.A. (2003). Increases in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta 217: 896–903 [DOI] [PubMed] [Google Scholar]
- Corneille S., Cournac L., Guedeney G., Havaux M., Peltier G. (1998). Reduction of the plastoquinone pool by exogenous NADH and NADPH in higher plant chloroplasts. Characterization of a NAD(P)H-plastoquinone oxidoreductase activity. Biochim. Biophys. Acta 1363: 59–69 [DOI] [PubMed] [Google Scholar]
- DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schunemann D., Finazzi G., Joliot P., Barbato R., Leister D. (2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Davuluri G.R., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23: 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C., Mus F., Cuiné S., Billon E., Cournac L., Peltier G. (2009). Characterization of Nda2, a plastoquinone-reducing Type II NAD(P) dehydrogenase in Chlamydomonas chloroplasts. J. Biol. Chem. 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Deikman J., Fischer R.L. (1998). Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 7: 3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Ishida S., Ishikawa N., Sato F. (2008). Chloroplastic NAD(P)H dehydrogenase complex and cyclic electron transport around photosystem I. Mol. Cells 25: 158–162 [PubMed] [Google Scholar]
- Enfissi E.M., Barneche F., Ahmed I., Lichtle C., Gerrish C., McQuinn R.P., Giovannoni J.J., Lopez-Juez E., Bowler C., Bramley P.M., Fraser P.D. (2010). Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A., Willmitzer L. (2004). Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Enfissi E.M., Bramley P.M. (2009). Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 15: 196–204 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Pinto M.E.S., Holloway D.E., Bramley P.M. (2000). Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Galpaz N., Wang Q., Menda N., Zamir D., Hirschberg J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Bläsing O.E., Palacios-Rojas N., Pankovic D., Hendriks J.H., Fisahn J., Höhne M., Günther M., Stitt M. (2004). Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y., Larher F. (1997). Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 251: 153–157 [DOI] [PubMed] [Google Scholar]
- Guera A., Sabater B. (2002). Changes in the protein and activity levels of the plastid NADH-plastoquinone-oxidoreductase complex during fruit development. Plant Physiol. Biochem. 40: 423–442 [Google Scholar]
- Hirschberg J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Horváth E.M., Peter S.O., Joët T., Rumeau D., Cournac L., Horváth G.V., Kavanagh T.A., Schäfer C., Peltier G., Medgyesy P. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. (2008). Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P., Romer S., Kuntz M., Camara B. (1992). Characterization and molecular cloning of a flavoprotein catalyzing the synthesis of phytofluene and ζ-carotene in Capsicum chromoplasts. Eur. J. Biochem. 209: 399–407 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T., Ohad I., Beyer P., Hirschberg J. (2004). Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol. 136: 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Satoh F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa N., Endo T., Sato F. (2008). Electron transport activities of Arabidopsis thaliana mutants with impaired chloroplastic NAD(P)H dehydrogenase. J. Plant Res. 121: 521–526 [DOI] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Johnson G.N. (2005). Cyclic electron transport in C3 plants: Fact or artefact? J. Exp. Bot. 56: 407–416 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Simkin A.J., Gaffe J., Laboure A.M., Kuntz M., Carol P. (2000). A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 123: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Lennon A.M., Prommeenate P., Nixon P.J. (2003). Location, expression and orientation of the putative chlororespiratory enzymes, Ndh and IMMUTANS, in higher-plant plastids. Planta 218: 254–260 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Matsumara H., Miyachi S. (1980). Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol. 69: 465–470 [Google Scholar]
- Mayer M.P., Beyer P., Kleinig H. (1990). Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur. J. Biochem. 191: 359–363 [DOI] [PubMed] [Google Scholar]
- Meissner R., Chague V., Zhu Q., Emmanuel E., Elkind Y., Levy A.A. (2000). Technical advance: A high throughput system for transposon tagging and promoter trapping in tomato. Plant J. 22: 265–274 [DOI] [PubMed] [Google Scholar]
- Meissner R., Jacobson Y., Melamed S., Levyatuv S., Shalev G., Ashri A., Elkind Y., Levy A.A. (1997). A new model system for tomato genetics. Plant J. 12: 1465–1472 [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., Wang Z., Jetter R., Venger I., Adato A., Aharoni A. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L.A., et al. (2005). The SOL Genomics Network: A comparative resource for Solanaceae biology and beyond. Plant Physiol. 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nakatsuka A., Murachi S., Okunishi H., Shiomi S., Nakano R., Kubo Y., Inaba A. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievelstein V.J., Vandekerckove J., Tadros M.H., vonLintig J., Nitschke W., Beyer P. (1995). Carotene desaturation is linked to a respiratory redox pathway in Narcissus pseudonarcissus chromoplast membranes. Involvement of a 23-kDa oxygen-evolving-complex-like protein. Eur. J. Biochem. 233: 864–872 [DOI] [PubMed] [Google Scholar]
