Floral scent and color play major roles in the plant's life cycle. Using petunia as a model system, a MYB-like factor was identified that transcriptionally regulates floral scent but not pigmentation. The multilayered regulation allows efficient control of metabolic flux in the phenylpropanoid pathway.
Abstract
Floral scent, which is determined by a complex mixture of low molecular weight volatile molecules, plays a major role in the plant's life cycle. Phenylpropanoid volatiles are the main determinants of floral scent in petunia (Petunia hybrida). A screen using virus-induced gene silencing for regulators of scent production in petunia flowers yielded a novel R2R3-MYB–like regulatory factor of phenylpropanoid volatile biosynthesis, EMISSION OF BENZENOIDS II (EOBII). This factor was localized to the nucleus and its expression was found to be flower specific and temporally and spatially associated with scent production/emission. Suppression of EOBII expression led to significant reduction in the levels of volatiles accumulating in and emitted by flowers, such as benzaldehyde, phenylethyl alcohol, benzylbenzoate, and isoeugenol. Up/downregulation of EOBII affected transcript levels of several biosynthetic floral scent-related genes encoding enzymes from the phenylpropanoid pathway that are directly involved in the production of these volatiles and enzymes from the shikimate pathway that determine substrate availability. Due to its coordinated wide-ranging effect on the production of floral volatiles, and its lack of effect on anthocyanin production, a central regulatory role is proposed for EOBII in the biosynthesis of phenylpropanoid volatiles.
INTRODUCTION
The survival of many plant species depends on their ability to attract pollinators to their flowers at anthesis (Pichersky and Gershenzon, 2002; Gang, 2005). To this end, various strategies have been developed by plants, involving flower architecture, color, and the production of nectar and scent, characters that determine the pollinator(s) for each species (Bradshaw and Schemske, 2003; Hoballah et al., 2007).
Flower scent is a highly dynamic trait that, in many instances, is rhythmic and coordinated with the diurnal activity of the pollinator (Dudareva and Pichersky, 2006). It is a composite trait, assembled from a variety of compounds (Dudareva and Negre, 2005; Knudsen et al., 2006; Pichersky et al., 2006; Schuurink et al., 2006). A particular flower's aroma is usually a compilation of dozens to hundreds of different molecules from diverse biosynthetic pathways, including phenylpropanoids, terpenes, and fatty acid derivatives (Gang, 2005; Knudsen et al., 2006; Gershenzon and Dudareva, 2007). Volatile phenylpropanoids derive from the highly diverse general phenylpropanoid pathway that starts with Phe metabolism via, for example, the action of l-phenylalanine ammonia lyase (PAL) (Boatright et al., 2004; Figure 1), phenylacetaldehyde synthase (PAAS) (Kaminaga et al., 2006), and aromatic amino acid decarboxylase (Tieman et al., 2006). Moreover, numerous essential substances, such as flavonoids, lignin, and other mono- and polyphenolic compounds, derive from this metabolic pathway (Boatright et al., 2004).
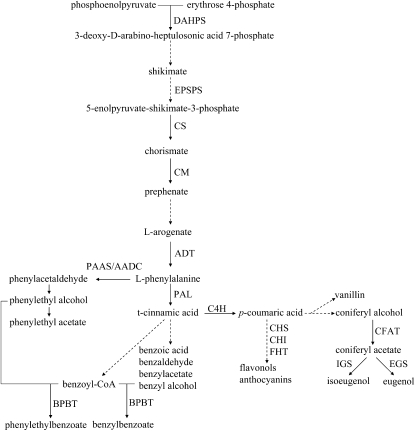
Figure 1.
Schematic Diagram of the Shikimate and Phenylpropanoid Pathways in Plants.
Continuous arrows represent a one-step enzymatic reaction, and dashed arrows represent several enzymatic reactions or as yet undescribed steps. DAHPS, 3-deoxy-d-arabino-heptulosonic acid 7-phosphate synthase; EPSPS, 5-enolpyruvate-shikimate-3-phosphate synthase; ADT/PDT, arogenate dehydratase/prephenate dehydratase; AADC, aromatic amino acid decarboxylase; C4H, cinnamic acid 4-hydroxylase; CHS, chalcone synthase; CHI, chalcone isomerase; CFAT, coniferyl alcohol acetyltransferase; IGS, isoeugenol synthase; EGS, eugenol synthase; BPBT, benzoyl-CoA:benzyl alcohol/2-phenylethanol benzoyltransferase.
Structural genes encoding enzymes responsible for the synthesis of several floral volatile phenylpropanoids via acetylation, methylation, decarboxylation, etc., have been identified and characterized (Dudareva et al., 2004). In contrast with the rapid progress in recent years in characterizing and mapping the reactions leading to the formation of volatile phenylpropanoids (Pichersky and Dudareva, 2007; Dudareva and Pichersky, 2008), very little is known about regulation at the molecular level of floral volatile phenylpropanoid production. To date, ODORANT1 (ODO1) is the only transcription factor that has been characterized as a regulator of scent production in flowers (Verdonk et al., 2005). ODO1 belongs to the MYB transcription factor family, and its suppression in petunia (Petunia hybrida) leads to decreased levels of emitted volatile phenylpropanoids (Verdonk et al., 2005).
Petunia (family: Solanaceae) possesses large flowers that produce a wealth of volatiles from diverse biochemical pathways/branches; it is one of only a handful of model plants that is commonly used for scent studies (Channeliere et al., 2002; Dudareva et al., 2004; van Schie et al., 2006; Dexter et al., 2008). Petunia is amenable to numerous high-throughput approaches for the characterization of DNA sequences, including the frequently used reverse-genetics approach of virus-induced gene silencing (VIGS) (Quattrocchio et al., 1998; Chen et al., 2004; Verdonk et al., 2005; Spitzer et al., 2007). Using Tobacco rattle virus (TRV) as the vector for VIGS, we recently showed the applicability of this approach to analyses of floral scent–related genes, including transcription factors, in petunia lines P720 and B1 (Spitzer et al., 2007). Suppression of the anthocyanin pathway via chalcone synthase (CHS) silencing was used as the reporter, allowing easy visual identification of anthocyanin-less silenced flowers/tissues with no effect on the level of volatile emissions. The reliability of this system for scent studies was apparent from the consistency of the VIGS phenotypes relative to those previously described following RNA interference (RNAi) silencing of the respective genes, as was exemplified with BSMT (encoding SAM:benzoic acid/salicylic acid carboxyl methyltransferase), PAAS, and ODO1 (Spitzer et al., 2007). Here, we present the isolation and characterization of a novel MYB-like factor, EMISSION OF BENZENOIDS II (EOBII), regulating scent production in petunia flowers. Since EOBII is flower specific, and since other traits, such as flower pigmentation and architecture, were not affected in flowers with suppressed EOBII, we suggest that this factor is a critical regulator of the machinery responsible for floral fragrance.
RESULTS
Isolation of EOBII
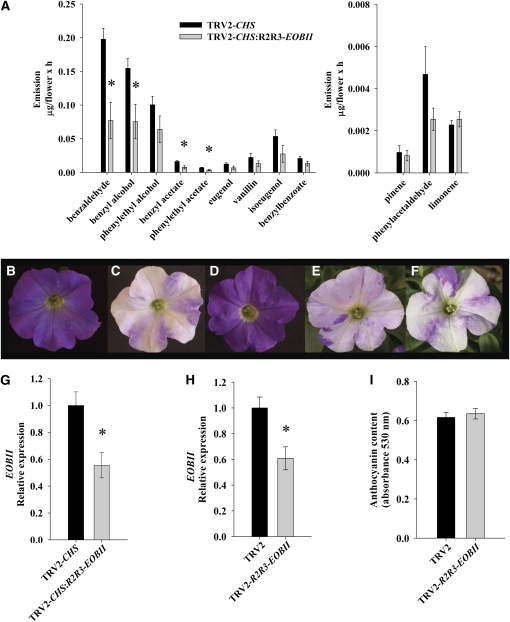
To generate a collection of putative regulatory factors from petunia flowers (P. hybrida line P720), we used degenerate primers to amplify, among others, the domains R2R3, bHLH, RWD, WRKY, and EPF, which are present in transcription factors belonging to the R2R3-Myb, basic helix-loop-helix, WD40, and zinc-finger protein families. As a template for PCR, we used reverse-transcribed RNA prepared from petunia corollas 1 and 2 d postanthesis (dpa), when scent production is elevated. This collection of sequenced PCR fragments was then screened for their relevance to scent production, using a VIGS system based on the TRV2-CHS vector, which enables easy identification of silenced tissues (Spitzer et al., 2007). One of the sequences, termed R2R3-EOBII, containing SANT/MYB DNA binding domains indicative of MYB-like transcription factors, was chosen for further analyses. Infection of line P720 petunia plants (volatile compounds of line P720 are shown in Supplemental Table 1 online) with the TRV2-CHS vector fused to R2R3-EOBII (TRV2-CHS:R2R3-EOBII) led to a significant reduction in the levels of several flower-emitted volatile phenylpropanoids relative to plants infected with control TRV2-CHS empty vector (Figure 2A). The levels of the emitted monoterpene volatiles limonene and pinene, as well as other floral characteristics, such as corolla shape, size, and color, were similar in TRV2-CHS– and TRV2-CHS:R2R3-EOBII–infected plants (Figures 2A, 2C, and 2F). Suppression of EOBII in the corollas of TRV2-CHS:R2R3-EOBII–infected petunia plants compared with those in control TRV2-CHS–infected plants was verified by real-time RT-PCR (Figure 2G). EOBII suppression resulting from infection with the CHS-lacking vector TRV2-R2R3-EOBII compared with infection with CHS-lacking TRV2 (Figure 2H) also did not affect floral shape or anthocyanin content (Figures 2B, 2D, and 2I).
