This article describes the molecular analysis of large contiguous sequences produced from the bread wheat genome. It provides novel insights into the number, distribution, and density of genes along chromosome 3B and reveals an unexpectedly high amount of noncollinear genes compared to model grass genomes.
Abstract
To improve our understanding of the organization and evolution of the wheat (Triticum aestivum) genome, we sequenced and annotated 13-Mb contigs (18.2 Mb) originating from different regions of its largest chromosome, 3B (1 Gb), and produced a 2x chromosome survey by shotgun Illumina/Solexa sequencing. All regions carried genes irrespective of their chromosomal location. However, gene distribution was not random, with 75% of them clustered into small islands containing three genes on average. A twofold increase of gene density was observed toward the telomeres likely due to high tandem and interchromosomal duplication events. A total of 3222 transposable elements were identified, including 800 new families. Most of them are complete but showed a highly nested structure spread over distances as large as 200 kb. A succession of amplification waves involving different transposable element families led to contrasted sequence compositions between the proximal and distal regions. Finally, with an estimate of 50,000 genes per diploid genome, our data suggest that wheat may have a higher gene number than other cereals. Indeed, comparisons with rice (Oryza sativa) and Brachypodium revealed that a high number of additional noncollinear genes are interspersed within a highly conserved ancestral grass gene backbone, supporting the idea of an accelerated evolution in the Triticeae lineages.
INTRODUCTION
Bread wheat (Triticum aestivum) is allohexaploid with three homoeologous genomes (2n=6x=AABBDD) and has one of the largest higher plant genomes (17 Gb, 40 times larger than the rice (Oryza sativa) genome [International Rice Genome Sequencing Project, 2005]). Because of its size and high repetitive sequence content (∼80%; Smith and Flavell, 1975), sequencing the wheat genome has been perceived as too challenging; consequently, its organization and composition remain largely unknown. Most of the wheat genomic sequences available to date have been obtained during map-based cloning projects or comparative studies at disease resistance, storage protein, grain hardness, domestication, or vernalization loci (for review, see Feuillet and Salse, 2009; Krattinger et al., 2009). In all cases, the sequences corresponded to single BAC clones or small BAC contigs of 129 kb on average, and only five contigs larger than 300 kb are available to date in the databanks (http://srs.ebi.ac.uk), the largest of which has a maximum contiguous size of 450 kb (Ogihara et al., 2005). Analyses of the 3.8 Mb representing all wheat genomic sequences available in the public databases in 2005 showed an average gene density of 1 gene per 24 kb with only ∼55% of transposable elements, thereby indicating a clear bias toward gene-rich regions in these samples (Sabot et al., 2005). Early studies based on EST mapping in cytogenetic bins suggested that the gene distribution is highly heterogeneous in wheat with >90% of the gene space clustered in 29% of the genome, mainly in the telomeric part of the chromosomes (Erayman et al., 2004; Qi et al., 2004). By contrast, sampling of the wheat genome performed through the sequencing of randomly chosen BAC clones or large samples of BAC end sequences (BES) provided evidence for a more homogeneous gene distribution in the genome. In a preliminary study of four BAC clones from a whole bread wheat genome (cv Chinese Spring) BAC library, Devos et al. (2005) found an average gene density of 1/75 kb, while Charles et al. (2008) reported a gene density of 1/100 kb homogeneously distributed after sequencing 10 BACs (1.43 Mb) randomly chosen from the 3B chromosome-specific BAC library. Finally, end sequencing of 10,000 BAC clones distributed along the physical map of chromosome 3B led to an estimate of 1 gene/165 kb (Paux et al., 2006). Recently, draft sequences of 217 additional BAC clones from the Chinese Spring BAC library have been deposited in the databanks (AC200765-851, AC207901-60, AC216550-85, AC232247-62, AC238983-88, and DQ767609-30) by Bennetzen et al., and annotation to refine these estimates is underway (http://www.nsf.gov/awardsearch/showAward.do?AwardNumber=0501814).
Further insights into the composition of the wheat genome have been provided by other random sequencing efforts. Analysis of 3 Mb of plasmid end sequences produced from Aegilops tauschii (the D genome progenitor of hexaploid bread wheat) indicated that 92% of repeated sequences (Li et al., 2004) contained 68% of transposable elements (TEs), whereas the annotation of 11 Mb of BESs from the 3B chromosome of hexaploid wheat (Paux et al., 2006) revealed a repeat content of 86% as well as an estimated gene number of 6000 for chromosome 3B (i.e., 36,000 genes per diploid genome). This contrasted dramatically with the predictions of Rabinowicz et al. (2005) who suggested as much as 98,000 genes per subgenome in bread wheat based on the analysis of 1597 plasmid ends from a methyl-filtration library. Similar variations in gene number were reported for maize (Zea mays) until its genome was fully sequenced (Schnable et al., 2009). Early genetic analyses suggested that maize genes are clustered primarily in high-coding density islands representing 10 to 20% of the genome (Carels et al., 1995). Sequencing of large sets of random BAC clones led to the revision of the gene island concept and suggested that the 42,000 to 59,000 estimated genes are largely spread in 78% of the genome (Haberer et al., 2005). Finally, the first improved maize genome sequence draft provided an estimate of 32,000 non-TE-related genes (Schnable et al., 2009) that are found with an increased density in the subtelomeric regions (Soderlund et al., 2009), a pattern that was also observed in sorghum (Sorghum bicolor; Paterson et al., 2009).
During its evolution, the hexaploid wheat genome has been shaped by different evolutionary forces. At the global level, its basic structure results from large rearrangements that range from an ancestral grass whole-genome duplication, to chromosome fusions and chromosome number reduction (Luo et al., 2009; Salse et al., 2009), and more recently to two polyploidization events that brought together the A, B, and D genomes into a single nucleus (McFadden and Sears, 1946). At the sequence level, its organization and composition can be depicted as two main compartments with different evolutionary dynamics and relative importance: a small conservative part that is subjected to selection pressure and mostly corresponds to the gene space and a much larger and more variable component that is under more dynamic evolution and comprises the TE space as well as duplicated genes and gene fragments. Evidence for some of the mechanisms underlying the dynamics of these two compartments (e.g., TE insertions, illegitimate and unequal recombination, and interchromosomal and tandem duplications) have been provided by comparative analyses of homeologous loci (for review, see Feuillet and Salse 2009) as well as by gene family studies (Akhunov et al., 2007), but, to date, little is known about the extent and relative impact of these mechanisms on the organization and distribution of the gene and TE spaces along the wheat chromosomes.
The current lack of knowledge about the complexity and organization of the hexaploid wheat genome sequence hampers the delineation of the most cost-effective and informative sequencing strategy even with the advent of the Next Generation Sequencing technologies (Metzker, 2009). To reduce the complexity of the analyses, the International Wheat Genome Sequencing Consortium (www.wheatgenome.org) embarked a few years ago on a chromosome-based approach (Dolezel et al., 2009). Recently, we provided a proof of concept for this approach with the construction of the physical map of the 1 Gb wheat chromosome 3B (Paux et al., 2008). In this study, we gained additional knowledge about the organization and composition of the wheat genome by the complete sequencing and annotation of 13 Mb-sized (0.3 to 3.1 Mb) BAC contigs selected from different regions of the chromosome and by whole chromosome shotgun sequencing. Our data show that (1) genes are present along the whole chromosome and are clustered mainly into numerous, very small islands separated by large blocks of repetitive elements, and (2) genome expansion occurred homogeneously along the chromosome through specific TE bursts. In addition, they reveal an accelerated evolution through tandem or interchromosomal gene duplications in the telomeric regions that led to an increase in the gene number in wheat compared with related grasses without disruption of the ancestral gene backbone. These gene rearrangements combined with the differential insertion or removal of specific TE families resulted in a contrasted sequence composition that is now observed between the proximal and distal regions of the wheat chromosomes.
RESULTS
Mb-Sized Contig and Whole Chromosome 3B Shotgun Sequencing
Thirteen contigs representing 152 BAC clones were selected for sequencing out of the 1036 established for the physical map of chromosome 3B (Paux et al., 2008). Twelve contigs originated from seven different deletion bins on the short (four contigs) and long (eight contigs) arms of the chromosome, while one contig was from the centromere (Table 1). Five of the long-arm contigs are part of the large (200 Mb) telomeric bin 3BL7-0.63-1.00 and were selected for their putative contrasted gene content estimated after screening of the 3B BAC library with a set of 399 ESTs previously assigned to this bin (see Methods). On the short arm, two contigs (ctg0954 and ctg0011) were selected from a region of 12 centimorgans located between the markers gwm389 and gwm493 that carries a high density of disease resistance genes (Paux et al., 2008). The 152 BAC clones were sequenced by Sanger or 454 Roche GSFLX technologies and completely assembled into single scaffolds. This resulted in 18.212 Mb, including eight contiguous sequences larger than 1 Mb (up to 3.1 Mb) that represent 2% of the 3B chromosome and 0.3% of the whole B genome (accession numbers FN564426-37 and FN645450). To support sequence annotation and obtain additional whole 3B chromosome sequence data, Illumina/Solexa sequencing was performed on sorted and amplified 3B chromosomes. A total of 54,808,646 short reads of 36 bp were generated, resulting in 1,973,111,256 nucleotides that represent 2X coverage of the chromosome (accession number ERA000182). This was used to establish a Mathematically Defined Repeats (MDR) index by counting occurrence of 17-mers and to evaluate the number of genes carried by chromosome 3B. High-quality sequence annotation was performed with a combination of automated procedures and manual curation based on sequence similarities and MDR patterns (see Methods).
Table 1.
