This work identifies the putative transcription factor gene VDD as a direct target of MADS box proteins that regulate ovule development. It finds that antipodal and synergid cell identity and/or differentiation are affected in vdd mutant female gametophytes, providing insights into the pathways regulated by the ovule identity factors.
Abstract
In Arabidopsis thaliana, the three MADS box genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1), and SHP2 redundantly regulate ovule development. Protein interaction studies have shown that a multimeric complex composed of the ovule identity proteins together with the SEPALLATA MADS domain proteins is necessary to determine ovule identity. Despite the extensive knowledge that has become available about these MADS domain transcription factors, little is known regarding the genes that they regulate. Here, we show that STK, SHP1, and SHP2 redundantly regulate VERDANDI (VDD), a putative transcription factor that belongs to the plant-specific B3 superfamily. The vdd mutant shows defects during the fertilization process resulting in semisterility. Analysis of the vdd mutant female gametophytes indicates that antipodal and synergid cell identity and/or differentiation are affected. Our results provide insights into the pathways regulated by the ovule identity factors and the role of the downstream target gene VDD in female gametophyte development.
INTRODUCTION
In Arabidopsis thaliana, ovule primordia appear from the placental tissue at stage 8 of flower development (Smyth et al., 1990). Ovule differentiation is complete at stage 13, when the mature embryo sac awaits fertilization (Schneitz et al., 1995). It has been shown that the MADS box genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1), and SHP2 redundantly regulate the identity of the ovule integuments that develop from the chalazal region, since in the stk shp1 shp2 triple mutant, the integuments are transformed into carpelloid structures leading to complete sterility (Pinyopich et al., 2003; Brambilla et al., 2007). Furthermore, genetic and protein interaction studies have shown that these ovule identity factors interact with SEPALLATA (SEP) MADS domain factors and that these interactions are essential for their function in ovule development (Favaro et al., 2003). It is clear that these MADS domain transcription factors are key regulators of ovule development. However, there is very limited information about the genes that are regulated by them. It could well be that they are involved in the regulation of both sporophyte and gametophyte development since not only integument development but also embryo sac development was arrested in the stk shp1 shp2 triple mutant (Brambilla et al., 2007; Battaglia et al., 2008).
Differentiation of the embryo sac occurs contemporarily and in coordination with the development of the diploid sporophytic tissues of the ovule. Megasporogenesis takes place in the nucellus when integument primordia elongate from the chalazal region. Shortly after, the functional megaspore undergoes three rounds of mitosis without cellularization to form the syncytial female gametophyte, or embryo sac, with eight haploid nuclei. Subsequently, nuclear migration and cellularization take place, and the mature female gametophyte consists of seven cells: three antipodal cells, two synergid cells, one egg cell, and one central cell containing two polar nuclei that fuse prior or during fertilization (Schneitz et al., 1995). The formation of the next sporophytic generation depends on long- and short-range interactions between male and female gametophytes. The male gametophyte, or pollen tube, follows chemotactic signals produced by the female gametophyte and is guided into the micropylar opening of the ovule (Hülskamp et al., 1995; Ray et al., 1997). As the pollen tube approaches the micropyle, one of the synergid cells initiates degeneration and is penetrated by the pollen tube, which arrests its growth, bursts, and releases the two sperm cells to ensure double fertilization. These processes are referred to as pollen tube guidance and reception (reviewed in Weterings and Russell, 2004). Both male and female gametophytes play fundamental roles in the control of male gamete delivery (Johnson and Lord, 2006).
Recently, some of the mechanisms underlying double fertilization in angiosperms have been dissected at the molecular level. The identification and characterization of female gametophytic mutants showing defects in embryo sac cell differentiation allowed the determination of the contributions of specific embryo sac cells to pollen tube guidance and reception (Higashiyama et al., 2001; Huck et al., 2003; Kasahara et al., 2005; Portereiko et al., 2006; Chen et al., 2007; Pagnussat et al., 2007; Punwani et al., 2007; Bemer et al., 2008; Shimizu et al., 2008; Steffen et al., 2008; Okuda et al., 2009; Srilunchang et al., 2010). A key role during the fertilization process is played by the synergid cells. These haploid cells are not only responsible for the production and secretion of a signal that guides the pollen tubes toward the embryo sac (Higashiyama et al., 2001; Kasahara et al., 2005; Okuda et al., 2009, Tsukamoto et al., 2010), but they also mediate pollen tube reception (Huck et al., 2003; Rotman et al., 2003; Escobar-Restrepo et al., 2007). Once pollen tubes correctly reach the micropyle, synergid-specific expression of the FERONIA (FER) receptor-like kinase is required for pollen tube growth arrest, rupture, and sperm cell discharge (Huck et al., 2003; Escobar-Restrepo et al., 2007). In fer mutants, the pollen tubes fail to arrest and keep growing within the embryo sac, leading to pollen tube overgrowth (Huck et al., 2003; Rotman et al., 2003). Such pollen tube overgrowth has also been observed in the absence of the LORELEI function (Capron et al., 2008), in self-fertilized absence of mutual consent mutants (Boisson-Dernier et al., 2008), and in scylla (syl) mutant embryo sacs (Rotman et al., 2008). Interestingly, at low frequency, syl/SYL heterozygous plants show proliferation of the central cell nucleus in the absence of fertilization, indicating that the pollen tube overgrowth phenotype may also depend on some central cell functions (Rotman et al., 2008).
Here, we report the functional characterization of VERDANDI (VDD), a direct target gene of the ovule identity factors STK, SHP1, and SHP2. VDD plays a role in female gametophyte development and fertilization. Since in the vdd mutant both synergids and antipodal cells lose their cellular identity, we named this mutant after one of the three Norns, the goddesses of fate in Norse mythology (Brodeur, 1916). Although pollen tubes get attracted, the transmission efficiency of the vdd mutant allele is drastically reduced through the female gametophyte due to a defect in the subsequent fertilization process.
RESULTS
Identification of Genes Expressed in Ovule Primordia
In the model plant Arabidopsis, ovule identity is redundantly regulated by the activity of the MADS box genes STK, SHP1, and SHP2 (Pinyopich et al., 2003). Genetic and biochemical evidence showed that ovule identity proteins interact with SEP factors in order to function (Favaro et al., 2003). These interactions might result in higher-order MADS domain complexes as has been suggested previously (Egea-Cortines et al., 1999). To identify target gene(s) of the ovule identity complex, we isolated cells from ovule primordia (stage 8-9 of flower development) using laser microdissection (see Supplemental Figure 1 online). RNA was extracted, amplified, and used for RNA profiling studies based on the Affymetrix ATH1 GeneChip. Using the GeneSpring 7.2 program, these expression data were analyzed, revealing that more than 14,000 genes are expressed in ovule primordia (see Supplemental Data Set 1 online). For our further analyses of these candidates, we considered only genes that were annotated as putative transcription factors. The reason for doing this is that we would like to study the network that is regulated by these MADS domain factors and because until now the majority of the direct target genes that have been identified for MADS domain proteins encode transcription factors (Sablowski and Meyerowitz, 1998; Wagner et al., 1999; Hepworth et al., 2002; Lamb et al., 2002; Ito et al., 2004; William et al., 2004; Gómez-Mena et al., 2005; Sundström et al., 2006). This reduced our set of genes to 1024 putative transcription factor–encoding genes (see Supplemental Data Set 2 online).
Since MADS domain proteins recognize and bind CArG boxes [CC(A/T)6GG] (Nurrish and Treisman, 1995), we further restricted our sample through the identification of ovule primordial-expressed transcription factors that contain CArG box consensus sequences in their genomic region (see Methods). For each gene, we considered a region comprising 3 kb upstream of its putative transcription start site, its complete coding sequence, including introns, and 1 kb downstream of the termination codon. Since the ovule identity proteins probably form a higher-order MADS domain complex that binds to multiple CArG boxes (Egea-Cortines et al., 1999; Favaro et al., 2003), we considered only those genes that had at least two MADS domain binding sites in their genomic region, with a distance between them of <300 bp. These selection criteria allowed us to identify a subset of 15 transcription factor–encoding genes as putative targets of the ovule identity complex (see Supplemental Table 1 online).
VDD Is a Direct Target of the Ovule Identity Complex
To investigate which of these 15 genes were indeed targets of the ovule identity MADS domain factors, chromatin immunoprecipitation (ChIP) experiments were performed using an STK antibody. Since STK is redundant for its ovule identity function with SHP1 and SHP2, we used flower tissue isolated from the shp1 shp2 double mutant for these experiments. This approach likely increased the amount of STK protein that binds to the target sites and optimized the ChIP analysis.
