A crucial step for diseases caused by phytopathogenic fungi is the penetration of the plant cuticle. Here, we highlight that two conserved plasma membrane proteins are required for plant surface sensing and regulate the penetration process via MAP kinase signaling.
Abstract
The dimorphic fungus Ustilago maydis switches from budding to hyphal growth on the plant surface. In response to hydrophobicity and hydroxy fatty acids, U. maydis develops infection structures called appressoria. Here, we report that, unlike in Saccharomyces cerevisiae and other fungi where Sho1 (synthetic high osmolarity sensitive) and Msb2 (multicopy suppressor of a budding defect) regulate stress responses and pseudohyphal growth, Sho1 and Msb2-like proteins play a key role during appressorium differentiation in U. maydis. Sho1 was identified through a two-hybrid screen as an interaction partner of the mitogen-activated protein (MAP) kinase Kpp6. Epistasis analysis revealed that sho1 and msb2 act upstream of the MAP kinases kpp2 and kpp6. Furthermore, Sho1 was shown to destabilize Kpp6 through direct interaction with the unique N-terminal domain in Kpp6, indicating a role of Sho1 in fine-tuning Kpp6 activity. Morphological differentiation in response to a hydrophobic surface was strongly attenuated in sho1 msb2 mutants, while hydroxy fatty acid–induced differentiation was unaffected. These data suggest that Sho1 and the transmembrane mucin Msb2 are involved in plant surface sensing in U. maydis.
INTRODUCTION
A crucial step for phytopathogenic fungi to initiate infection of their hosts is the penetration of the plant cuticle. The most common strategy for entry into the plant tissue is the development of specialized infection structures known as appressoria (Tucker and Talbot, 2001). Appressorium formation is induced upon contact with the plant surface. The cues responsible for this differentiation range from chemical signals, such as ethylene, epicuticular waxes, and cutin monomers to the physical nature of the surface, such as hydrophobicity, hardness, and topography (Tucker and Talbot, 2001; Kumamoto, 2008). Processing of these signals depends on conserved signaling cascades. In Magnaporthe oryzae and Botrytis cinerea, cAMP signaling and mitogen-activated protein (MAP) kinase signaling are required for appressorium differentiation (Lee and Dean, 1993; Xu and Hamer, 1996; Choi and Dean, 1997; Zhao et al., 2005; Doehlemann et al., 2006). The involvement of MAP kinases in appressorium development has also been demonstrated in Cochliolobus heterostrophus, Pyrenophora teres, and Colletotrichum species (Lev et al., 1999; Kim et al., 2000; Takano et al., 2000; Ruiz-Roldan et al., 2001). While intracellular signaling cascades have been linked to appressorium development in many phytopathogenic fungi, there is still poor knowledge about upstream receptors that regulate appressorial differentiation (Kumamoto, 2008). The only example so far is Pth11 (gene that plays a role in pathogenicity), a seven-transmembrane protein from M. oryzae. Pth11 has been suggested to be a surface sensor acting upstream of the cAMP pathway. However, pth11 mutants are impaired in appressorium formation in response to both cutin monomers and hydrophobic surface cues (DeZwaan et al., 1999).
Ustilago maydis is the causative agent of maize (Zea mays) smut disease. A prerequisite for infection of its host plant maize is the fusion of two compatible cells, and this is achieved through a pheromone-receptor system (Bölker et al., 1992). The pheromone signal is transmitted through a MAP kinase cascade consisting of the MEKK (mitogen-activated protein kinase kinase kinase) Kpp4 (Andrews et al., 2000), the MEK (mitogen-activated kinase kinase) Fuz7 (Banuett and Herskowitz, 1994), and the MAP kinase Kpp2 (Mayorga and Gold, 1999; Müller et al., 1999, 2003). After cell fusion, a dikaryotic filament is established, which represents the infective form of U. maydis (Feldbrügge et al., 2004). In such filaments, the formation of septa leads to the accumulation of empty sections in the older parts of the filament. Only the growing tip cell is filled with cytoplasm and differentiates into an appressorium at appropriate sites on the plant surface (Snetselaar and Mims, 1992). In contrast with the dome-shaped, melanine-pigmented appressoria of M. oryzae and Colletotrichum graminicola, where high turgor pressure allows plant penetration predominantly by mechanical force (Howard et al., 1991; Bechinger et al., 1999), appressoria in U. maydis are only slightly swollen tips of the filaments, and penetration is thought to occur by secretion of lytic enzymes (Schirawski et al., 2005). Recent work demonstrated that a hydrophobic surface is essential and sufficient to induce not only septated filaments, but also appressorium formation in U. maydis, and addition of hydroxy fatty acids strongly enhances appressorium formation efficiency (Mendoza-Mendoza et al., 2009b). The MAP kinase Kpp2 is involved in appressorium development (Müller et al., 2003), while another MAP kinase, Kpp6, which also acts downstream of Fuz7 (Di Stasio et al., 2009), is required for the appressorial penetration step (Brachmann et al., 2003; see Supplemental Figure 1A online). The morphological switch from budding to filamentous growth in response to the hydrophobic surface also depends on the MAP kinase Kpp2 (Mendoza-Mendoza et al., 2009b). However, the upstream receptors for the MAP kinase cascade that lead to appressorium development and penetration of the plant cuticle have not been identified so far.
The plasma membrane spanning protein Sho1 and the transmembrane mucin Msb2 are conserved proteins that serve as stress sensors in many fungal systems (Roman et al., 2005, 2009; Krantz et al., 2006; Norice et al., 2007; Ma et al., 2008; Boisnard et al., 2008) and function upstream of MAP kinase cascades (Seet and Pawson, 2004; Cullen, 2007). In Saccharomyces cerevisiae, Sho1p and Msb2p were shown to interact and regulate signaling cascades involved in osmotic stress response and pseudohyphal growth (Chen and Thorner, 2007). In the high osmolarity glycerol (HOG) pathway, a SH3 (Src homology 3) domain-mediated interaction between Sho1p and the MEK Pbs2p leads to activation of the MAP kinase Hog1p (Brewster et al., 1993; Maeda et al., 1995). In the filamentous growth (FG) pathway, the MAP kinase Kss1p is phosphorylated by the MEK Ste7p (Liu et al., 1993; Roberts and Fink, 1994). In both pathways, upstream components are shared: The MEKK Ste11p activates Ste7p in the FG pathway as well as Pbs2p in the HOG pathway (Liu et al., 1993; Roberts and Fink, 1994; Posas and Saito, 1997). The tetraspan membrane protein Sho1p and the single transmembrane mucin Msb2p act at the head of the HOG and the FG pathway. While Sho1p interacts directly with Ste11p and Pbs2p, Msb2p is thought to function through the small GTPase Cdc42p and the PAK kinase Ste20p to activate Ste11p (Maeda et al., 1995; O'Rourke and Herskowitz, 1998; Cullen et al., 2004; Tatebayashi et al., 2007; see Supplemental Figure 1B online). S. cerevisiae possesses a second transmembrane mucin, Hkr1p, that acts in parallel with Msb2p in the HOG pathway but is dispensable for the FG pathway (Tatebayashi et al., 2007; Pitoniak et al., 2009).
In a previous study, a homolog of Sho1p was found to interact with the MAP kinase Kpp6 from U. maydis (Mendoza-Mendoza et al., 2009a). We also identified a transmembrane mucin with similarity to Msb2p from S. cerevisiae in the genome of U. maydis but no ortholog of Hkr1p. In this article, we demonstrate that Sho1 and Msb2 are crucial virulence factors that localize to the plasma membrane. Intriguingly, Sho1 and Msb2 proteins in U. maydis are not involved in stress responses but specifically regulate appressorium development in response to a hydrophobic surface by acting upstream of the MAP kinases Kpp2 and Kpp6.
RESULTS
The Plasma Membrane Protein Sho1 Interacts Specifically with the MAP Kinase Kpp6
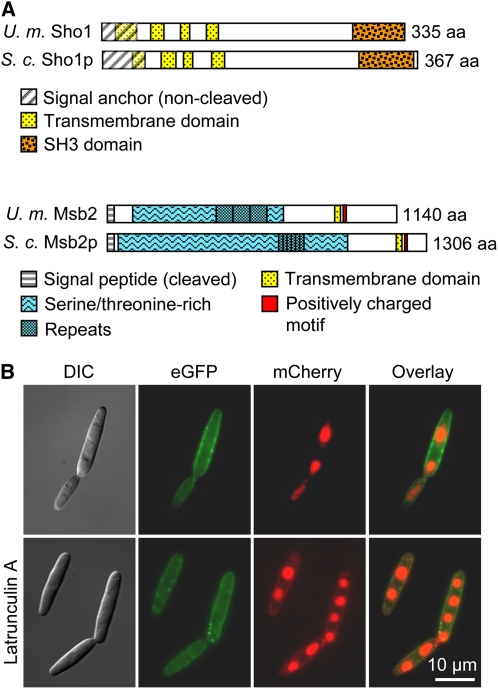
By yeast two-hybrid screening, using the MAP kinase Kpp6 from U. maydis as bait, we identified the C-terminal domain of Um03156 as putative interactor (Mendoza-Mendoza et al., 2009a). Um03156 encodes a 335–amino acid protein (http://mips.gsf.de/genre/proj/ustilago/) with similarity to the osmosensor Sho1p from S. cerevisiae (25% identity; Figure 1A). Sho1 and its orthologs in other fungi display an N-terminal secretion signal, four characteristic transmembrane domains, and a C-terminal SH3 domain. The Um03156 protein from U. maydis also displays such a domain structure and shows highest identity (61%) to Sho1p from yeast in the SH3 domain. Therefore, we designated um03156 as sho1.
Figure 1.
Domain Architecture and Localization of U. maydis Sho1 and Msb2.
(A)Top rows: Schematic representation of the domain structure of U. maydis Sho1 and S. cerevisiae Sho1p. Both proteins contain a noncleaved secretion signal (SignalP), four transmembrane domains, of which the first is part of the secretion signal, and a C-terminal SH3 domain. Bottom rows:Schematic representation of the domain structure of U. maydis Msb2 and S. cerevisiae Msb2p. Msb2 of U. maydis and Msb2p of S. cerevisiae share common features of signaling mucins. They have a cleaved signal peptide (SignalP) and one transmembrane domain close to the N terminus. The large extracellular part is Ser/Thr rich and includes tandem repeats. The short cytoplasmic tail contains a positively charged motif (RKHRK in U. maydis Msb2; RRR in S. cerevisiae Msb2p). aa, amino acids.
(B) Localization of Sho1-GFP and Msb2-mCherry in U. maydis. SG200sho1GFP/msb2mCherry cells were grown to mid log phase in YEPSL and analyzed microscopically without addition (top row) or after addition of latrunculin (bottom row). The fluorescent signals corresponding to Sho1-GFP and Msb2-mCherry (middle columns) were merged (right column). Bright-field images are shown on the left.
To determine the localization of Sho1 protein in U. maydis, a green fluorescent protein (gfp) tag was C-terminally fused to sho1 in the native locus in the solopathogenic strain SG200. This fusion protein was biologically active (see below and Supplemental Figure 2A online). Sho1-GFP showed an asymmetric patchy distribution in the plasma membrane of the mother cells and only faint staining in the plasma membrane of daughter cells (Figure 1B). The subcellular localization of Sho1-GFP was also determined by cell fractionation. Sho1-GFP was exclusively found in the membrane fraction, confirming that Sho1 is a plasma membrane protein (see Supplemental Figure 3 online).
The interaction between Sho1 and Kpp6 was verified by in vivo coimmunoprecipitation. Since kpp6 is not expressed in budding cells (Brachmann et al., 2003), a myc-kpp6 fusion construct was placed under the control of the constitutively active otef promoter. This construct was integrated in multiple copies into the ip locus of SG200Δkpp6 and SG200sho1GFP. Antibodies against the Myc-tag were used to precipitate Myc-Kpp6, and this weakly coprecipitated Sho1-GFP in SG200sho1GFP/otef:kpp6-mc (see Supplemental Figure 4A online).