Features of the 13-Mb Contig Sequences from Chromosome 3B
| Bin | Contig Name | Contig Size (bp) | No. BACs | No. Genes | No. Pseudogenes | No. Gene Fragments | Gene Density (kb−1) | No. Genes per Mb | TE Content (%) | GC Content (%) |
| 3BS8-0.78-0.87 | ctg0011 | 1266078 | 16 | 21 | 5 | 10 | 48.7 | 20.5 | 49.7 | 44.8 |
| 3BS8-0.78-0.87 | ctg0954 | 3109948 | 26 | 41 | 8 | 4 | 63.5 | 15.8 | 63.2 | 45.6 |
| 3BS1-0.33-0.55 | ctg1030 | 619476 | 6 | 0 | 0 | 0 | <619.5 | 0.0 | 97.8 | 48.8 |
| C-3BS5-0.07 | ctg1035 | 711534 | 5 | 1 | 1 | 0 | 355.8 | 2.8 | 89.9 | 45.6 |
| Centromere | 100L17 | 268551 | 1 | 0 | 0 | 0 | <268.6 | 0.0 | 96.1 | 43.0 |
| 3BL2-0.22-0.28 | ctg0616 | 786544 | 6 | 6 | 0 | 0 | 131.1 | 7.6 | 90.7 | 46.7 |
| 3BL2-0.22-0.28 | ctg0382 | 1610902 | 12 | 9 | 4 | 0 | 123.9 | 8.1 | 88.7 | 46.0 |
| 3BL1-0.31-0.38 | ctg0005 | 1715514 | 12 | 9 | 1 | 1 | 171.6 | 5.8 | 92.1 | 46.1 |
| 3BL7-0.63-1.00 | ctg0528 | 1033236 | 8 | 4 | 0 | 0 | 258.3 | 3.9 | 91.2 | 46.3 |
| 3BL7-0.63-1.00 | ctg0464 | 2543369 | 21 | 22 | 3 | 7 | 101.7 | 9.8 | 82.4 | 46.9 |
| 3BL7-0.63-1.00 | ctg0091 | 2776447 | 23 | 16 | 4 | 0 | 138.8 | 7.2 | 89.4 | 46.4 |
| 3BL7-0.63-1.00 | ctg0079 | 1305738 | 10 | 13 | 0 | 0 | 100.4 | 10.0 | 88.0 | 46.4 |
| 3BL7-0.63-1.00 | ctg0661 | 465250 | 6 | 6 | 1 | 2 | 66.5 | 15.0 | 74.5 | 45.5 |
| 18212587a | 152a | 148a | 27a | 24a | 104.1b | 9.6b | 81.5b | 46.2b |
Contig localization in deletion bins is indicated from the top to the bottom of the chromosome. The contig size, number of BACs per contig, as well as the number gene, pseudogenes, and gene fragments are indicated. Only genes and pseudogenes were taken into account for calculating the gene density and the number of genes per megabase.
Sum of the column values.
Average gene density (in kb−1 and gene per Mb), TE, and GC contents (in %) calculated by considering the sizes of the 13 contigs relative to the total contig length (18 Mb).
General Features of the Sequence Composition
Annotation of the 18.2 Mb of contig sequences revealed 199 non-TE genic features that were classified into three categories (see Methods for definitions): 148 protein coding genes, 27 pseudogenes, and 24 gene fragments (Table 1; see Supplemental Table 1 online for gene functions). The gene assignments were all supported by at least a full-length cDNA, an EST, and/or a homolog in another genome. Among these assignments, 76% showed a hit with one of the 40,349 T. aestivum unigenes (National Center for Biotechnology Information [NCBI] build#55). The coding fraction of the sample represents 242 kb (i.e., 1.5%) of the sequences, while the TE content is 81.4% (graphical view of the annotations in Supplemental Figure 1 online). An average guanine-cytosine (GC) content of 46.2% was found for all BACs (sd =1.6%), whereas the 1973 Mb of Solexa reads generated from sorted chromosomes and previous analysis of 11 Mb of BES (Paux et al., 2006) indicated significantly lower values of 42.8 and 44.5%, respectively. In contrast with the constant GC content, the composition in genes and TEs was highly variable between the different BAC sequences. The proportion of TEs ranged from 19 to 100%, while the gene content ranged from 0 to 10 genes per BAC. Coding exons show a high GC content: 59.0% ± 8.5%, which decreases to 54.7 ± 8.1 for pseudogenes and to 51.5 ± 9.5 for gene fragments, indicating that loss of function is followed by a period of relaxed selection for codon usage.
Gene Structures and Intron-Associated TEs
Analysis of the 175 gene and pseudogene models revealed a gene size ranging from 309 bp to 15.8 kb with an average of 3300 ± 2900 bp, an average number of 5.6 ± 5.5 exons per gene (median = 4), and 20% of genes without introns (see Supplemental Figure 2 online). One-third of the genes contained only one or two exons, whereas,18% had 10 or more exons comparable to what has been described for maize (Haberer et al., 2005). The average coding sequence (CDS) size of 1382 ± 852 bp (N50 = 1260 bp) was close to the average value (1143 bp) obtained from 6137 full-length cDNAs from wheat (Mochida et al., 2009), indicating that our sample provides a good representation for the wheat genes. Intron sizes were highly variable with a median size of 130 bp, similar to that observed in rice (median 138 bp; Yu et al., 2002), and slightly lower than maize introns (median 166 bp; Haberer et al., 2005). In total, 118 TEs (or truncated derivatives) were found in introns for 33% of the genes. Most of them (82) corresponded to miniature inverted-repeat transposable elements (MITEs), and there was a clear correlation between gene islands and MITE islands along the contigs (see Supplemental Figure 3 online), thereby confirming the preferential association of these small elements with genes (Wessler et al., 1995; Sabot et al., 2005). In addition, long interspersed nuclear elements (LINEs) (Stasy family) were also preferentially associated with genes similar to what was observed in rice (International Rice Genome Sequencing Project, 2005) and grapevine (Vitis vinifera; Jaillon et al., 2007). Very few complete LINEs (10/170) were found in the data set, confirming that retrotransposition of these elements often leads to the insertion of a small partial element by aborted reverse transcription (Jurka, 1997). By contrast, long terminal repeat (LTR) retrotransposons, which are the most represented TEs in the wheat genome, were almost completely excluded from the genes with only eight LTR retrotransposon fragments detected in introns.
Gene Density and Gene Distribution along Chromosome 3B
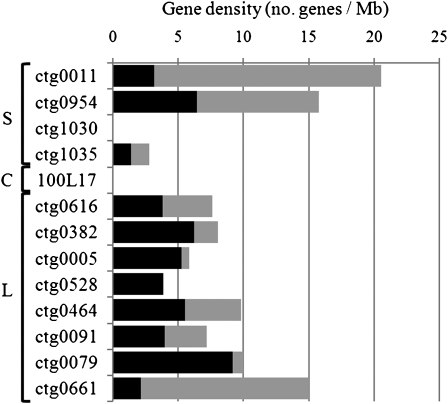
The average gene density was 1 gene per 104 kb. Interestingly, comparisons between the distal and proximal contig sequences revealed a twofold increase in gene density toward the telomeres with 1 gene per 184 kb in the proximal regions (ctg1030, 1035, 100L17, 0616, 0382, and 0005) versus 1 per 86 kb in the more distal regions (ctg0011, 0954, 0528, 0464, 0091, 0079, and 0661), thereby suggesting differential gene density distribution along chromosome 3B (Figure 1). At the contig scale, genes were found in 95% of the sample sequences (11/13 contigs), with the exception of two contigs originating from the centromeric region (100L17) and the middle of the short arm (ctg1030) that both exhibited a TE content of 97% (Table 1). Twenty-eight blocks of TEs larger than 200 kb and representing 8.77 Mb in total were found, the largest of which was 709 kb (in ctg0091). Thus, our data indicate that genes are present in the majority (73%) of the BAC clones and that Mb-sized regions without genes are rare. This observation was further supported by hybridization experiments of macroarrays, comprising the 7440 BACs of the 3B physical map minimal tiling path (MTP) (Paux et al., 2008), with 13 different wheat cDNA samples originating from mRNAs extracted at different growth stages and organs. Although it was not possible to determine the exact number of genes present on a BAC with this method, the results showed that 48% (3563) of the MTP BACs carry at least one expressed gene. This ratio is slightly lower than the prediction from the contig sequence annotation, probably since it is based solely on gene expression and is limited by the sensitivity of hybridizations on macroarrays. By combining the hybridization results with the position of each BAC within contigs, we found on average at least one expressed gene every 220 kb. The largest region without detected genes was 840 kb, which is in the range of the 709 kb calculated on the sequenced contigs. In addition, no significant differences in the density of positive BAC clones were observed between the different bins (χ2 test, P value = 0.39), confirming that genes are distributed across the entire chromosome 3B.
Figure 1.
Gene Density and Level of Synteny along the 13 Contigs.
Gene density calculated for the 13 sequenced contigs displayed according to their chromosomal location from the top to the bottom (S, short arm; C, centromeric region; L, long arm). Densities of the syntenic (black) and locus-specific (gray) genes are represented and expressed in number of genes per megabases.
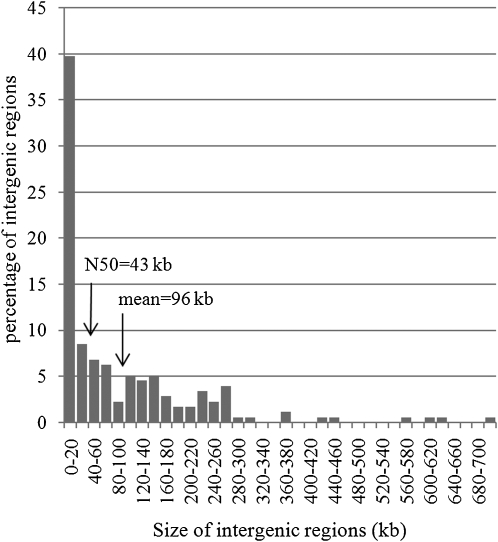
Previous work suggested that genes in wheat are preferentially found in gene islands, but the length and density of such islands had not been defined so far primarily because random single BAC analyses are limited in the ability to estimate average distances between genes. Here, we took advantage of having access to large and contrasted regions to better define the proportion and features of gene islands by calculating first the size distribution of intergenic intervals (Figure 2). One hundred and seventy six intergenic distances (IGDs) were calculated on the basis of the 175 genes and pseudogenes identified within the 13 sequenced contigs. The average IGD was 96 ± 128 kb and the median 43 kb. The IGD distribution pattern (Figure 2) indicated that very few genes are separated by a distance of 75 to 100 kb, a size that would correspond to an even distribution of genes along the chromosome (Figure 2). By contrast, short and large IGDs were overrepresented with 40% smaller than 20 kb and 24% larger than 150 kb. Based on these results, we propose to choose the N50 value as a threshold to define gene islands in wheat as clusters of at least two genes separated by <43 kb. Using this threshold, 75% (132) of the annotated genes/pseudogenes belonged to 42 gene islands, whereas 23% (41) could be considered as isolated genes. Thus, these results suggest that a majority of wheat genes are clustered into numerous islands of very small size that contain less than four genes (3.2 ± 1.6 genes ranging from 2 to 10) on average.