Figure 2.
Suppression of Volatile Emission in Flowers of Petunia Plants Infected with TRV2-CHS:R2R3-EOBII.
(A) Dynamic headspace analyses, followed by GC-MS, were performed from 17 h00 to 09 h00. Flowers of plants infected with TRV2-CHS were used as controls. Graphs represent the average of four to seven independent experiments with standard errors indicated by vertical lines. Bars with asterisks are significantly different (P ≤ 0.05) from their corresponding controls. The significance of differences between treatment and control samples (asterisks) was calculated using Student's t test.
(B) to (F) Flowers of petunia line P720 infected with TRV2, TRV2-CHS, TRV2-R2R3-EOBII, TRV2-CHS:EOBII, and TRV2-CHS:R2R3-EOBII, respectively.
(G) and (H) Quantitative real-time PCR analysis of EOBII transcript levels in petunia corollas infected with TRV2-CHS and TRV2-CHS:R2R3-EOBII (G) or with TRV2 and TRV2-R2R3-EOBII (H). Presented data were normalized to that from TRV2-CHS– or TRV2-infected corollas with standard errors indicated by vertical lines. Significance of differences (P ≤ 0.05; n = 4) between treatments (asterisks) was calculated (Student's t test) based on the raw transcript levels' data normalized to Actin.
(I) Anthocyanin levels were determined in corollas of TRV2-R2R3-EOBII– and control TRV2-infected petunia. Each graph represents the average of five independent experiments; se is indicated by vertical lines.
[See online article for color version of this figure.]
Involvement of EOBII in Volatile Phenylpropanoid Production
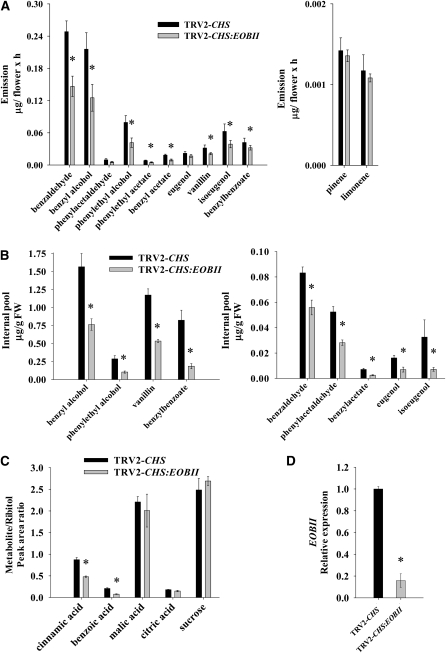
The full-length sequence of EOBII was cloned (see Methods) and used to construct a TRV2 vector containing 205 bp of the EOBII 3′ untranslated region (TRV2-CHS:EOBII). The untranslated region was harnessed to allow higher specificity of VIGS-derived suppression of EOBII in petunia corollas. Infection of plants with the TRV2-CHS:EOBII construct, compared with empty TRV2-CHS, had no effect on flower shape or size (Figures 2C and 2E) but led to a strong decrease in the levels of several volatile phenylpropanoids emitted from corollas (Figure 3A), similar to that which had been found in the TRV2-CHS:R2R3-EOBII–infected plants (Figure 2A). Suppression of EOBII in corollas of plants infected with TRV2-CHS:EOBII was confirmed using real-time RT-PCR. As shown in Figure 3D, the EOBII transcript levels in corollas of TRV2-CHS:EOBII–infected plants was ∼15% of that in control, TRV2-CHS–infected plants. Note that we did not detect any reduction in the levels of MYB-like transcript, which shows high similarity to EOBII (82% identity in nucleotide sequences), in TRV2-CHS:EOBII–silenced plants (see Supplemental Figure 1A online).
Figure 3.
Suppression of EOBII Expression Reduces Levels of Volatile Phenylpropanoids in Petunia Flowers.
(A) Dynamic headspace analyses, followed by GC-MS, were performed (from 17 h00 to 09 h00) on TRV2-CHS:EOBII–infected flowers. Flowers of plants infected with TRV2-CHS were used as controls. Graphs represent the average of seven to nine independent experiments with standard errors indicated by vertical lines. Bars with asterisks are significantly different (P ≤ 0.05) from their corresponding controls. The significance of differences between treatment and control samples (asterisks) was calculated using Student's t test.
(B) Internal pools of volatile compounds accumulated in corollas 1 dpa (collected at 07 h00) of EOBII-silenced plants compared with corollas of TRV2-CHS–infected plants. Columns represent the mean values of three to five independent experiments. Standard errors are indicated by vertical lines. Bars with asterisks are significantly different (P ≤ 0.05).
(C) Levels of metabolites collected from EOBII-silenced and TRV2-CHS–infected corolla limbs at anthesis (collected at 07 h00) as determined by GC-MS analysis of samples derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide using ribitol as an internal standard. Columns represent the mean values of three independent experiments. Standard errors are indicated by vertical lines. Bars with asterisks are significantly different (P ≤ 0.05) from their corresponding controls.
(D) Quantitative real-time PCR analysis was used to determine EOBII transcript levels in petunia flowers infected with TRV2-CHS and TRV2-CHS:EOBII. Presented data were normalized to that from TRV2-CHS–infected corollas, with standard errors indicated by vertical lines. Significance of differences (P ≤ 0.05; n = 3) between treatments (asterisks) was calculated (Student's t test) based on the raw transcript levels' data normalized to Actin.
To further detail the effect of EOBII suppression on corolla-produced phenylpropanoid volatiles, their internal pools were analyzed in TRV2-CHS:EOBII–silenced petunia flowers and compared with those in TRV2-CHS–silenced ones. The internal pools of phenylpropanoid volatiles that could be detected in TRV2-CHS:EOBII–infected plants, namely, benzyl alcohol, phenylethyl alcohol, vanillin, benzylbenzoate, benzaldehyde, phenylacetaldehyde, benzylacetate, eugenol, and isoeugenol, were significantly lower than in control TRV2-CHS–infected flowers (Figure 3B), indicating EOBII's involvement in the production of these phenylpropanoid volatiles in petunia corollas. Analyses of the levels of cinnamic and benzoic acid, representing key branch points in the production of volatile phenylpropanoids, revealed a significant reduction in the TRV2-CHS:EOBII corollas compared with TRV2-CHS ones (Figure 3C). Levels of the primary metabolism compounds malic acid, citric acid, and sucrose were unaffected in corollas of plants infected with TRV2-CHS:EOBII versus TRV2-CHS (Figure 3C).
Temporal and Spatial Expression Profiles of EOBII
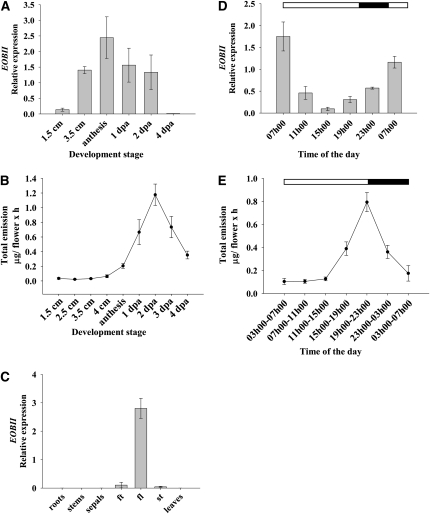
To characterize EOBII expression, its transcript levels were analyzed in petunia flowers at different stages of development, at various time points during the day and night, and in different organs and tissues using real-time RT-PCR. EOBII transcript levels in corollas increased in parallel to flower development from the young 1.5-cm bud to flowers at anthesis, and it was almost undetectable in senescing flowers at 4 dpa (Figure 4A). Transcript levels of genes involved in the biosynthesis of floral scent compounds (e.g., CM, PAL2, BPBT, CFAT, and IGS) also increased in parallel to flower development (see Supplemental Figures 2A, 2C, 2E, 2G, and 2I online). Gas chromatography–mass spectrometry (GC-MS) headspace analyses of the emission patterns of volatile phenylpropanoids during flower development revealed an increase in their levels in parallel to flower development as well, peaking at 2 dpa and then dropping in senescing flowers (Figure 4B).
Figure 4.
Spatial, Temporal, and Developmental Regulation of EOBII Transcript Levels and Scent Emission in Petunia Corollas.