To analyze whether the observed interaction between Sho1 and Kpp6 is specific, the yeast two-hybrid system was used. The coding region of four MAP kinases from U. maydis were cloned in the bait vector pGBKT7 and cotransformed with pGADT7-Sho1131-335 (see Supplemental Figure 4B online) into the S. cerevisiae strain AH109. The MAP kinases tested were the previously characterized kinases Kpp2 and Kpp6 (Mayorga and Gold, 1999; Müller et al., 1999; Brachmann et al., 2003), as well as Um02357, a MAP kinase related to Hog1p, and Um10107, a MAP kinase related to Slt2p. All strains showed similar expression levels of the respective MAP kinase proteins (see Supplemental Figure 4C online). The resulting strains were able to grow on selection plates; however, on high stringency plates, only the strain transformed with pGBKT7-Kpp6 and pGADT7-Sho1131-335 was able to grow (see Supplemental Figure 4D online), indicating that Sho1 and Kpp6 interact specifically.
U. maydis Encodes a Mucin-Like Protein
In S. cerevisiae, Sho1p regulates filamentous growth and osmotic stress responses together with Msb2p (O'Rourke and Herskowitz, 2002; Cullen et al., 2004; Tatebayashi et al., 2007). The U. maydis um00480 gene product showed significant similarity to yeast Msb2p (28% identity) as well as to proteins annotated as Msb2 orthologs in other fungi (Krantz et al., 2006) and was therefore designated Msb2. msb2 encodes a putative protein of 1140 amino acids and contains all the classical characteristics described for transmembrane mucin proteins (Singh and Hollingsworth, 2006), such as an N-terminal signal sequence (amino acids 1 to 30), a Ser/Thr-rich domain (amino acids 108 to 692) that includes in the case of U. maydis Msb2 three identical repetitive domains of 69 amino acids each (amino acids 430 to 636), and one transmembrane domain (amino acids 895 to 916) followed by a positively charged Lys/Arg-rich motif (RKHRK between amino acids 916 and 920; Figure 1A).
A biologically active C-terminal fusion of Msb2 (see below and Supplemental Figure 2A online) with the cherry fluorescent protein (mCherry) was generated in SG200sho1GFP to investigate putative colocalization of Msb2 and Sho1. The Msb2-mCherry fusion protein was concentrated in vacuoles but was absent from the plasma membrane (Figure 1B). We reasoned that this might result from a highly efficient actin-dependent endocytosis pathway and treated cells with latrunculin A, an inhibitor of actin polymerization. Latrunculin A has been demonstrated to efficiently inhibit endocytosis of the U. maydis pheromone receptor Pra1 (Fuchs et al., 2006). In latrunculin A–treated cells, the majority of Msb2-mCherry still localized to vacuoles, but under such conditions, a weak signal could additionally be detected in the plasma membrane and this signal colocalized with Sho1-GFP (Figure 1B).
Typically, cell surface–associated mucins are highly glycosylated and proteolytically cleaved, which separates the large extracellular part from the membrane-bound C terminus (Hollingsworth and Swanson, 2004). The predicted size of the Msb2-mCherry fusion protein is 145 kD. Immunoblotting revealed that the Msb2-mCherry protein is larger than 170 kD (see Supplemental Figure 3 online). Furthermore, a 65-kD fragment that could represent a specific C-terminal cleavage product of Msb2-mCherry was detected. The full-length protein as well as the C-terminal fragment were mainly found in the membrane fraction and only to a minor extent in the cytoplasm (see Supplemental Figure 3 online). These results indicate that U. maydis Msb2 is a membrane-associated mucin.
sho1 and msb2 Are Not Required for Mating and Do Not Affect Stress Responses
To investigate the function of sho1 and msb2 in U. maydis, deletion mutants and double deletion mutants were generated in the compatible haploid strains FB1 and FB2 as well as in the solopathogenic strain SG200 using a one-step gene replacement procedure (Kämper, 2004). Haploid deletion derivatives were tested for mating by cospotting cultures on potato dextrose (PD) charcoal plates. After 48 h, all strain combinations had developed vigorous dikaryotic hyphae that gave the colonies a fuzzy appearance. However, the mating response of crosses with Δsho1 Δmsb2 strains was slightly less efficient than in combinations with sho1 and msb2 single mutants or wild-type strains (see Supplemental Figure 5A online). When SG200 and the SG200 mutant derivatives were spotted on PD charcoal plates, SG200Δsho1 Δmsb2 showed slightly reduced filamentation after 24 h (Figure 2A). These results likely reflect that sho1 and msb2 are not required for a successful cell fusion reaction while the development of dikaryotic hyphae is slightly attenuated in the absence of both genes.
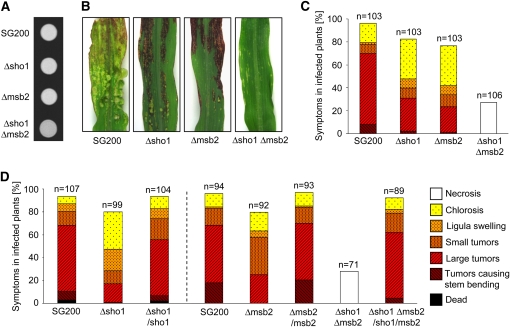
Figure 2.
Pathogenicity of sho1 and msb2 Mutant Strains in the SG200 Background.
(A) The solopathogenic strain SG200 and its derivatives indicated below were spotted on PD charcoal plates and incubated for 24 h at 28°C. The white fuzzy colonies reflect the formation of b-dependent filaments.
(B) Representative leaves 12 d after infection with the indicated strains.
(C) Disease symptoms caused by SG200, sho1, and msb2 single mutants and sho1 msb2 double mutants in the SG200 background. The indicated strains were injected into maize seedlings and symptoms were scored 12 d after infection. Based on the severity of symptoms observed on each plant, symptoms were grouped into color-coded categories according to Kämper et al. (2006). Colors and patterns for disease scores are indicated to the right of (D). Three independent experiments were performed and the average values are expressed as a percentage of the total number of infected plants (n), which is given above each column. Tested strains are listed below each column.
(D) Complementation of SG200-derived sho1 and msb2 single and double mutants. Plants were infected with the indicated strains and evaluated as described in (C).
In S. cerevisiae, Candida albicans, and Aspergillus fumigatus, deletion of putative orthologs of sho1 resulted in strains with increased sensitivity to oxidative and osmotic stress as well to cell wall interfering compounds like congo red and calcofluor (Maeda et al., 1995; Roman et al., 2005; Ma et al., 2008). To investigate whether sho1 and msb2 in U. maydis contribute to growth under stress conditions, serial dilutions of SG200, the sho1 and msb2 mutant derivatives, as well as the sho1 msb2 double mutant were plated on agar containing the respective stressors. Growth on these plates was indistinguishable between mutant strains and SG200 on media containing congo red (70 μg/mL), calcofluor (50 μM), H2O2 (1.5 mM), NaCl (1 M), sorbitol (2 M), and water agar containing 1% glucose (see Supplemental Figure 6A online). Under nitrogen limiting conditions, S. cerevisiae switches from bipolar budding to pseudohyphal growth, and this process depends on Sho1p and Msb2p (Gimeno et al., 1992; Cullen et al., 2004). U. maydis also grows filamentously on low ammonium medium (Smith et al., 2003). To test if Sho1 and Msb2 have a function during this process, the colony morphology of SG200, the sho1 and msb2 single mutant derivatives as well as the sho1 msb2 double mutant were analyzed on nitrogen limiting medium (SLAHD). All strains showed a filamentous colony morphology after 36 h (see Supplemental Figure 6B online). This indicates that contrary to the situation in S. cerevisiae and other fungi, sho1 and msb2 in U. maydis do not participate in stress sensing.
sho1 and msb2 Are Required for Pathogenic Development in U. maydis
To analyze the role of sho1 and msb2 during pathogenic development, 7-d-old maize plants were infected with SG200, SG200Δsho1, SG200Δmsb2, and SG200Δsho1 Δmsb2. Disease symptoms were scored after 12 d according to severity (Kämper et al., 2006), and averages from three independent experiments are shown (Figure 2). Compared with SG200, a decreased tumor rate was observed in infections with SG200Δsho1 and SG200Δmsb2, and in both cases, the percentage of plants with severest disease symptoms was most strongly reduced (Figures 2B and 2C). Interestingly, SG200Δsho1 Δmsb2 was unable to cause tumors and instead necrotic lesions were observed (Figures 2B and 2C). To verify the reduced virulence independently, maize plants were infected with compatible mixtures of wild-type and sho1 or msb2 deletion strains, respectively (see Supplemental Figure 5B online). While the overall symptom severity was higher than in SG200 and its derivatives, single sho1 or single msb2 mutants were again attenuated in virulence. In crosses between FB1Δsho1 Δmsb2 and FB2Δsho1 Δmsb2, neither tumor formation nor chlorosis was observed and instead necrotic lesions developed (see Supplemental Figure 5B online), which corroborates the findings for SG200Δsho1 Δmsb2.
To complement the sho1 and msb2 single mutants, either sho1 or msb2 was reintroduced in single copy in the ip locus of the respective SG200-derived deletion strains. In the resulting strain SG200Δmsb2/msb2, virulence could be completely restored, while in SG200Δsho1/sho1, complementation levels were slightly lower (Figure 2D). To explain this, we speculate that the sho1 locus contains regulatory upstream or downstream elements that are missing in the complementation construct. To demonstrate that the loss of virulence in SG200Δsho1 Δmsb2 resulted only from disruption of sho1 and msb2, a sho1 msb2 double construct was integrated in single copy in the ip locus of the sho1 msb2 double mutant. In this strain, virulence was almost completely restored (Figure 2D). The slight attenuation compared with SG200 is likely due to incomplete complementation by the sho1 gene (see above).
To investigate what is underlying the induction of necrotic lesions after infection with SG200Δsho1 Δmsb2, infected leaves were harvested after 3 d and stained with propidium iodide and calcofluor. Calcofluor allows detection of the fungal material on the leaf surface, while the non-membrane-permeable propidium iodide stains plant cell walls and DNA, if the membrane is ruptured (i.e., in dead plant cells). In infections with SG200, the propidium iodide stain was almost exclusively found in the plant cell walls, while after infection with SG200Δsho1 Δmsb2, plant cell walls in the infected area were more strongly stained, plant cell nuclei were brightly stained, and these cells showed internal blue staining presumed to reflect autoflourescence (see Supplemental Figure 7 online). This indicates that the Δsho1 Δmsb2 mutant elicits plant defense responses associated with plant cell death.
