Brassinosteroid (BR) homeostasis is established by the regulatory circuit between receptor BRI1-mediated signaling and BR synthesis. RAVL1 modulates the strength of the circuit by activating expression of both BRI1 and synthetic genes and is necessary for feedback responses to BR levels.
Abstract
Temporal and spatial variation in the levels of and sensitivity to hormones are essential for the development of higher organisms. Traditionally, end-product feedback regulation has been considered as the key mechanism for the achievement of cellular homeostasis. Brassinosteroids (BRs) are plant steroid hormones that are perceived by the cell surface receptor kinase Brassinosteroid Insensitive1. Binding of these hormones to the receptor activates BR signaling and eventually suppresses BR synthesis. This report shows that RAVL1 regulates the expression of the BR receptor. Furthermore, RAVL1 is also required for the expression of the BR biosynthetic genes D2, D11, and BRD1 that are subject to BR negative feedback. Activation by RAVL1 was coordinated via E-box cis-elements in the promoters of the receptor and biosynthetic genes. Also, RAVL1 is necessary for the response of these genes to changes in cellular BR homeostasis. Genetic evidence is presented to strengthen the observation that the primary action of RAVL1 mediates the expression of genes involved in BR signaling and biosynthesis. This study thus describes a regulatory circuit modulating the homeostasis of BR in which RAVL1 ensures the basal activity of both the signaling and the biosynthetic pathways.
INTRODUCTION
Brassinosteroids (BRs) are plant hormones that are structurally similar to animal steroid hormones (Grove et al., 1979). BRs are perceived on the cell surface via their binding to the transmembrane receptor kinase, Brassinosteroid Insensitive1 (BRI1) (Li and Chory, 1997; Kinoshita et al., 2005). BRs are essential for the elongation of stems and roots and the differentiation of vascular bundles, as well as for senescence, stress response, photomorphogenesis, and tropisms (Mitchell et al., 1970; Clouse, 1996; Li and Chory, 1999; Li et al., 2005). Recently, it has been shown that biomass production and grain yield can be enhanced by controlling the level of endogenous BR in rice (Oryza sativa) plants (Sakamoto et al., 2006; Wu et al., 2008).
BR-mediated developmental processes are largely determined by endogenous levels of and sensitivity to these hormones. BR distribution is widespread in most plant tissues (Bajguz and Tretyn, 2003); however, the distribution of BRs exhibits temporal and spatial variations. Bioactive BRs are much more abundant in young actively growing tissues than in mature vegetative tissues (Shimada et al., 2003), and the levels of bioactive BRs are higher in shoots than in roots (Bancos et al., 2002). The expression levels of endogenous BRs are determined by biosynthesis, degradation, and conjugation. Its levels correlate with the expression patterns of the BR biosynthetic genes BR-6-oxidase and DWARF4 and with the BR synthesis–related P450 genes (Bancos et al., 2002; Shimada et al., 2003). The inactivation of BRs by degradation and conjugation mechanisms contributes to the homeostasis of BRs (Neff et al., 1999; Rouleau et al., 1999; Nakamura et al., 2005; Poppenberger et al., 2005).
Hormonal sensitivity of plant tissues may be largely influenced by the activity of receptors and signal transduction chain compounds (Davies, 2004). Optimal homeostasis of BRs is achieved by the balance of its signaling and biosynthesis. Signaling is initiated by binding of BR to the receptor kinase, BRI1, which eventually leads to the repression of BR biosynthetic genes. BRs activate the BRI1 receptor by stimulating its autophosphorylation (Wang et al., 2005a). The active BRI1 transmits the BR signal to the downstream genes Bri1 Suppressor1 (BSU1) and BR INSENSITIVE2 (BIN2). BSU1 encodes a phosphatase that acts as a positive regulator (Mora-García et al., 2004). By contrast, BIN2 encodes a GSK3/Shaggy-like kinase and acts as a negative regulator of transcription factors (Li and Nam, 2002). Moreover, Bri1 Ems Suppressor 1 (BES1) and Brassinazole-Resistant 1 (BZR1) are substrates of BIN2. Recently, it was shown that BSU1 inactivates BIN2 (Kim et al., 2009). BES1 interacts with a basic helix-loop-helix transcription factor BES1-interacting Myc-like 1 (BIM1). The BES1/BIM1 complex synergistically binds to the E-box element CANNTG and activates BR-induced gene promoters (Yin et al., 2005). BZR1 plays a dual role in feedback regulation of BR synthesis and downstream growth response (Wang et al., 2002; He et al., 2005); it enhances the expression of genes involved in cellular growth and suppresses the expression of genes involved in BR synthesis, thus producing the negative feedback suppression of BR biosynthesis.