The RNA was extracted from corollas at different development stages (A), from different organs (C), and from corollas at different time points during the day/night (D). EOBII transcript levels were determined by quantitative real-time PCR analysis using Actin as the reference. Graphs represent the average of three independent experiments, with standard errors indicated by vertical lines. ft, flower tube; fl, flower limb; st, stamen. Dynamic headspace analyses, followed by GC-MS, were performed (from 17 h00 to 09 h00) with flowers at different development stages (B) and with flowers (2 dpa) at different time points throughout the day/night (E). Black part of horizontal bar indicates nighttime hours. Each time point represents the average of three to five independent experiments, with standard errors indicated by vertical lines.
Analysis of EOBII as well as of biosynthetic floral scent–related transcript levels in different organs further supported the involvement of EOBII in floral scent production. Expression of EOBII was flower specific: no or only negligible levels of its transcript were detected in the roots, stems, leaves, and sepals. A comparison of EOBII expression levels in various floral organs revealed its highest transcript level in flower limbs and much lower levels in the stamen and flower tube (Figure 4C). Tissue-specific expression patterns of the floral scent–related genes CM, PAL2, BPBT, CFAT, and IGS were essentially identical to that of EOBII (see Supplemental Figures 2B, 2D, 2F, 2H, and 2J online). The diurnal expression pattern of EOBII was found to be rhythmic, with transcript levels being highest in the early morning hours and lowest during the late afternoon hours (Figure 4D). Analysis of the pattern of floral scent emission during the day/night hours showed that volatile emission peaks at night (Figure 4E).
EOBII Belongs to the Family of R2R3-MYB Transcription Factors
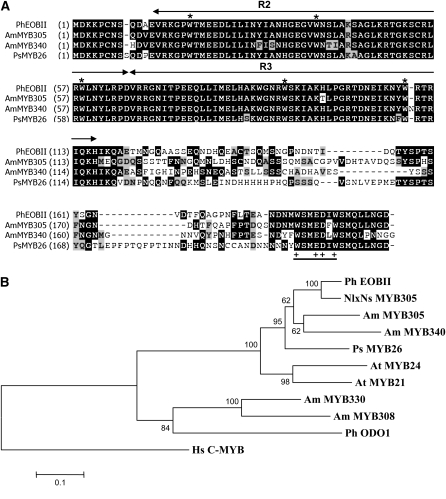
EOBII cDNA contains an open reading frame (ORF) encoding a 197–amino acid protein (Figure 5A) with a conserved R2R3 binding domain (important for interaction with promoter elements) (Kranz et al., 1998) near its N terminus and a W/Y-MDDIW motif (of major importance for transactivation; Li et al., 2006) at its C terminus (Figure 5A). Based on the motifs outside of the DNA binding R2R3 domain, EOBII can be classified into subgroup 19 of the R2R3-MYB transcription factors based on Kranz et al. (1998). Phylogenetically, EOBII is rather distant from Ph ODO1 (which belongs to a separate subgroup), while it shares high homology with subgroup 19 members from other plants, such as Ps MYB26, NlxNs MYB305, Am MYB305, Am MYB340, At MYB24, and At MYB21 (Figure 5B; see Supplemental Data Set 1). These members have been shown to be predominantly expressed in their respective flowers and involved in regulation of the phenylpropanoid pathway (Sablowski et al., 1994; Moyano et al., 1996; Uimari and Strommer, 1997; Shin et al., 2002; Li et al., 2006; Liu et al., 2009). The latter five genes have also been shown to activate PAL expression ectopically. To test whether EOBII also upregulates PAL, we expressed the EOBII gene in tobacco (Nicotiana tabacum) (see Supplemental Figure 3A online). Real-time RT-PCR analyses revealed a strong (1.5- to 2.5-fold) increase in PALB transcript levels in tobacco flowers ectopically expressing EOBII (see Supplemental Figures 3B and 3D online). Levels of transcript coding for PALA, CHS, chalcone isomerase (CHI), and flavanone 3-hydroxylase (FHT), as well as anthocyanin levels, were not affected in these tobacco flowers (see Supplemental Figures 3C and 3E to 3H online). Levels of volatiles emitted by tobacco flowers were also not affected by ectopic expression of EOBII (see Supplemental Figure 3I online). Note, however, that terpenes, and not phenylpropanoid volatiles, are emitted by tobacco flowers.
Figure 5.
EOBII Belongs to Subgroup 19 of the MYB Proteins.
(A) Sequence alignment of P. hybrida EOBII (Ph OBII), A. majus MYB305 (Am MYB305), A. majus MYB340 (Am MYB340), and Pisum sativum MYB26 (Ps MYB26). Identical amino acids are shaded in black and conserved changes in gray. The two MYB repeats (R2 and R3) are indicated with sets of arrows, and the critical Trp residues are indicated by asterisks. The W/Y-MDDIW motif region is underlined (+ indicates conserved amino acids). Numbers in parentheses indicate amino acid positions.
(B) Phylogenetic tree displaying the similarity of EOBII to other R2R3-MYB proteins. The dendogram was constructed using MEGA4 software with the neighbor-joining algorithm; unrelated human (Hs) C-MYB protein was used as the outgroup. Bootstrap values are indicated at branch nodes, and the scale bar indicates the number of amino acid substitutions per site. Names of proteins are given in uppercase letters, and their origin is indicated by a two letter prefix: Am, Antirrhinum majus; Ph, Petunia hybrida; NlxNs, Nicotiana langsdorffii × Nicotiana sanderae; Ps, Pisum sativum; At, Arabidopsis thaliana. GenBank accession numbers are given in Methods.
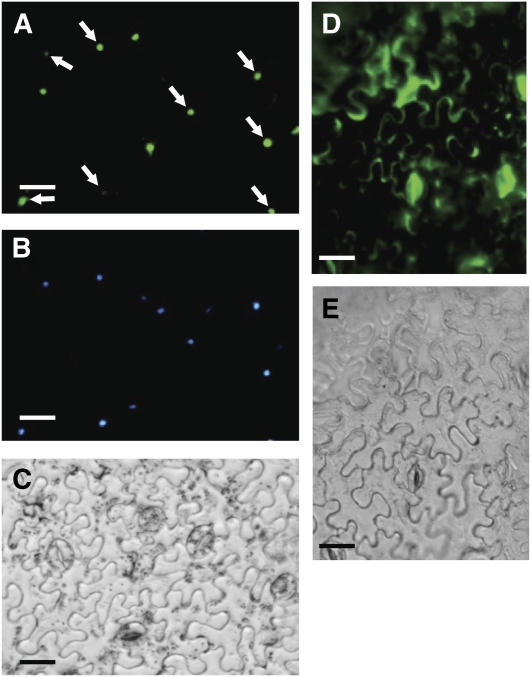
To establish EOBII's nuclear localization, we transiently expressed an EOBII:GFP (green fluorescent protein) fusion protein in leaves and followed its compartmentalization within the cell. Microscopy analysis localized the GFP fluorescence to the nucleus (Figures 6A to 6C), while a GFP control (not fused to EOBII) accumulated in the cytosol, as expected (Figures 6D and 6E).
Figure 6.
Nuclear Localization of EOBII.
(A) Localization of the EOBII:GFP fusion protein in the leaf epidermis.
(B) Nucleus stained with 4',6-diamidino-2-phenylindole.
(C) Bright field.
(D) and (E) Fluorescence of nonfused GFP used as a control and bright field of leaf epidermis, respectively. Arrows indicate nuclei with 4',6-diamidino-2-phenylindole and GFP cofluorescence.
Bars = 25 μm.
[See online article for color version of this figure.]
EOBII Is Required for the Expression of Genes Involved in Floral Phenylpropanoid Scent Production
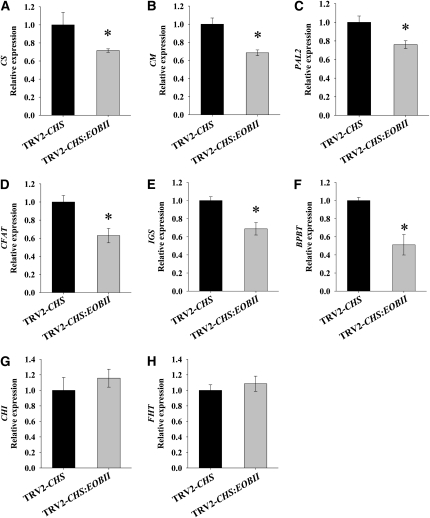
To further detail the effect of EOBII silencing on scent production, we analyzed the expression levels of genes involved in floral scent production in TRV2-CHS:EOBII–suppressed petunia flowers compared with control TRV2-CHS–suppressed flowers. As can be seen from Figures 7A and 7B, the level of transcripts encoding the shikimate pathway biosynthetic enzymes chorismate synthase (CS) and chorismate mutase (CM), respectively, was significantly downregulated in EOBII-silenced corollas compared with controls silenced with the empty vector. The level of PAL2 transcript, representing the first committed step in the phenylpropanoid pathway, was also significantly downregulated in EOBII-suppressed flowers (Figure 7C). The levels of transcripts encoding enzymes that direct the production of the specific phenylpropanoid scent volatiles CFAT, IGS, and BPBT were also significantly lower in EOBII-silenced flowers than in controls (Figures 7D to 7F). On the other hand, accumulation of transcripts coding for 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase, 5-enol-pyruvylshikimate-3-phosphate synthase, PAL1, and PAAS were not affected by EOBII suppression (see Supplemental Figures 1B to 1E online). The levels of CHI and FHT transcripts, coding for key enzymes in the early segment of the flavonoid biosynthetic pathway, were also similar in both control and EOBII-suppressed petunia corollas (Figures 7G and 7H), further supporting the specificity of EOBII to volatile phenylpropanoid formation. Interestingly, the level of transcript encoding ODO1 was significantly downregulated in EOBII-suppressed flowers (Figures 8A and 8B). By contrast, in analyses of petunia (cv Mitchell; W115) flowers with RNAi-suppressed ODO1 expression (Verdonk et al., 2005), EOBII transcript level was not affected: its level was similar in ODO1-suppressed and control nontransgenic cv Mitchell flowers (Figures 8C and 8D).