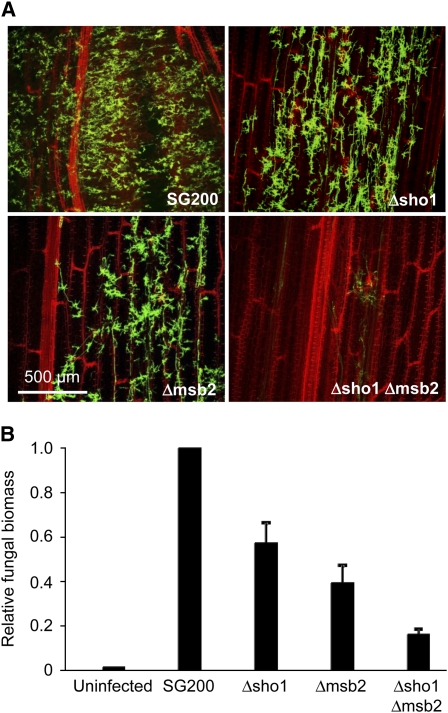
To elucidate at which developmental stage the mutants are affected, the infected plant areas were stained with carbohydrate binding wheat germ agglutinin–alexa fluor conjugate (WGA-AF 488) and analyzed by confocal microscopy 6 d after inoculation. Massive intracellular fungal proliferation was observed in leaves infected with SG200, while leaves infected with SG200Δsho1 or SG200Δmsb2 showed reduced fungal material (Figure 3A). By contrast, after infection with SG200Δsho1 Δmsb2, only very few intracellular hyphae could be detected (Figure 3A). To quantify fungal biomass, leaves of infected plants were detached 3 d after infection, fungal cells on the surface were removed by latex treatment and total DNA was extracted. Relative fungal biomass was determined by quantitative real-time PCR using the fungal mfa1 gene and the plant GAPDH gene as internal standards (Figure 3B). sho1 and msb2 single mutants were about twofold and threefold reduced in fungal biomass, respectively, compared with SG200, while the double mutant showed a reduction of about fivefold (Figure 3B). Because of the severely reduced amount of intracellular fungal material, we consider it likely that the sho1 msb2 double mutant is affected in a step prior to or during penetration.
Figure 3.
Host Colonization Is Impaired in sho1 and msb2 Mutants.
(A) Confocal projections of representative leaves 6 d after infection with the indicated strains. Plant tissue is shown at the layer of vascular bundles. Fungal material was stained by WGA-AF488 (green). Plant material was stained by propidium iodide (red).
(B) Quantification of fungal biomass by quantitative PCR. U. maydis–specific and plant-specific primers were used to determine the relative fungal biomass in plants 3 d after infection with the indicated strains. Columns give ratios of fungal DNA to plant DNA, and the ratio in SG200-infected plants was set to 1.0. Means of three independent experiments with 30 leaves per strain are shown, and error bars indicate standard deviations.
Appressorium Development Is Regulated by sho1 and msb2
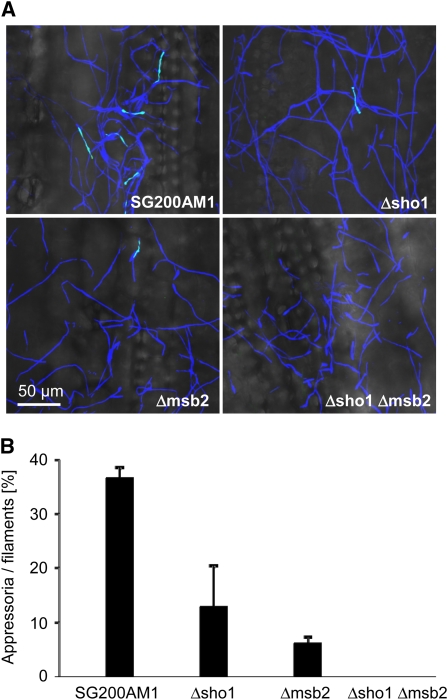
To address whether filamentation on the leaf surface and appressorium formation were affected by sho1 and/or msb2 deletion, we introduced the AM1 marker, which is a promoter fused to a triple gfp that is specifically upregulated in those hyphal cells that differentiate appressoria (Mendoza-Mendoza et al., 2009b). Calcofluor staining revealed that SG200AM1, sho1, and msb2 single as well as double mutant strains showed comparable filamentation on the leaf surface (Figure 4A). However, when the percentage of hyphae that had developed appressoria was determined 18 h after inoculation, both SG200AM1Δsho1 and SG200AM1Δmsb2 were significantly reduced in appressorium formation compared with SG200AM1, while SG200AM1Δsho1 Δmsb2 did not develop any appressoria (Figures 4A and 4B).
Figure 4.
Sho1 and Msb2 Affect Appressorium Formation on the Plant Surface.
(A) Maize seedlings were infected with SG200AM1 and its derivatives as indicated. Eighteen hours after infection, the surface of the third oldest leaf was analyzed by confocal microscopy. Fungal hyphae were stained by calcofluor (blue), and expression of the AM1 marker (green) indicates appressorium formation. The overlays of maximum projections of both channels with the corresponding bright-field images are depicted.
(B) Quantification of appressoria on the plant surface. Using the same strains as in (A), the average percentage of filaments that had formed appressoria to the total number of filaments was determined. For each strain (indicated below each column), >900 filaments were analyzed in three independent experiments. Error bars indicate standard deviations.
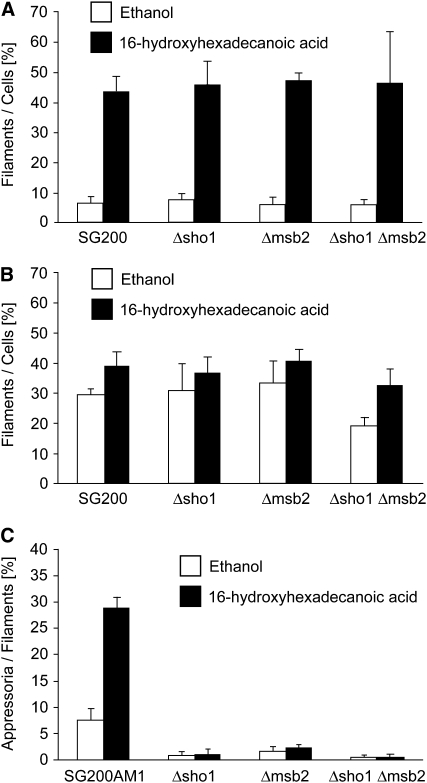
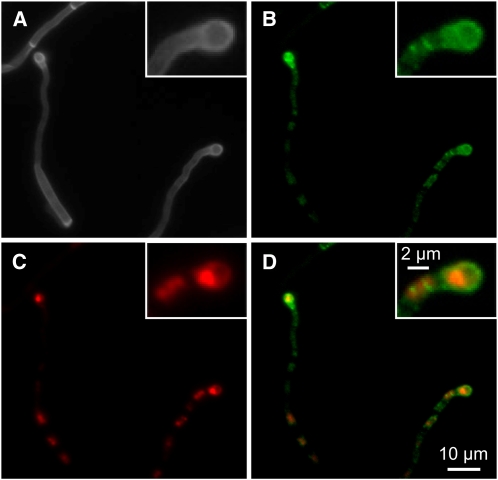
We recently established in vitro conditions for filamentation and appressorium development: filaments resembling conjugation tubes can be induced in SG200 by 16-hydroxyhexadecanoic acid, while hydrophobicity induces filaments with empty sections. On the hydrophobic surface, a few appressoria are induced and this level is strongly enhanced when 16-hydroxyhexadecanoic acid is added (Mendoza-Mendoza et al., 2009b). The sho1 and msb2 single mutant as well as the double mutant responded to 16-hydroxyhexadecanoic acid like the parental strain SG200 (Figure 5A; see Supplemental Figure 8 online). On the hydrophobic surface, SG200Δsho1 Δmsb2 was affected in filamentation, while the sho1 and msb2 single mutants reacted like the progenitor strain (Figure 5B; see Supplemental Figure 9 online). The addition of 16-hydroxyhexadecanoic acid enhanced filamentation in all mutant strains as described previously for SG200 (Mendoza-Mendoza et al., 2009b). However, with respect to appressorium development on the hydrophobic surface, the sho1 and msb2 single as well as the double mutant were severely reduced, and the addition of 16-hydroxyhexadecanoic acid had only a slight enhancing effect in the single mutants and no effect in the double mutant (Figure 5C; see Supplemental Figure 9 online). This shows that sho1 and msb2 specifically affect development of appressoria. Therefore, we determined the localization of Sho1-GFP during appressorium formation. Sho1-GFP showed a patchy distribution in filaments but accumulated strongly in appressoria (Figures 6A and 6B). Attempts to visualize Msb2-mCherry in filaments failed due to the background fluorescence of the inducing surface. To enhance expression of msb2-mCherry, we placed the gene under the control of the constitutively active otef promoter (Spellig et al., 1996) and integrated this construct into the ip locus in strains SG200Δmsb2 and SG200sho1GFP. The construct was able to complement SG200Δmsb2 (see Supplemental Figure 2B online). In filaments, Msb2-mCherry accumulated in vacuoles that were distributed throughout the filament. In appressoria, Msb2-mCherry strongly accumulated in vacuoles (Figures 6A and 6C), corroborating a function of Msb2 during this stage of development. Colocalization with Sho1-GFP was not apparent (Figure 6D).
Figure 5.
sho1 and msb2 Mutants Respond to Hydroxy Fatty Acid but Are Impaired in Their Response to Hydrophobic Surfaces.
(A) Filamentation in liquid: The indicated strains (below each column) were tested for their response to 16-hydroxyhexadecanoic acid. Cells were incubated in liquid culture (2% YEPSL) supplemented with either 100 μM 16-hydroxyhexadecanoic acid dissolved in ethanol (black columns) or with ethanol (white columns) for 18 h at 28°C. The average percentage of cells that grew filamentously was determined by microscopy analysis.
(B) Filamentation on a hydrophobic surface: Cell suspensions of the indicated strains in 2% YEPSL were sprayed on Parafilm M with 100 μM 16-hydroxyhexadecanoic acid dissolved in ethanol (black columns) or with ethanol (white columns) and incubated for 18 h at 28°C. After staining of fungal cells with calcofluor, the average percentage of cells that have formed filaments was determined microscopically.
(C) Appressorium formation on a hydrophobic surface: SG200AM1 and its derivatives indicated below the columns were sprayed on a hydrophobic surface as described in (B), incubated for 18 h at 28°C, and stained with calcofluor. After microscopy analysis, the average percentage of cells that expressed the AM1 marker was determined relative to the cells that had formed filaments.
In three independent experiments, >900 cells ([A] and [B]) or filaments (C) per strain were analyzed, and error bars indicate standard deviations.
Figure 6.
Localization of Sho1-GFP and Msb2-mCherry in Appressoria.
(A)to (D) SG200sho1GFP/otef:msb2mCherryHA was sprayed with 100 μM 16-hydroxyhexadecanoic acid on paraffin wax and incubated as described in Figure 5B. A section displaying two appressoria is analyzed. The appressorium on the right-hand side is magnified in the inset.
(A) Calcofluor staining.
(B) Visualization of Sho1-GFP (green).
(C) Visualization of Msb2-mCherry (red).
(D) Overlay of (B) and (C).