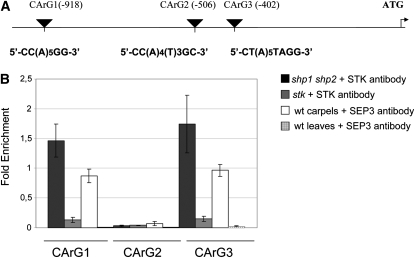
These experiments demonstrated the binding of STK to CArG box-containing regions in three of the 15 putative targets (see Supplemental Table 1 online). Here, we focus on the VDD gene encoding a transcription factor belonging to a poorly characterized gene family, namely, the reproductive meristem (REM) family (Franco-Zorrilla et al., 2002; Swaminathan et al., 2008; Romanel et al., 2009). Sequence analysis of the VDD genomic region revealed the presence of three putative CArG boxes (Figure 1A). Quantitative real-time PCR performed on chromatin immunoprecipitated using the anti-STK antibody showed an enrichment of the genomic regions containing CArG boxes 1 and 3, while the putative CArG box 2 sequence was not bound by STK (Figure 1B). Chromatin immunoprecipitated from the stk single mutant was used as negative control. These data strongly indicate that the STK protein directly interacts with the promoter region of the VDD gene.
Figure 1.
Quantitative Real-Time PCR on Chromatin Immunoprecipitated with STK and SEP3 Antibodies.
(A) Schematic representation of the position of the CArG boxes in the promoter region of the VDD gene.
(B) ChIP enrichment tests by quantitative real-time PCR show STK- and SEP3-specific binding to the CArG boxes 1 and 3. The stk single mutant was used as a negative control in the STK-ChIP and wild-type leaves as negative control for the SEP3-ChIP assays. Fold enrichment was calculated over the negative controls. Error bars represent the propagated error value using three replicates (see Methods).
Since SEP proteins are necessary for the formation of ovule identity protein complexes (Favaro et al., 2003), we also tested the binding of SEP3 to the CArG box–containing region of VDD. Our results showed an enrichment of fragments containing the same CArG boxes (1 and 3) that are bound by the STK protein (Figure 1B). These results demonstrate that the ovule identity protein complex composed of SEP3 and STK proteins directly interacts with the promoter region of the VDD gene.
VDD Is Not Expressed during Ovule Development in the stk shp1 shp2 Triple Mutant
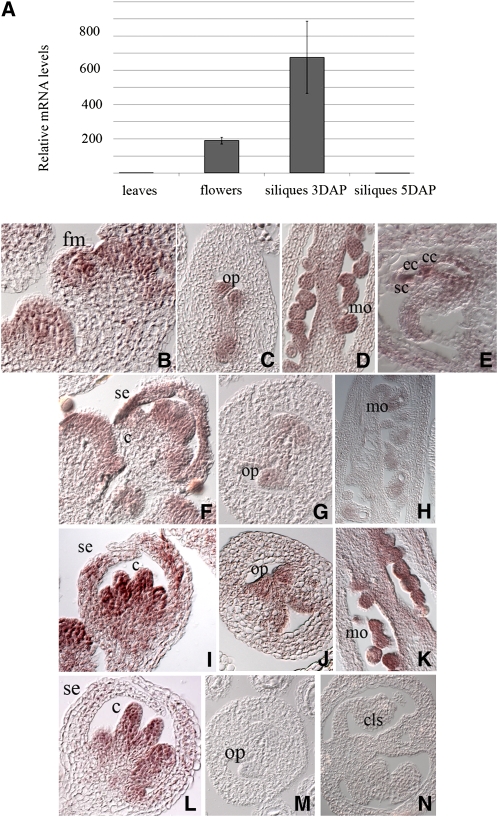
We studied the spatial and temporal expression pattern of VDD through quantitative real-time PCR (Figure 2A). This experiment showed that VDD is highly expressed in the reproductive tissues and during early stages of seed development. The amount of VDD transcript strongly decreased during late stages of seed formation (Figure 2A). To analyze better the VDD expression profile at the cellular level, in situ hybridization experiments were performed (Figures 2B to 2E). This analysis revealed that VDD is expressed in the inflorescence and floral meristems (Figure 2B). During ovule formation, VDD is expressed during all the stages of ovule development, including the developing embryo sac (Figures 2C to 2E).
Figure 2.
Spatial and Temporal Expression Pattern of VDD in Wild-Type, stk, shp1shp2 Double, and stk shp1 shp2 Triple Mutant Backgrounds.
(A) Quantitative real-time RT-PCR performed on cDNA obtained from leaves, flower, and siliques at 3 d after pollination (DAP) and siliques at 5 DAP. The relative mRNA levels indicate that VDD is strongly expressed in the reproductive tissues before fertilization and in the early stages of seed development. Error bars represent the propagated error value using three replicates.
(B) to (E) In situ hybridization experiment performed in wild-type plants.
(B) VDD is expressed in the floral meristem and in developing carpels and stamens.
(C) During ovule formation VDD mRNA is present in ovule primordia.
(D) VDD expression is detectable during later stages of ovule formation.
(E) In the mature embryo sac, VDD transcripts are found in the synergids, egg, and central cells.
(F) to (H) In situ hybridization experiment performed in the stk single mutant background.
(F) VDD is expressed in young flowers.
(G) Within ovule primordia, the signal is reduced compared with wild-type plants.
(H) Mature ovules show a decreased signal compared with wild-type plants.
(I) to (K) In situ hybridization experiment performed in the shp1 shp2 double mutant background.
(I) VDD is expressed in young flowers as in wild-type plants.
(J) Ovule primordia in the shp1 shp2 mutant plants express VDD.
(K) The hybridization signal is visible in mature ovules.
(L) and (M) In situ hybridization experiment performed in the stk shp1 shp2 triple mutant background.
(L) VDD is expressed in developing stamens and carpels.
(M) and (N) VDD transcripts are not detected in the ovule primordia (N). No hybridization signal is visible in the carpel-like structures that develop inside the stk shp1 shp2 mutant carpel.
c, carpel; cls, carpel-like structures; cc, central cell; ec, egg cell; fm, floral meristem; mo, mature ovules; op, ovule primordia; sc, synergid cells; se, sepal.
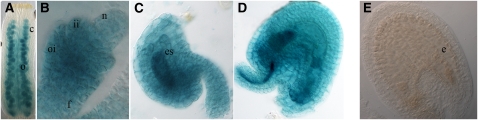
The VDD expression profile in the developing ovule and megagametophyte was confirmed by analyzing reporter gene expression of a construct consisting of 1221 bp upstream of the VDD translation start site, the complete genomic VDD coding region fused to the uidA reporter gene that encodes β-glucuronidase (GUS), and 389 bp corresponding to the VDD 3′ untranslated region. Transgenic plants transformed with this construct showed GUS activity in the same tissues where we observed VDD expression in our in situ hybridization experiments before fertilization (Figures 3A to 3D). However, we could not detect GUS expression in the developing seeds, indicating that this construct did not include the regulatory regions that are necessary for VDD transcription following fertilization (Figure 3E). We did not test longer constructs because the neighboring genes immediately flank the sequences we used: At5g18005 maps 1222 bp upstream of the VDD start site, while At5g17990 is located immediately downstream of the 3′ untranslated region.
Figure 3.
Expression of the pVDD:VDD-GUS Reporter Gene.
(A) GUS expression (blue color) is detectable during ovule formation.
(B) At stage 12 of flower development, GUS transcript is present in the funiculus, integuments, and nucellus.
(C) Within the ovule, the VDD promoter drives the expression of the reporter gene in the gametophytic and sporophytic tissues.
(D) The GUS reporter gene is transcribed in the mature female gametophyte and in the integuments.
(E) Following fertilization, the selected VDD promoter was not active in the developing endosperm and embryo.
c, carpel; e, embryo; es, embryo sac; f, funiculus; ii, inner integument; n, nucellus; o, ovules; oi, outer integument.
Since VDD is a direct target of STK, we were interested in investigating whether the expression of VDD is regulated by the ovule identity factors. Therefore, we analyzed VDD expression in the stk, shp1 shp2, and stk shp1 shp2 mutants (Figures 2F to 2N). Compared with the expression in wild-type ovules, a reduction in expression of VDD was observed in the stk single mutant (Figures 2G and 2H). Interestingly, no hybridization signal could be detected in stk shp1 shp2 triple mutant ovules, indicating that VDD expression within the ovule is strictly dependent on the activity of the ovule identity factors STK, SHP1, and SHP2 (Figures 2M and 2N). VDD was normally expressed in other floral tissues of the triple mutant, suggesting that other factors may regulate VDD expression in these organs (Figure 2L).