In contrast with what is observed for other plant hormones, BRs appear to be synthesized in all tissues and organs and to function where they are synthesized (Symons et al., 2008); therefore, different levels of BR homeostasis need to be established, depending on developmental stage and environmental inputs. It is thus crucial to understand how plants control the endogenous levels of BR during development. Here, we show that the rice RAVL1 gene participates in the maintenance of BR homeostasis by enhancing the expression of BRI1 and several BR biosynthetic genes. The RAVL1-mediated mechanism plays a significant role in ensuring the basal activities of the signaling and synthesis processes that are part of the antagonistic relation.
RESULTS
Identification and Expression of Rice RAVL1
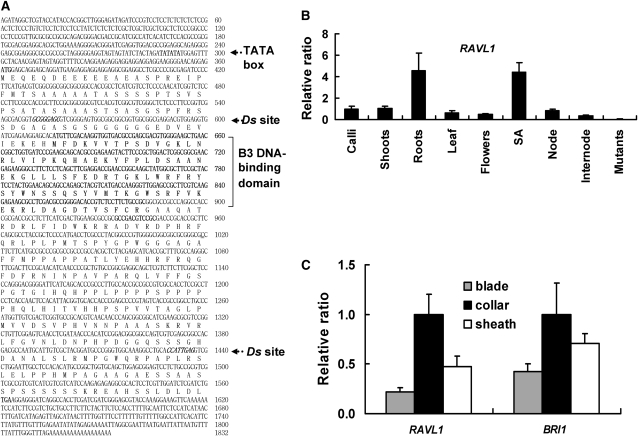
RAVL1 was identified by examination of two mutant alleles identified in a Ds transposant rice population (Kim et al., 2004) as dark-green and semidwarf plants. The mutants were named ravl1-1 and ravl1-2 (see below), and the trap Ds elements were located in the N-terminal region of ravl1-1 and the C-terminal region of ravl1-2. RAVL1 contains one exon and encodes a protein containing a B3 DNA binding domain (Figure 1A). A B3 or B3-like DNA binding domain is present in Viviparous 1/Abscisic Acid-Insensitive 3 (VP1/ABI3), auxin response factors (ARFs), and Related to ABI3/VP1 (RAV). Unlike RAV, RAVL1 does not carry an AP2 domain (Riechmann et al., 2000). A revertant RAVL1-R allele was obtained from regenerated plants of ravl1-1 calli carrying Ac. The revertant allele carries a 6-bp insertion at the Ds excision site and exhibits wild-type phenotypes.
Figure 1.
DNA and Protein Sequences and Expression of RAVL1.
(A) The B3-DNA binding domain spans from amino acids 86 to 195. The C-terminal region is rich in Pro and Ser residues. The two italicized 8-bp stretches indicated by arrows in the N- and the C-terminal regions represent duplicated sequences of Ds insertions of ravl1-1 and ravl1-2, respectively.
(B) qRT-PCR measured the relative amounts of RAVL1 mRNA. RAVL1 is expressed in all tissues examined. SA, shoot apices. Tissues were extracted from 1- to 2-week-old seedlings.