Figure 7.
EOBII Silencing Leads to Downregulation of the Expression of Genes in the Shikimate and Phenylpropanoid Pathways.
Quantitative real-time PCR analysis of CS (A), CM (B), PAL2 (C), CFAT (D), IGS (E), BPBT (F), CHI (G), and FHT (H) transcript levels in EOBII-silenced petunia corollas compared with corollas infected with TRV2-CHS. Samples were collected from corollas 1 dpa at 08 h00. Presented data were normalized to that from TRV2-CHS–infected corollas, with standard errors indicated by vertical lines. Significance of differences (P ≤ 0.05; n = 4) between treatments (asterisks) was calculated (Student's t test) based on the raw transcript levels' data normalized to Actin.
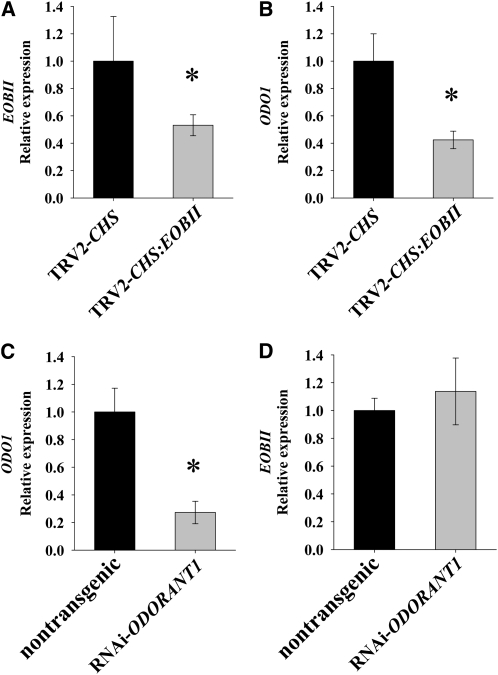
Figure 8.
Interrelationship Between EOBII and ODO1.
(A) and (B) EOBII silencing leads to downregulation of ODO1 expression. Quantitative real-time PCR analysis of EOBII (A) and ODO1 (B) transcript levels in EOBII-silenced petunia corollas compared with corollas infected with TRV2-CHS. Samples were collected from corollas 1 dpa at 21 h00.
(C) and (D) EOBII transcript level is not affected by downregulation of ODO1. Quantitative real-time PCR analysis of ODO1 (C) and EOBII (D) transcript levels in corollas of ODO1-RNAi–suppressed petunia plants (W115) compared with those in control nontransgenic plants. Samples were collected from corollas 1 dpa at 07 h00. Presented data were normalized to control corollas, with standard errors indicated by vertical lines. Significance of differences (P ≤ 0.05; n = 6) between treatments (asterisks) was calculated (Student's t test) based on the raw transcript levels' data normalized to Actin.
Overexpression of EOBII Upregulates Transcript Levels of Genes Involved in Floral Phenylpropanoid Scent Production
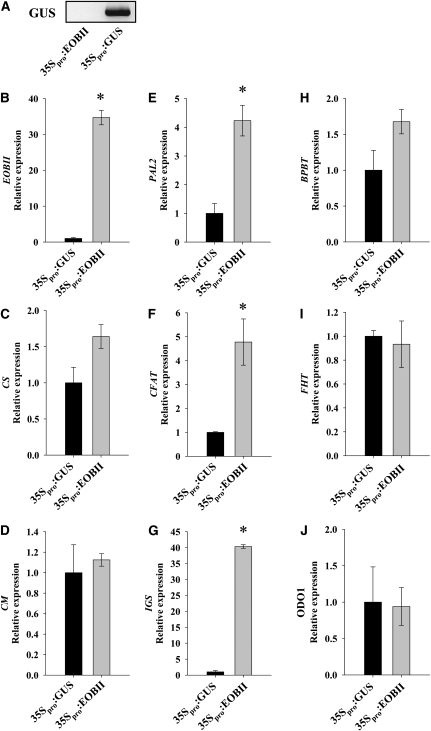
To determine EOBII's ability to upregulate scent-related genes, EOBII was transiently overexpressed in petunia buds. Analyses of 35Spro:EOBII Agrobacterium tumefaciens–infiltrated flowers relative to control 35Spro:β-glucuronidase (GUS)-infiltrated ones (at 15 h00) revealed an ∼35-fold increase in EOBII transcript levels in the former (Figures 9A and 9B). Transcript levels of IGS were most strongly affected by ectopic expression of EOBII, with a 40-fold increase in expression in EOBII-overexpressing corollas compared with controls. The levels of PAL2 and CFAT transcripts were increased approximately fourfold in EOBII-overexpressing flowers, while the levels of other analyzed genes were not significantly affected by ectopic expression of EOBII (Figures 9C to 9J).
Figure 9.
EOBII Overexpression Leads to Upregulation of Genes in the Shikimate and Phenylpropanoid Pathways.
(A) RT-PCR analysis of GUS levels in cDNA synthesized from mRNA extracted from petunia corollas 48 h after infiltration with Agrobacterium carrying the 35Spro:EOBII construct or 35Spro:GUS as a control. Ethidium bromide–stained agarose gel with the GUS product is shown.
(B) to (J) Quantitative real-time PCR analysis of EOBII, CS, CM, PAL2, CFAT, IGS, BPBT, FHT, and ODO1 transcript levels in petunia corollas 48 h after infiltration (at 15 h00) with Agrobacterium carrying the 35Spro:EOBII construct or 35Spro:GUS as a control. Presented data were normalized to that from 35Spro:GUS-infiltrated corollas, with standard errors indicated by vertical lines. Significance of differences (P ≤ 0.05; n = 3) between treatments (asterisks) was calculated (Student's t test) based on the raw transcript levels' data normalized to Actin.
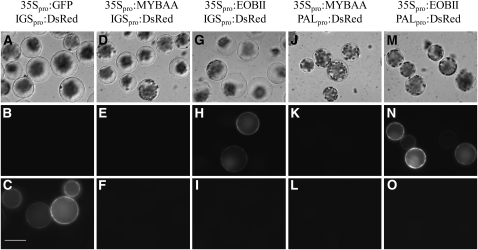
To further characterize EOBII's ability to activate the expression of early and late scent-related genes, we first generated constructs containing tobacco PALB and petunia IGS promoter sequences fused to DsRed: PALpro:DsRed and IGSpro:DsRed, respectively. Next, we cotransformed 35Spro:EOBII and PALpro:DsRed or IGSpro:DsRed constructs into protoplasts extracted from Arabidopsis leaves (Figure 10). An unrelated MYB regulatory factor (MYBAA) or GFP under the control of the 35S promoter (35Spro:MYBAA and 35Spro:GFP, respectively) were used as controls for promoter activation analyses. Only protoplasts cotransformed with 35Spro:EOBII and IGSpro:DsRed or with 35Spro:EOBII and PALpro:DsRed expressed DsRed (Figures 10H and 10N). Control protoplasts cotransformed with 35Spro:MYBAA or 35Spro:GFP instead of 35Spro:EOBII did not yield DsRed fluorescent signal (Figures 10B, 10E, and 10K).
Figure 10.
EOBII Activates IGS and PAL Promoters.
Arabidopsis protoplasts were cotransformed with 35S-driven EOBII (35Spro:EOBII) together with DsRed driven by the petunia IGS promoter (IGSpro:DsRed) ([G] to [I]) or with DsRed driven by tobacco PALB promoter (PALpro:DsRed) ([M] to [O]). As controls, protoplasts were cotransformed with IGS:DsRed together with 35S:GFP ([A] to [C]), with Solanum lycopersicum MYBAA driven by 35S (35Spro:MYBAA) ([D] to [F]), or with PALpro:DsRed together with 35Spro:MYBAA ([J] to [L]). Bright-field images are shown in panels (A), (D), (G), (J), and (M); DsRed signal is shown in (B), (E), (H), (K), and (N); and GFP signal in (C), (F), (I), (L), and (O). Bar = 50 μm.