Sho1 and Msb2 Act Upstream of the MAP Kinases Kpp2 and Kpp6
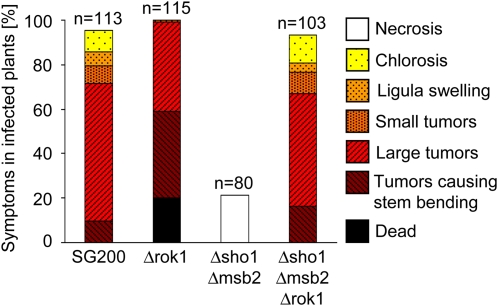
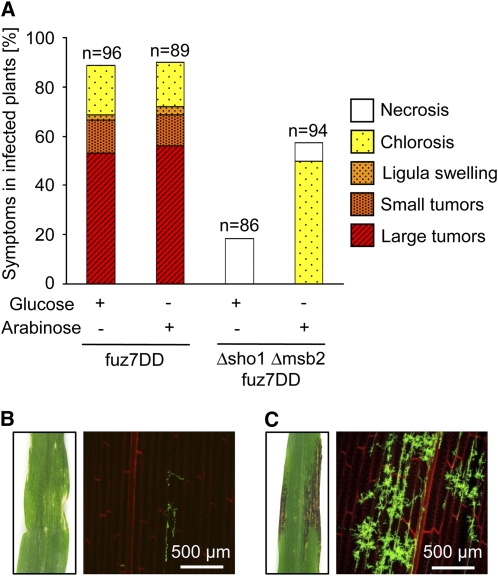
In U. maydis, the MAP kinase Kpp2 is implicated in appressorium development induced by the hydrophobic stimulus (Müller et al., 2003; Mendoza-Mendoza et al., 2009b), while the related MAP kinase Kpp6 is specifically needed for appressorium function (Brachmann et al., 2003). The dual specificity phosphatase Rok1 is a negative regulator of the MAP kinases Kpp2 and Kpp6, and deletion of rok1 leads to hypervirulence (Di Stasio et al., 2009). To test whether activated Kpp2 and Kpp6 restore tumor development in SG200Δsho1 Δmsb2, we deleted rok1 in this strain. While plants infected with SG200Δsho1 Δmsb2 showed no tumor development 12 d after infection, SG200Δsho1 Δmsb2 Δrok1–infected plants developed tumors and symptom severity was comparable to SG200 infections (Figure 7). To substantiate that Sho1 and Msb2 activate the MAP kinase module containing Kpp2 and Kpp6 and to rule out the possibility that rok1 suppresses the virulence phenotype of the Δsho1 Δmsb2 mutant by a mechanism that is independent of Kpp2 and Kpp6, we integrated the constitutively active version of the MEK fuz7 (fuz7DD; Müller et al., 2003) under the control of the arabinose-inducible crg1 promoter in the genome of SG200Δsho1 Δmsb2 as well as in the genome of SG200. Expression of fuz7DD was induced by adding arabinose to the inoculum prior to infection of maize plants. As a control, arabinose was replaced by glucose, which represses the activity of the crg1 promoter (Bottin et al., 1996). When SG200fuz7DD was used to infect maize plants, symptom development was comparable irrespective of whether glucose or arabinose was added to the inoculum (Figure 8A). SG200Δsho1 Δmsb2 fuz7DD induced necrosis when glucose was added (Figures 8A and 8B). However, when arabinose was added as an inducer for fuz7DD, we observed chlorotic areas and anthocyanin production, which is indicative of successful penetration (Figures 8A and 8C). Microscopy analysis of leaves 6 d after infection confirmed that penetration had occurred and the amount of fungal material inside the leaf was much more abundant when fuz7DD was induced compared with the repressed situation (Figures 8B and 8C). These data indicate that Sho1 and Msb2 act upstream of the MAP kinases Kpp2 and Kpp6.
Figure 7.
The Deletion of rok1 Suppresses the Virulence Phenotype of SG200Δsho1 Δmsb2.
Maize plants were infected with SG200 and the indicated derivatives. Disease rating was done as described in the legend to Figure 2. Tested strains are listed below each column, the numbers of infected plants (n) are given above each column, and the color and pattern code for disease rating is depicted on the right.
Figure 8.
Expression of a Constitutively Active Allele of fuz7 Partially Bypasses the Need for Sho1 and Msb2.
(A) The indicated SG200 derivatives were inoculated into maize seedlings. Prior to infection, either arabinose (to induce fuz7DD expression) or glucose (to repress fuz7DD expression) was added to the inoculums to a final concentration of 1%. Twelve days after infection, symptoms were scored as described in the legend to Figure 2. Inoculated strains and glucose/arabinose supplements are listed below each column, numbers of infected plants (n) are given above each column, and the color and pattern code for disease rating is depicted on the right.
(B) Macroscopy and microscopy symptoms after infection with SG200Δsho1 Δmsb2 fuz7DD in the presence of glucose. A representative infected leaf is shown on the left 12 d after infection. Confocal microscopy performed as described in the legend to Figure 3A reveals poor colonization of plant tissue 6 d after infection (right panel).
(C) Macroscopy and microscopy symptoms after infection with SG200Δsho1 Δmsb2 fuz7DD in the presence of arabinose. A representative infected leaf 12 d after infection is shown on the left. Confocal microscopy performed as described in legend to Figure 3A reveals strongly enhanced colonization of plant tissue 6 d after infection (right panel).
Functional Analysis of Msb2
If Msb2 functions as a cell surface receptor, the transmembrane domain should be important for function. To test this hypothesis, the transmembrane domain within the otef:msb2-mCherryHA construct was deleted and the construct integrated in the ip locus of SG200Δmsb2. Plant infections revealed that msb2ΔTM-mCherry-ha could not complement the virulence defect of the msb2 deletion strain (see Supplemental Figure 10A online). Furthermore, only the full-length protein could be detected in immunoblot analysis, whereas the C-terminal Msb2-mCherryHA fragment was absent (see Supplemental Figure 10B online). These results indicate that the transmembrane domain of Msb2 is essential for processing and function of Msb2.
U. maydis Msb2 and S. cerevisiae Msb2p have similar domain structures but display only 28% amino acid identity (Figure 1A). To address whether U. maydis msb2 is able to complement the S. cerevisiae msb2 mutant, we made use of the yeast strain PC538 that is defective in the mating pathway and has a FUS1:HIS3 reporter whose expression is driven only by the FG pathway (Cullen et al., 2004). The PC538 derivative PC948 carries the msb2 deletion and is hence unable to grow on medium without His (Cullen et al., 2004). U. maydis msb2 and S. cerevisiae MSB2 were inserted under the control of the constitutively active ADH1 promoter into the free replicating plasmid pVTU260 and transformed into PC948. Expression of U. maydis Msb2 was verified by immunoblot analysis (see Supplemental Figure 11A online). S. cerevisiae strain PC948, expressing U. maydis msb2, was unable to grow on medium lacking His, while S. cerevisiae MSB2 could complement the growth phenotype of the msb2 deletion strain (see Supplemental Figure 11B online). This demonstrates that, although characteristic features are shared between the Msb2 proteins in U. maydis and S. cerevisiae, they do not represent functional homologs.
The SH3 Domain of Sho1 Interacts with a Negative Regulatory Domain of Kpp6
To establish the biological significance of the observed interaction between Sho1 and Kpp6, we delineated the region responsible for the interaction between Kpp6 and Sho1. Different segments of Sho1 and Kpp6 were tested by two-hybrid interaction for their ability to interact with each other. In all cases, the amounts of fusion proteins expressed were comparable (see Supplemental Figure 12A online). We found that Kpp6 interacted specifically with the SH3 domain of Sho1 (amino acids 279 to 335), while the linker domain (amino acids 131 to 278) that is situated between the SH3 domain and the fourth transmembrane domain showed no interaction with Kpp6. With respect to Kpp6, we mapped the interaction domain to amino acids 1 to 170, which constitutes the unique N-terminal domain of Kpp6 (see Supplemental Figures 12B and 12C online). Based on the knowledge that SH3 domains interact with Pro-rich domains (PRDs; Li, 2005), we searched for such a motif in the N terminus of Kpp6. This led to the identification of the motif KPLPPSP (corresponding to amino acids 127 to 133), which conforms to type I PRDs (R/KxxPxxP; Li, 2005). To test the functional significance of this motif in Kpp6, we generated a mutant in which two Pro residues are substituted by Ala (KPLAASP). Two-hybrid interactions demonstrated that Kpp6P130A P131A is no longer able to interact with the SH3 domain of Sho1 despite similar expression levels of the respective fusion proteins (see Supplemental Figure 12 online).
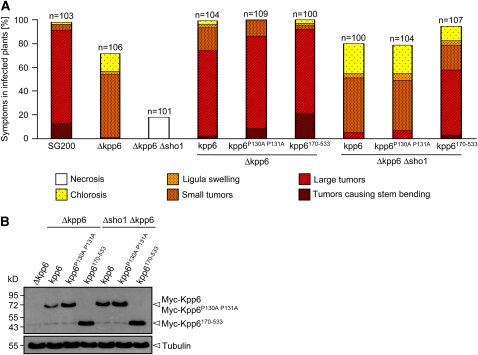
To test whether the observed interaction of Sho1 and Kpp6 is relevant for pathogenic development of U. maydis, a sho1 allele lacking the SH3 domain (sho1Δ280-335) was generated in the endogenous locus in SG200. Pathogenicity assays revealed that this mutant was attenuated in virulence to a level slightly above that observed in the sho1 deletion mutant (see Supplemental Figure 13 online). With respect to the functional relevance of the interaction domain in Kpp6, we generated alleles in which either the N-terminal domain was deleted or the PR motif was mutated. To avoid problems associated with the complex regulation of kpp6 (Brachmann, 2001), these alleles were expressed from the constitutive otef promoter and carried an N-terminal myc tag. A derivative of SG200Δkpp6 expressing N-terminally myc-tagged wild-type kpp6 from the otef promoter (otef:kpp6) served as control. In this control strain, partial complementation of the kpp6 deletion phenotype was observed (Figure 9A). In SG200Δkpp6 derivatives expressing kpp6P130A P131A, virulence was elevated compared with SG200Δkpp6/otef:kpp6, and the expression of kpp6Δ1-169 in SG200Δkpp6 led to a further increase in virulence (Figure 9A). However, when the same kpp6 alleles were introduced in SG200Δsho1 Δkpp6, which is unable to cause disease (Figure 9A), symptom development was restored to the same extent with both the wild-type allele and the kpp6P130A P131A allele (Figure 9A). On the other hand, introduction of the kpp6Δ1-169 allele into SG200Δsho1 Δkpp6 led again to increased virulence compared with that of kpp6 or kpp6P130A P131A. The virulence-enhancing effect of the kpp6Δ1-169 allele was significantly higher in the Δsho1 Δkpp6 background than in the Δkpp6 background (Figure 9A). This illustrates that the N terminus of Kpp6 as well as the PRD alone have a negative role during virulence. In the absence of sho1, the negative effect of the PRD alone was no longer visible. To elucidate the basis for these findings, we determined the Kpp6 protein levels of the various strains (Figure 9B). This revealed that Kpp6P130A P131A levels were significantly higher than Kpp6 levels in the Δkpp6 strain background. By contrast, in the Δsho1 Δkpp6 strain background, Kpp6 and Kpp6P130A P131A protein levels were comparable and as high as the Kpp6P130A P131A levels in the Δkpp6 strain background (Figure 9B). Compared with the Kpp6P130A P131A level, the amount of the truncated version Kpp6Δ1-169 was further elevated and was indistinguishable in the Δkpp6 and Δsho1 Δkpp6 strain background (Figure 9B). This suggests that Sho1 negatively influences the stability of Kpp6 through direct interaction with the N terminus of Kpp6.
Figure 9.
The Interaction of Sho1 and Kpp6 Influences Pathogenicity.
(A) Wild-type kpp6, kpp6P130A P131A, and N-terminally truncated kpp6Δ1-169 were constitutively expressed as N-terminal Myc-fusion proteins in either SG200Δkpp6 or SG200Δsho1 Δkpp6. The SG200 derivatives listed below each column were inoculated into maize seedlings, and symptoms were scored 12 d after infection as described in the legend to Figure 2.
(B) The indicated SG200Δkpp6- and SG200Δsho1 Δkpp6-derived strains were grown in liquid YEPSL to an OD600 of 1.0, and proteins were extracted and subjected to SDS-PAGE. After blotting, anti-Myc was used to detect Myc-Kpp6 as well as its mutated and truncated alleles (top panel). The Myc antibody detects with low signal intensity one unspecific cross-hybridizing protein at the size of Myc-Kpp6Δ1-169 that was disregarded. Tubulin served as loading control and was detected with antitubulin (bottom panel). The fusion proteins and tubulin are indicated by arrowheads on the right. The molecular mass marker is depicted on the left.
DISCUSSION
In this article, we showed that Sho1 and Msb2 in U. maydis are essential virulence factors. sho1 and msb2 mutants are specifically reduced in pathogenic development and are not significantly affected in mating and stress responses. The virulence defects could be traced back to a failure in developing appressoria.