- Nixon P.J. (2000). Chlororespiration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1541–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Barrette T.R., DellaPenna D. (1995). Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Takami T., Shikanaï T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCL in Arabidopsis. Plant Cell 21: 3623–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G., Cournac L. (2002). Chlororespiration. Annu. Rev. Plant Biol. 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. (2003). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8: 33–41 [DOI] [PubMed] [Google Scholar]
- Picton S., Gray J., Barton S., AbuBakar U., Lowe A., Grierson D. (1993). cDNA cloning and characterisation of novel ripening-related mRNAs with altered patterns of accumulation in the ripening inhibitor (rin) tomato ripening mutant. Plant Mol. Biol. 23: 193–207 [DOI] [PubMed] [Google Scholar]
- Rick C.M., Butler L. (1956). Cytogenetics of the tomato. Adv. Genet. 8: 267–382 [Google Scholar]
- Rodermel S. (2001). Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 6: 471–478 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G.N., Nijkamp H.J., Hille J. (1994). Ds read-out transcription in transgenic tomato plants. Mol. Gen. Genet. 243: 426–433 [DOI] [PubMed] [Google Scholar]
- Rumeau D., Bécuwe-Linka N., Beyly A., Louwagie M., Garin J., Peltier G. (2005). New subunits NDH-M, N and O encoded by nuclear genes are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sandmann G., Malkin R. (1984). Light inhibition of respiration is dueto a dual function of the cytochrome b6/f complex and the plastocyanin/cytochrome c-553 pool. Arch. Biochem. Biophys. 234: 105–111 [DOI] [PubMed] [Google Scholar]
- Sazanov L.A., Burrows P.A., Nixon P.J. (1998a). The chloroplast Ndh complex mediates the dark reduction of the plastoquinone pool in response to heat stress in tobacco leaves. FEBS Lett. 429: 115–118 [DOI] [PubMed] [Google Scholar]
- Sazanov L.A., Burrows P.A., Nixon P.J. (1998b). The plastid ndh genes code for an NADH-specific dehydrogenase: Isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. USA 95: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Scheible R., Stitt M. (1988). Comparison of NADP-malate dehydrogenase activation QA reduction and O2 evolution in spinach leaves. Plant Physiol. Biochem. 26: 473–481 [Google Scholar]
- Schnurr G., Misawa N., Sandmann G. (1996). Expression, purification and properties of lycopene cyclase from Erwinia uredovora. Biochem. J. 315: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M., Gilbert M., Laboure A.M., Kuntz M. (2007). Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 145: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanaï T. (2007a). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanaï T. (2007b). The NAD(P)H dehydrogenase complex in photosynthetic organisms: Subunit composition and physiological function. Funct. Plant Sci. Biotechnol. 1: 129–137 [Google Scholar]
- Shikanaï T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Shikanai T. (2007). Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52: 539–547 [DOI] [PubMed] [Google Scholar]
- Sirpiö S., Allahverdiyeva Y., Holmström M., Khrouchtchova A., Haldrup A., Battchikova N., Aro E.-M. (2009). Novel nuclear encoded subunits of the chloroplast NAD(PH dehydrogenase complex. J. Biol. Chem. 284: 905–912 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Steinbrenner J., Linden H. (2003). Light induction of carotenoid biosynthesis genes in the green alga Haemotococcus pluvialis: Regulation by photosynthetic redox control. Plant Mol. Biol. 52: 343–356 [DOI] [PubMed] [Google Scholar]
- Suorsa M., Sand S., Aro E.-M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2: 1127–1140 [DOI] [PubMed] [Google Scholar]
- Takabayashi A., Ishikawa N., Obayashi T., Ishida S., Obokata J., Endo T., Sato F. (2009). Three novel subunits of the chloroplastic NAD(P)H dehydrogenase identified by bioinformatics and reverse genetic approaches. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Tiessen A., Hendriks J.H.M., Stitt M., Branscheid A., Gibon Y., Farre E.M., Geigenberger P. (2002). Starch synthesis in potato tubers is regulated byposttranslational redox modification of ADP-glucose pyrophosphorylase: A novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E., et al. (2006). Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: Effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol. Biol. 60: 773–792 [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T.C., Dekker B.M.M., Hoekema A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Woitsch S., Romer S. (2003). Expression of xanthophyll biosynthetic genes during light-dependent chloroplast differentiation. Plant Physiol. 132: 1508–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wright D.A., Wetzel C., Voytas D.F., Rodermel S. (1999). The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Schaub P., Ghisla S., Al-Babili S., Krieger-Liszkay A., Beyer P. (2010). The lycopene cyclase CrtY from Pantoea ananatis (formerly Erwinia uredovora) catalyzes an FADred-dependent non-redox reaction. J. Biol. Chem. 285: 12109–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata J.M., Guera A., Esteban-Carrasco A., Martin M., Sabater B. (2005). Chloroplasts regulate leaf senescence: delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ. 12: 1277–1284 [DOI] [PubMed] [Google Scholar]
- Zhang H., Whitelegge J.P., Cramer W.A. (2001). Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 276: 38159–38165 [DOI] [PubMed] [Google Scholar]