DISCUSSION
Floral scent composition and intensity are determined by the plant's genetic background (Iijima et al., 2004), developmental status (Dudareva et al., 2000), and physiological condition (Underwood et al., 2005; Colquhoun et al., 2009), as well as time of day (Kolosova et al., 2001; Simkin et al., 2004), light (Hendel-Rahmanim et al., 2007), and other environmental factors (Verdonk et al., 2003; Schuurink et al., 2006; van Schie et al., 2006). In many flowering plants, including petunia, the difference in the levels of volatile compounds emitted by mature flowers after anthesis versus flowers before anthesis, or by flowers during the night versus the daytime, is striking (Boatright et al., 2004; Figures 4B and 4E). At present, we have only very limited knowledge of the molecular factors regulating the expression of genes involved in this process (Verdonk et al., 2005; Schuurink et al., 2006). Among the main factors slowing our progress in delineating the molecular regulatory mechanism of floral scent biosynthesis are the complexities of the character and of the analytical methods used for its measurement and the lack of extensive genetic data on plants used as models for scent studies.
The VIGS approach, which makes use of posttranscriptional gene silencing to downregulate target sequences, has been used for the isolation and characterization of various genes in diverse plant species, including petunia (Chen et al., 2004; Robertson, 2004; Spitzer et al., 2007). TRV is widely used as a VIGS vector because the infection is very efficient and produces only mild symptoms in the host plant (Ratcliff et al., 2001; Spitzer et al., 2007). We recently demonstrated the applicability of the TRV-based VIGS system for studying floral scent in petunia using CHS silencing as a marker (Spitzer et al., 2007). The approach was found to be applicable to both structural and regulatory genes responsible for volatile production (Spitzer et al., 2007). This high-throughput reverse genetics tool was harnessed to identify EOBII following large-scale screening for floral scent–related regulators. Silencing of EOBII in petunia resulted in reduced levels of phenylpropanoid volatiles emitted from corollas, while it had no effect on the levels of two terpenoid scent volatiles, analyzed for comparison. The levels of all analyzed internal pools of phenylpropanoid scent compounds were significantly reduced following EOBII suppression (Figure 3), indicating that biosynthetic steps, rather than the volatile release machinery, are affected.
The spatial pattern of EOBII expression (Figure 4C) was directly correlated with that of phenylpropanoid scent production and the expression of scent-related genes, which occurred predominantly in flower limbs (see Supplemental Figure 2 online; Underwood et al., 2005; Colquhoun et al., 2009). For example, expression of eugenol synthase (EGS), CFAT, BSMT, IGS, BPBT, PAAS, and ODO1 is highest in limbs of petunia corollas; relatively low, but nevertheless detectable, expression of the latter five genes has also been found in tubes and/or ovaries of petunia flowers (Negre et al., 2003; Verdonk et al., 2005; Kaminaga et al., 2006; Koeduka et al., 2006, 2008; Dexter et al., 2007, 2008). Analyses of the developmental pattern of EOBII expression revealed that it peaks at anthesis (Figure 4A), similar to some other scent-related genes (see Supplemental Figure 2 online; Verdonk et al., 2005; Dexter et al., 2007; Colquhoun et al., 2009). High levels of expression of EOBII and most biosynthetic scent-related genes precede peak scent emission (anthesis versus 2 dpa, respectively). This may allow, for example, building up an internal pool of scent volatiles; alternatively, the lack of high levels of volatile emission at anthesis may be explained by limitations in substrate availability (Boatright et al., 2004; Schuurink et al., 2006; Hendel-Rahmanim et al., 2007; Pichersky and Dudareva, 2007; Moyal Ben Zvi et al., 2008). In this respect, a lack of correlation has been shown between the activities of several scent-related enzymes (e.g., PAAS, BSMT and BPBT) and the production patterns of their respective volatiles (Kolosova et al., 2001; Boatright et al., 2004; Colquhoun et al., 2009).
The diurnal accumulation pattern of transcripts coding for scent-related biosynthetic genes is complex, while the emission of most scent compounds is synchronized (Kolosova et al., 2001; Schuurink et al., 2006; Hendel-Rahmanim et al., 2007; Farhi et al., 2009). It has been suggested that circadian and light regulation of scent production was adapted by plants, from an evolutionary standpoint, relatively recently (Hendel-Rahmanim et al., 2007) and that a combination of various mechanisms was harnessed by plants for rhythmic production/emission of scent compounds (Kolosova et al., 2001; Simkin et al., 2004; Hendel-Rahmanim et al., 2007). EOBII expression is rhythmic with a phase preceding that of biosynthetic floral scent–related genes and that of scent production, as might be expected for a scent regulator: maximal levels of the transcript were detected in the early morning hours, preceding, for example, peak BPBT and IGS transcript accumulation (Boatright et al., 2004; Dexter et al., 2008; Colquhoun et al., 2009) by ∼6 h, and peak emission of benzyl benzoate and isoeugenol (products of BPBT and IGS, respectively; see Supplemental Figure 4 online) by ∼12 h. By contrast, diurnal expression of ODO1 has been shown to coincide with scent production (Verdonk et al., 2005). This difference in EOBII and ODO1 expression patterns could potentially be explained by the finding that ODO1 expression is affected by that of EOBII (Figure 8).
Based on the sequence analyses, EOBII belongs to a large family of MYB transcription factors. It is localized in the nucleus, its up- and downregulation affect transcript levels of several biosynthetic floral scent-related genes, and it can transcriptionally activate PAL and IGS promoters (Figures 6, 9, and 10). These findings and the observation that its expression in tobacco also leads to increased levels of PALB transcript (see Supplemental Figure 3 online) strongly support EOBII's regulatory function in floral scent production. Several close homologs of EOBII from various plants, such as MYB305 and MYB340 from snapdragon (Antirrhinum majus) and NlxNs MYB305 from Nicotiana langsdorffii × Nicotiana sanderae (ornamental tobacco), have been characterized. These homologs target flavonoid biosynthetic genes, and NlxNs MYB305 has been recently shown to transcriptionally regulate nectarin genes in the ornamental tobacco floral nectary as well (Sablowski et al., 1994; Moyano et al., 1996; Liu et al., 2009). ODO1, also shown to regulate floral scent production, is phylogenetically distant from EOBII (Figure 5B). Furthermore, while suppression of ODO1 leads to downregulation of several genes from the shikimate pathway (Verdonk et al., 2005), suppression of EOBII downregulated the expression of ODO1 and of the volatile phenylpropanoid-related biosynthetic genes BPBT, IGS, and CFAT, in addition to some of the shikimate pathway genes (Figure 7). The lack of full overlap in target shikimate genes between EOBII and ODO1 may be due to the different genetic backgrounds of the analyzed petunia plants, differences in their diurnal expression patterns, or differences in the extent of suppression when ODO1 is downregulated directly compared with its downregulation through EOBII; the possibility that in flowers with RNAi-suppressed ODO1, an additional ODO1 homolog is also affected cannot be ruled out at present. Once the ODO1 promoter becomes available, it will be informative to establish whether EOBII interacts with it directly or via an additional factor(s). The lack of complete overlap in genes affected by EOBII and ODO1 would support the latter scenario.
Interaction between the branches of the phenylpropanoid pathway leading to volatile phenylpropanoids and flavonoids can be anticipated on the basis of their common biochemical origin, as well as their similar biological roles (Hoballah et al., 2007; Moyal Ben Zvi et al., 2008). Diversion of metabolic flux from one branch of the phenylpropanoid pathway to another has been demonstrated in carnation (Dianthus caryophyllus) flowers (Zuker et al., 2002). In addition, ectopic expression of the Arabidopsis MYB factor PAP1 in petunia has been found to enhance both anthocyanin and volatile phenylpropanoid levels in flowers due to elevated metabolic flux in both branches (Moyal Ben Zvi et al., 2008). On the other hand, overexpression of AN2, the petunia homolog of PAP1, did not affect scent emission in Petunia axillaris, while strongly enhancing floral pigmentation (Hoballah et al., 2007). The effect of EOBII suppression, similar to that of ODO1, was specific to volatile phenylpropanoid production and not flavonoid accumulation (Figures 2, 7G, and 7H; Verdonk et al., 2005). Multilayer regulation of different branches of the phenylpropanoid pathway is also apparent from the different patterns of their temporal activation: while pigments accumulate during early stages of flower development, production of floral scent volatiles reaches maximum levels in mature flowers following anthesis (Quattrocchio et al., 1998; Spelt et al., 2000; Schuurink et al., 2006). Thus, the expression patterns of EOBII and ODO1 (Verdonk et al., 2005), with low transcript levels in young flowers that peak at anthesis, may indicate differences in the activation time of the various branches of the phenylpropanoid pathway.