Using bioinformatic tools, Msb2 of U. maydis was recently classified as an ortholog of S. cerevisiae Msb2p (Krantz et al., 2006). Complementation studies have now shown that these proteins are not functional homologs, which is not unexpected, in view of the light that they regulate distinct processes in both organisms (see below). Nevertheless, with respect to domain structure, Msb2p from S. cerevisiae and Msb2 from U. maydis are clearly related (Figure 1A). For Msb2p in S. cerevisiae, it has been demonstrated that the extracellular domain is highly N- as well as O-glycosylated (Cullen et al., 2004; Yang et al., 2009), and using the program NetNGlyc 1.0 (http://cbs.dtu.dk/services/NetNGlyc/), seven N-glycosylation sites in U. maydis Msb2 (eight in S. cerevisiae Msb2p) were predicted. Furthermore, the combination of all six recommended constraints of the program OGPET 1.0 (http://ogpet.utep.edu/OGPET/) predicts an average of 12 O-glycosylation sites for Msb2 in U. maydis (nine for S. cerevisiae Msb2p). We consider it likely that Msb2 in U. maydis is a glycosylated transmembrane mucin. This assertion is supported by the finding that its apparent molecular mass of >170 kD is significantly higher than its expected mass of 147 kD as well as the finding that the transmembrane domain is crucial for Msb2 function.
While Sho1- and Msb2-related proteins serve as stress sensors in all fungal systems analyzed to date (Maeda et al., 1995; O'Rourke and Herskowitz, 2002; Roman et al., 2005, 2009; Norice et al., 2007; Boisnard et al., 2008; Ma et al., 2008), sho1 and msb2 are not needed for general stress responses in U. maydis. Responses to osmotic stress, oxidative stress, or cell wall stress were not altered in sho1 and msb2 single or double mutants, suggesting that the function of Sho1 and Msb2 in U. maydis is uncoupled from the HOG pathway and serves in pathogenicity only.
In U. maydis, pathogenic development is initiated by b-dependent filaments, which are septated and accumulate empty sections. On the plant surface, sho1 and msb2 mutants were able to form these filaments as efficiently as the progenitor strain. However, single deletion mutants in sho1 or msb2 showed reduced colonization of the plant tissue, and the sho1 msb2 double deletion mutant had an almost complete colonization defect. The quantification of appressoria on the plant surface revealed that single deletion strains of sho1 or msb2 form fewer appressoria than SG200, and the sho1 msb2 double deletion strain was not able to form appressoria. Thus, Sho1 and Msb2 have cooperative and essential functions during appressorium development. Interestingly, mutants lacking an ortholog to MSB2 in Fusarium oxysporum showed delayed invasive growth and were strongly attenuated in virulence (E. Perez-Nadales and A. Di Pietro, personal communication), suggesting that mucins may have a general function during the early infection stages of phytopathogenic fungi.
We observed strong plant defense responses when the sho1 msb2 double mutant grows on the leaf surface. This most likely reflects pathogen-associated molecular pattern-induced defense responses that are suppressed after wild-type hyphae penetrate the epidermal layer (Doehlemann et al., 2008a). Due to the penetration defect, the sho1 msb2 double mutant would be unable to suppress this reaction. The failure of the sho1 msb2 mutant to develop appressoria is at variance with results that rely on fungal biomass determinations. In these experiments, only a fivefold reduction in colonization was observed relative to the progenitor strain SG200. In addition, confocal microscopy allowed us to detect rare colonization events by the sho1 msb2 mutant. We consider intracellular fungal biomass determination during early infection stages to be error prone as it relies on complete removal of fungal hyphae from the leaf surface by latex treatment. To explain the rare colonization successes of the sho1 msb2 mutant, we speculate that this reflects the ability of U. maydis to enter plant tissue as a saprophyte without the formation of appressoria. This could actually be facilitated by dead plant cells as they are found underneath SG200Δsho1 Δmsb2 cells growing on the leaf surface.
In yeast, Msb2p has been shown to interact with Sho1p (Cullen et al., 2004). Attempts to visualize an interaction between Sho1 and Msb2 in U. maydis relying on epitope-tagged proteins and coimmunoprecipitations were unsuccessful. This could suggest that interactions are only transient and may be restricted to appressoria (i.e., a stage not yet accessible to biochemical analysis). Msb2p of yeast has also been demonstrated to interact with Cdc42p, and this complex is hypothesized to provide sensory capacity in the FG pathway that is transmitted via the PAK kinase Ste20p (Cullen et al., 2004). In U. maydis, Cdc42 is part of a signaling module consisting of the Rho-GEF Don1 and the Ste20-like kinase Don3 (Weinzierl et al., 2002). While don3 mutants are not affected in pathogenic development (Weinzierl et al., 2002), cdc42 mutants are nonpathogenic (Mahlert et al., 2006). A second PAK in U. maydis, Cla4, a member of the Cla4 subfamily of Ste20-like kinases, is involved in the regulation of cell polarity during budding and filamentation and cla4 mutants are nonpathogenic (Leveleki et al., 2004). It will be interesting to determine whether pathogenicity defects of cdc42 and cla4 mutants occur at the stage of appressorium formation as this could provide a direct link to Sho1 and Msb2.
Recently, the dual specificity phosphatase Rok1 has been characterized as a negative regulator of Kpp2 and Kpp6. Deletion of rok1 results in hypervirulence, which can be partially explained by increased appressorium formation (Di Stasio et al., 2009). When the rok1 gene was deleted in the Δsho1 Δmsb2 background, the resulting strain became fully pathogenic, indicating that Sho1 and Msb2 feed into the Kpp2/Kpp6 MAP kinase cascade, and a lack of this input can be compensated for by the deletion of rok1. This is corroborated by demonstrating that expression of the constitutively active allele of the MEK Fuz7 that activates the MAP kinases Kpp2 and Kpp6 (Müller et al., 2003; Di Stasio et al., 2009) allowed to bypass the colonization defect of the sho1 msb2 deletion strain. Since tumor formation was not observed when fuzDD was induced by arabinose prior to infection with SG200Δsho1 Δmsb2 fuz7DD, this most likely indicates that fuz7DD expression ceases after penetration (due to insufficient amounts of arabinose in the plant tissue) and that Sho1 and Msb2 have additional, Fuz7-dependent functions during the biotrophic phase. Since U. maydis appears to require the formation of appressoria for cell-to-cell passages (Doehlemann et al., 2009), it is conceivable that the reinitiation of this program is affected in the sho1 msb2 double mutant. Overall, these results place Sho1 and Msb2 upstream of Kpp2 and Kpp6, where they function as sensors to activate MAP kinase signaling.
An intriguing feature of this signaling pathway is that Sho1 protein was identified as a two-hybrid interactor of Kpp6 (Mendoza-Mendoza et al., 2009a). We demonstrated here that this interaction occurs via the SH3 domain of Sho1 and the PR-motif in Kpp6. This motif is located in the N-terminal extension of Kpp6, which is absent from Kpp2 (Brachmann et al., 2003). kpp6 is strongly upregulated in b-dependent filaments and is specifically needed for appressoria to penetrate (Brachmann et al., 2003). The finding that a kpp6 allele that lacks the N-terminal domain is significantly stronger in complementing the pathogenicity defect of kpp6 mutants than wild-type kpp6 suggests that the N terminus of Kpp6 has an inhibitory function. A Kpp6 variant that no longer interacts with Sho1 due to mutations in the PR domain also causes enhanced virulence and this effect requires Sho1. The demonstration that Kpp6 protein levels are elevated when the PRD is mutated or sho1 is deleted makes it likely that the interaction with Sho1 destabilizes Kpp6. We speculate that the interaction with Sho1 could impose a negative feedback loop that might fine-tune Kpp6 levels during the differentiation of appressoria. In S. cerevisiae, it has been demonstrated that a negative feedback loop exists between Sho1p and Hog1p that is based on phosphorylation of Sho1p by Hog1p (Hao et al., 2007). The cytoplasmic domain of U. maydis Sho1 contains three putative MAP kinase phosphorylation sites. However, simultaneous mutation of all three sites did not affect virulence of the respective strain (D. Lanver and R. Kahmann, unpublished data), making a phosphorylation-based feedback loop unlikely. Despite the negative function of the PRD in the N terminus of Kpp6, this domain must additionally confer regulatory information. This is based on the observation that the deletion of the entire N terminus causes stronger virulence than mutating only the PRD in Kpp6. Furthermore, in this case, the enhanced virulence does not depend on Sho1.
The deletion of the SH3 domain in sho1 reduces virulence to a similar extent as disruption of the entire sho1 gene. Thus, the negative effect of the SH3 domain of Sho1 on Kpp6 levels cannot be the only function of this domain. In yeast, the MEK Pbs2p is recruited to the plasma membrane via interaction of the Sho1p SH3 domain with the Pro-rich motif KPLPPLP in the N terminus of Pbs2p (Maeda et al., 1995; Raitt et al., 2000; Reiser et al., 2000). In U. maydis, the MEK Fuz7 acts downstream of Sho1. Since Fuz7 lacks such a Pro-rich motif and we currently have no evidence that Sho1 and Fuz7 interact, further experimentation will be necessary to link the SH3 domain in Sho1 to components of the MAP kinase module.
In S. cerevisiae, activation of Msb2 is regulated by starvation-dependent induction of its cognate aspartyl protease Yps1p (Vadaie et al., 2008). Yps1p processes Msb2p into a secreted extracellular form and a cell-associated form. This cleavage releases the inhibitory mucin domain and generates the active form of Msb2 (Vadaie et al., 2008). Our finding of two forms of Msb2 in U. maydis, a high molecular mass form and a form representing the C terminus also suggests a processing event. However, we currently have no indication that the extracellular domain in Msb2 assumes a negative regulatory function. In S. cerevisiae, Msb2-GFP localizes primarily to vacuoles, and this localization depends on efficient processing of Msb2p (Vadaie et al., 2008). Given that U. maydis Msb2-mCherry also accumulates in vacuoles in the appressorium, we speculate that Msb2 could be activated when appressoria are formed.
Recently, Msb2p in S. cerevisiae has been identified as a target for the mannosyltransferase Pmt4p and deletion of pmt4 leads to underglycosylated Msb2p, as well as to enhanced activity of the FG pathway (Yang et al., 2009). Interestingly, the deletion of pmt4 in U. maydis causes a specific defect in appressorium development, while vegetative growth and filament formation are not affected (Fernandez-Alvarez et al., 2009). Since Msb2 is an upstream component of the MAP kinase cascade regulating appressorium development, Msb2 is likely to be a mannosylated target of Pmt4 in U. maydis.
In U. maydis, appressoria can be efficiently induced on a hydrophobic surface when hydroxy fatty acids are added (Mendoza-Mendoza et al., 2009b). In contrast with the situation on the leaf surface, in vitro appressorium formation was not only abolished in sho1 msb2 double mutants, but already in sho1 and msb2 single mutants. This could indicate that on the plant surface additional signals are provided that allow sho1 and msb2 single mutants to form appressoria. Under the in vitro conditions, sho1 and msb2 mutants were able to develop filaments in response to hydroxy fatty acids alone, indicating that this pathway is unaffected. Formation of septated filaments by the sho1 msb2 double mutant occurred inefficiently on the hydrophobic surface but was increased when hydroxy fatty acids were added. We take this to indicate that the hydrophobic surface is only poorly sensed. Moreover, with respect to the in vitro differentiation of appressoria, the sho1 and msb2 mutants are defective. We speculate that the reduced response to hydrophobicity of sho1 and msb2 mutants in vitro results from inefficient perception of the hydrophobic stimulus. As this is the crucial signal for appressorial differentiation (Mendoza-Mendoza et al., 2009b), the reduced perception could translate into the complete failure to form these infection structures. So far, it is not entirely clear what is sensed by Sho1p/Msb2p in S. cerevisiae (Cullen, 2007). The physical properties of the heavily glycosylated extracellular domains of mucins change dramatically in response to changes in extracellular milieu, and in higher eukaryotes, mucins have been shown to bind to and detect the presence of eukaryotic cells, proteins, and microorganisms (De Nadal et al., 2007). We consider these properties to be extendable to abiotic surface recognition, although formal proof for this needs further experimentation.