Figure 2.
Distribution of the Size of the 176 IGDs.
The x axis displays the different size intervals for the IGDs, while the y axis presents the percentage of intergenic regions found for each size interval. The mean (96 ± 128 kb) and median (42.5 kb) values are indicated by arrows.
Assessing the 3B Gene Catalog Set
The average gene density of 1 gene/104 kb leads to an estimate of 9560 genes on chromosome 3B, 60% more genes than previously estimated by BAC end sequencing (6000 genes in Paux et al. 2006). Such a high number of genes could be due partly to an overrepresentation of distal contigs with 69% of the sample sequences having two times the gene density of that observed in proximal regions. Considering that distal regions represent half of the chromosome (500 Mb), a weighted value of ∼8400 genes on 3B can be estimated (5700 telomeric and 2700 centromeric) for a total of 50,000 per diploid genome.
To get another estimate for the gene number on this chromosome, we used the 1973 Mb of sequences obtained by Solexa/Illumina sequencing of sorted chromosome 3B. First, the 55 million reads were mapped against the 510,160 bp of sequence that corresponds to the 199 low-copy genic regions identified during the annotation of the 18 Mb of contig sequences. The results revealed an average coverage of 1.2x per gene with large variations ranging from 0 to 3.7x, whereas ∼25% of the genic regions were covered <0.2 times. The N50 was 0.8x, indicating that despite a theoretical coverage of 2x, only half of the 3B chromosome sequence is covered more than 0.8 times by the Solexa sequence reads. The low GC content (42.8%) of the Solexa sequences compared with the average value (46%) observed in wheat BAC sequences may indicate a preferential amplification of the repeated fraction during the preparation of sorted 3B DNA before sequencing. This would result in an underrepresentation of the genic fraction and the observed discrepancy. The 0.8x value was applied in a second analysis whereby the 1973 Mb of Solexa reads were aligned against the 40,349 sequences present in the wheat unigenes database (NCBI build#55). A total of 2748 unigenes were identified with a coverage of at least 0.8x. Taking into account the N50 value (i.e., 5496 unigenes in total) and the fact that ∼24% of the annotated genic regions from the Mb-sized contigs were not found in the wheat unigene set, a total of 7230 genic regions was estimated for chromosome 3B by this whole-chromosome shotgun approach. As it is not possible to distinguish complete genes from gene fragments in the short reads data set, the gene number estimate can be refined by taking into account the proportion of gene fragments observed during the Mb-sized contig annotation (12.1% of the genic regions). This results in an estimate of ∼6360 genes for chromosome 3B and, therefore, ∼40,000 for the B genome.
Composition and Distribution of the TEs
To obtain novel information on the relative distribution and evolution patterns of TEs along wheat chromosomes and provide a foundation for the future annotation of the wheat genome, particular attention was given to the identification and annotation of TEs. In total, 3222 complete or truncated elements were identified, including 126 known and 818 new families. TEs represented 81.4% of the genomic sample sequences (Table 2; see Supplemental Figure 4 online) and ranged from 46 bp in size for a MITE (a nonautonomous transposon Mariner Athos) to 31,157 bp for a Jorge CACTA transposon. To correct potential bias in the representation of the sequences, the most gene-rich regions (ctg0954 and ctg0011) and the most repetitive centromeric region (100L17) were removed from the statistical analyses. The corrected value indicated 88.2% of TEs, a level higher than the 76.3% estimated previously from the analysis of 11 Mb of BES from the 3B chromosome library (Paux et al., 2006). Class 1 retrotransposons and class 2 DNA transposons accounted for 65.9 and 14.5% of the sequences, respectively (Table 2). This ratio is in between rice, in which class 2 outnumber class 1 elements (International Rice Genome Sequencing Project, 2005), and maize where class 2 represents 8.6% of the genome (Schnable et al., 2009). Interestingly, while the class 1 TE proportion was similar to the 68.7% observed in BESs (Paux et al., 2006), class 2 elements were 3 times more abundant than in the previous estimate (5.6%). The difference was mostly due to the CACTA transposons that represented 13.6% of the 13 contigs sequences (Table 2) versus 4.9% of the BESs (Paux et al., 2006). The detection of CACTA transposons is important not only for understanding genome composition but also because of their role in capturing and shuffling exons (Wicker et al., 2003) as shown for the Pack-MULES and helitrons in rice and maize (Jiang et al., 2004; Morgante et al., 2005). Here, we identified four CACTAs containing three different host gene fragments and one gene likely to be complete. Thus, extrapolating this value to the whole chromosome, CACTAs would be involved potentially in capturing ∼200 gene fragments on chromosome 3B, the same number reported recently for the Sorghum bicolor genome (Paterson et al., 2009). In addition, five helitron-derived elements were found in contigs ctg0011 and ctg0954, but none contained host gene fragments.
Table 2.
Classification of the 3222 TEs Annotated in the Whole 18.2 Mb of Sequence Produced from the 13-Mb Contigs of Chromosome 3B
| Type of TE | No. Copies | % of TE Copies | No. Bases | % of TE Fraction | % of Genome Fraction | |
| LTR retrotransposons | ||||||
| Gypsy | 1,219 | 37.83 | 7,939,750 | 53.23 | 43.59 | 64.10 |
| Complete | 688 | 56.44 | 6,264,453 | 78.90 | 34.40 | |
| Solo LTR | 91 | 7.47 | 175,245 | 2.21 | 0.96 | |
| Truncated | 367 | 30.11 | 1,195,157 | 15.05 | 6.56 | |
| Unknown | 73 | 5.99 | 304,895 | 3.84 | 1.67 | |
| Copia | 504 | 15.64 | 3,036,081 | 20.35 | 16.67 | |
| Complete | 264 | 52.38 | 2,374,144 | 78.20 | 13.04 | |
| Solo LTR | 35 | 6.94 | 65,294 | 2.15 | 0.36 | |
| Truncated | 160 | 31.75 | 435,234 | 14.34 | 2.39 | |
| Unknown | 45 | 8.93 | 161,409 | 5.32 | 0.89 | |
| Unknown | 127 | 3.94 | 698,013 | 4.68 | 3.83 | |
| Complete | 75 | 59.06 | 583,189 | 83.55 | 3.20 | |
| Solo LTR | 20 | 15.75 | 33,150 | 4.75 | 0.18 | |
| Truncated | 29 | 22.83 | 73,461 | 10.52 | 0.40 | |
| Unknown | 3 | 2.36 | 8,213 | 1.18 | 0.05 | |
| Non-LTR retrotransposons | ||||||
| LINE | 170 | 5.28 | 317,598 | 2.13 | 1.74 | 1.75 |
| SINE | 3 | 0.09 | 716 | 0.00 | 0.00 | |
| Transposons | ||||||
| CACTA | 332 | 10.30 | 2,479,618 | 16.62 | 13.61 | 14.53 |
| Complete | 137 | 41.2 | 1,521,871 | 61.38 | 8.36 | |
| Truncated | 156 | 46.99 | 639,258 | 25.78 | 3.51 | |
| Unknown | 39 | 11.75 | 318,489 | 12.84 | 1.75 | |
| Harbinger | 35 | 1.09 | 26,479 | 0.18 | 0.15 | |
| Mariner | 294 | 9.12 | 38,550 | 0.26 | 0.21 | |
| Mutator | 74 | 2.30 | 43,724 | 0.29 | 0.24 | |
| Hat | 7 | 0.22 | 9,444 | 0.06 | 0.05 | |
| Others | 193 | 5.99 | 49,180 | 0.33 | 0.27 | |
| Helitrons | ||||||
| Helitron | 5 | 0.16 | 13,562 | 0.09 | 0.07 | 0.07 |
| Unclassified | 259 | 8.04 | 264,184 | 1.77 | 1.45 | 1.45 |
| Total | 3,222 | 14,916,899 | 81.90 | |||
The distribution of complete, solo LTR, truncated, and unknown (i.e., undefined or partially sequenced) is indicated for the LTR retrotransposons and the CACTA transposons. For these later subcategories, the proportions of copy numbers and TE fraction are expressed as a percentage within each superfamily.
Similarity searches against the TREP10 databank (http://wheat.pw.usda.gov/ITMI/Repeats/) and clustering of newly discovered elements enabled the classification of the 3222 TEs into 944 different families containing 1 to 226 members. Despite this wide diversity, only eight families (Fatima, Jorge, Angela, Laura, Sabrina, WIS, Wilma, and Nusif) account for >50% of the TE fraction (see Supplemental Figure 5A online). Interestingly, 61% of the repeat families showed a very low MDRN90 (<10), confirming that the wide majority of TEs is weakly repeated. The elements found in the 818 new families accounted for 13.9% of the total TE fraction. They will greatly enrich the TE databanks and improve future high throughput TE annotations of the wheat genome using automated pipelines.
To investigate evolutionary forces that shaped the wheat genome, we determined precisely the proportions of complete (with two complete TSDs and/or LTRs/TIRs) versus truncated elements. A total of 1027 (59%) LTR retrotransposons and 137 (47%) CACTAs were identified as complete with quite similar ratios between the different sequenced regions (sd =11 and 14%, respectively; Table 2). This result indicates that most of the TEs are still complete in the wheat genome but are highly nested and that target site duplications of an ancient element may be found far apart from each other (e.g., >200 kb away in some cases). When individual BACs were considered within each contig, the proportion of complete LTR retrotransposons and CACTA transposons decreased significantly because in 18% of the cases (up to 50% for small BACs), the rest of the sequence was not present on the same BAC.