The importance of substrate availability for efficient metabolic flow toward the production of floral volatiles has been illustrated in several studies, and both positive and negative feedback loops have been characterized (Blount et al., 2000; Boatright et al., 2004; Dexter et al., 2007; Moyal Ben Zvi et al., 2008). For example, RNAi-driven downregulation of CFAT in petunia was found to result in reduced levels of isoeugenol (synthesized from coniferyl acetate) as well as of several other volatile phenylpropanoid compounds, such as benzyl acetate, phenylacetaldehyde, and phenylethyl alcohol (Dexter et al., 2007). No changes were observed in the levels of other volatile compounds, such as benzaldehyde, benzyl alcohol, or benzylbenzoate, or in the transcript levels of scent-related genes, such as IGS and BPBT. Based on in vitro assays, it was suggested that this differential effect on volatile production is due to overaccumulation of coniferyl aldehyde and homovanillic acid in CFAT-RNAi–silenced corollas, which in turn inhibits the activity of PAAS and BPBT enzymes. Silencing of EOBII, while also leading to a reduction in the levels of volatiles observed in CFAT-RNAi flowers, had a much wider-ranging effect on volatile production and the expression of related genes (Figures 1, 3, and 7). Genes from the shikimate pathway (CS and CM) and those coding for enzymes intimately involved in the formation of volatile phenylpropanoids (CFAT, IGS, and BPBT) were affected by EOBII. The observation that expression of some of the genes (e.g., CM and ODO1) were not affected by EOBII overexpression, while they were downregulated in EOBII-suppressed corollas, suggests that additional factors are involved in determining their expression. The simultaneous regulation of both CM and PAL by EOBII should enable efficient modulation of metabolic flux into the phenylpropanoid pathway since Phe is a feedback inhibitor of CM (Eberhard et al., 1993; Mobley et al., 1999). The coordinated wide-ranging effect of EOBII on the production of floral volatiles may suggest that it plays a central regulatory role in the biosynthesis of phenylpropanoid volatiles from the different branches. Moreover, a characterization of factors upstream of EOBII, as well as those interacting with EOBII and ODO1, should open the way to deciphering the developmental regulation of interconnections between specialized and primary metabolism.
METHODS
Plant Material
Rooted petunia plantlets (Petunia hybrida line P720) were obtained from Danziger-Dan Flower Farm (Mishmar Hashiva, Israel). Transgenic cv Mitchell (W115) with RNAi-suppressed ODO1 (Verdonk et al., 2005) was kindly provided by Robert C. Schuurink, University of Amsterdam, The Netherlands. Plants were grown in the greenhouse under 25°C/20°C day/night temperatures and natural photoperiod.
Cloning of EOBII and Construction of Vectors
The MYB-like DNA fragments were isolated by RT-PCR using as a template RNA from flowers of P. hybrida line P720 and degenerate primers. Primers were designed complementary to the conserved amino acid sequences KKGPWTPEE (5′-AAGAAAGCNSCWTGGCANSMNGMAGAA-3′) and DNEIKNYWN (5′-TTCCARTRRTTNTTNAYNTCRTTRTC-3′) of the R2R3-MYB motifs. The PCR products were cloned into pGEMT-Easy (Promega) and sequenced. One of these fragments, containing a 216-bp ORF homologous to Ps MYB26, was termed R2R3-EOBII and transferred to pTRV2 and pTRV2-CHS constructs (Spitzer et al., 2007) to generate TRV2-R2R3-EOBII and TRV2-CHS:R2R3-EOBII, respectively. To clone the EOBII gene, PCR was performed on a petunia cDNA library using primers based on the sequence of R2R3-EOBII (forward primer, 5′-GAACTGCATGCTAAGTGGG-3′, and reverse primer: 5′-GACCACCTGTTTCCCCACTTAGC-3′), and forward and reverse primers for the lambda GEM4 vector (5′-GATTTAGGTGACACTATAGGGAGACC-3′ and 5′-GTAATACGACTCACTATAGGGC-3′). The full-length EOBII was cloned using primers to the 5′ and 3′ ends of the in silico–assembled EOBII sequence. To generate pTRV2-CHS containing the 3′ region of EOBII (TRV2-CHS:EOBII), 205 bp (669 to 873) of EOBII was PCR amplified using forward primer, 5′-TTGAGTTAAACCCTAGATTGAACC-3′, and reverse primer, 5′-AACCATAGGCACCTCCATGCATC-3′, and inserted upstream of CHS. To construct pTRV2 with EOBII fused upstream of GFP (pTRV2-EOBII:GFP), the 591-bp ORF of EOBII was PCR amplified using forward primer, 5′-AACGAGATGGATAAAAAACCATGCAACTCTCA-3′, and reverse primer, 5′-TCCTGGTCCATCACCATTAAGCAATTGCATGG-3′, and ligated (in frame with GFP) into HpaI-restricted pTRV2-GFP. The last nine nucleotides at the 5′ end of the reverse primer allowed generation of a Gly-Pro-Gly amino acid bridge between EOBII and GFP. To generate a binary vector containing EOBII under the control of the cauliflower mosaic virus 35S promoter (35S:EOBII), the full-length EOBII was PCR amplified using forward primer, 5′-ATGGATAAAAAACCATGCAACTCTCAAGATG-3′, and reverse primer, 5′-CGTACTCACTCATGAGATGGTTCAATC-3′. The PCR product was cloned into a pCd shuttle vector between the cauliflower mosaic virus 35S promoter and octopine synthase terminator; the resultant plasmid was then restricted with KpnI/XbaI, and the generated restriction fragment was inserted into binary vector pCGN1559 containing the neomycin phosphotransferase II gene (Moyal Ben Zvi et al., 2008).
Agroinoculation of TRV Vectors
Agrobacterium tumefaciens (strain AGLO) transformed with pTRV1 and pTRV2 derivatives were prepared as described previously (Liu et al., 2002). The Agrobacterium culture was grown overnight at 28°C in Luria-Bertani medium with 50 mg L−1 kanamycin and 200 μM acetosyringone. The cells were harvested and resuspended in inoculation buffer containing 10 mM MES, pH 5.5, 200 μM acetosyringone, and 10 mM MgCl2 to an OD550 of 10. Following an additional 3 h of incubation at 28°C, the bacteria containing pTRV1 were mixed with those containing the pTRV2 derivatives in a 1:1 ratio; 200 to 400 μL of this mixture was applied to the cut surface, after removing the apical meristems, of petunia plantlets. For the pTRV2-EOBII:GFP infection, the plasmid was electroporated into A. tumefaciens (strain AGLO). Following overnight culture, bacteria were harvested and used to infect leaves of 3-week-old seedlings of Nicotiana benthamiana by infiltration using a syringe (Liu et al., 2002). Plants were grown for 36 h in a growth chamber at 22°C. Transient expression of GFP fusions in the tobacco epidermis was evaluated 36 h after infection using a fully motorized epifluorescence inverted microscope (Olympus-IX8 Cell-R) with a 12-bit Orca-AG CCD camera (Hamamatsu).
Collection and GC-MS Analysis of Volatile Compounds
For dynamic headspace analysis, flowers were collected 1 dpa at 17 h00, unless otherwise indicated. Volatiles emitted from detached petunia flowers were collected for the indicated periods of time using an adsorbent trap consisting of a glass tube containing 100 mg Porapak Type Q polymer (80/100 mesh; Alltech) and 100 mg 20/40 mesh activated charcoal (Supelco), held in place with plugs of silanized glass wool (Guterman et al., 2006). Trapped volatiles were eluted using 1.5 mL hexane, and 2 μg isobutylbenzene was added to each sample as an internal standard.
To determine the pool sizes of volatile compounds in corolla limbs, petal tissues (0.5 g fresh weight) were collected at 07 h00, ground in liquid nitrogen, and extracted in hexane (4 mL g−1 tissue) containing 2 μg isobutylbenzene as the internal standard. Following overnight incubation with shaking at 150 rpm, extracts were centrifuged at 10,500g for 10 min, and the supernatant was filtered through a 25-mL syringe with a 0.2-μM sterile nylon filter.
GC-MS analysis (1 μL sample) was performed using a device composed of a Pal autosampler (CTC Analytic), a TRACE GC 2000 equipped with an Rtx-5SIL MS (Restek; i.d. 0.25 μm, 30 m × 0.25 mm) fused-silica capillary column, and a TRACE DSQ quadrupole mass spectrometer (ThermoFinnigan). Helium was used as the carrier gas at a flow rate of 0.9 mL min−1. The injection temperature was set to 250°C (splitless mode) and the interface to 280°C, and the ion source was adjusted to 200°C. The analysis was performed under the following temperature program: 2 min of isothermal heating at 40°C followed by a 10°C min−1 oven temperature ramp to 250°C. The transfer line temperature was 280°C. The system was equilibrated for 1 min at 70°C before injection of the next sample. Mass spectra were recorded at 2.65 scan s−1 with a scanning range of 40 to 450 mass-to-charge ratio and an electron energy of 70 eV. Compounds were tentatively identified (>95% match) based on NIST/EPA/NIH Mass Spectral Library (Data Version: NIST 05, Software Version 2.0d) using the XCALIBUR v1.3 program (ThermoFinnigan). Further identification of major compounds was based on comparison of mass spectra and retention times with those of authentic standards (Sigma-Aldrich) analyzed under similar conditions.