(C) Mature leaves of normal plants were separated into blades, collars, and sheaths. The relative amounts of RAVL1 (left graph) and BRI1 (right graph) mRNA were measured by qRT-PCR. 25S rRNA was used as control to normalize the expression data. Error bars are se of the means of three qPCR replicates.
The expression pattern of RAVL1 was determined using the β-glucuronidase (GUS) reporter gene inserted in the trap Ds. GUS staining was detected in all the young and immature tissues. Strong expression was detected in lateral roots, coleoptiles, and stigmas and lodicules of flowers (see Supplemental Figure 1 online). Quantitative real-time PCR (qRT-PCR) analysis confirmed the observation that the RAVL1 mRNA was present in all the tissues analyzed (Figure 1B). Expression of RAVL1 in lateral root primordia and immature leaves was confirmed by in situ RNA hybridization (see Supplemental Figure 1 online).
In mature leaves, low GUS activity was observed with the exception of lamina joints (see Supplemental Figure 1 online). The levels of RAVL1 mRNA were examined in leaf blades, lamina joints (collar), and sheaths of 1-month-old plants (Figure 1C): its expression was highest in joints and lowest in blades. It should be noted that the expression patterns of RAVL1 were almost identical to those of BRI1 (Figure 1C).
BR-Related Phenotypes of RAVL1 Mutants and Overexpressors
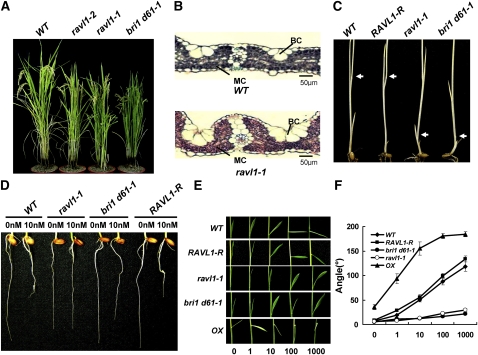
Under normal growth conditions, ravl1 mutants show delay in germination, retardation in seedling growth, a semidwarf appearance (Figure 2A), and dark-green leaves. The semidwarf phenotype results from reduced elongation of upper internodes (see Supplemental Figure 1 online). Microscopy inspection of dark-green leaves revealed that the number of bulliform cells was increased in the mutants when compared with wild-type plants (Figure 2B). An increase in the number and size of bulliform cells is also observed in BR-deficient dwarf1 (brd1) rice mutants (Hong et al., 2002). In addition, the other ravl1 phenotypes (i.e., late germination, retarded shoot growth, and dark-green leaves) are also often found in BR-related mutants (Yamamuro et al., 2000; Hong et al., 2003).
Figure 2.
Phenotypes of ravl1 Plants.
(A) Mature 4-month-old plants are shown. Plants grown in the field were transferred to pots for photographing. WT, wild type.
(B) Cross sections of leaves of 3-month-old plants grown in the field. MC, mesophyll cells; BC, bulliform cells. Samples were stained with Safranin O and Fast Green.
(C) Skotomorphogenesis of ravl1-1. Plants were grown in soil for 2 weeks in the dark. The arrows indicate nodes.
(D) Root growth was compared among ravl1-1, bri1 d61-1, and revertant plants, in the presence of 10 nM epiBL.
(E) Lamina inclination to epiBL. The indicated amounts of epiBL in 1 μL of ethanol were spotted on the tip of the second lamina of 6-d-old seedlings. OX indicates the RAVL1 overexpressor.
(F) Measurements of lamina inclination in response to epiBL. Mean values are derived from measurements of 10 plants. Error bars are se of the mean values.