METHODS
Strain Construction and Growth Conditions
The Escherichia coli strains DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes. The Saccharomyces cerevisiae strain AH109 (Clonetech) was used for two-hybrid interaction studies. The S. cerevisiae strains PC538 and PC948 (Cullen et al., 2004) were used for msb2 complementation studies. All S. cerevisiae strains are listed in Supplemental Table 1 online.
Ustilago maydis strains were grown in liquid YEPSL (0.4% yeast extract, 0.4% peptone, and 2% sucrose) or on solid potato dextrose (PD) plates. For mating assays and filament induction, PD plates containing 1% activated charcoal were used (Holliday, 1974).
Haploid U. maydis strains FB1 and FB2, and solopathogenic strains SG200, SG200AM1, SG200Δkpp6, and SG200Δrok1 have been described previously (Banuett and Herskowitz, 1989; Brachmann et al., 2003; Kämper et al., 2006; Di Stasio et al., 2009; Mendoza-Mendoza et al., 2009b).
For gene disruptions, the PCR strategy described by Kämper (2004) and the SfiI insertion cassette system (Brachmann et al., 2004) were used. All gene replacement constructs were sequenced after cloning. After transformation, DNA gel blot analysis was performed to confirm gene replacements or insertions. All U. maydis strains are listed in Supplemental Table 2 online.
For generation of sho1 (um03156) deletion mutants, two 1.0-kb fragments containing the 5′ flanking region and the 3′ flanking region of the sho1 gene were amplified by PCR using FB1 genomic DNA as template with the primer combinations oAM150/oAM151 (5′-GAGTCTTGGACTGTTCGCGATAC-3′/5′-CACGGCCTGAGTGGCCCAGCATGCCTTGGTTCAGCCAAGCTG-3′) for the left border and oAM152/oAM153 (5′-GTGGGCCATCTAGGCCGAGATCGAGCTGGTCTATAC-3′/5′-CGACGACTTGTTTGAGTTTGGCC-3′) for the right border. The PCR fragments were digested with SfiI and ligated to the hygromycin resistance cassette isolated as a 2.7-kb SfiI fragment from plasmid pMF1-h (Brachmann et al., 2004). The ligation product was cloned into pCRII-TOPO (Invitrogen) to obtain pΔsho1-Hyg. The plasmid was digested with Eco0109I and HindIII prior to transformation in SG200. The 2.7-kb hygromycin cassette of pΔsho1-Hyg was replaced by the 1.4-kb nourseothricin resistance cassette from pMF1-n (Brachmann et al., 2004), resulting in pΔsho1-Nat. This plasmid was digested with PstI and BamHI before transformation and was used for sho1 disruption in FB1, FB2, and SG200Δkpp6.
For deletion of only the SH3 domain of sho1, the primer combination oDL02/oDL03 (5′-TAGAAGCTTCTGTTCCACAGCTTGGCTG-3′/5′-TATGGCGCGCCCTAGTAGCCGTAGTCGGGCAG-3′) was used to amplify the 5′ region of sho1. Primer oDL03 introduces a stop codon at amino acid position 280, just before the SH3 domain starts. The PCR product was digested with AscI. The 3′ flanking region of sho1 was amplified using the primers oDL04 (5′- GATGAGCTCCGAGATCGAGCTGGTCTATACC-3′) and oDL05 (5′-ATCAAGCTTCGACGACTTGTTTGAGTTTGG-3′), following digestion with SacI. A three-fragment ligation including a 3.0-kb AscI/SacI fragment containing the nos terminator and the hygromycin resistance cassette, the PCR fragments corresponding to the 3′ flanking region of sho1 and to the C-terminally truncated sho1 gene was performed. The ligation product was cloned into pCRII-TOPO to obtain psho1Δ280-335. The plasmid was digested with HindIII and transformed in SG200, resulting in strain SG200sho1Δ280-335.
To generate msb2 (um00480) deletion mutants, two 1.0-kb fragments comprising the 5′ flank and the 3′ flank of the msb2 gene were generated by PCR with the primer pairs oOM1/oOM2 (5′-TACACCTCATCATTCACGCTAACGC-3′/5′-CACGGCCTGAGTGGCCAAAGAGACAAGTGGGAGGCTGACG-3′) and oOM3/oOM4 (5′-GTGGGCCATCTAGGCCTGTTTGCTTTGGTTGTAACGGAACG-3′/5′-TGTCTGGCTGCACCACTCTATTTACG-3′). The PCR products were digested with SfiI and ligated with the 1.9-kb SfiI hygromycin resistance cassette from pBS-hhn (Kämper, 2004). The resulting product was cloned into pCRII-TOPO to generate pΔmsb2-Hyg, which was digested with HindIII and XbaI and transformed in SG200, FB1, and FB2. To generate sho1 and msb2 double deletions strains, the 1.9-kb hygromycin resistance cassette of pΔmsb2-Hyg was replaced by the 1.9-kb carboxin resistance cassette of pBS-Cbx (Kämper, 2004) to yield pΔmsb2-Cbx. Prior to transformation in FB1Δsho1 and FB2Δsho1, the plasmid was digested with HindIII and XbaI. For generation of SG200Δsho1 Δmsb2, the PCR-amplified msb2 borders were ligated with the 1.4-kb nourseothricin resistance cassette from pMF1-n and the ligation product cloned into pJet1 (Fermentas). Prior to transformation in SG200Δsho1, the plasmid was digested with BspEI and PvuII.
To complement SG200Δsho1 with sho1, a 1.9-kb fragment corresponding to the promoter and the open reading frame (ORF) of sho1 was amplified using primers oAM154 (5′-CGCTCGGTACCCGGTGATTTGTGATTAACACGTC-3′) and oDL01 (5′-CTAGGCGCGCCCTATAGGAGCTGCATGTAGTTGCTG-3′), following digestion with AscI. p123 (Aichinger et al., 2003) was linearized with NotI and refilled with Klenow according to the recommendations of the manufacturer (New England Biolabs). Then, the linearized plasmid was cut with Acc65I and the 4.7-kb Acc65I/blunt fragment was ligated with the 1.9-kb Acc65I/blunt sho1 fragment. The resulting plasmid, p123Psho1:sho1, contains the ORF of sho1 under the control of its own promoter and the nos terminator. p123Psho1:sho1 was linearized with SspI and transformed into SG200Δsho1. Single-copy integrations in the ip locus were identified as described previously (Loubradou et al., 2001).
For construction of the msb2 complementation plasmid, p123 was digested with NdeI and NotI, and the 4.5-kb fragment was ligated with the 6.9-kb NdeI/NotI fragment corresponding to the promoter and the ORF of msb2, which was amplified using the primers oAM288 (5′-GCCCCGCATATGGCTGATGAAGAAAGAGCACT-3′) and oAM290 (5′- GCCTGCGGCCGCATTTAAAGGAGAACCGAGTTG-3′). p123Pmsb2:msb2 contains the ORF of msb2 under the control of its own promoter and the nos terminator. For complementation, plasmid p123Pmsb2:msb2 was linearized with SspI and integrated in single copy in the ip locus of SG200Δmsb2. For complementation of the sho1 msb2 double mutant, p123Psho1:sho1 was digested with PvuII and HpaI, and the 2.3-kb fragment corresponding to the promoter, the sho1 gene, and the nos terminator were ligated into HpaI-linearized p123Pmsb2:msb2. The resulting plasmid p123Psho1:sho1-Pmsb2:msb2 was integrated in single copy into the ip locus of SG200Δsho1 Δmsb2.
To generate SG200sho1GFP, a 1.0-kb left border corresponding to the coding region of sho1 was amplified using the primers oDL08 (5′-GGTGGCCGCGTTGGCCCGCTCGATCGCCACCGGTAGGAGCTGCATGTAGTTGCTGGG-3′) and oDL02. The right border was amplified using the primers oDL05 and oDL09 (5′-ATAGGCCTGAGTGGCCGAGATCGAGCTGGTCTATAC-3′). Both fragments were digested with SfiI and ligated with the 3.7-kb SfiI fragment from pBS-eGFP, which contains the egfp gene, the nos terminator, and the hygromycin resistance cassette (Brachmann, et al., 2004). The ligation product was cloned in pCRII-TOPO to yield psho1-eGFP. In this plasmid, the sho1 gene is fused to egfp via a linker encoding the amino acids PVAIERANAAT. Prior to transformation and integration in the sho1 locus, the plasmid was digested with HindIII.
To construct a C-terminal fusion of Msb2 to the red fluorescent protein mCherry (Shaner et al., 2004), the 0.7-kb NcoI/BsrgI fragment of pMF5-n (Becht et al., 2006) corresponding to egfp was replaced with a 0.7-kb NcoI/BsrgI mcherry fragment (kindly provided by M. Bölker) resulting in pMF5-mCherry. A 1.0-kb fragment corresponding to the 3′ coding region of msb2 was amplified with the primer pair oAM331/oAM284 (5′-CACGGCCGCGTTGGCCCCGGTGGCGATCGAGCGAAGGAGAACCGAGTTGCTCATC-3′/5′-GACGGCGCAAATCTTTGCAT-3′), and a second 1.0-kb fragment was generated by PCR with the primers oAM332 (5′-GTTGGCCTGAGTGGCCATCTAGTTTGGTGCTTCTTTT-3′) and oAM333 (5′-GCATTCAGTCGGCGTCCCATCCAGC-3′). Both fragments were digested with SfiI and ligated with the 2.4-kb SfiI fragment from pMF5-mCherry. After cloning this construct into pCR4-TOPO (Invitrogen), the resulting plasmid pmsb2-mCherry was digested with SnaBI and PmeI and transformed in SG200sho1GFP. In the resulting strain SG200sho1GFP/msb2mCherry, the native msb2 gene is fused to mcherry via a linker encoding the amino acids RSIATGANAAT, and this is followed by the nos terminator and the nourseothricin resistance cassette.
For overexpression of msb2-ha-mcherry-ha, first pONG was constructed. Using primers oDL114 (5′-AATACCATGGTGAGCAAGGGCGAGGAGG-3′), oDL115 (5′-TATGCGGCCGCTTTAAGCGTAATCTGGAACATCGTATGGGTA-3′), and pPotef:vcp1-mCherryHA (A. Djamei and R. Kahmann, unpublished data) as template, a 0.7-kb product containing the mcherry-ha gene was amplified and digested with NcoI and NotI. pPotef:vcp1-mCherryHA is a plasmid in which the secreted effector gene vcp1 is fused to the otef promoter and a C-terminal mcherry-ha tag. A second PCR with the primer pair oD111/oDL112 (5′-ATAGGATCCAGGCCTGAGTGGCCATGACAGAGGAGGACTCTGTGCTTTATCCG-3′/5′-TTATCCATGGTGGCCGCGTTGGCCCCTAGGAGCTGCATGTAGTTGCTGGG-3′) using p123Psho1:sho1 as template amplified a part of the sho1 gene. The 0.6-kb product was digested with BamHI and NcoI. A three-fragment ligation with the two fragments mentioned above and the 5.5-kb BamHI/NotI fragment of p123 was performed to yield pONG. In pONG, the sho1 part flanked by SfiI sites serves as stuffer for the integration of genes to be fused to mcherry-ha. To insert msb2-ha into pONG, the primer combinations oDL79 (5′-[P]GTGCCCGACTATGCCGGCGCCAGTCCCTCCACTGCTCCCTCGT-3′)/oDL125 (5′-TATGGCCGCGTTGGCCGCAAGGAGAACCGAGTTGCTCATC-3′) and oDL80 (5′-[P]GTCGTAGGGGTAGGCTGCGCCGCTGTCATCATCGCTGC-3′)/oDL124 (5′-ATAGGCCTGAGTGGCCATGGTTCTGTTTCGACCCAAC-3′) using p123Pmsb2:msb2 as template generated two PCR products, 2.2 and 1.3 kb in length, respectively. Both fragments were cut with SfiI and ligated with the 6.3-kb SfiI fragment of pONG, resulting in pPotef:msb2HA-mCherryHA. In this plasmid, the msb2 gene with an internal ha tag (corresponding to amino acid 421) is C-terminally fused to mcherry-ha. Expression of the fusion gene is driven by the otef promoter. pPotef:msb2HA-mCherryHA was linearized with Ssp1 and integrated into the ip locus of SG200Δmsb2 and SG200sho1GFP. To delete the transmembrane domain of msb2, a 9.7-kb fragment was amplified with the primer pair oDL197 (5′-[P]TGGCGCAAGCATCGCAAGG-3′)/oDL198 (5′-[P]ACTGTTGCGCAGCGTCGAGTC-3′) using pPotef:msb2HA-mCherryHA as template. The resulting fragment was circularized to yield pPotef:msb2HAΔTM-mCherryHA. This plasmid was linearized with SspI and integrated in single copy in the ip locus of SG200Δmsb2.