The vdd-1 Mutation Causes a Female Gametophytic Defect
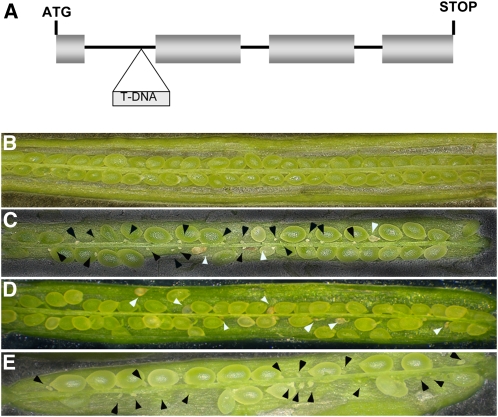
To understand the function of VDD, we characterized the vdd-1 mutant allele that carries a T-DNA insertion in the first intron (Figure 4A). Segregation analysis of the progeny of plants heterozygous for the vdd-1 allele revealed a distorted segregation from the expected 1:2:1 ratio (wild type:vdd-1/VDD:vdd-1). We obtained wild-type and heterozygous plants in a segregation ratio 1.0:1.3 but did not recover any plant homozygous for the vdd-1 mutation (Table 1).
Figure 4.
Seed Set in Wild-Type, vdd-1/VDD Heterozygous Plants, Complemented vdd-1 Heterozygous Plants, and pSTK:amiR-vdd Plants.
(A) Schematic representation of the vdd-1 mutant allele. T-DNA is inserted in the first intron, 44 bp upstream the 3′ splicing acceptor site.
(B) Wild-type silique showing full seed set.
(C) Siliques of vdd-1/VDD plants containing aborted ovules (black arrows) and aborted seeds (white arrows).
(D) Siliques of vdd-1/VDD plants complemented with the genomic region of the VDD gene. The complementation construct is able to rescue VDD expression in the female gametophyte. Aborted seeds (white arrowheads), but not aborted ovules, are present.
(E) Siliques of wild-type plants transformed with the pSTK:amiRvdd construct. Aborted ovules only (black arrowheads) are present.
Table 1.
Segregation Analysis of the vdd-1 Mutant Allele
| Progeny Genotype |
na Transmission Efficiencyb |
|||||||
| Male Genotype | Female Genotype | VDD/VDD | vdd-1/VDD | vdd-1/vdd-1 | Total | TEmale | TEfemale | P Valuec |
| vdd-1/VDD | vdd-1/VDD | 92 | 120 | 0 | 212 | n.a.d | n.a. | 0.054 |
| vdd-1/VDD | VDD/VDD | 151 | 128 | n.a. | 279 | 84.8% | n.a. | 0.169 |
| VDD/VDD | vdd-1/VDD | 207 | 57 | n.a. | 264 | n.a. | 27.5% | 2.7e-20 |
Total number of plants scored.
Transmission efficiencies (TE) as calculated by Howden et al. (1998).
P value obtained using the χ2 test under the hypothesis of a 1:1 segregation ratio.
n.a., not applicable.
To test whether female, male, or both gametophytes were affected due to the lack of VDD activity, we performed reciprocal crosses of vdd-1/VDD and wild-type plants. Crossing wild-type female with heterozygous pollen showed that transmission efficiency (TE; Howden et al., 1998) via the male gametophyte was only slightly reduced (TEmale= 84.8%) and not significantly different from the expected 1:1 segregation (χ2 test, P value = 0.17). By contrast, crossing a heterozygous female with wild-type pollen showed a strongly reduced TE through the female gametophyte (TEfemale= 27.5%), significantly deviating from the expectation (P value = 2.7 e-20) (Table 1). Assuming a normal TE through the male gametophyte, we expected to obtain 6.9% vdd-1/vdd-1 homozygous seedlings in the progeny of a selfed plant heterozygous for the vdd-1 mutation. However, such homozygous plants were not identified (n = 212), indicating that homozygous vdd-1 is also zygotically lethal.
Analysis of the siliques of plants heterozygous for the VDD T-DNA insertion showed that two different ovule abortion phenotypes could be distinguished (Figures 4B and 4C): abortions of ovules that were not fertilized (35%), which are expected to be due to the female gametophyte defect, and seed abortion postfertilization (10%, n = 5451). The latter phenotype probably explains why we did not observe homozygous vdd-1 plants. In siliques of wild-type sibling plants grown under the same conditions, ovule abortion was around 2%, which is in agreement with previously reported observations (Acosta-García and Vielle-Calzada, 2004). In this article, we will focus our attention on the role of VDD during female gametophyte formation. Concerning the seed phenotype, differential interference contrast (DIC) microscopy analysis showed that 8% of the developing seeds in the vdd-1 heterozygous siliques were arrested at the globular stage, indicating that VDD plays a role during the early stages of seed formation (see Supplemental Figure 2 online).
To confirm that the female gametophytic defect was caused by the loss of VDD activity, we performed complementation experiments using plants heterozygous for the vdd-1 mutation. Into these plants, we introduced a binary vector carrying the genomic region of the VDD gene, which included 1221 bp upstream of the translation start site, the complete genomic coding region, and 389 bp downstream of the stop codon. As already described, the reporter gene construct containing the same VDD sequences showed GUS expression in the young ovules and during female gametophyte development, whereas it was not expressed during seed formation (Figure 3). The T-DNA also encodes a visible selection marker (enhanced yellow fluorescent protein [EYFP]) under the control of the strong napin seed-specific promoter, allowing visible selection of transformant seeds (Stuitje et al., 2003; Battaglia et al., 2006). Selected T1 transformants were tested for the presence of the complementation construct and the presence of the vdd-1 allele. Analysis of the siliques in six of these T1 lines showed a reduced seed abortion rate in three plants. Since the T-DNA was hemizygous in T1 plants, we isolated EYFP positive seeds and selected four T2 plants that had only EYFP-positive seeds, indicating that the complementation construct was homozygous and did not segregate. When we analyzed the siliques of these plants, we did not detect abortion due to unfertilized ovules; as expected, we still observed early seed abortion (Figure 4D). Furthermore, segregation analysis of the vdd-1 allele in plants (n = 48) obtained from these seeds showed a segregation ratio of 1:2 (wild type:vdd-1/VDD), and no homozygous vdd-1 mutants were identified. This suggests that we successfully complemented the female gametophytic defect caused by the absence of VDD activity. Because at the time of manuscript preparation there were no other vdd mutant alleles available, we also investigated VDD's role during embryo sac formation by silencing VDD in wild-type plants via an artificial microRNA (Schwab et al., 2006) specific for the VDD gene. We expressed this artificial microRNA under the control of the STK promoter (pSTK; Kooiker et al., 2005). Analysis of the siliques of transgenic plants (six siliques for each plant) expressing the pSTK:amiR-vdd construct showed ovule abortion due to nonfertilization, whereas we did not observe increased early seed abortion in comparison to wild-type plants (Figure 4E). Since the pSTK promoter is active before fertilization, these results support the hypothesis that the vdd-1 mutation caused the female gametophytic defects. Due to the presence of wild-type VDD activity during seed formation, transgenic plants carrying the pSTK:amiR-vdd construct did not show seed abortion (Figure 4E).
Pollen Tube Guidance Is Normal but Fertilization Does Not Occur in vdd-1 Mutant Ovules
To examine whether the observed fertilization defect arose from defective embryo sac development, DIC microscopy analysis was performed on heterozygous vdd-1 plants. Interestingly, analysis of unpollinated mature ovules showed that all ovules in heterozygous vdd-1 plants were morphologically indistinguishable from each other and embryo sacs were composed of seven cells as in the wild type (see Supplemental Figure 2 online).
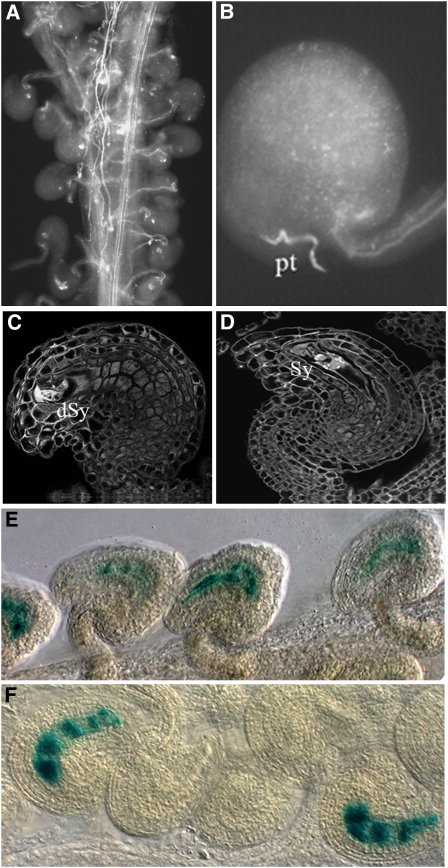
Subsequently, we analyzed by aniline blue staining whether the vdd-1 phenotype is due to a defect in pollen tube guidance (Figures 5A and 5B). In wild-type plants, pollen tubes grow along the transmitting tract toward the funiculus and subsequently to the micropyle and into the female gametophyte. When we pollinated vdd-1 heterozygous plants with wild-type pollen, 86% of pollen tubes successfully reached the micropyle (n = 545). These results were not statistically different compared with wild-type plants in which 88% of pollen tubes reached the micropyle (n = 229).