Extraction and GC-MS Analysis of Nonvolatile Metabolites
The extraction protocol was modified from Brosche et al. (2005). Briefly, frozen petal tissues (100 mg fresh weight) were ground in liquid nitrogen, incubated for 15 min at 70°C, and lightly shaken in 1.4 mL of 90% (v/v) aqueous methanol with ribitol (120 μL of 0.2 mg/mL) as an internal standard. Following centrifugation at 18,500g for 10 min, the extract was mixed with 1500 μL water and 750 μL chloroform and subsequently centrifuged for 15 min at 30,000g, 4°C. Aliquots of the methanol/water phase (150 μL) were dried in a vacuum overnight. The dry residue was modified for GC-MS analysis according to Brosche et al. (2005). Residues after reduction were redissolved and derivatized for 120 min at 37°C in 40 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine followed by a 30-min treatment with 70 μL N-methyl-N-[trimethylsilyl]trifluoroacetamide at 37°C. A retention time standard mixture (10 μL of 0.029% v/v n-dodecane, n-pentadecane, n-nonadecane, n-docosane, noctacosane, n-dotracontane, and n-hexatriacontane dissolved in pyridine) was added before trimethylsilylation. Sample volumes of 1 μL were then injected into the GC column in splitless mode. The GC-MS system was run as described above with some modifications. The injection temperature was set at 300°C, the interface at 300°C, and the ion source adjusted to 270°C. The analysis was performed under the following temperature program: 5 min of isothermal heating at 70°C, followed by a 5°C min−1 oven temperature ramp to 350°C, and a final 5-min heating at 300°C. Mass spectra were recorded at 3 scan s−1 with a scanning range of 40 to 600 m/z. Both chromatograms and mass spectra were evaluated using the XCALIBUR v1.3 program. A retention time and MS library for automatic peak quantification of metabolite derivatives were implemented within the NIST 2.0 method format. Substances were identified by comparison with authentic standards, as described by Roessner-Tunali et al. (2003).
To determine anthocyanin levels, petunia corollas were extracted with methanol containing 1% (v/v) HCl (50 mg fresh tissue per 1 mL acidic methanol). Absorption values of the extract at A530 and A657 were measured using the formula A530 − 0.25(A657), which allows for subtraction of chlorophyll interference.
Real-Time PCR Analysis
Petunia total RNA was extracted with a Tri-reagent kit (Molecular Research Center) and treated with RNase-free DNase (Fermentas). First-strand cDNA was synthesized using 1 μg total RNA, oligo d(T) primer, and Reverse Transcriptase ImProm-II (Promega). PCR was performed for 40 cycles (94°C for 15 min and then cycling at 94°C for 10 s, 60°C for 30 s, and 72°C for 20 s). Real-time PCR primers for EOBII amplification were forward primer, 5′-CAAGCAGGCCCTAATTTTCTC-3′, and reverse primer, 5′-TCATGAGATGGTTCAATCTAGGG-3′. These primers are unable to amplify EOBII fragments cloned in pTRV2 since the first primer is complementary to a region of EOBII that was not used in the pTRV2 constructs. Primers (forward and reverse, respectively) for BPBT were 5′-TGTTGAAGGGTGATGCTCAA-3′ and 5′- GGATTTGGCATTTCAAACAAA-3′; for PAL2, 5′- TGCTAATGGTGAACTTCATCCA-3′ and 5′-TGACATTCTTCTCACTTTCACCA-3′; for FHT, 5′-GCCTTAACCAAGGCATGTGT-3′ and 5′-TAGCTTGAAGCCCACCAACT-3′; for CHI, 5′-TGAAAGAGTAGCGGAACTGCT-3′ and 5′-GGTAACAGTTTACAACATCAGGC-3′; for CS, 5′-TGTTCCAATGGTTGAAGCAA-3′ and 5′-GTTTCAAAGGGCAACCTCAG-3′; for CM, 5′-GATGCACGGTTCTACGCTTC-3′ and 5′-AATTGAACCGCCAAGTGAGT-3′; for CFAT, 5′-CCAATGCCTAGCCCTAACAA-3′ and 5′-GGACGCTTCTTCACATCACA-3′; for IGS, 5′-CCACGTCAAAAGAGTGAGCA-3′ and 5′-CCAGTGGTTTTCTCCCAAGA-3′; for ODO1, 5′-ACCAACCTACCAACCAACCA-3′ and 5′-ATGATGACCCCTCCAACAAG-3′; Actin, used as reference for standardization of cDNA amounts, 5′-TGCTGATCGTATGAGCAAGGAA-3′ and 5′-GGTGGAGCAACAACCTTAATCTTC-3′. Real-time quantitative PCR was performed in the presence of SYBR Green I dye (ABgene) on a Corbett Research Rotor-Gene 6000 cycler, and data analysis was performed using Rotor-Gene 6000 series software 1.7.
Protoplast Transformation
To clone the IGS promoter regions from petunia line W115, a GenomeWalker Kit (Clontech) was used according to the manufacturer's instructions. The promoter region was PCR amplified using the kit's forward primer and IGS reverse primer 5′-CACCAGATGAAGGAGTAAGGGACTGCA-3′ and cloned into pGEMT (Promega; pGEMT-IGSpro). To clone 590 bp of the IGS promoter upstream of DsRed (IGSpro:DsRed), we first removed the 35S promoter from the pSAT6A-DsRed2-N1 vector (Tzfira et al., 2005). The resultant promoterless vector was used to clone the PCR-amplified (5′-TATTTAGGTGACACTATAG-3′ and 5′-GGATCCTCTCTCTTGGATTTGCATGGA-3′) promoter region of IGS upstream of DsRed. Similarly, the tobacco PALB promoter region (GB: AB008200) was first PCR amplified using primers 5′-AAGCTTGATCCGGACAAGAATGCA-3′ and 5′-GGATCCTGTTAAAGGTTGTGAGGA-3′ and then cloned upstream of DsRed into the promoterless pSAT6A-DsRed2-N1 vector, yielding the PALpro:DsRed construct.
Protoplasts were isolated from Arabidopsis leaf mesophyll, and 35Spro:EOBII and IGSpro:DsRed, or 35Spro:EOBII and PALpro:DsRed, were transiently expressed using the polyethylene glycol transformation method (Locatelli et al., 2003); 8 × 104 protoplasts were used for transformation with IGS:DsRed and pSAT6-EGFP-C1 (35S:GFP; Tzfira et al., 2005), IGS:DsRed and 35S:MYBAA (TIGR:TC208520; kindly provided by Asaph Aharoni, Weizmann Institute, Israel), and PAL:DsRed and 35S:MYBAA used as controls. Experiments were repeated three times, with a transformation efficiency of 70 to 80%. Imaging of GFP was performed by fully motorized epifluorescence inverted microscope (Olympus-IX8 Cell-R) 48 h after transformation, with the 12-bit Orca-AG CCD camera.
Transient Overexpression of EOBII in Petunia Flowers
Transient overexpression of EOBII in petunia buds was performed according to Long et al. (2009). Petunia buds 3 d before anthesis were vacuum infiltrated with A. tumefaciens containing 35S:EOBII or GUS under the same promoter (35S:GUS). Following incubation for 48 h in the dark, RNA was extracted from infiltrated corollas and the transcript levels of selected genes were analyzed using real-time RT-PCR.
Phylogenetic Analysis
The dendogram was constructed using the neighbor-joining algorithm with MEGA4 software (Tamura et al., 2007). Bootstrapping was performed with 1000 replicates. Unrelated human (Hs) C-MYB protein was used as the outgroup. The multiple alignment was created with the ClustalW algorithm using AlignX of the Vector NTI Advance 9.0 program.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under the following accession numbers: P. hybrida EOBII (EU360893), CHS (X14599), CHI (AB213651.1), BPBT (AY611496), PAL2 (CO805160), FHT (X60512), CS (TIGR accession TC2915), CM (CO805161), CFAT (DQ767969), IGS (DQ372813), IGS promoter (GU983699), ODO1 (AY705977), Actin (U60495), and ODO1 (Q50EX6); Solanum lycopersicum MYBAA (TIGR accession TC208520); Nicotiana tabacum PALB (AB008200); Nicotiana langsdorffii × Nicotiana sanderae MYB305 (ABU97107); Antirrhinum majus MYB305 (P81391), MYB340 (P81396), MYB330 (P81395), and MYB308 (P81393); Arabidopsis MYB21 (Q9LK95) and MYB24 (AAM63674); Pisum sativum MYB26 (CAA71992); and Homo sapiens C-MYB (M15024).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transcript Levels of EU360892, DAHPS, EPSPS, PAL1, and PAAS Are Not Affected in EOBII-Silenced Corollas.
Supplemental Figure 2. Developmental and Spatial Expression of Floral Scent–Related Genes from the Shikimate and Phenylpropanoid Pathways.
Supplemental Figure 3. Tobacco Flowers Ectopically Expressing EOBII Accumulate Increased Levels of PALB Transcript.
Supplemental Figure 4. Levels of Benzylbenzoate and Isoeugenol Emitted from Petunia Flowers During the Day/Night.
Supplemental Table 1. The Predominant Volatiles Emitted by Wild-Type Petunia P720.
Supplemental Data Set 1. FASTA File of ClustalW Alignment, Using MEGA4 Software, for Dendogram Construction.
Acknowledgments
We thank the Danziger-Dan Flower Farm for providing the plant material and Hillary Voet for assistance in the statistical analyses. This work was funded by Israel Science Foundation Grants 505/05 and 269/09 to A.V. A.V. is an incumbent of the Wolfson Chair in Floriculture.