To express kpp6 in U. maydis, pPotef:kpp6NA (Brachmann, 2001) was used as progenitor plasmid. This plasmid is a derivative of pkpp6NA (Brachmann et al., 2003), constructed by exchange of almost the complete kpp6 promoter (region −2371 to −82) with the 873-bp otef promoter from pOTEF-SG (Spellig et al., 1996). pPotef:kpp6NA contains the otef promoter, followed by 81-bp 5′ untranslated region, the complete ORF, and 435-bp 3′ untranslated region of kpp6. To integrate myc-tagged versions of kpp6, three PCR reactions with the primer pair oDL131 (5′-ATACCCGGGATGGAGGAGCAGAAGCTGATCTC-3′)/oDL132 (5-TATGCGGCCGCTCAACGAAGAAGCGGCTGAAATTC-3′) were performed, using pGBKT7-kpp6, pGBKT7-kpp6170-533, and pGBKT7-kpp6P130A P131A (see Supplemental Methods online) as templates. The PCR products (1.1, 1.1, and 0.6 kb, respectively) were digested with SmaI and BspEI and separately ligated with the 6.0 BspEI/SmaI fragment of pPotef:kpp6NA, resulting in pPotef:myc-kpp6, pPotef:myc-kpp6Δ1-169, and pPotef:myc-kpp6P130A P131A. The plasmids were digested with SspI and integrated in single copy into the ip locus of SG200Δkpp6 (Brachmann et al., 2003) and SG200Δsho1 Δkpp6, respectively. The plasmid pPotef:myc-kpp6 was also integrated in the ip locus of SG200Δkpp6 and SG200sho1-GFP in multiple copy.
The triple deletion strain of sho1, msb2, and rok1 was constructed by linearization of pΔrok1-Cbx (Di Stasio et al., 2009) with PvuI and subsequent transformation into SG200Δsho1 Δmsb2.
To generate strains carrying the fuz7DD allele under the control of the crg1 promoter, p123Pcrg1:fuz7DD (Müller et al., 2003) was linearized with AgeI and integrated in single copy in the ip locus of SG200 and SG200Δsho1 Δmsb2, respectively.
For construction of sho1 and msb2 mutants containing the AM1 reporter gene, pAM1 (Mendoza-Mendoza et al., 2009b) was linearized with AgeI and transformed in SG200Δsho1, SG200Δmsb2, and SG200Δsho1 Δmsb2, respectively. Single-copy integrations of the AM1 marker in the ip locus were selected.
Plant Infections
Solopathogenic strains were grown in YEPSL medium to an OD600 of 0.8 and concentrated in water to a final OD600 of 1.0. This suspension was inoculated into 7-d-old seedlings of Early Golden Bantam (Olds Seeds). Compatible haploid strains were mixed (1:1) prior to infection. Disease symptoms were evaluated according to the disease rating criteria reported by Kämper et al. (2006). Three independent experiments were performed, and the average values (for each symptom category) were expressed as a percentage of the total number of infected plants.
Induction of Filaments and Appressoria
The in vitro system for inducing filaments and appressoria in U. maydis was applied as described previously (Mendoza-Mendoza et al., 2009b) with minor modifications. Briefly, SG200 and derivatives were grown in YEPSL at 28°C to an OD600 of 0.6 to 0.8. The cells were resuspended in 2% YEPSL to an OD600 of 0.2 and supplemented with either 100 μM (f.c.) 16-hydroxyhexadecanoic acid (Sigma-Aldrich) or an appropriate amount of ethanol (1%, f.c.), the solvent of 16-hydroxyhexadecanoic acid. For filament induction in liquid, the samples were incubated at 28°C on a rotating wheel, and after 18 h the number of cells that had developed filaments relative to the number of total cells was determined using light microscopy. To quantify filament formation on a hydrophobic surface, cells were sprayed (EcoSpray Labo Chimie) on Parafilm M and incubated at 100% humidity at 28°C for 18 h. The samples were stained with calcofluor to visualize fungal cells, and the percentage of cells that had developed filaments relative to total cells was determined using fluorescence microscopy. For appressoria quantification, SG200AM1 derivatives were inoculated into 7-d-old maize seedlings 2 cm above ground or were sprayed on Parafilm M, as described above. After 18 h, the third leaf of infected plants was prepared, washed with water, and stained with calcofluor (Sigma-Aldrich). Likewise, the Parafilm M samples were washed with water and stained. Using fluorescence microscopy, filaments expressing the AM1 marker that is expressed in cells forming appressoria were visualized by their GFP fluorescence, and the ratio of appressoria to filamentous cells was determined. All experiments were performed in three biological replicates.
To visualize Sho1-GFP and Msb2-mCherryHA in appressoria, a flat hydrophobic surface lacking background fluorescence was generated by melting and casting Granopent P (Carl Roth).
Microscopy
For analysis of U. maydis sporidia, cells were grown at 28°C to an OD600 of 0.6. Latrunculin A (Sigma-Aldrich) was applied at a final concentration of 10 μM following a 2-h incubation at room temperature. To stain fungal material, samples were incubated in calcofluor Fluorescent Brightner 28 (100 μg/mL in 0.2 M Tris/HCl, pH 8.0; Sigma-Aldrich) for 30 s. For staining of plant cells with propidium iodide (Sigma-Aldrich), fresh leaves were incubated in 10 μg/mL propidium iodide (in PBS, pH 7.4) for 20 min. To examine fungal colonization inside the leaf tissue 6 d after infection, the third oldest leaf was destained in ethanol, transferred to 10% KOH, incubated at 95°C overnight, washed once with PBS buffer (140 mM NaCl, 16 mM Na2HPO4, 2 mM KH2PO4, 3.5 mM KCl, and 1 mM Na2-EDTA, pH 7.4), and incubated under vacuum in staining solution (10 μg/mL propidium iodide and 10 μg/mL WGA-AF 488 in PBS, pH 7.4) according to Doehlemann et al. (2008b). WGA-AF 488 was purchased from Invitrogen.
For microscopy, an Axioplan II microscope (Zeiss) with differential interference contrast optics was used. Fluorescence of GFP, mCherry, and calcofluor was observed using GFP (ET470/40BP, ET495LP, and ET525/50BP), TexasRed (HC562/40BP, HC593LP, and HC624/40BP), and 4',6-diamidino-2-phenylindole (HC375/11BP, HC409BS, and HC447/60BP) filter sets (Semrock). Pictures were taken with a CoolSNAP-HQ charge-coupled device camera (Photometrics). Image processing was done with MetaMorph software (Universal Imaging).
Confocal microscopy was performed using a TCS-SP5 confocal microscope (Leica Microsystems). For GFP fluorescence, an excitation of 488 nm and detection at 495-530 nm was used. Propidium iodide and mCherry fluorescence was excited with 561 nm and detected at 580-630 nm. To visualize WGA-AF 488, an excitation of 488 nm and subsequent detection at 500-540 nm was employed. Calcofluor and autofluorescence were excited with a 405 nm laser and detected at 415-460 nm. Images were processed using LAS-AF software (Leica Microsystems).
Quantitative Real-Time PCR
An iCycler-machine (Bio-Rad) in combination with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) was used. For data analysis, iCycler software (Bio-Rad) was used.
To determine fungal biomass after infection, we followed the procedure described by Di Stasio et al. (2009), with some modifications. Briefly, plants were inoculated with the solopathogenic strains SG200, SG200Δsho1, SG200Δmsb2, and SG200Δsho1 Δmsb2, and 3 d after inoculation, the third oldest leaf was prepared. Fungal cells on the leaf surface were peeled off by applying liquid latex (Orion). Subsequently, 4-cm-long sections below the injection holes were excised, and 10 excised sections were pooled. From this material, total DNA was extracted as described by Di Stasio et al. (2009). Quantitative real-time PCR analysis was conducted with primer pairs mfa1-RT-FW (5′-GCTTTCGATCTTCGCTCAGAC-3′)/mfa1-RT-RV (5′-CAACAACACAGCTGGAGTAGC-3′) for detecting U. maydis DNA and Gapdh-FW (5′-CTTCGGCATTGTTGAGGGTTTG-3′)/Gapdh-RV (5′-TCCTTGGCTGAGGGTCCGTC-3′) for detection of maize DNA. The experiment was done in three biological replicates, and the ratio of SG200 DNA to plant DNA was set to 1.0.
Molecular Techniques
For molecular analysis, standard procedures were followed (Sambrook et al., 1989). Transformation of U. maydis was performed as published previously (Schulz et al., 1990), and DNA was isolated according to the protocol of Hoffman and Winston (1987). For extraction of proteins from S. cerevisiae, 10 mL of cells (OD600 of 1.0) were washed with 50 mM Tris/HCl, pH 7.5, and resuspended in 100 μL ESB buffer (80 mM Tris-HCl, pH 8.0, 2% SDS, 10% glycerol, 100 mM DTT, and 0.01% bromophenol blue). Samples were boiled for 3 min at 95°C and supplemented with glass beads, and cells were disrupted by incubation for 3 min on a Vibrax shaker (IKA). After additional boiling (1 min at 95°C), samples were subjected to SDS polyacrylamide electrophoresis. For isolation of proteins from U. maydis, 2 mL of cells (OD600 of 1.0) were washed in water and resuspended in 100 μL Thorner buffer (8 M urea, 5% SDS, 0.1 mM EDTA, 0.01% bromophenol blue, 100 mM DTT, and 100 mM Tris-HCl, pH 6.8). Samples were boiled for 10 min at 95°C, supplemented with glass beads, and incubated 5 min on a Vibrax shaker (IKA). After a second boiling step (10 min at 95°C), 10 μL was loaded for SDS polyacrylamide gel electrophoresis. Transfer of proteins to PVDF nitrocellulose membranes (Amersham Pharmacia Biotech) was conducted in a semidry blot chamber (UniEquip). Blots were probed with monoclonal GAL4-DBD, GAL4-TA (both Santa Cruz Biotechnology), anti-HA (Sigma-Aldrich), anti-c-Myc (Sigma-Aldrich), anti-GFP (Roche), and antitubulin (Merck) antibodies. To detect mCherry, a rabbit serum against RFP was used (kindly provided by M. Thanbichler). Horseradish peroxidase–conjugated anti-mouse or anti-rabbit IgG (Cell Signaling) was used as secondary antibody, and the ECL Plus System (Amersham Pharmacia Biotech) was used for protein detection.