Figure 5.
Fertilization Analysis in vdd-1 Heterozygous Plants.
(A) Pollen tube staining with aniline blue shows that all embryo sacs in the vdd-1 heterozygous background are reached by pollen tubes.
(B) Detailed image of aniline blue–stained pollen tube reaching the micropyle in the vdd-1 heterozygous carpel.
(C) CLSM image of a wild-type fertilized female gametophyte. Observation performed 12 h after pollination showed one degenerating synergid in all the embryo sacs. The strong fluorescence signal indicates the degeneration of a synergid cell.
(D) Detailed image of a female gametophyte in the vdd-1 heterozygous pistil. In this genetic background, not all the embryo sacs show synergid degeneration at 12 h after pollination.
(E) Following pollination of wild-type pistils with pollen carrying the MINI3:GUS reporter construct, all seeds showed GUS activity in the developing endosperm.
(F) Polllination of heterozygous vdd-1 pistils with MINI3:GUS pollen. GUS activity is not observed in all of the ovules.
dSy, degenerated synergid; pt, pollen tube; Sy, synergid.
Despite the fact that vdd-1 mutant embryo sacs retained the capability to attract and guide pollen tubes, we investigated whether fertilization occurs once that pollen tube enters the vdd-1 mutant female gametophyte. Therefore, we pollinated pistils of wild-type and heterozygous vdd-1 mutant plants with pollen obtained from plants homozygous for the MINISEED3:GUS construct (Luo et al., 2005). As expected, following double fertilization, 90% of the ovules in wild-type pistils expressed the GUS reporter gene in the endosperm (Figure 5E). By contrast, ∼30% of the ovules in heterozygous vdd-1 mutant plants did not show GUS activity (Figure 5F), suggesting that fertilization did not occur in these ovules. Since we did not observe pollen tube overgrowth in the vdd-1 female gametophyte, it is most likely that a subsequent step, such as sperm cell delivery or gamete fusion, is affected in vdd-1 female gametophytes.
To investigate this further, we analyzed whether the synergid cells degenerated in heterozygous vdd-1 plants. In wild-type plants, pollen tube entrance in the micropyle is followed by degeneration of one synergid cell, which is revealed by intense fluorescence using confocal laser scanning microscopy (CLSM) (Christensen et al., 1997). As expected, when we pollinated wild-type pistils, high fluorescence was visible in almost all the embryo sacs at 12 h after pollination, suggesting that synergid degeneration occurred (Figure 5C). Interestingly, when we pollinated pistils of vdd-1 heterozygous plants using wild-type pollen, we observed frequently no synergid degeneration (Figure 5D), which might explain the observed defect in fertilization.
VDD Affects Accessory Cell Differentiation in the Embryo Sac
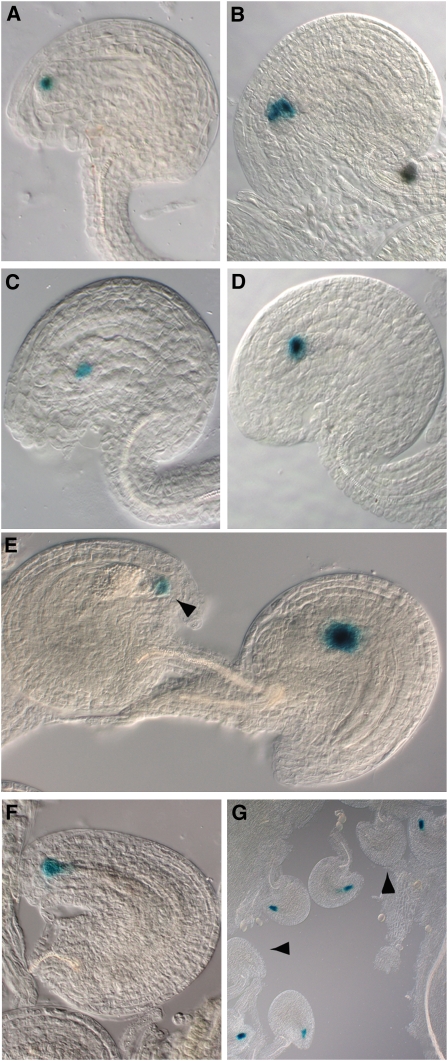
To investigate whether the identities of the vdd-1 embryo sac cells, in particular the synergids, were altered, the expressions of different cell-specific molecular markers were analyzed (Figure 6; see Supplemental Table 2 online). The marker lines were crossed with the heterozygous vdd-1 mutant and reporter gene expression was analyzed in the F2 generation. The expressions of the egg cell-specific marker EG1 (Gross-Hardt et al., 2007) and a central cell-specific marker (Chaudhury et al., 1997) were not altered in any of the embryo sacs (Figures 6A to 6D), indicating that gametic cell fate was not affected.
Figure 6.
Expression Pattern of Gametophytic Cell-Specific Markers in Wild-Type and vdd-1 Heterozygous Plants.
Plants homozygous for the gametophytic marker constructs were analyzed 48 HAE if not otherwise indicated.
(A) and (B) Egg cell–specific marker expression.
(A) Wild-type plants showed GUS expression in 98% of the female gametophytes (n = 289).
(B) vdd-1 heterozygous plants showed blue staining in 97% of the megagametophytes (n = 304).
(C) and (D) Central cell marker expression in wild-type plants (C) (98% of female gametophytes; n = 282) and in the vdd-1 heterozygous plants (D) (99% of megagametophytes; n = 327).
(E) Antipodal cell marker expression in the vdd-1/VDD plants 72 HAE. At this time point, vdd-1/VDD heterozygous siliques showed GUS expression in the synergids cells (arrowhead) (49%; n = 244). In the remaining 51% of megagametophytes (n = 254), blue staining was visible in the antipodal cells.
(F) and (G) Expression profile of the ET2634 synergid cell marker.
(F) In wild-type plants, the ET2634 synergid cell marker is visible in almost all of the mature embryo sacs (96%; n = 665).
(G) In the vdd-1 heterozygous plants, 32% of the mature embryo sacs (n = 740) did not express the synergid-specific cell marker (arrowheads).
When we analyzed accessory cell (synergid and antipodal) specification, we observed an abnormal expression profile for antipodal and synergid cell markers when compared with wild-type female gametophytes (Figures 6E to 6G). The expression of the antipodal cell marker (Yu et al., 2005) was analyzed at 24, 48, and 72 h after emasculation (HAE) in wild-type and vdd-1/VDD heterozygous siliques. We analyzed the expression at 24 HAE to exclude the possibility that our observations were influenced by the degeneration of antipodal cells. Unlike previously reported (Murgia et al., 1993; Christensen et al., 1997), we did not observe antipodal cell death prior to fertilization, and they were still clearly present at this time point. In heterozygous plants, at 24 HAE, only 55% of the ovules expressed the antipodal cell marker, whereas we did not detect GUS expression in the remaining ovules (n = 309). Interestingly, at 48 HAE, 6% of the ovules expressed the antipodal cell marker in the synergids instead of the antipodal cells, whereas in 38% of the megagametophytes, we were not able to detect GUS expression (n = 480). The rest of the ovules showed antipodal cell expression. At 72 HAE, the number of megagametophytes that showed GUS expression in the synergids further increased to almost 50% (n = 498) (Figure 6E). At 24, 48, and 72 HAE wild-type sibling plants showed GUS expression in the antipodal cells in 98, 97, and 98% of the megagametophytes, respectively (n = 324, 403, and 456, respectively).
Analysis of the expression of the synergid cell marker line ET2634 (Gross-Hardt et al., 2007) in vdd-1 heterozygous plants 48 HAE revealed that 32% of the megagametophytes did not express the synergid-specific marker (n = 740). For this marker also, an inversion of the expression was occasionally observed: 4% of the megagametophytes expressed the synergid marker in the antipodals instead of the synergid cells, which was not observed in wild-type plants. In comparison, wild-type sibling plants homozygous for the reporter construct showed GUS expression in 96% of the synergids at 48 HAE (n = 665) (Figures 6F and 6G).
Our data indicate that cell identity of antipodal and synergid cells in the vdd-1 mutant embryo sac is not correctly specified and/or that their differentiated state is not maintained. Interestingly, the 32% of female gametophytes that did not express the synergid cell marker corresponds to the percentage of ovule abortion observed in vdd-1/VDD heterozygous plants, whereas we found wrong specification of the antipodal cells in almost 50% of the gametophytes in vdd-1 heterozygous siliques. These data suggest that the fertilization defects described in the vdd-1 mutant gametophyte correlates with defects in the development or differentiation of synergid cells.