TE content, both in terms of amount and type, was highly variable between the different chromosomal locations (see Supplemental Tables 2 and 3 online). The two distal ctg0011 and ctg0954 show a very low TE content (59.2%) compared with more proximal ones (100L17 and ctg1030), which contain close to 100% of TEs without any gene. The composition of the centromeric sequence was very different from the other regions. It consisted exclusively of three types of LTR retrotransposons: Cereba, Quinta, and a newly identified Copia (Unnamedfam6) repeated in 27, 18, and 4 highly nested copies, respectively. The Quinta and Cereba elements were also found in other proximal contigs (see Supplemental Table 2 online) but at a lower density and with much older insertion dates (1.6 versus 0.6 million years) than those identified at the centromere. Indeed, the youngest average insertion time (0.490 million years ago) among all LTR retrotransposon activities was found for the centromeric BAC elements. These results suggest that the current wheat centromeres have been shaped by the recent reactivation of TEs ancestrally present in the proximal regions. Alternatively, the recent insertion time estimates may originate from a reduced substitution rate around the centromere compared with the other regions. It may also be due to an increased rate of sequence conversion related to a high level of unequal homologous recombination between LTRs as observed previously at rice centromeres (Ma and Bennetzen, 2006).
Other TE families exhibited contrasted distribution patterns: Barbara retrotransposons were found mainly in the proximal regions, whereas some CACTAs (Caspar, Clifford, and Boris) were repeated preferentially in the distal parts of the chromosome. Finally, Laura retrotransposons were found in significantly higher density in the geneless contig ctg1030 than in the other contigs (24.0 versus 4.7%). This may reflect preferential insertion of this element in a region (bin 3BS1-0.33-0.55) that contains heterochromatin (Gill et al., 1991), thereby suggesting a potential Laura signature for heterochromatic DNA.
Different Waves of TE Amplification Have Shaped the Wheat Genome
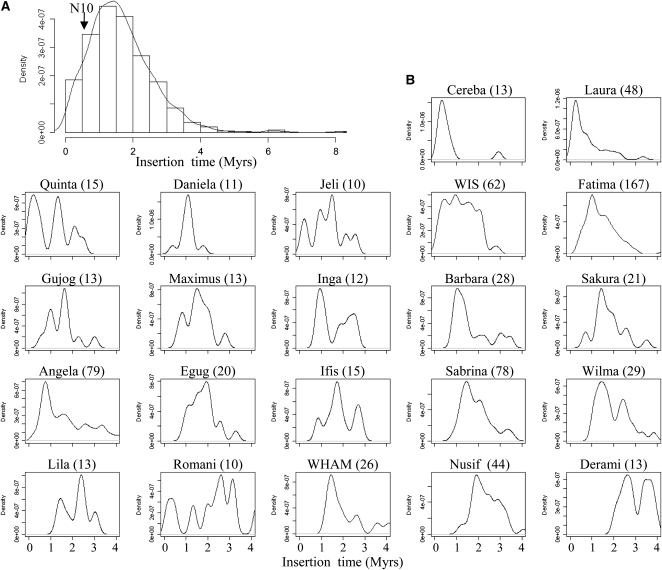
LTR nucleotide divergence indicated that 90% of the LTR retroelements transposed <3 million years ago with the oldest element (a Gypsy Lisa) inserted 7.5 million years ago and the most recent (two Gypsy Quinta elements with 100% identical LTRs) inserted <40,000 years ago (Figure 3A). The burst of amplification peaks at 1.4 million years ago (i.e., before the allopolyploidization event 0.5 million years ago [Huang et al., 2002; Dvorak et al., 2006] at the origin of the tetraploid ancestor Triticum turgidum). Furthermore, each family of LTR retrotransposons exhibited a specific pattern of activity (Figure 3B). Most of the highly repeated families showed a single narrow peak (e.g., Laura, Cereba, and Fatima), suggesting that amplification occurred mainly during a short period of time (<1 million years) possibly followed by silencing. By contrast, some families, such as WIS, seem to have been amplified over a long period of time (over 2.5 million years). The TE transposition time was also specific to each family with some showing recent transposition activity (e.g., Cereba, Quinta, and Laura; <1 million years ago), while others have been inactive since ∼2 million years ago (e.g., Derami, Nusif, and WHAM), indicating that transposition bursts did not involve all TE families at once and that the wheat genome was shaped by a succession of amplification waves involving different families. Moreover, the results show that, except for the centromeric regions, no significant LTR retrotransposon activity has affected the wheat genome in the past 0.5 million years (N10, Figure 3A).
Figure 3.
Ages of LTR-Retrotransposon Insertions.
Distribution of the age of the insertion of 880 complete LTR retrotransposons (A) and the 13 most abundant LTR retrotransposon families (B). The insertion time was calculated based on the LTR sequence divergence using a substitution rate of 1.3 × 10−8 substitutions/site/year (Ma and Bennetzen, 2004). For each family, the number of complete copies used to calculate the insertion pattern is indicated in brackets. The cutoff value of 0.5 million years (Myrs) distinguishing the 10% of youngest elements (N10) is indicated with an arrow.
Homogeneous Expansion and Accelerated Evolution of the Wheat Genome
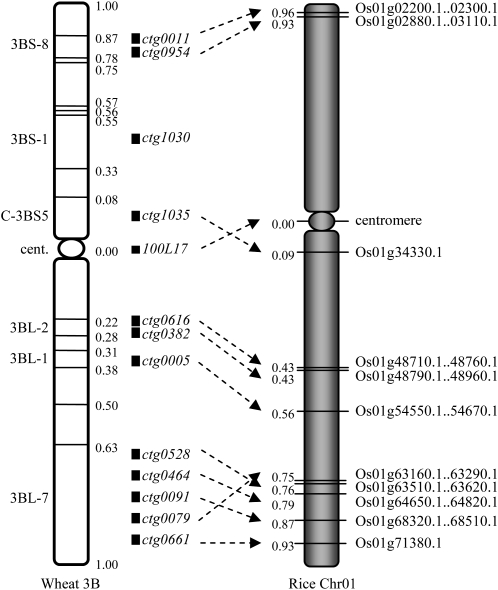
Wheat chromosome 3B is highly collinear to rice chromosome 1 (Sorrells et al., 2003; Salse et al., 2008). Alignment of the 13 contig sequences revealed that they are mostly collinear between the two genomes (Figure 4). We estimated the relative physical distances between the contigs by their position within the deletion bins and compared it with the relative distance of their orthologous regions on rice chromosome 1. This did not reveal any significant difference along the two chromosomes (Figure 4). Since genes generally are evenly distributed along rice chromosome 1, our findings suggest that the proximal cytogenetic bins of wheat chromosome 3B contain as many orthologous genes as the distal ones and that the distal and proximal regions have been expanded to the same extent in wheat. Furthermore, at the scale of the BAC contigs, the size ratio of wheat and rice orthologous regions (14x; 13.1 Mb in wheat versus 0.91 Mb in rice) corresponds to the ratio of size between the wheat B and rice genomes (15x). All regions have expanded at a similar level (14 ± 5x) whatever their proximal or distal location. Thus, despite different burst times and contrasted patterns of insertion, globally, the amplification of TEs occurred at the same intensity across the wheat chromosomes, resulting in an apparently homogeneous expansion of the genome. To confirm this result at the sequence level, we compared the distances between all available adjacent pairs of orthologous genes in wheat and rice. For the 74 analyzed IGDs, we found that only 30% (22/74) had a ratio between 5 and 30x, indicating an expansion level around the average value. By contrast, 40% (30/74) showed little or no expansion (ratio <5), thereby contributing to the formation of gene islands, while 30% (22/74) have massively expanded (ratio >30). We did not observe any differences for this pattern between the distal and proximal regions. Thus, we conclude that the average expansion factor of 14x observed between the wheat and rice genomes does not correspond to a globally homogeneous expansion but encompasses great local variations that can be of several orders of magnitude.
Figure 4.
Chromosomal Location of the 13 Sequenced Contigs from the Wheat 3B Chromosome and Their Orthologous Regions on Chromosome 1 of Rice.
The boundaries of 16 deletion bins are indicated by horizontal lines across the 3B chromosome, and their distances from the centromere are expressed as a fraction of length of chromosomal arms. In rice chromosome 1, the first and last genes carried by the orthologous regions are indicated. Their distances from the centromere were calculated using a centromere position at 16.8 Mb (http://rice.plantbiology.msu.edu/pseudomolecules/centromere.shtml). The relative order of the contigs in bin 3BL7-0.63-1.00 was determined by genetic mapping (data not shown).
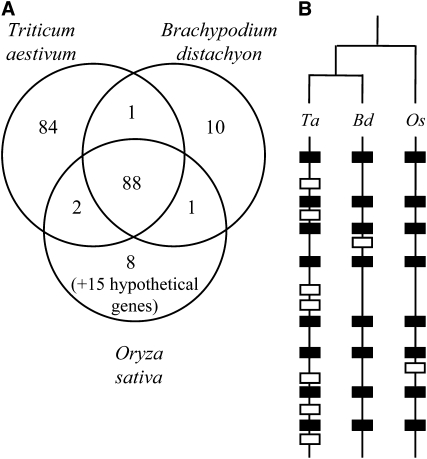
To further investigate the evolution patterns along wheat chromosome 3B, we compared systematically the 175 wheat predicted genes and pseudogenes with the rice and Brachypodium distachyon genome sequences. Only half (52%, 91 genes including six pseudogenes) were strictly orthologous with genes of rice chromosome 1 or of B. distachyon chromosome 2 (Figure 5A). Their products shared 77% ± 12% and 82% ± 12% amino acid identity (see Supplemental Table 1 online), respectively. Among the 91 orthologous genes, 88 were conserved and syntenic between the three genomes, two were common with rice but absent in B. distachyon, and one was conserved with B. distachyon while absent in rice (Figure 5A). These 91 genes represent an ancestral Poaceae genic backbone that exhibits a constant gene density of 1 gene per 200 kb (Figure 1) across the proximal and distal contigs. This is reflected in the conserved homogeneous distribution of genes observed through the alignment of the wheat 3B and rice 1 chromosomes (Figure 4).