Accession Numbers
Sequence data from this article can be found at GenBank/EMBL databases under accession numbers XP_759303 (sho1) and XP_756627 (msb2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Illustration of Signaling Cascades in U. maydis and S. cerevisiae.
Supplemental Figure 2. Sho1-GFP and Msb2-mCherry Fusions Are Functional.
Supplemental Figure 3. Sho1 and Msb2 Are Membrane Associated.
Supplemental Figure 4. Sho1 Interacts Specifically with Kpp6.
Supplemental Figure 5. Pathogenicity of Haploid sho1 and msb2 Mutant Strains after Mating.
Supplemental Figure 6. Stress Response in sho1 and msb2 Mutants.
Supplemental Figure 7. SG200Δsho1 Δmsb2 Induces Plant Cell Death.
Supplemental Figure 8. Hydroxy Fatty Acids Induce Filamentation in sho1 and msb2 Mutants.
Supplemental Figure 9. Hydrophoic Surface-Induced Differentiation in sho1 and msb2 Mutants.
Supplemental Figure 10. The Transmembrane Domain of Msb2 Is Essential for Function.
Supplemental Figure 11. U. maydis Msb2 Cannot Functionally Replace S. cerevisiae Msb2p.
Supplemental Figure 12. The SH3 Domain of Sho1 Interacts with the N Terminus of Kpp6.
Supplemental Figure 13. The SH3 Domain of Sho1 Is Essential for Function.
Supplemental Table 1. S. cerevisiae Strains Used in This Study.
Supplemental Table 2. U. maydis Strains Used in This Study.
Supplemental Methods.Two-Hybrid Interaction Studies, Complementation of the S. cerevisiae msb2 Mutant, Stress Assays, Coimmunoprecipitation Analysis, and Subcellular Fractionation of Proteins.
Supplemental References.
Acknowledgments
We thank X. Xia for help with plasmid and strain constructions, A. Djamei and G. Doehlemann for help with confocal microscopy, and P. Berndt and J. Freitag for stimulating discussions. We thank E. Perez-Nadales and A. Di Pietro for communicating results prior to publication, P. J. Cullen for providing yeast strains, M. Bölker for providing the mcherry gene, and M. Thanbichler for antibodies. D.L. was supported through the International Max Planck Research School and the GRK1216. We acknowledge support through the EU network Signalpath.
References
- Aichinger C., Hansson K., Eichhorn H., Lessing F., Mannhaupt G., Mewes W., Kahmann R. (2003). Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics 270: 303–314 [DOI] [PubMed] [Google Scholar]
- Andrews D.L., Egan J.D., Mayorga M.E., Gold S.E. (2000). The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant Microbe Interact. 13: 781–786 [DOI] [PubMed] [Google Scholar]
- Banuett F., Herskowitz I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F., Herskowitz I. (1994). Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8: 1367–1378 [DOI] [PubMed] [Google Scholar]
- Bechinger C., Giebel K.F., Schnell M., Leiderer P., Deising H.B., Bastmeyer M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285: 1896–1899 [DOI] [PubMed] [Google Scholar]
- Becht P., Konig J., Feldbrugge M. (2006). The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 119: 4964–4973 [DOI] [PubMed] [Google Scholar]
- Boisnard S., Ruprich-Robert G., Florent M., Da Silva B., Chapeland-Leclerc F., Papon N. (2008). Role of Sho1p adaptor in the pseudohyphal development, drugs sensitivity, osmotolerance and oxidant stress adaptation in the opportunistic yeast Candida lusitaniae. Yeast 25: 849–859 [DOI] [PubMed] [Google Scholar]
- Bölker M., Urban M., Kahmann R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68: 441–450 [DOI] [PubMed] [Google Scholar]
- Bottin A., Kämper J., Kahmann R. (1996). Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 13: 342–352 [DOI] [PubMed] [Google Scholar]
- Brachmann A. (2001). Die frühe Infektionsphase von Ustilago maydis: Genregulation durch das bW/bE-Heterodimer. PhD dissertation (Munich, Germany: Ludwig-Maximilians-University; ). [Google Scholar]
- Brachmann A., Konig J., Julius C., Feldbrugge M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272: 216–226 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Schirawski J., Muller P., Kahmann R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster J.L., de Valoir T., Dwyer N.D., Winter E., Gustin M.C. (1993). An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Chen R.E., Thorner J. (2007). Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Dean R.A. (1997). The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J. (2007). Signaling mucins: The new kids on the MAPK block. Crit. Rev. Eukaryot. Gene Expr. 17: 241–257 [DOI] [PubMed] [Google Scholar]
- Cullen P.J., Sabbagh W., Jr, Graham E., Irick M.M., van Olden E.K., Neal C., Delrow J., Bardwell L., Sprague G.F., Jr (2004). A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. (1999). Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadal E., Real F.X., Posas F. (2007). Mucins, osmosensors in eukaryotic cells? Trends Cell Biol. 17: 571–574 [DOI] [PubMed] [Google Scholar]
- Di Stasio M., Brefort T., Mendoza-Mendoza A., Munch K., Kahmann R. (2009). The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 73: 73–88 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Berndt P., Hahn M. (2006). Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59: 821–835 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kuhnemann J., Sonnewald U., Kahmann R., Kamper J. (2008a). Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56: 181–195 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Vranes M., de Vries R.P., Kamper J., Kahmann R. (2008b). Establishment of compatibility in the Ustilago maydis/maize pathosystem. J. Plant Physiol. 165: 29–40 [DOI] [PubMed] [Google Scholar]
- Feldbrügge M., Kamper J., Steinberg G., Kahmann R. (2004). Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7: 666–672 [DOI] [PubMed] [Google Scholar]
- Fernandez-Alvarez A., Elias-Villalobos A., Ibeas J.I. (2009). The O-mannosyltransferase PMT4 is essential for normal appressorium formation and penetration in Ustilago maydis. Plant Cell 21: 3397–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs U., Hause G., Schuchardt I., Steinberg G. (2006). Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell 18: 2066–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C.J., Ljungdahl P.O., Styles C.A., Fink G.R. (1992). Unipolar cell division in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Hao N., Behar M., Parnell S.C., Torres M.P., Borchers C.H., Elston T.C., Dohlman H.G. (2007). A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr. Biol. 17: 659–667 [DOI] [PubMed] [Google Scholar]
- Hoffman C.S., Winston F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of E. coli. Gene 57: 267–272 [DOI] [PubMed] [Google Scholar]
- Holliday R. (1974). Ustilago maydis. Handbook of Genetics, King R.C., ed (New York: Plenum Press; ), pp. 575–595 [Google Scholar]
- Hollingsworth M.A., Swanson B.J. (2004). Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 4: 45–60 [DOI] [PubMed] [Google Scholar]
- Howard R.J., Ferrari M.A., Roach D.H., Money N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88: 11281–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271: 103–110 [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Liu Z.M., Li D., Kolattukudy P.E. (2000). Two novel genes induced by hard-surface contact of Colletotrichum gloeosporioides conidia. J. Bacteriol. 182: 4688–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz M., Becit E., Hohmann S. (2006). Comparative genomics of the HOG-signalling system in fungi. Curr. Genet. 49: 137–151 [DOI] [PubMed] [Google Scholar]
- Kumamoto C.A. (2008). Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat. Rev. Microbiol. 6: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.H., Dean R.A. (1993). cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell 5: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Sharon A., Hadar R., Ma H., Horwitz B.A. (1999). A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: Diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96: 13542–13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveleki L., Mahlert M., Sandrock B., Bolker M. (2004). The PAK family kinase Cla4 is required for budding and morphogenesis in Ustilago maydis. Mol. Microbiol. 54: 396–406 [DOI] [PubMed] [Google Scholar]
- Li S.S. (2005). Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 390: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C.A., Fink G.R. (1993). Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741–1744 [DOI] [PubMed] [Google Scholar]
- Loubradou G., Brachmann A., Feldbrugge M., Kahmann R. (2001). A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40: 719–730 [DOI] [PubMed] [Google Scholar]
- Ma Y., Qiao J., Liu W., Wan Z., Wang X., Calderone R., Li R. (2008). The sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infect. Immun. 76: 1695–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Takekawa M., Saito H. (1995). Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558 [DOI] [PubMed] [Google Scholar]
- Mahlert M., Leveleki L., Hlubek A., Sandrock B., Bolker M. (2006). Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 59: 567–578 [DOI] [PubMed] [Google Scholar]
- Mayorga M.E., Gold S.E. (1999). A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34: 485–497 [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Berndt P., Djamei A., Weise C., Linne U., Marahiel M., Vranes M., Kamper J., Kahmann R. (2009b). Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71: 895–911 [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Eskova A., Weise C., Czajkowski R., Kahmann R. (2009a). Hap2 regulates the pheromone response transcription factor prf1 in Ustilago maydis. Mol. Microbiol. 72: 683–698 [DOI] [PubMed] [Google Scholar]
- Müller P., Aichinger C., Feldbrugge M., Kahmann R. (1999). The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Müller P., Weinzierl G., Brachmann A., Feldbrugge M., Kahmann R. (2003). Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2: 1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norice C.T., Smith F.J., Jr, Solis N., Filler S.G., Mitchell A.P. (2007). Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 6: 2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S.M., Herskowitz I. (1998). The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12: 2874–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S.M., Herskowitz I. (2002). A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22: 4739–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitoniak A., Birkaya B., Dionne H.M., Vadaie N., Cullen P.J. (2009). The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol. Biol. Cell 13: 3101–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Saito H. (1997). Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science 276: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Raitt D.C., Posas F., Saito H. (2000). Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19: 4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Salah S.M., Ammerer G. (2000). Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2: 620–627 [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Fink G.R. (1994). Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- Roman E., Cottier F., Ernst J.F., Pla J. (2009). Msb2 signaling mucin controls activation of Cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot. Cell 8: 1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman E., Nombela C., Pla J. (2005). The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25: 10611–10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Roldan M.C., Maier F.J., Schafer W. (2001). PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant Microbe Interact. 14: 116–125 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Frisch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schirawski J., Bohnert H.U., Steinberg G., Snetselaar K., Adamikowa L., Kahmann R. (2005). Endoplasmic reticulum glucosidase II is required for pathogenicity of Ustilago maydis. Plant Cell 17: 3532–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306 [DOI] [PubMed] [Google Scholar]
- Seet B.T., Pawson T. (2004). MAPK signaling: Sho business. Curr. Biol. 14: R708–R710 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Singh P.K., Hollingsworth M.A. (2006). Cell surface-associated mucins in signal transduction. Trends Cell Biol. 16: 467–476 [DOI] [PubMed] [Google Scholar]
- Smith D.G., Garcia-Pedrajas M.D., Gold S.E., Perlin M.H. (2003). Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorhism. Mol. Microbiol. 50: 259–275 [DOI] [PubMed] [Google Scholar]
- Snetselaar K.M., Mims C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84: 193–203 [Google Scholar]
- Spellig T., Bottin A., Kahmann R. (1996). Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252: 503–509 [DOI] [PubMed] [Google Scholar]
- Takano Y., Kikuchi T., Kubo Y., Hamer J.E., Mise K., Furusawa I. (2000). The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant Microbe Interact. 13: 374–383 [DOI] [PubMed] [Google Scholar]
- Tatebayashi K., Tanaka K., Yang H.Y., Yamamoto K., Matsushita Y., Tomida T., Imai M., Saito H. (2007). Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26: 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.L., Talbot N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39: 385–417 [DOI] [PubMed] [Google Scholar]
- Vadaie N., Dionne H., Akajagbor D.S., Nickerson S.R., Krysan D.J., Cullen P.J. (2008). Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181: 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl G., Leveleki L., Hassel A., Kost G., Wanner G., Bolker M. (2002). Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 45: 219–231 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Hamer J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Tatebayashi K., Yamamoto K., Saito H. (2009). Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. EMBO J. 28: 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J.R. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17: 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]