DISCUSSION
VDD Is a Direct Target of the Ovule Identity MADS Domain Transcription Factor Complex
MADS box genes belonging to the AG subfamily play a redundant role in the regulation of ovule development (Pinyopich et al., 2003; Brambilla et al., 2007), and their function has been well conserved during the course of plant evolution (Colombo et al., 2008). This was underlined not only by genetic studies, but also through the analysis of protein–protein interactions, which were found to be conserved between distantly related species (Favaro et al., 2002; Ferrario et al., 2003; Immink et al., 2003; Dreni et al., 2007). Despite the efforts to describe the molecular pathways regulated by MADS domain factors during flower development, very few direct target genes have been identified and characterized to date (Lamb et al., 2002; Hepworth et al., 2002; Ito et al., 2004; William et al., 2004; Gómez-Mena et al., 2005; Sundström et al., 2006), and this knowledge is completely missing for the ovule identity MADS domain factors.
Most efforts to identify direct target genes used transgenic plants in which a transcription factor was under external control, allowing the controlled induction of its activity. Subsequent genome-wide transcript profiling shows that genes are significantly changed in expression upon induction (Ito et al., 2004, 2007; Gómez-Mena et al., 2005). Other approaches include the comparison of differences in the transcriptome between wild-type and mutant plant tissues (Zik and Irish, 2003; Wellmer et al., 2004). Only recently, the ability to perform high-throughput sequencing on immunoprecipitated chromatin allowed the description of AP1 and SEP3 binding sites at the genome-wide level and the correlation of SEP3 function with plant hormone signaling pathways in Arabidopsis inflorescences (Kaufmann et al., 2009, 2010). Here, we present an alternative method to identify targets of the MADS domain ovule identity factors. This method starts with defining a subset of genes that are expressed in a specific cell type. In this study, we used ovule primordia at a very early stage of development (the 8-cell stage). Because we look here at expressed genes, we exclude the identification of genes that are completely silenced by the ovule identity factors. Subsequently, we selected from this subset of genes only those encoding putative transcription factors. We made this selection because until now most of the MADS domain target genes that have been identified encode transcription factors. The next step was to identify those genes that contain a putative MADS domain binding site. Since these CArG sequences are rather frequent in the Arabidopsis genome (Gómez-Mena et al., 2005), we considered only those genes that contain in their genomic region at least two putative CArG boxes within a distance <300 bp. This reduced the number of candidate targets to a subset of 15 genes. ChIP experiments using antibodies against STK confirmed 4 of these 15 genes as in vivo targets of STK. Although the criteria used to select these genes may seem a bit arbitrary, the results that we obtained in this study (along with those of other studies) support the model that MADS domain protein complexes often interact with DNA by contacting multiple nearby CArG sequences (Egea-Cortines et al., 1999; Gregis et al., 2008; Liu et al., 2008).
Ovule identity determination in Arabidopsis is dependent on the interaction between STK (or SHP1 or SHP2) and the SEP proteins (Favaro et al., 2003; Pinyopich et al., 2003; Brambilla et al., 2007). Therefore, we tested whether the CArG boxes in the VDD genomic region that are bound by STK were also interacting with SEP3. Interestingly, CArG boxes 1 and 3, which are directly contacted by STK, are also direct targets bound by SEP3.
This result suggests that SEP3-STK heterodimers could interact with DNA through the formation of tetramers as previously proposed for other MADS box complexes (Theissen and Saedler, 2001). Moreover, our binding analyses indicate that MADS domain protein dimers display affinity for specific CArG elements, since CArG box 2, which maps only 95 bp from CArG 3, is not contacted by the ovule identity protein complex composed of STK and SEP3. It will be interesting to investigate by site specific mutagenesis whether in the absence of a high affinity binding site, like CArG 1 or 3, the STK-SEP3 protein complex is able to interact with the VDD promoter using the lower-affinity CArG box 2.
VDD Expression Requires the Activity of the Ovule Identity Factors STK, SHP1, and SHP2
The ability to isolate ovule primordia and to analyze the tissue-specific transcriptome through the combination of laser microdissection and microarray hybridization allowed us to identify which genes are coexpressed with the ovule identity gene STK. As already discussed, this approach led to the identification of VDD as the first direct target of the ovule identity complex. This gene belongs to a plant-specific transcription factor family, namely, the REM family, which appears to be highly expanded in Arabidopsis (Swaminathan et al., 2008; Romanel et al., 2009). Despite the presence of at least 76 REM genes in the Arabidopsis genome, to date, VERNALIZATION1 is the only REM for which a function has been determined (Levy et al., 2002). This lack of functional information regarding the role of REM genes during plant growth might be due to high functional redundancy within this gene family and/or may be due to the fact that the mutant phenotypes are difficult to observe.
In general, AtGenExpress data (Schmid et al., 2005) shows that most of the REM-encoding genes are tissue-specifically expressed (Swaminathan et al., 2008; Romanel et al., 2009). VDD transcripts are present in the same tissues as the ovule identity genes STK, SHP1, SHP2, AG, and SEP (Yanofsky et al., 1990; Ma et al., 1991; Rounsley et al., 1995; Flanagan et al., 1996; Mandel and Yanofsky, 1998). VDD expression studies by in situ hybridization using different mutant backgrounds highlighted the redundancy of the ovule identity factors STK, SHP1, and SHP2 in the regulation of VDD expression within the ovule, since the expression of VDD is strictly dependent on the activity of the three MADS domain ovule identity factors. Only the absence of all three ovule identity proteins leads to complete absence of VDD expression during ovule development. This suggests that the ovule identity complex that binds the CArG boxes 1 and 3 in the VDD regulatory region is composed of STK, SHP1, SHP2, and SEP proteins. However, since our ChIP experiments do not include IP experiments using SHP antibodies, we cannot at this point exclude that STK binds preferentially to these sites and that only in the absence of STK, the SHP proteins will replace STK. The hypothesis that STK may be the main player regulating VDD expression is supported by the fact that loss of STK activity results in a significant reduction in VDD expression. Other transcription factors mediate the expression of VDD in the inflorescence and floral meristems, suggesting that other MADS domain factors might regulate this gene during other phases of flower development.
VDD Is Required for Cell Differentiation in the Female Gametophyte
Morphological and genetic analyses of plants carrying the vdd-1 allele showed that this mutation was transmitted through the female gametophyte at a significantly reduced efficiency. This reduction in vdd-1 transmission is due to a defect in fertilization after the pollen tube reaches the female gametophyte. In particular, pollen tubes correctly find their way along the funiculus into the micropyle and arrest their growth, but fertilization does not occur. Understanding the mechanisms that control the fertilization process represents an intriguing aspect of plant reproductive biology. After entering the micropylar opening, sperm cell discharge is under the control of both male and female gametophytes. In the female, the synergid-specific expression of FER is required for pollen tube growth arrest, rupture, and sperm cell release (Huck et al., 2003; Escobar-Restrepo et al., 2007). Continuous pollen tube overgrowth into fer mutant female gametophytes is linked to the absence of the signaling cascade regulated by the FER receptor-like Ser-Thr kinase in the receptive synergid cell (Escobar-Restrepo et al., 2007). The recent characterization of the syl mutant has indicated that additional embryo sac cells may also be involved in pollen tube reception (Rotman et al., 2008). It is thus possible that aspects of the molecular dialogue during fertilization takes place not only between male and female gametophytes but also among the haploid cells of the megagametophyte. The functional characterization of the FER homologs ANXUR1 (ANX1) and ANX2 allowed the identification of a male signaling cascade necessary to prevent pollen tube rupture before entering the micropyle. A precise molecular dialogue between the FER-dependent and ANX-dependent signaling cascades may be necessary for proper pollen tube reception (Boisson-Dernier et al., 2009; Miyazaki et al., 2009).
Silencing of the VDD gene segregates with defects in embryo sac cell specification and a lack of fertilization. Expression analysis of antipodal- and synergid-specific marker genes showed that the identity of these cells is compromised in plants heterozygous for the vdd mutation. The frequency with which we observe the loss of synergid cell identity correlates well with the reduced transmission efficiency of the vdd-1 allele through the female gametophyte. Therefore, this defect in synergid differentiation explains the lack of fertilization in the vdd-1 mutant embryo sacs. This is even further supported by the fact that we frequently do not observe synergid degeneration in the vdd-1 heterozygous mutant.