Figure 5.
Level of Synteny between Wheat, Rice and B. distachyon Genomes.
(A) Venn diagram of the syntenic and nonsyntenic genes between wheat, rice, and B. distachyon. Fifteen additional hypothetical rice genes that do not share any similarity within the sequence databanks and represent putative prediction errors are mentioned on the diagram. All other nonsyntenic genes identified (84 in wheat, 10 in B. distachyon, and 8 in rice) have homologs in the compared species.
(B) Schematic representation of orthologous chromosomes displaying orthologous (black) and nonsyntenic (white) genes in wheat (Ta), B. distachyon (Bd), and rice (Os).
In addition to this ancestral backbone, wheat contigs carry an unexpectedly high proportion (48% of the gene content; Figure 5) of non collinear genes. While 21 out of the 84 noncollinear genes are likely pseudogenes, the majority (75%) shows complete open reading frames (ORFs) and significant similarities over >70% of their length with a homologous gene product in rice and B. distachyon, indicating that they do not correspond to wrongly annotated repetitive elements. Noncollinear genes were found in all contigs but were more abundant in the subtelomeric contigs (Figure 1). Ctg0954 and ctg0011 showed the highest degree of noncollinearity with 62 and 87% (33/53 and 27/31), respectively, of their coding fraction not found in orthologous regions of the other genomes. By contrast, the whole set of 13 genes carried by ctg0079 was collinear in B. distachyon (only one gene missing in rice). Thus, this result strongly suggests that the twofold increase in gene density observed in the subtelomeric regions is due mainly to the presence of a high amount of noncollinear genes. Interestingly, nonsyntenic genes were not found in clusters; rather, they were interspersed along the ancestral gene backbone, thus disrupting collinearity with rice and B. distachyon at many locations (Figure 5B). One of the most striking examples is contig ctg0954 in which 29 wheat locus-specific genes are interspersed along the ancestral conserved backbone composed of 18 orthologous genes (see Supplemental Figure 6 online). Comparison with the orthologous region that was partially sequenced at the Rph7 locus in barley (Hordeum vulgare; Brunner et al., 2003) showed that noncollinear wheat genes are conserved in barley. In addition, comparison of the 84 noncollinear wheat genes with the barley mapped ESTs (http://www.harvest-web.org; assembly #35) revealed that half of them are on chromosome 3H and are likely syntenic with wheat. Thus, our results suggest that the high level of gene rearrangements observed in wheat compared with rice and B. distachyon is a common feature of the Triticeae.
The noncollinear wheat genes shared similarities with genes located on several other chromosomes in rice and B. distachyon, suggesting that they originate mainly from independent events of translocation or interchromosomal duplication in the wheat genome. Moreover, for these genes, the best BLAST hits were collinear in the rice and B. distachyon genomes in 80% of the cases revealing ancestral loci that have been duplicated and/or translocated to new chromosomal locations in wheat specifically. Interestingly, a significant correlation (r2 = 0.744) was observed between the level of collinearity disruption and the level of TE activity (see Supplemental Figure 7 online) with the most rearranged contigs (ctg0011, ctg0954, and ctg0661) showing the most recent (1.0 to 1.4 million years on average) TE activity, while the most conserved regions (ctg0528, ctg0079, and ctg0005) contained the oldest TEs (1.9 to 2.1 million years on average). Thus, together, these results suggest that interchromosomal duplications mediated by transposable elements had a major impact on increasing the number of genes and in relocating genes at nonsyntenic regions in the wheat and most probably the ancestral Triticeae genome.
We also examined the ratio of genes found in the rice and B. distachyon orthologous regions but not present on chromosome 3B: only nine nonconserved genes were found in rice and 11 in B. distachyon (Figure 5A). All of the nonconserved genes, except the pair Os01g48810/Bradi2g46610, were not orthologous between rice and B. distachyon, indicating that their absence in wheat does not reflect a deletion from an ancestral gene in the Triticum lineage but rather a gene insertion specifically in the rice or B. distachyon lineages (Figure 5B). The analysis of the complete genomes of rice and B. distachyon showed that these locus-specific genes are duplicated within their respective genomes, suggesting a predominant role for interchromosomal duplications in gene movement in these genomes. Further analyses are underway to determine whether this is the same in wheat, in particular whether homologs of the noncollinear genes are also found and in which proportion on other wheat chromosomes.
Tandem duplicated genes represented 33% of the gene content in our data set with 26 groups of duplicated genes composed of two to seven copies. Tandem gene duplications occurred primarily in the telomeric regions with 64 of the 66 duplicated genes found in six distal contigs. Only three pairs of duplicated genes were found duplicated in rice and B. distachyon as well, revealing that they were ancestrally duplicated and that several copies are maintained by selection following ancestral sub- or neofunctionalization. The 23 remaining tandem duplicated groups are found in a single copy at syntenic positions in rice and B. distachyon, indicating that most of the gene duplications occurred recently in the Triticeae evolution. These results demonstrate that, in addition to interchromosomal duplications, tandem gene duplication in distal regions is a major force driving gene number increases in wheat with a strong potential for the creation of new functions. Most (18/26) of the duplicated groups have at least one gene copy that is a pseudogene, revealing that, in most of cases, gene duplication does not increase fitness sufficiently enough to result in both copies being maintained by selection. Interestingly, among the 51 pseudogenes and gene fragments identified, the vast majority (88%; 45/51) were nonsyntenic, indicating that pseudogenization concerned mainly the additional noncollinear genes.
In conclusion, considering that the ancestral gene content remains highly conserved and that half of the coding potential consists of genes inserted recently, our data support the idea that the wheat genome contains more genes than the rice and B. distachyon genomes.
DISCUSSION
Mb Level Sequencing Provides Insights into the Wheat Genome Composition
Following the establishment of the first physical map of a wheat chromosome (Paux et al., 2008), we produced and analyzed 18 Mb of contiguous sequences and 2 Gb of whole chromosome survey short reads from chromosome 3B of hexaploid wheat. Together with the 11 Mb of BES obtained previously on the same chromosome (Paux et al., 2006), this unique sample represents a large and diverse sampling of wheat sequences compared with other sequences produced thus far, which include only a few contigs larger than 300 kb. Comparison of the features from the three different sequence data sets indicated putative bias induced by different approaches in assessing genome composition. Evidence for bias was determined first with GC content estimates. While contigs displayed a similar GC content (46%) as the Triticum/Aegilops BAC sequences present in the public databases, BES and Solexa sequences showed significantly lower values, suggesting a potential bias for AT-rich regions in these samples. This was particularly true for the Solexa sequencing sample in which amplification of sorted chromosomal DNA prior to sequencing seems to be less efficient for the genic regions (GC-rich) than for the TE fraction (AT-rich). For the BESs, the reduced GC% is likely related to an intrinsic feature of the wheat genome in which HindIII restriction sites (used to produce the wheat BAC libraries) are overrepresented in eight of the most repeated TE families that display a low GC content (F. Choulet, unpublished data).
Another discrepancy was found in the estimation of the percentage of CACTA elements. In this study and in a recent analysis of 10 single wheat BAC clones from chromosome 3B (Charles et al., 2008), 3 times more CACTA elements were identified than in the BES survey of the same chromosome (Paux et al., 2006). CACTAs are particularly prone to underestimation because their sequences are highly variable (Wicker et al., 2008), and it is more difficult to identify them by similarity from short sequences than from long stretches of DNA. Furthermore, having the ability to analyze large contiguous sequences enabled us to find that even if they are highly nested, the majority of CACTAs and LTR retrotransposons are still complete in the wheat genome. This feature was not observed in previous studies based on individual BAC sequencing (Charles et al., 2008) because the nested clusters generally span distances that are larger than a BAC. Combined with the fact that most TEs transposed more than 0.5 million years ago (i.e., before the two polyploidization events), these results support the idea that deletion forces have been low in comparison to those of amplification during evolution, thereby contributing to the large size of the wheat genome (Wicker and Keller, 2007).
Finally, significant variations between the different samples were found when estimating the number of genes. Depending on the sequence sample, between 6000 and 8400 genes (+1000 gene fragments) were predicted for chromosome 3B; this would translate to 36,000 to 50,000 for the B genome of hexaploid wheat. This is slightly higher than the 32,000 to 40,000 annotated genes obtained from the rice, sorghum, and maize whole-genome sequences (International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; Schnable et al., 2009). The lower estimate for wheat is in the range of the number of unigenes (40,349) deduced from the EST collection, which by definition cannot be complete (NCBI build#55). On the other hand, the upper estimate is still lower than the 98,000 genes per diploid genomes estimated through random sequencing of wheat methyl-filtrated libraries (Rabinowicz et al., 2005; Paux et al., 2006). These discrepancies can have different origins. The lower estimate was obtained from BESs and is most likely an underestimation. Indeed, we found evidence that due to the increased frequency of HindIII sites in some TE families, BESs obtained from the HindIII 3B-specific BAC library (Safar et al., 2004) originate preferentially from the repeated fraction of the chromosome, thereby leading to an underestimation of the gene content. We also believe that the whole 3B survey using Solexa reads (7230 genes) slightly underestimates the gene number because the coverage is highly uneven when amplification is used prior to sequencing and genic regions tend to be less represented. Thus, assessing the gene content by this approach is rather unreliable. Finally, we believe that the precise annotation of the 13 Mb-sized contigs provides the best estimate at this time because complete gene structures with ORF, pseudogenes, and gene fragments as well as different members of tandemly repeated gene families could be distinguished and taken into account in the gene number calculation. This level of resolution cannot be achieved with short sequence surveys that collapse paralogs and cannot distinguish gene fragments from complete genes. Similar conclusions were reached for maize, where the characterization of the gene and repeat spaces based on genome survey sequences (Messing et al., 2004) was further refined by sequencing large BAC or contig sequences (Haberer et al., 2005; Kronmiller and Wise, 2009).