To test whether RAVL1 is involved in BR-mediated developmental processes, we measured the mutants' growth and morphological responses by observing (1) internode elongation in the dark, (2) growth response of seminal roots to epibrassinolide (epiBL), and (3) inclination of lamina joints after exposure to exogenous epiBL. A weak allele of rice bri1, d61-1, and the RAVL1-R revertant of ravl1-1 were used as control plants. The bri1 d61-1 mutation arose from a single amino acid change (from Thr to Ile at residue 989) in the kinase domain of BRI1 (Yamamuro et al., 2000). In dark growth conditions, BR-defective rice mutants fail to elongate low internodes (Yamamuro et al., 2000; Tanabe et al., 2005). Skotomorphogenesis was examined in ravl1-1 seedlings. As shown in Figure 2C, etiolated seedlings of ravl1-1 mutant plants exhibited short internodes; similar results were obtained for bri1 d61-1 seedlings. Growth conditions with ample light lead to an increase in the number of bulliform cells and mesophyll cell layers in leaf blades of rice plants (Chonan, 1967); therefore, it is tempting to speculate that the changes in internal structure of ravl1-1 leaves described in Figure 2B may be a consequence of an aberrant response to light. However, further work is required to examine this question.
Seminal root growth was measured in the presence of several phytohormones (i.e., 1-amino-cyclopropane-1-carboxylic acid [ACC], abscisic acid [ABA], gibberellin [GA3], naphthalene-1-acetic acid [NAA], and epiBL) (see Supplemental Figure 2 online). Among these hormones, epiBL had the most severe effect on root growth. Differences in root growth patterns were also detected between mutant and wild-type plants treated with ACC and NAA. However, under the same conditions, the BR signal mutant bri1 d61-1 also showed responses to ACC and NAA very similar to those of the ravl1-1 mutant. Therefore, it is plausible that these altered responses to ACC and NAA might also occur in BR mutants. Furthermore, unlike their response to epiBL treatment, mutants were sensitive to ACC and NAA. The growth of ravl1-1 roots was insensitive to 10 nM epiBL, a dosage at which bri1 d61-1 roots were slightly inhibited (Figure 2D). In wild-type plants, root growth was not affected up to a concentration of 1 pM epiBL, but it was gradually inhibited with increasing concentration of epiBL, starting at 10 pM (see Supplemental Figure 3 online). Higher concentrations (100 nM and 1 μM) of epiBL inhibited root growth of ravl1-1 and bri1 d61-1 plants, although much more severe effects were observed for wild-type plants. Root curling was detected at 10 nM epiBL in wild-type plants. By contrast, root curling of ravl1-1 plants was less sensitive to epiBL treatment. Even at 100 nM epiBL, which induced root curling in bri1 d61-1 plants, ravl1-1 mutant roots grew straight without any morphological distortion (Figure 2D; see Supplemental Figure 3 online). This result suggests that the ravl1 mutant is less sensitive to epiBL than are wild-type plants.
The lamina inclination assay has been used in rice as the typical bioassay of BR sensitivity (Fujioka et al., 1998). Lamina inclination assays were performed by spotting 1 μL of ethanol containing epiBL on the tip of laminae of 6-d-old seedlings. Wild-type plants exhibited epiBL dosage-dependent lamina inclination. By contrast, ravl1-1 plants did not show obvious inclination of lamina joints after epiBL treatment, even though a very weak response to epiBL was detected at high concentrations (i.e., 1000 ng/μL) (Figures 2E and 2F). Taken together, these results suggest that ravl1 mutant plants have reduced responses to epiBL.
For further assessment of the BR-specific functions of RAVL1, we generated two different types of RAVL1 overexpressor lines driven by the 35S promoter. One was overexpression of a full-length RAVL1 and the other was one of RAVL1 fused with green fluorescent protein (GFP) at the C terminus. In normal growth conditions, RAVL1 and RAVL1-GFP overexpressors exhibited strong lamina inclination (Figure 3C; see Supplemental Figure 4 online). Analysis of the RAVL1 mRNA levels of these overexpressors showed that the intensity of the lamina inclination in transgenic plants correlated with the abundance of RAVL1 mRNA (Figure 3D; see Supplemental Figure 4 online). RAVL1 overexpressors also showed a hypersensitive inclination of lamina joints after epiBL treatment (Figures 2E and 2F). Because BRs induce dose-dependent swelling of the adaxial cells of the lamina joint (Marumo et al., 1968), we examined the adaxial cell layers of lamina joints using perpendicular sections. There were no distinct differences in the width of the adaxial cells; however, their length was at least twofold longer in RAVL1 overexpressors compared with wild-type plants (see Supplemental Figure 4 online).