Until now, most of the female gametophytic mutants affecting pollen tube reception led to the identification of factors that are potentially involved in the signal transduction cascade (Escobar-Restrepo et al., 2007; Boisson-Dernier et al., 2008; Capron et al., 2008). It is not clear how VDD fits into this pathway. Since VDD encodes a putative transcription factor, it likely regulates the expression of downstream genes involved in the specification of the two accessory cell types, which is reflected by the lack of fertilization in vdd-1 mutant embryo sacs. The antipodal and synergid cells are located at opposite poles of the embryo sac, and the common origin of these cells can be traced back to the one-nucleate stage of embryo sac development, when the two nuclei migrate to opposite poles (Christensen et al., 1997). Therefore, it might be that VDD is already influencing gene expression before the migration of these nuclei, subsequently affecting their specification and differentiation, leading to defects in male gamete discharge. However, it could also be that VDD is involved in a pathway that establishes accessory cell identity based on differences in signals at the two poles of the embryo sac. It is important to point out that synergid identity is not completely lost in vdd mutant embryo sacs as they are still competent to attract the pollen tube and induce its growth arrest. It is therefore possible that vdd mutant synergids have a mixed identity, losing some aspects of synergid function while gaining others, as indicated by the gain of and loss of expression of antipodal and synergid cell markers, respectively. One of our challenges for the future will be to identify the genes that are regulated by VDD.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (ecotype Columbia) and mutant plants were grown at 22°C under short-day (8 h light/16 h dark) or long-day (16 h light/8 h dark) conditions. The Arabidopsis stk, shp1shp2, and stk shp1 shp2 mutants were kindly provided by M. Yanofsky (Pinyopich et al., 2003). Gametophytic cell marker line corresponding to the egg cell (Gross-Hardt et al., 2007), central cell (promoter of the gene At1g02580; Chaudhury et al., 1997), and antipodal cell (promoter of the gene At1g36340; Yu et al., 2005) marker lines were kindly provided by R. Gross-Hardt. The synergid cell marker line (ET2634) was generated in the lab of U. Grossniklaus. All the gametophytic marker lines analyzed encode for a nuclear localization signal that is in frame with the GUS reporter gene. The MINI3:GUS reporter line was kindly provided by M. Luo. Arabidopsis (ecotype Columbia) seeds carrying the vdd-1 mutant allele were obtained from the Syngenta Arabidopsis Insertion Library (SAIL 50_C03) collection (www.Arabidopsis.org/abrc/sail.jsp). T-DNA is inserted in the first intron, 44 bp upstream the second exon.
Laser Microdissection
Young inflorescences from wild-type plants were prepared as previously described (Kerk et al., 2003). Ovules corresponding to stage 8-9 (referred as ovule primordia) were dissected using a Leica laser microdissection system (LMD 6000; Leica Microsystems). The selected cells were cut using a UV laser (337-nm wavelength). The dissection conditions were optimized as follows: L40x objective at power 35 to 45 and speed 3 to 4. Samples from ∼30 sections (at least 1000 cells) were collected in a single tube. RNA was extracted from three different tubes. RNA from laser microdissection cells was extracted using the PicoPure RNA isolation kit (Arcturus) according to the manufacturer's instructions. RNA obtained from three independent extractions was pooled and amplified as describe by Van Gelder et al. (1990).
Identification of Putative CArG Sequences
The genomic regions located 3 kb upstream of the ATG, 1 kb downstream of the stop codon and in the exons and introns of the ovule primordia expressed transcription factors were analyzed to identify CArG sequences. The Transfac bioinformatic program available at the Biobase website (http://www.biobase-international.com) allowed us to identify perfect CArG boxes, CArG sequences with one mismatch, and AG, AGL15, SHP1, and SEP binding sites deduced by a probability matrix. To restrict the sample further, we selected genes containing two putative CArG sequences within a distance of ∼300 bp.
ChIP and Quantitative Real-Time PCR Analysis
ChIP experiments were performed as a modified version of a previously reported protocol (Gregis et al., 2008); a detailed protocol is available in the Supplemental Methods online. STK polyclonal antibody was obtained against the synthetic peptide: NH2-RTKVAEVERYQHH-COOH. The polyclonal SEP3 antibody was obtained against the following synthetic peptides: NH2-EVDHYGRHHHQQQQHSQA-COOH and NH2-SQQEYLKLKERYDALQRCOOH. Antibodies were produce by Primm.
Enrichment of the target region was determined using a Sybr Green Assay (iQ_ SYBR Green Supermix; Bio-Rad). The quantitative real-time PCR assay was conducted in triplicate and was performed in a Bio-Rad iCycler iQ optical system (software version 3.0a). Relative enrichment was calculated normalizing the amount of immunoprecipitated DNA against an ACTIN2/7 (ACT2/7) fragment and against total INPUT DNA. In particular, for the binding of STK to the selected genomic regions, the affinity of the purified sample obtained in the shp1 shp2 mutant background was compared with the affinity-purified sample obtained in the stk single mutant background, which was used as negative control. For the binding of SEP3 to the selected genomic regions, the affinity of the purified sample obtained from wild-type carpel tissue was compared with the affinity-purified sample obtained from wild-type leaf tissue, which was used as negative control.
Fold enrichment was calculated using the following formulas, where Ct.tg is target gene mean value, Ct.i is input DNA mean value, and Ct.nc is actin (negative control) mean value: dCT.tg = CT.i-CT.tg and dCT.nc =CT.i-CT.nc. The propagated error values of these CTs are calculated: dSD.tg = sqrt((sd.i)^2+ (sd.tg^2)/sqrt(n) and dSD.nc = sqrt((sd.i)^2+ (sd.nc^2)/sqrt(n), where n = number of replicate per sample.
Fold-change over negative control (actin and wild-type plants) was calculated finding the “delta delta CT” of the target region as follows: ddCT = dCT.tg- dCT.nc and ddSD = sqrt((dSD.tg)^2+ (dSD.nc)^2. The transformation to linear “fold-change” values is obtained as follows: FC = 2^(ddCT) and FC.error = ln(2)*ddSD*FC.
Oligo sequences are as follows: ACTIN2/7 forward, 5′-CCAATCGTGAGAAAATGACTCAG-3′; ACTIN2/7 reverse, 5′-CCAAACGCAGAATAGCATGTGG-3′); CArG 1 forward, 5′-AACATTGCTTTCTCCTTCCAAA-3′; CArG1 reverse, 5′-CAAAAGGGAGTTCAAGTGAAAGAAC-3′; CArG2 forward, 5′-CTACATTCTACAGACTAGCTAG-3′; CArG2 reverse, 5′-CTAAAAAGACAGCGTCATATTTCC-3′; CArG3 forward, 5′-GGAAATATGACGCTTGTCTTTTTAG-3′; CArG3 reverse, 5′- CAGAAACAGCAATATGCTCGTG-3′.
Expression Analysis
For the microarray hybridization experiment, RNA purified from LCM cells was amplified, labeled, and hybridized on the ATH1 GeneChip at the Affymetrix Microarray Unit at the Molecular Oncology Foundation Institute (IFOM; http://www.ifom-firc.it). Two hybridization replicates were performed to reduce the technical variability; each replicate was measured twice. Expression levels were calculated as an average of the closest three values out of the four measurements. All the samples were normalized together to a per-chip and per-gene median value. Clustering analysis was performed using condition tree clustering on all samples. Similarity was measured using Spearman correlation (GeneSpring version 7.2). Microarray data were normalized and analyzed using GeneSpring software. Our analysis identified 14,575 genes significantly expressed in the ovule primordia with a P value < 0.05 (see Supplemental Data Set 1 online). As reported in the text, we focused our attention on transcription factor encoding genes. For these genes, the q-value was calculated, which allowed us to measure the minimum false discovery rate (FDR) that is incurred when calling that test significant. In our analysis, q-values were measured from the corresponding P values using a freely available database (genomics.princeton.edu/storeylab/qvalue). Q-value estimation was done using a specific FDR level of 0.05, a lambda range from 0.0 to 0.90, and a smoother pi_0 method as main parameters. FDR analysis allowed the identification of 1024 transcription factor genes that are significantly expressed in ovule primordia (Storey et al., 2004) (see Supplemental Data Set 2 online). A comparison of the replicates for each tissue type was done and the mean signal values were ranked, revealing the number of genes that passed the cutoff and furthermore are considered as expressed genes.