Thus, even if our sample may not be completely representative of the whole genome, we conclude that, in addition to faster and more random approaches, megabase-level sequencing provides essential knowledge of the genome composition and organization for the delineation of the best strategy for sequencing the entire genome. Individual chromosome sequencing efforts that are currently underway at the international level (see www.wheatgenome.org) will help to confirm and refine our estimates for the whole genome in the near future.
Genes Are Mainly Clustered into Small Islands Spread Out along the 3B Chromosome
Previous work based on EST mapping into deletion bins concluded that 94% of wheat genes are clustered into gene-rich regions spanning 29% of the chromosomes (Erayman et al., 2004). In addition, since crossing-over frequency is related to gene density (Akhunov et al., 2003b), it was suggested that centromeric regions, where recombination is largely suppressed in wheat, could be devoid of genes. Here, Mb-sized contig sequences and MTP macroarray hybridizations show that regions larger than 800 kb without genes are rare on chromosome 3B, even in the proximal regions. This refines the findings of Devos et al. (2005) and Charles et al. (2008) who also observed the presence of one or two genes per BAC in a majority of clones randomly chosen from whole-genome and chromosome 3B-specific libraries. This finding has great implications for the future sequencing of the wheat genome since accessing the entire wheat gene space will require sequencing at least 90% of the BAC contigs obtained for the minimal tiling paths of the different wheat chromosomes. Therefore, cost-efficient strategies need to be established to ensure complete sequencing of these contigs. Spatial gene distribution and sequencing strategy were refined on the same model for the maize genome after the first large sequences were analyzed (Haberer et al., 2005; Liu et al., 2007; Kronmiller and Wise, 2009) and revealed a much more homogeneous distribution than proposed previously (Carels et al., 1995; Barakat et al., 1997).
The possibility of examining large contiguous sequences from contrasted regions also enabled us to better define the gene island concept in wheat. Gene islands reflect inhomogeneous expansion of the genome and are not found in compact genome species such as rice, Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000), and Brachypodium (Huo et al., 2009). They are, however, common features of large and repetitive genomes, such as the 2.5 Gb maize genome. Interestingly, wheat and maize share similar genome structures despite divergent evolutionary histories (Salse et al., 2009). In maize, the gene distribution is quite similar to wheat with small islands (one to four genes/island) separated by blocks of repetitive elements and only 22% of the annotated BACs without genes (compared with 27% in our sample; Kronmiller and Wise, 2009). This suggests that common evolutionary forces govern the dynamics of large plant genomes. Gene islands may originate from two different evolutionary scenarios: (1) selection against the separation of genes by TE insertions that would be deleterious for gene expression or regulation and (2) homogeneous expansion combined with preferential deletions in gene-rich regions. The latter was proposed as the explanation for increased gene density in the distal regions of the sorghum chromosomes (Paterson et al., 2009). In maize, old TEs were found more frequently in BACs without genes than in gene-rich BACs, suggesting that the elimination rate of LTR retroelement is higher in gene-rich regions (Liu et al., 2007). In fact, since old elements are enlarged by subsequent insertions of younger TEs, the probability of finding old elements is lower in gene-rich than in gene-free BACs simply because the intergenic spaces studied are smaller. This is an intrinsic limit of sequencing isolated BAC clones as opposed to large stretches of contiguous sequence. In our sample, old LTR retroelements were found at every locus (except the centromeric BAC). TE deletion also can be monitored partly through the identification of solo-LTRs (Devos et al., 2002). Here, they were found mainly in the large blocks of TEs rather than in the gene islands, suggesting that TE removal is not the main driver for the formation of gene islands in wheat (Tian et al., 2009). The apparently homogeneous expansion of the wheat genome resulted from a combination of massive expansion of some regions that accumulated TEs and may have served as a nucleation point for additional TE insertions with selection against TE insertion maintaining a majority of genes in close vicinity from each other. To investigate further the dynamics of gene islands in wheat, we are developing a transcriptional map of the wheat 3B chromosome, which will permit us to determine whether genes located in gene islands are preferentially expressed or coregulated when compared with more isolated genes.
Additional Noncollinear Genes Are Interspersed within a Very Conserved Ancestral Grass Gene Backbone
Our results indicate a gene density that is two times higher in the distal (1 gene/87 kb) than in the proximal contigs (1 gene/184 kb). At the same time, we observed that the relative distances of the genes to the centromeres were similar on the wheat 3B and rice 1 chromosomes, indicating that the distribution of genes belonging to the ancestral Poaceae backbone is homogeneous in the two species. In addition, the size ratio of wheat and rice orthologous regions (14x) revealed that despite different burst times and nonrandom patterns of insertion, the amplification of TEs occurred at the same intensity across the wheat chromosomes, resulting in a homogeneous expansion of the proximal and distal regions. Thus, differential efficiency of transposition cannot explain the increased gene density toward the telomeres. By contrast, our findings suggest that TE-mediated interchromosomal and tandem gene duplications are primarily responsible for the higher gene density and higher disruption of collinearity in the telomeric regions of wheat chromosome 3B. Comparative sequence analyses revealed that 99% of the genes that were orthologous between rice and B. distachyon were also found at syntenic locations on chromosome 3B, revealing that no rearrangement of the ancestral gene order has occurred over the whole 18 Mb of wheat contigs. However, a high proportion of nonsyntenic genes was observed in the distal regions, especially on 3BS. All nonsyntenic genes shared significant similarity with genes in rice or B. distachyon genomes located on different chromosomes, indicating that they may originate from interchromosomal duplications of DNA fragments carrying complete or partial genes as previously suggested by Li and Gill (2002) and more recently by Akhunov et al. (2007). The fact that the nonsyntenic genes are not clustered but rather interspersed along the ancestral backbone also supports the idea of multiple events of long-distance duplications. This is in perfect agreement with previous results based on EST hybridizations that showed that 25% of gene loci are duplicated (either interchromosomal or intrachromosomal) in wheat, especially in the distal regions (Akhunov et al., 2003b). In addition, one-third on the genes were found tandemly duplicated within the distal contigs with a majority of nonsyntenic genes, suggesting that, in addition to translocations, tandem gene duplications played a significant role in increasing the number of genes at the telomeric ends of the chromosomes during wheat evolution.
High duplication and translocation activities that occurred in the wheat and most likely barley lineages resulted in an increased level of synteny perturbations, which was suggested previously by EST mapping for the A and D genomes (Akhunov et al., 2003a) and in the distal bin of 4BL (See et al., 2006). Here, we observed that the rice and B. distachyon genomes are much more similar to each other, in terms of gene content, than they are to wheat. This confirms at the sequence level the accelerated evolution in the Triticeae that was suggested very recently through comparison of an EST genetic map of A. tauschii with the rice and sorghum genomes (Luo et al., 2009). Thus, although Brachypodium is phylogenetically closer to wheat than rice (Griffiths et al., 2006) and sequence homology is higher, the lineage-specific rearrangements that occurred in wheat have disrupted the synteny as much with Brachypodium as with rice. Therefore, for structural, synteny-based genomic analyses, Brachypodium will not provide significantly better support than rice for estimating the gene order and content of the wheat chromosomes.
Evidence for TE-mediated interchromosomal duplications was obtained from two types of results. First, there was a clear correlation between the levels of nonsyntenic genes and TE activity found in the telomeric regions. Second, we identified four genes that were included in CACTA transposons. CACTAs are rare in the rice (0.3%) and maize genomes (3%) where gene capture has been driven massively by Pack-Mules and helitrons (Jiang et al., 2004; Morgante et al., 2005), although CACTAs carrying gene fragments were already observed in maize (Li et al., 2009). By contrast, they were estimated to account for 14% of the wheat B genome, suggesting that, as was shown recently for sorghum (Paterson et al., 2009), CACTA-mediated gene capture may be one of the main mechanisms for gene amplification and mobilization in wheat (Akhunov et al., 2007).
Following a normal evolutionary pattern, we would expect that the 91 genes conserved at regions syntenic between wheat, rice, and B. distachyon were present in each of the A, B, and D diploid ancestral homeologous genomes. Interestingly, only a very small fraction (7%) of these genes indicated pseudogenization, suggesting that despite different episodes of polyploidization and segmental duplications, the wheat genes were not affected by major structural rearrangements as previously observed by sequencing homeologous loci in wheat (Chalupska et al., 2008). Thus, in contrast with paleopolyploids such as maize, gene loss has not been that extensive in wheat since its polyploid origins, confirming that the mechanisms of coping with polyploidization have varied significantly for different genomes (Hufton and Panopoulou, 2009). Polyploidization events may have occurred too recently (∼500,000 years) to observe massive loss of redundant functions by genetic drift; however, a more probable explanation is that most of the duplicated gene copies have been maintained by selection since they represent more than just redundant gene functions. Indeed, there is accumulating evidence for differential expression of homoeologous genes in wheat (Bottley et al., 2006; Pumphrey et al., 2009), suggesting that subfunctionalization is playing a major role in maintaining the structural integrity of duplicated genes along the wheat chromosomes.
Here, by analyzing a unique set of Mb-sized contiguous sequences and of whole chromosome survey sequences from a wheat chromosome, we gained extensive insights into the composition, organization, and evolution of the wheat genome that will permit us to better define the genome sequencing and annotation strategies. With the development of additional physical maps and the sequencing of the first chromosomes in the near future, we will be able to refine our knowledge and develop even better tools to access and exploit the wheat genome. With one Insertion Site-Based Polymorphism marker every 3.8 kb and one Simple Sequence Repeat every 13 kb (Paux et al., 2010), the wheat genome sequence holds the potential for unlimited marker development and for a paradigm shift in breeding. The preliminary work presented here is paving the way toward enabling this transformative technology for wheat.