References
- Blount J.W., Korth K.L., Masoud S.A., Rasmussen S., Lamb C., Dixon R.A. (2000). Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 122: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright J., Negre F., Chen X., Kish C.M., Wood B., Peel G., Orlova I., Gang D., Rhodes D., Dudareva N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135: 1993–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H.D., Schemske D.W. (2003). Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178 [DOI] [PubMed] [Google Scholar]
- Brosche M., et al. (2005). Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol. 6: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channeliere S., Riviere S., Scalliet G., Jullien F., Szecsi J., Dolle C., Vergne P., Dumas C., Bendahmane M., Hugueney P., Cock J.M. (2002). Analysis of gene expression in rose petals using expressed sequence tags. FEBS Lett. 515: 35–38 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Jiang C.Z., Gookin T.E., Hunter D.A., Clark D.G., Reid M.S. (2004). Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol. Biol. 55: 521–530 [DOI] [PubMed] [Google Scholar]
- Colquhoun T.A., Verdonk J.C., Schimmel B.C.J., Tieman D.M., Underwood B.A., Clark D.G. (2009). Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry : . [DOI] [PubMed] [Google Scholar]
- Dexter R., Qualley A., Kish C.M., Ma C.J., Koeduka T., Nagegowda D.A., Dudareva N., Pichersky E., Clark D. (2007). Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 49: 265–275 [DOI] [PubMed] [Google Scholar]
- Dexter R.J., Verdonk J.C., Underwood B.A., Shibuya K., Schmelz E.A., Clark D.G. (2008). Tissue-specific PhBPBT expression is differentially regulated in response to endogenous ethylene. J. Exp. Bot. 59: 609–618 [DOI] [PubMed] [Google Scholar]
- Dudareva N., Murfitt L.M., Mann C.J., Gorenstein N., Kolosova N., Kish C.M., Bonham C., Wood K. (2000). Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Negre F. (2005). Practical applications of research into the regulation of plant volatile emission. Curr. Opin. Plant Biol. 8: 113–118 [DOI] [PubMed] [Google Scholar]
- Dudareva N., Pichersky E. (2006). Floral scent metabolic pathways: Their regulation and evolution. Biology of Floral Scent, Dudareva N., Pichersky E., (Boca Raton, FL: CRC Press; ), pp. 55–78 [Google Scholar]
- Dudareva N., Pichersky E. (2008). Metabolic engineering of plant volatiles. Curr. Opin. Biotechnol. 19: 181–189 [DOI] [PubMed] [Google Scholar]
- Dudareva N., Pichersky E., Gershenzon J. (2004). Biochemistry of plant volatiles. Plant Physiol. 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard J., Raesecke H.R., Schmid J., Amrhein N. (1993). Cloning and expression in yeast of a higher plant chorismate mutase. Molecular cloning, sequencing of the cDNA and characterization of the Arabidopsis thaliana enzyme expressed in yeast. FEBS Lett. 334: 233–236 [DOI] [PubMed] [Google Scholar]
- Farhi M., Lavie O., Masci T., Hendel-Rahmanim K., Weiss D., Abeliovich H., Vainstein A. (2009). Identification of rose phenylacetaldehyde synthase by functional complementation in yeast. Plant Mol. Biol. . [DOI] [PubMed] [Google Scholar]
- Gang D.R. (2005). Evolution of flavors and scents. Annu. Rev. Plant Biol. 56: 301–325 [DOI] [PubMed] [Google Scholar]
- Gershenzon J., Dudareva N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Guterman I., Masci T., Chen X., Negre F., Pichersky E., Dudareva N., Weiss D., Vainstein A. (2006). Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: Rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Mol. Biol. 60: 555–563 [DOI] [PubMed] [Google Scholar]
- Hendel-Rahmanim K., Masci T., Vainstein A., Weiss D. (2007). Diurnal regulation of scent emission in rose flowers. Planta 226: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Hoballah M.E., Gubitz T., Stuurman J., Broger L., Barone M., Mandel T., Dell'Olivo A., Arnold M., Kuhlemeier C. (2007). Single gene-mediated shift in pollinator attraction in petunia. Plant Cell 19: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y., Davidovich-Rikanati R., Fridman E., Gang D.R., Bar E., Lewinsohn E., Pichersky E. (2004). The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol. 136: 3724–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaga Y., et al. (2006). Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281: 23357–23366 [DOI] [PubMed] [Google Scholar]
- Knudsen J.T., Eriksson R., Gershenzon J., Stahl B. (2006). Diversity and distribution of floral scent. Bot. Rev. 72: 1–120 [Google Scholar]
- Koeduka T., et al. (2006). Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 103: 10128–10133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka T., Louie G.V., Orlova I., Kish C.M., Ibdah M., Wilkerson C.G., Bowman M.E., Baiga T.J., Noel J.P., Dudareva N., Pichersky E. (2008). The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant J. 54: 362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N., Gorenstein N., Kish C.M., Dudareva N. (2001). Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13: 2333–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Li J.G., Yang X.Y., Wang Y., Li X.J., Gao Z.F., Pei M., Chen Z.L., Qu L.J., Gu H.Y. (2006). Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem. Biophys. Res. Commun. 341: 1155–1163 [DOI] [PubMed] [Google Scholar]
- Liu G., Ren G., Guirgis A., Thornburg R.W. (2009). The MYB305 transcription factor regulates expression of nectarin genes in ornamental tobacco floral nectary. Plant Cell 21: 2672–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Locatelli F., Vannini C., Magnani E., Coraggio I., Bracale M. (2003). Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. Plant Cell Rep. 21: 865–871 [DOI] [PubMed] [Google Scholar]
- Long M.C., Nagegowda D.A., Kaminaga Y., Ho K.K., Kish C.M., Schnepp J., Sherman D., Weiner H., Rhodes D., Dudareva N. (2009). Involvement of snapdragon benzaldehyde dehydrogenase in benzoic acid biosynthesis. Plant J. 59: 256–265 [DOI] [PubMed] [Google Scholar]
- Mobley E.M., Kunkel B.N., Keith B. (1999). Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene 240: 115–123 [DOI] [PubMed] [Google Scholar]
- Moyal Ben Zvi M., Negre-Zakharov F., Masci T., Ovadis M., Shklarman E., Ben-Meir H., Tzfira T., Dudareva N., Vainstein A. (2008). Interlinking showy traits: Co-engineering of scent and colour biosynthesis in flowers. Plant Biotechnol. J. 6: 403–415 [DOI] [PubMed] [Google Scholar]
- Moyano E., Martinez-Garcia J.F., Martin C. (1996). Apparent redundancy in Myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell 8: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre F., Kish C.M., Boatright J., Underwood B., Shibuya K., Wagner C., Clark D.G., Dudareva N. (2003). Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15: 2992–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Dudareva N. (2007). Scent engineering: Toward the goal of controlling how flowers smell. Trends Biotechnol. 25: 105–110 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Gershenzon J. (2002). The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Noel J.P., Dudareva N. (2006). Biosynthesis of plant volatiles: Nature's diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J.F., van der Woude K., Mol J.N., Koes R. (1998). Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C. (2001). Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Robertson D. (2004). VIGS vectors for gene silencing: Many targets, many tools. Annu. Rev. Plant Biol. 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R.W.M., Moyano E., Culianezmacia F.A., Schuch W., Martin C., Bevan M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink R.C., Haring M.A., Clark D.G. (2006). Regulation of volatile benzenoid biosynthesis in petunia flowers. Trends Plant Sci. 11: 20–25 [DOI] [PubMed] [Google Scholar]
- Shin B., Choi G., Yi H., Yang S., Cho I., Kim J., Lee S., Paek N.-C., Kim J.-H., Song P.-S., Choi G. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30: 23–32 [DOI] [PubMed] [Google Scholar]
- Simkin A.J., Underwood B.A., Auldridge M., Loucas H.M., Shibuya K., Schmelz E., Clark D.G., Klee H.J. (2004). Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of {beta}-Ionone, a fragrance volatile of petunia flowers. Plant Physiol. 136: 3504–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J., Koes R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer B., et al. (2007). Reverse genetics of floral scent: Application of Tobacco rattle virus-based gene silencing in petunia. Plant Physiol. 145: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tieman D., Taylor M., Schauer N., Fernie A.R., Hanson A.D., Klee H.J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Tian G.-W., Lacroix B.T., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Uimari A., Strommer J. (1997). Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 12: 1273–1284 [DOI] [PubMed] [Google Scholar]
- Underwood B.A., Tieman D.M., Shibuya K., Dexter R.J., Loucas H.M., Simkin A.J., Sims C.A., Schmelz E.A., Klee H.J., Clark D.G. (2005). Ethylene-regulated floral volatile synthesis in petunia corollas. Plant Physiol. 138: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie C.C.N., Haring M.A., Schuurink R.C. (2006). Regulation of terpenoid and benzenoid production in flowers. Curr. Opin. Plant Biol. 9: 203–208 [DOI] [PubMed] [Google Scholar]
- Verdonk J.C., Haring M.A., van Tunen A.J., Schuurink R.C. (2005). ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk J.C., Ric de Vos C.H., Verhoeven H.A., Haring M.A., van Tunen A.J., Schuurink R.C. (2003). Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62: 997–1008 [DOI] [PubMed] [Google Scholar]
- Zuker A., Tzfira T., Ben-Meir H., Ovadis M., Shklarman E., Itzhaki H., Forkmann G., Martens S., Neta-Sharir I., Weiss D., Vainstein A. (2002). Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene. Mol. Breed. 9: 33–41 [Google Scholar]