Quantitative real-time RT-PCR experiments were performed on cDNA obtained from leaves, flower, and siliques at 3 d after pollination and siliques at 5 d after pollination. Total RNA was extracted using the LiCl method (Verwoerd et al., 1989). DNA contamination was removed using the Ambion TURBO DNA-free DNase kit according to the manufacturer's instructions (http://www.ambion.com/). The treated RNA was subjected to reverse transcription using the ImProm-IITM reverse transcription system (Promega). VDD transcripts were detected using a Sybr Green Assay (iQ SYBR Green Supermix; Bio-Rad) with the reference gene UBIQUITIN. The real-time PCR assay was conducted in triplicate and was performed in a Bio-Rad iCycler iQ Optical System (software version 3.0a). Relative enrichment of VDD transcripts was calculated normalizing the amount of mRNA against a UBIQUITIN fragment. Diluted aliquots of the reverse-transcribed cDNAs were used as templates in quantitative PCR reactions containing the iQ SYBR Green Supermix (Bio-Rad). The difference between the cycle threshold (Ct) of VDD and that of UBIQUITIN (ΔCt = CtVDD − CtUBIQUITIN) was used to obtain the normalized expression of VDD, which corresponds to 2−ΔCt. The expression of VDD was analyzed by the following primers: VDD forward, 5′-TGGATGGAACCAGTTTGTGA-3′, and VDD reverse, 5′-CTTCACATCTTTGTAGATGCTC-3′. The expression of UBIQUITIN was analyzed using the following primers: UB forward, 5′-CTGTTCACGGAACCCAATTC-3′, and UB reverse, 5′-GGAAAAAGGTCTGACCGACA-3′.
For in situ hybridization analysis, Arabidopsis flowers were fixed and embedded in paraffin as described previously (Huijser et al., 1992). Sections of plant tissue were probed with digoxigenin-labeled VDD antisense RNA corresponding to nucleotides 240 to 557. Hybridization and immunological detection were performed as described previously (Coen et al., 1990).
For the GUS assays, gametophytic cell-specific marker lines were used as female and pollinated with pollen obtained from vdd-1 heterozygous plants to introduce the reporter constructs into the vdd-1 mutant background. The F2 progeny obtained from self-fertilization of F1 plants heterozygous for the vdd-1 allele and the reporter constructs were analyzed for GUS expression to identify vdd-1/VDD plants homozygous for the reporter constructs. Flowers were emasculated and harvested 48 h following emasculation for GUS staining. MINI3:GUS pattern in the vdd-1 heterozygous background was analyzed 26 h after pollination. All GUS assays were performed overnight as described previously (Liljegren et al., 2000). Samples were incubated in clearing solution, dissected, and observed using a Zeiss Axiophot D1 microscope equipped with DIC optics. Images were captured on an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1).
Microscopy
To analyze ovule development in vdd-1 heterozygous plants, flowers at different developmental stages were cleared and analyzed as described previously (Brambilla et al., 2007).
For the aniline blue staining experiments, vdd-1 heterozygous plants were emasculated and pollinated 24 h after the emasculation. Pollen tube growth was analyzed 24 h after pollination. Aniline blue staining was performed as described by Huck et al. (2003).
For the synergid degeneration analysis, wild type and vdd-1 heterozygous flowers were emasculated and pollinated using wild-type pollen 24 h after emasculation. Pistils were fixed 12 h after pollination (hap) and observed by CLSM following the Braselton et al. (1996) protocol. Samples were excited using a 532-nm laser. Emission was selected between 570 and 740 nm.
Plasmid Construction and Arabidopsis Transformation
For the VDD promoter analysis, wild-type Arabidopsis plants (ecotype Columbia) were transformed with a construct containing 1221 bp upstream of the VDD translation start site, the complete VDD genomic coding region fused to the GUS reporter gene, and 389 bp corresponding to the VDD 3′ untranslated region. The pBGWFS7 vector (Karimi et al., 2002) was modified to substitute the T35S fragment with the VDD 3′ untranslated region sequence. The PCR product obtained with the primer 5′- CCATGGACACCATGACGATGATGATATTTTA-3′ in combination with the primer 5′-GACGTCCCAGAAGAGGCTTATGATA-3′ was cloned into the pBGWFS7 vector using the NcoI and AatII restriction sites. Following this, the VDD genomic region was amplified using the forward oligo 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCCGAACTTTATTCCGGATA-3′ in combination with the reverse oligo 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTCTTCTTTGGAGACTTTCACAC-3′. This PCR product was cloned in the pBGWFS7 vector modified as described above.
For the molecular complementation experiment, a 3-kb genomic region containing the VDD gene was amplified by PCR using the oligo Atp1669 (5′-TCTAGACCCGAACTTTATTCCGGATA-3′) located 1221 bp upstream of the ATG and the oligo Atp1670 (5′-TCTAGACCAGAAGAGGCTTATGATA-3′) located 389 bp downstream of the stop codon. The obtained PCR product was cloned in the XbaI site of the pFLUAR binary vector (Stuitje et al., 2003), which contains the YFP coding sequence under the control of the NAPIN promoter. Constructs were verified by sequencing and used to transform vdd-1 heterozygous plants using the floral dip method (Clough and Bent, 1998). Transformant seeds were visually selected by fluorescence microscopy.
For the pSTK:amiRvdd construct, the genomic fragment corresponding to the STK promoter was amplified using the primers Atp1509 (5′-TCTGACGTCAGGCGTTTTTGTTGGGTATGTTCTCAC-3′) and Atp 1508 (5′-TCTGACGTCAGGCATCCTTCATTTTAAACATC-3′) (Kooiker et al., 2005) and was cloned in the binary vector pBGW0 (http://www.psb.ugent.be/gateway) upstream of the recombination Gateway cassette using the AatII restriction site (Invitrogen). The artificial RNA directed against the VDD gene was prepared according to the information available at the Web microRNA Designer website (WMD wmd2.weigelworld.org/cgi-bin/mirnatools.pl?page=8#experimentalProcedure). Site-directed mutagenesis on a template plasmid containing the MIR319a precursor was performed using the following primers: oligo I (5′-GATTTACTAACAGTTTCCACCCTTCTCTCTTTTGTATTCC-3′), oligo II (5′-GAAGGGTGGAAACTGTTAGTAAATCAAAGAGAATCAATGA-3′), oligo III (5′-GAAGAGTGGAAACTGATAGTAATTCACAGGTCGTGATATG-3′), and oligo IV (5′-GAATTACTATCAGTTTCCACTCTTCTACATATATATTCCT-3′). The obtained PCR product was cloned into the TOPO vector (Invitrogen) and further recombined into the pBGW0 binary vector containing the STK promoter. Wild-type plants were transformed using the floral dip method (Clough and Bent, 1998), and transformants were identified through BASTA selection.
PCR-Based Genotyping
Identification of the vdd-1 mutant allele was performed by PCR analysis using the oligo Atp1220 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′) on the T-DNA left border and the oligo Atp1219 (5′-CGAAGGAGAGAAGCAGAGATG-3′). The VDD wild-type allele was identified using the oligo Atp 1219 in combination with oligo Atp1218 (5′-TGAAGTACCGGCTTCAGAGTC-3′).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ACTIN2/7, AT5G09810; AGAMOUS, AT4G18960; ANXUR1, AT3G04690; ANXUR2, AT5G28680; APETALA1, AT1G69120; FERONIA, AT3G51550; MINISEED3, AT1G55600; SEEDSTICK, AT4G09960; SEPALLATA1, AT5G15800; SEPALLATA2, AT3G02310; SEPALLATA3, AT1G24260; SEPALLATA4, AT2G03710; SHATTERPROOF1, AT3G58780; SHATTERPROOF2, AT2G42830; VERNALIZATION, AT3G18990; and VERDANDI, AT5G18000.
Author Contributions
L.M.-H. and R.B. are the main contributors to the experimental part of this manuscript. R.B. drafted the manuscript. F.G. performed ChIP and real-time RT-PCR experiments. M.R. performed complementation tests. C.E. and U.G. performed pollen tube guidance experiments and provided advice on experimental approaches. M.M.K. and U.G. provided advice on experimental approaches and helped with writing of the manuscript. The work was generally performed in the group of L.C. who provided funding and supervised the research and the writing of the manuscript.
Supplemental Data
The following materials are available in the online version of this manuscript.
SupplementalFigure 1. Laser Microdissection of Ovule Primordia.
Supplemental Figure 2. vdd-1 Mutant Female Gametophyte.
Supplemental Table 1. List of the 15 Transcription Factor-Encoding Genes Selected for the ChIP Analysis.
Supplemental Table 2. Cell-Specific Marker Gene Expression.
Supplemental Data Set 1. List of Ovule Primordia Expressed Genes According to Microarray Analysis.
Supplemental Data Set 2. List of Transcription Factors Encoding Genes Expressed in the Ovule Primordia According to Microarray Analysis.
Supplemental Methods.
Acknowledgments
We thank Rita Gross-Hardt and Thomas Dresselhaus for providing the female gametophyte-specific marker lines, Timothy Nelson for information concerning the laser microdissection technique, Robert Sablowski for helpful discussion regarding the analysis of microarray hybridization data, and Alice Bianchi for technical help. This research was supported by the European Union Marie Curie Research and Training Network Project Transistor and by the Ministero dell'Istruzione, dell'Universitá e della Ricerca CisCode project (RBER062B5L), the Networking the European Research Area-Plant Genomics Project (ERA-PG): Seeds for Growth––Identification of transcriptional programs controlling seeds growth and development from Arabidopsis to rice (RBER06NRMT), and the University of Zürich.