METHODS
BAC Screening and Contig Selection
In total, 152 BACs (see Supplemental Table 4 online) from the Minimal Tiling Path of 13 contigs selected from the physical map of chromosome 3B (Paux et al., 2008) were used for complete sequencing. Sequences and annotation are available through the Wheat 3B Physical Map Genome Browser at http://urgi.versailles.inra.fr/cgi-bin/gbrowse/wheat_FPC_pub/. The contigs were chosen to cover different regions of the 3B chromosome. Two contigs (ctg0011 and ctg0954) originated from the subtelomeric deletion bin (3BS8-0.778-0.87) on the short arm of the chromosome and were selected from a 12-centimorgan region carrying disease resistance gene. One contig (ctg1030) was located in a bin (3BS1-0.33-0.55) positioned at half of the 3BS chromosome arm. A contig (ctg1035) and a single BAC clone (TaaCsp3BFhA_0100L17 referred as 100L17) identified in pericentromeric and centromeric regions were selected to investigate more specifically the composition of centromeric sequences. On the long arm, two contigs (ctg0616 and ctg0382) originated from a subcentromeric region (bin 3BL2-0.22-0.28), and another one (ctg0005) was assigned to a more distal deletion bin (3BL1-0.31-0.38) at 1/3 of the chromosome arm. Finally, five contigs were chosen based on their EST content after screening the 3B BAC library with 399 ESTs assigned to the most distal deletion bin 3BL7-0.63-1.00 (229 ESTs previously assigned; Qi et al., 2004) and an additional 170 markers identified using synteny with rice chromosome 1. Three contigs (ctg0464, ctg0079, and ctg0661) carried two or more ESTs, one carried one EST (ctg0528), and one did not carry any EST (ctg0091).
BAC Sequencing and Assembly
Ten of the 13 contigs (ctg0005, ctg0079, ctg0091, ctg0382, ctg0464, 100L17, ctg0528, ctg0616, ctg0661, and ctg1035) were sequenced at the Centre National de Séquençage (Evry, France). First, 101 BACs were sequenced using Sanger technology with libraries obtained after mechanical shearing of BAC DNA and 5-kb fragment cloning into pcdna 2.1 plasmid vector (Invitrogen). DNA was purified and end-sequenced using dye terminator chemistry on ABI 3730 sequencers (Applied Biosystems) at 12× read coverage. Three other BACs were sequenced using 454-GS-FLX sequencer at 25× read coverage (Roche). Contigs ctg1030 and ctg0954 were sequenced by GATC Biotech (Konstanz, Germany) using Sanger technology at 8× read coverage. Finally, ctg0011 was sequenced using Sanger at 6× read coverage. BAC sequences were assembled using Phred/Phrap/Consed (Ewing and Green, 1998; Ewing et al., 1998; Gordon et al., 1998) for Sanger reads and Newbler (Roche) for 454 GS-FLX reads. Finishing sequencing reactions were performed for all clones until all contigs could be ordered and orientated for building a single supercontig for each BAC contig.
Annotation and Sequence Analyses
The TriAnnot pipeline (http://urgi.versailles.inra.fr/projects/TriAnnot/) was used for automatic annotation of genes. CDS predictions were combined with similarity searches using NCBI-BLAST (Altschul et al., 1997) against full-length cDNAs, unigenes, and ESTs from wheat (Triticum aestivum), Triticeae, and other Poaceae and against SwissProt and the rice (Orzya sativa) proteome specifically. Matching transcripts were mapped on the genomic DNA using Gmap (Wu and Watanabe, 2005). Predicted CDSs that do not share any significant similarity with any sequence in the databanks were discarded from the annotation. Gene models displaying translational stop codons, frameshift mutations, or small deletions (up to 30% of a complete homolog) within the ORF were considered as pseudogenes. Genes showing similarity over <50% of the length of their best homolog in databanks were considered as gene fragments. tRNA genes were identified using tRNAscan-SE (Lowe and Eddy, 1997).
For TE annotation, RepeatMasker Open (Smit et al., 1996–2004; http://www.repeatmasker.org) was used to find similarities against the TREP databank (http://wheat.pw.usda.gov/ITMI/Repeats/), and the dotter program (Sonnhammer and Durbin, 1995) was used to precisely annotate the exact borders of each TE by identifying long terminal repeats or terminal inverted repeats and the target site duplication. Reconstruction of the nested structures of TEs was manually curated under Artemis (Rutherford et al., 2000). Curated annotations were then inserted into our local relational database. Classification of TE was performed by following the procedure described by Wicker et al. (2007). For elements larger than 1 kb, a strong hit (≥80% identity) with an element from TREP and covering at least 500 bp was considered as threshold to assign a family. For elements smaller than 1 kb, a strong hit (≥80% identity) covering at least 50% of the query length was considered. A clustering of unknown elements was performed using exactly the same criteria to identify members of the same new family. Insertion dates of LTR retrotransposons were estimated by aligning both 5′ and 3′ LTRs using ClustalW (Larkin et al., 2007) and considering a mutation rate of 1.3 × 10−8 substitutions/site/year (SanMiguel et al., 1998; Ma and Bennetzen, 2004). This estimation was performed on 880 complete LTR retrotransposons containing no sequencing gap and for which both LTRs do not differ by >200 bp in size (caused by large insertion/deletion).
Solexa Sequencing of Sorted Chromosome 3B, Computing of an MDR Index, and Mapping Short Reads on Reference Sequences
The 3B chromosomes were sorted by flow cytometry and then amplified using Phi29 polymerase. DNA was then sequenced on a flow cell of Illumina/Solexa Genome Analyzer II using paired-end reads (average fragment size: 626 bp). Sequencing generated 54,808,646 paired-end reads of 36 nucleotides each, thus providing 1,973,111,256 nucleotides corresponding to theoretical 2X coverage of the complete chromosome. The complete sample was used for generating a MDR index using the Tallymer program (Kurtz et al., 2008). The optimal k-mer size was evaluated to 17 nucleotides, thus providing more than 1 billion 17-mers from the complete sample of which 81% are not unique. Tallymer was used to count the occurrence of each 17-mer (and its reverse complement) along the sequenced contigs according to the 3B MDR index. We developed a program (PlotMdr.pl) (available on request) that parses Tallymer results to generate MDR plots and compute the MDRN90 (i.e., the cutoff for the highest 10% of MDR values).
To estimate the real coverage of Solexa read data set, we selected 596 kb low-copy DNA regions corresponding to the 199 genic regions (introns and exons) annotated in the 13 completely sequenced contigs. To avoid unspecific alignment of 36-mers on repeated DNA, we first masked the regions showing MDR ≥5 (representing 86 kb). The addSolexareads script from the Consed suite was then used to map the 55 million of Solexa reads on the resulting 510 kb of reference sequences. The number of reads that correctly map to the template sequences was calculated to estimate the coverage of the Solexa data set. Then, the same procedure was applied to the 40,349 wheat unigenes (NCBI build#55) to identify and estimate the number of genes carried by chromosome 3B.
Hybridization on MTP-3B Macroarray
The 7440 BACs of the wheat chromosome 3B MTP (Paux et al., 2008) were gridded in duplicate on 22 × 22 nylon membranes (Proteigene) in 5 × 5 spot arrays with the control plate. The spotted membranes were incubated at 37°C on Luria-Bertani agar with 12.5 μg/mL chloramphenicol and were treated as described by Paux et al. (2004). Total RNAs of hexaploid wheat cv Chinese Spring were extracted from 500 mg of five organs (root, leaf, stem, spike, and grain) at two or three developmental stages each as described by Cossegal et al. (2008). DNA was removed using RQ1 RNase-free DNase (Promega). The RNeasy MinElute Cleanup kit (Qiagen) was used for purification, and cDNA synthesis was performed with the SMART PCR cDNA synthesis kit (Clontech) followed by purification with the QIAquick PCR purification kit (Qiagen).
DNA probes were labeled with [α-32P]dCTP (Perkin-Elmer; 50 ng SMART PCR cDNA and 50 pg Desmin cDNA) using the Megaprime DNA labeling system (GE Healthcare). Genomic DNA of wheat cv Chinese Spring was partially digested with RQ1 RNase-free DNase (Promega) at 37°C for 30 s, and the reaction was stopped by adding 2 μL of Stop Solution and incubating at 85°C for 15 min. Resulting DNA fragments (between 200 and 400 bp) were used as blocking DNA. Labeled cDNA probes were mixed with 2.5 μg blocking DNA (50× more than cDNA). The mix was denatured at 100°C for 5 min and incubated at 37°C for 30 min before hybridization. Hybridizations were performed as described by Paux et al. (2004). The wrapped filters were exposed for 1 to 2 d (Imaging Screen K; Bio-Rad). A hybridization step with labeled pIndigoBAC-5 polylinker was performed to validate filters quality and to normalize hybridization data.
Imaging screens were scanned with a Pharos FX Plus molecular imager (Bio-Rad) at 50-μm resolution, and ArrayVision 8.0 imaging software (Imaging Research) was used for signal detection and quantification. MTM density quantification method was chosen, and background for each grid was calculated based on the empty spots using median density method. The raw hybridization signals were normalized as described by Paux et al. (2004). Normalized intensity of each spot was divided by the normalized intensity of the same spot from the pIndigoBAC-5 polylinker hybridization to correct for bacteria growth bias. The median was calculated with the four growth-corrected normalized intensities of each BAC. Intensities above 4 times and below one-quarter of the median were excluded from the analysis. The corrected median was calculated with the remaining growth-corrected normalized intensities of each BAC. Unpaired Welch's two samples t tests were performed to compare the intensities of each BAC to the intensities of the negative controls using R (http://www.r-project.org/). A BAC was considered as positive if its P value was below 0.05 and its corrected median above the maximal corrected median of expressed transposable elements controls.
Accession Numbers
BAC contig sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers FN564426-37 and FN645450. Solexa sequence data can be found in the EMBL Sequence Read Archive under accession number ERA000182.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Graphical Representation of the Annotation of the 13-Mb Contig Sequences from Wheat Chromosome 3B.
Supplemental Figure 2. Distribution of the Number of Exons per Gene.
Supplemental Figure 3. Distribution of the Gene and MITE Densities along the 3.1-Mb ctg0954.
Supplemental Figure 4. Histograms of the Composition in Transposable Elements of the 13 Sequenced Contigs.