References
- Acosta-García G., Vielle-Calzada J.P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia R., Brambilla V., Colombo L. (2008). Morphological analysis of female gametophyte development in the bel1 stk shp1 shp2 mutant. Plant Biosyst. 142: 643–649 [Google Scholar]
- Battaglia R., Brambilla V., Colombo L., Stuitje A.R., Kater M.M. (2006). Functional analysis of MADS-box protein complexes controlling ovule development in Arabidopsis using the ethanol-inducible alc gene-expression system. Mech. Dev. 123: 267–276 [DOI] [PubMed] [Google Scholar]
- Bemer M., Wolters-Arts M., Grossniklaus U., Angenent G.C. (2008). The MADS domain protein DIANA acts together with AGAMOUS-LIKE 80 to specify the central cell in Arabidopsis ovules. Plant Cell 208: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Frietsch S., Kim T.H., Dizon M.B., Schroeder J.I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 8: 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Roy S., Kritsas K., Grobei M.A., Jaciubek M., Schroeder J.I., Grossniklaus U. (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V., Battaglia R., Colombo M., Masiero S., Bencivenga S., Kater M.M., Colombo L. (2007). Genetic and molecular interactions between BELL1 and MADS-box factors support ovule development in Arabidopsis. Plant Cell 19: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselton J.P., Wilkinson M.J., Clulow S.A. (1996). Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 71: 84–87 [DOI] [PubMed] [Google Scholar]
- Brodeur A.G. (1916). The Prose Edda by Snorri Struluson Translated from the Icelandic with an Introduction. (New York: American-Scandinavian Foundation; ). [Google Scholar]
- Capron A., Gourgues M., Neiva L.S., Faure J.E., Berger F., Pagnussat G., Krishnan A., Alvarez-Mejia C., Vielle-Calzada J.P., Lee Y.R., Liu B., Sundaresan V. (2008). Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. Plant Cell 20: 3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A.M., Ming L., Miller C., Craig S., Dennis E.S., Peacock W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Li H.J., Shi D.Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.A., King E.J., Jordan J.R., Drews G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10: 49–64 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliot R., Murphy G., Carpenter R. (1990). Floricaula a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Colombo L., Battaglia R., Kater M.M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13: 444–450 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P.B., An G., Colombo L., Kater M.M. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52: 690–699 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 3: 656–660 [DOI] [PubMed] [Google Scholar]
- Favaro R., Immink R.G.H., Ferioli V., Bernasconi B., Byzova M., Angenent G.C., Kater M.M., Colombo L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genomics 268: 152–159 [DOI] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L., Ditta G., Yanofsky M.F., Kater M.M., Colombo L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S., Immink R.G., Shchennikova A., Busscher-Lange J., Angenent G.C. (2003). The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan C.A., Hu Y., Ma H. (1996). Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 10: 343–353 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Cubas P., Jarrillo J.A., Fernandez-Calvín B., Salinas J., Martínez-Zapater J.M. (2002). AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol. 128: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M.M. (2008). AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J. 56: 891–902 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R., Kägi C., Baumann N., Moore J.M., Baskar R., Gagliano W.B., Jürgens G., Grossniklaus U. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S.R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S., Kuroiwa H., Kuroiwa T. (2001). Pollen tube attraction by the synergid cell. Science 293: 1480–1483 [DOI] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N., Moore J.M., Federer M., Grossniklaus U. (2003). The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- Huijser P., Klien J., Lonnig W.E., Meijer H., Saedler H., Sommer H. (1992). Bracteomania, an inflorescence anomaly is caused by the loss of function of the MADS-box gene SQUAMOSA in Antirrhinum majus. EMBO J. 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M., Schneitz K., Pruitt R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink R.G., Ferrario S., Busscher-Lange J., Kooiker M., Busscher M., Angenent G.C. (2003). Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genomics 268: 598–606 [DOI] [PubMed] [Google Scholar]
- Ito T., Ng K.H., Lim T.S., Yu H., Meyerowitz E.M. (2007). The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19: 3516–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., Lord M.E. (2006). Extracellular guidance cues and extracellular signalling pathways that direct pollen tube growth. The Pollen Tube: A Cellular and Molecular Perspective (Plant Cell Monographs), Malhó R., (Berlin, Heidelberg: Springer-Verlag; ), pp. 223–242 [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kerk N.M., Ceserani T., Tausta S.L., Sussex I.M., Nelson T.M. (2003). Laser capture microdissection of cells from plant tissues. Plant Physiol. 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiker M., Airoldi C.A., Losa A., Manzotti P.S., Finzi L., Kater M.M., Colombo L. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.S., Hill T.A., Tan Q.K., Irish V.F. (2002). Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129: 2079–2086 [DOI] [PubMed] [Google Scholar]
- Levy Y.Y., Mesnage S., Mylne J.S., Gendall A.R., Dean C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297: 243–246 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liu C., Chen H., Er H.L., Soo H.M., Kumar P.P., Han J.H., Liou Y.C., Yu H. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Yanofsky M.F., Meyerowitz E.M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Mandel M.A., Yanofsky M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11: 22–28 [Google Scholar]
- Miyazaki S., Murata T., Sakurai-Ozato N., Kubo M., Demura T., Fukuda H., Hasebe M. (2009). ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19: 1327–1331 [DOI] [PubMed] [Google Scholar]
- Murgia M., Huang B.-Q., Tucker S.C., Musgrave M.E. (1993). Embryo sac lacking antipodal cells in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 80: 824–838 [Google Scholar]
- Nurrish S.J., Treisman R. (1995). DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 115: 4076–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Sundaresan V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani J.A., Rabiger D.S., Drews G.N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19: 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S.M., Park S.S., Ray A. (1997). Pollen tube guidance by the female gametophyte. Development 124: 2489–2498 [DOI] [PubMed] [Google Scholar]
- Romanel E.A.C., Schrago C.G., Counago R.M., Russo C.A.M., Alves-Ferreira M. (2009). Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman N., Gourgues M., Guitton A.E., Faure J.E., Berger F. (2008). A dialogue between the sirene pathway in synergids and fertilization independent seed pathway in the central cell controls male gamete release during double fertilization in Arabidopsis. Mol. Plant 1: 659–666 [DOI] [PubMed] [Google Scholar]
- Rotman N., Rozier F., Boavida L., Dumas C., Berger F., Faure J.E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 4: 432–436 [DOI] [PubMed] [Google Scholar]
- Rounsley S.D., Ditta G.S., Yanofsky M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R., Meyerowitz E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749 [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 185: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.K., Ito T., Ishiguro S., Okada K. (2008). MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana. Plant Cell Physiol. 49: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srilunchang K.O., Krohn N.G., Dresselhaus T. (2010). DiSUMO-like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize. Development 137: 333–345 [DOI] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Portereiko M.F., Lloyd A., Drews G.N. (2008). AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 48: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Taylor J.E., Siegmund D. (2004). Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J. R. Stat. Soc. Ser. A Stat. Soc. 66: 187–205 [Google Scholar]
- Stuitje A.R., Verbree E.C., van der Linden K., Mietkiewska E.M., Nap J.P., Kneppers T.J.A. (2003). Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol. J. 1: 301–309 [DOI] [PubMed] [Google Scholar]
- Sundström J.F., Nakayama N., Glimelius K., an Irish V.F. (2006). Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 46: 593–600 [DOI] [PubMed] [Google Scholar]
- Swaminathan K., Peterson K., Jack T. (2008). The plant B3 superfamily. Trends Plant Sci. 13: 647–655 [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Qin Y., Huang Y., Dunatunga D., Palanivelu R. (2010). A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J. 62: 571–588 [DOI] [PubMed] [Google Scholar]
- Van Gelder R.N., von Zastrow M.E., Yool A., Dement W.C., Barchas J.D., Eberwine J.H. (1990). Amplified RNA synthesized from limited quantities of heterogenous cDNA. Proc. Natl. Acad. Sci. USA 87: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T.C., Dekker B.M., Hoekema A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Sablowski R.W., Meyerowitz E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 28: 582–584 [DOI] [PubMed] [Google Scholar]
- Wellmer F., Riechmann J.L., Alves-Ferreira M., Meyerowitz E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings K., Russell S.D. (2004). Experimental analysis of the fertilization process. Plant Cell 16 (suppl): S107–S118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William D.A., Su Y., Smith M.R., Lu M., Baldwin D.A., Wagner D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 101: 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yu H.J., Hogan P., Sundaresan V. (2005). Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139: 1853–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M., Irish V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]