Supplemental Figure 5. Proportion and MDR Analysis of the TE Families.
Supplemental Figure 6. Sequence Comparison of ctg0954 with the Rice Orthologous Region on Chromosome 1.
Supplemental Figure 7. Correlation between the Level of Synteny and the Level of TE Activity.
Supplemental Table 1. List of the Genes, Pseudogenes, and Gene Fragments Identified in the 13 Sequenced Contigs of Wheat Chromosome 3B.
Supplemental Table 2. Proportions of the 10 Most Represented TE Families within the 13 Contigs.
Supplemental Table 3. Proportions of the Known TE Families within Distal and Proximal Contigs.
Supplemental Table 4. List of the 152 Sequenced BAC Clones.
Acknowledgments
We thank D. Boyer from the Institut National de la Recherche Agronomique, Genetics Diversity and Ecophysiology of Cereals, for excellent technical assistance. We also thank P. Wincker, S. Samain, and V. Barbe from the Centre National de Séquençage (Evry, France) for their work on BAC sequencing. We thank I. Gut, C. Plançon, and Y. Duffourd from the Centre National de Génotypage (Evry, France) for their work on Illumina/Solexa sequencing and J. Doležel and H. Šimková (Institute of Experimental Botany, Olomouc, Czech Republic) for the sorted chromosome 3B DNA. This project was supported by grants from the Commissariat à l'Energie Atomique-Genoscope (AP2008), the Agence Nationale de la Recherche ANR-GPLA06001G SMART in the frame of the national Genoplante program, the Institut National de la Recherche Agronomique (AIP Sequencing “Plant ReSeq”), a Turkish Academy of Sciences - Outstanding Young Scientists Award, and the Swiss National Science Foundation (Grant 105620).
References
- Akhunov E.D., Akhunova A.R., Dvorak J. (2007). Mechanisms and rates of birth and death of dispersed duplicated genes during the evolution of a multigene family in diploid and tetraploid wheats. Mol. Biol. Evol. 24: 539–550 [DOI] [PubMed] [Google Scholar]
- Akhunov E.D., et al. (2003a). Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. USA 100: 10836–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov E.D., et al. (2003b). The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barakat A., Carels N., Bernardi G. (1997). The distribution of genes in the genomes of Gramineae. Proc. Natl. Acad. Sci. USA 94: 6857–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A., Xia G.M., Koebner R.M. (2006). Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906 [DOI] [PubMed] [Google Scholar]
- Brunner S., Keller B., Feuillet C. (2003). A large rearrangement involving genes and low-copy DNA interrupts the microcollinearity between rice and barley at the Rph7 locus. Genetics 164: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels N., Barakat A., Bernardi G. (1995). The gene distribution of the maize genome. Proc. Natl. Acad. Sci. USA 92: 11057–11060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupska D., Lee H.Y., Faris J.D., Evrard A., Chalhoub B., Haselkorn R., Gornicki P. (2008). Acc homoeoloci and the evolution of wheat genomes. Proc. Natl. Acad. Sci. USA 105: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles M., et al. (2008). Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180: 1071–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossegal M., Chambrier P., Mbelo S., Balzergue S., Martin-Magniette M.L., Moing A., Deborde C., Guyon V., Perez P., Rogowsky P. (2008). Transcriptional and metabolic adjustments in ADP-glucose pyrophosphorylase-deficient bt2 maize kernels. Plant Physiol. 146: 1553–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K., Bennetzen J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Ma J., Pontaroli A.C., Pratt L.H., Bennetzen J.L. (2005). Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc. Natl. Acad. Sci. USA 102: 19243–19248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J., Simkova H., Kubalakova M., Safar J., Suchankova P., Cihalikova J., Bartos J., Valarik M. (2009). Chromosome genomics in the Triticeae. Plant Genetics and Genomics, Feuillet C., Muehlbauer G.J., (New York: Springer; ), pp. 285–316 [Google Scholar]
- Dvorak J., Akhunov E.D., Akhunov A.R., Deal K.R., Luo M.C. (2006). Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23: 1386–1396 [DOI] [PubMed] [Google Scholar]
- Erayman M., Sandhu D., Sidhu D., Dilbirligi M., Baenziger P.S., Gill K.S. (2004). Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res. 32: 3546–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feuillet C., Salse J. (2009). Comparative genomics in the Triticeae. Plant Genetics and Genomics, Feuillet C., Muehlbauer G.J., (New York: Springer; ), pp. 451–477 [Google Scholar]
- Gill B.S., Friebe B., Endo T.R. (1991). Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830–839 [Google Scholar]
- Gordon D., Abajian C., Green P. (1998). Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Haberer G., et al. (2005). Structure and architecture of the maize genome. Plant Physiol. 139: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., Gornicki P. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton A.L., Panopoulou G. (2009). Polyploidy and genome restructuring: A variety of outcomes. Curr. Opin. Genet. Dev. 19: 600–606 [DOI] [PubMed] [Google Scholar]
- Huo N., Vogel J.P., Lazo G.R., You F.M., Ma Y., McMahon S., Dvorak J., Anderson O.D., Luo M.C., Gu Y.Q. (2009). Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol. Biol. 70: 47–61 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al. ; French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jurka J. (1997). Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 94: 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger S., Wicker T., Keller B. (2009). Map-based cloning of genes in Triticeae (wheat and barley). Plant Genetics and Genomics, Feuillet C., Muehlbauer G.J., (New York: Springer; ), pp. 337–358 [Google Scholar]
- Kronmiller B.A., Wise R.P. (2009). Computational finishing of large sequence contigs reveals interspersed nested repeats and gene islands in the rf1-associated region of maize. Plant Physiol. 151: 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Narechania A., Stein J.C., Ware D. (2008). A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC Genomics 9: 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Dai J., Li J., Yan J. (2009). Identification and characterization of CACTA transposable elements capturing gene fragments in maize. Chin. Sci. Bull. 54: 642–651 [Google Scholar]
- Li W., Gill B.S. (2002). The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the triticeae. Genetics 160: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang P., Fellers J.P., Friebe B., Gill B.S. (2004). Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40: 500–511 [DOI] [PubMed] [Google Scholar]
- Liu R., Vitte C., Ma J., Mahama A.A., Dhliwayo T., Lee M., Bennetzen J.L. (2007). A GeneTrek analysis of the maize genome. Proc. Natl. Acad. Sci. USA 104: 11844–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.C., et al. (2009). Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl. Acad. Sci. USA 106: 15780–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2006). Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden E., Sears E. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–89 107–116 [DOI] [PubMed] [Google Scholar]
- Messing J., Bharti A.K., Karlowski W.M., Gundlach H., Kim H.R., Yu Y., Wei F., Fuks G., Soderlund C.A., Mayer K.F., Wing R.A. (2004). Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101: 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M.L. (2009). Sequencing technologies - The next generation. Nat. Rev. Genet. 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Mochida K., Yoshida T., Sakurai T., Ogihara Y., Shinozaki K. (2009). TriFLDB: A database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 150: 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. (2005). Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37: 997–1002 [DOI] [PubMed] [Google Scholar]
- Ogihara Y., et al. (2005). Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33: 6235–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paux E., et al. (2010). Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted selection in wheat. Plant Biotechnol. J. 8: 196–210 [DOI] [PubMed] [Google Scholar]
- Paux E., Roger D., Badaeva E., Gay G., Bernard M., Sourdille P., Feuillet C. (2006). Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 48: 463–474 [DOI] [PubMed] [Google Scholar]
- Paux E., et al. (2008). A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322: 101–104 [DOI] [PubMed] [Google Scholar]
- Paux E., Tamasloukht M., Ladouce N., Sivadon P., Grima-Pettenati J. (2004). Identification of genes preferentially expressed during wood formation in Eucalyptus. Plant Mol. Biol. 55: 263–280 [DOI] [PubMed] [Google Scholar]
- Pumphrey M., Bai J., Laudencia-Chingcuanco D., Anderson O., Gill B.S. (2009). Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 181: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.L., et al. (2004). A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz P.D., Citek R., Budiman M.A., Nunberg A., Bedell J.A., Lakey N., O'Shaughnessy A.L., Nascimento L.U., McCombie W.R., Martienssen R.A. (2005). Differential methylation of genes and repeats in land plants. Genome Res. 15: 1431–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. (2000). Artemis: Sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Sabot F., Guyot R., Wicker T., Chantret N., Laubin B., Chalhoub B., Leroy P., Sourdille P., Bernard M. (2005). Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol. Genet. Genomics 274: 119–130 [DOI] [PubMed] [Google Scholar]
- Safar J., et al. (2004). Dissecting large and complex genomes: Flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 39: 960–968 [DOI] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Bolot S., Guilhot N., Courcelle E., Faraut T., Waugh R., Close T.J., Messing J., Feuillet C. (2009). Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl. Acad. Sci. USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U.M., Calcagno T., Cooke R., Delseny M., Feuillet C. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P., Gaut B.S., Tikhonov A., Nakajima Y., Bennetzen J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- See D.R., Brooks S., Nelson J.C., Brown-Guedira G., Friebe B., Gill B.S. (2006). Gene evolution at the ends of wheat chromosomes. Proc. Natl. Acad. Sci. USA 103: 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B., Flavell R.B. (1975). Characterisation of the wheat genome by renaturation kinetics. Chromosoma 50: 223–242 [Google Scholar]
- Soderlund C., et al. (2009). Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., Durbin R. (1995). A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10 [DOI] [PubMed] [Google Scholar]
- Sorrells M.E., et al. (2003). Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Rizzon C., Du J., Zhu L., Bennetzen J.L., Jackson S.A., Gaut B.S., Ma J. (2009). Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19: 2221–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S.R., Bureau T.E., White S.E. (1995). LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5: 814–821 [DOI] [PubMed] [Google Scholar]
- Wicker T., Guyot R., Yahiaoui N., Keller B. (2003). CACTA transposons in Triticeae. A diverse family of high-copy repetitive elements. Plant Physiol. 132: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Keller B. (2007). Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 17: 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Narechania A., Sabot F., Stein J., Vu G.T., Graner A., Ware D., Stein N. (2008). Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. (2005). GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Yu J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]