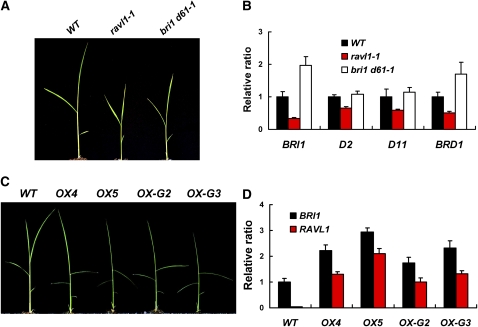
Figure 3.
Seedling Phenotypes of ravl1-1, bri1 d61-1, and RAVL1 Overexpressors and Correlated Transcript Levels of RAVL1, BRI1, D2, D11, and BRD1.
(A) Ten-day-old ravl1-1 and bri1 d61-1 seedlings are shown. WT, wild type.
(B) The expression levels of BRI1, D2, D11, and BRD1 were examined in ravl1-1 and bri1 d61-1 plants by qRT-PCR. 25S rRNA was used control to normalize the expression data. Error bars are se of the means of three qPCR replicates.
(C) Two-week-old seedling overexpressors of RAVL1 (OX) and the RAVL1-GFP fusion (OX-G) are shown. Based on phenotypic intensity, two independent lines (OX4 and OX5; OX-G2 and OX-G3) were chosen as representatives for the RAVL1 and RAVL1-GFP transformants, respectively.
(D) Expression levels of RAVL1 and BRI1 were determined in OX4, OX5, OX-G2, OX-G3, and wild-type seedlings by qRT-PCR. 25S rRNA was used control to normalize the expression data. Error bars are se of the means of three qPCR replicates.
Correlation of the Expression of RAVL1, BRI1, and BR Synthetic Genes
To test whether the expression of BRI1 is dependent on the expression of the RAVL1 gene, we examined BRI1 levels of expression in seedlings of ravl1-1, bri1 d61-1, and RAVL1 overexpressors. Like mature plants, ravl1-1 and bri1 d61-1 seedlings showed weaker growth when compared with the wild type (Figure 3A). qRT-PCR analysis revealed that BRI1 mRNA was decreased in the ravl1-1 mutant (Figure 3B), while the bri1 d61-1 mutation increased the level of BRI1 mRNA compared with that of wild-type seedlings. RAVL1 and the GFP fusion overexpressors were examined for the expression of BRI1. Both accumulated higher levels of BRI1 mRNA than wild types (Figure 3D). Furthermore, in the overexpressor lines, the upregulation of BRI1 was correlated with the levels of RAVL1 mRNA (Figure 3D).
To examine whether RAVL1 influences the BR biosynthetic pathway, we measured the mRNA levels of D2 (rice Ebisu dwarf/Dwarf2), D11 (rice DWARF11), and BRD1 in ravl1-1 and bri1 d61-1 mutants. The D2, D11, and BRD1 genes encode the BR biosynthetic enzymes CYP90D2, CYP724B1, and CYP85A1/2 (BR-C6-oxidase), respectively (Hong et al., 2002, 2003; Tanabe et al., 2005). The data showed that expression of BR synthetic genes was decreased in the ravl1-1 mutant (Figure 3B). By contrast, the signal-defective mutant bri1 d61-1 maintained normal or slightly higher levels of D2, D11, and BRD1 mRNA. This result was unexpected, as a decrease in BRI1 expression is hypothesized to weaken BR signaling pathways and consequently to enhance the expression of BR synthetic genes. These data suggest that RAVL1 simultaneously affects the expression of genes involved in BR signaling and BR biosynthesis.
Nuclear Localization and E-Box Binding Affinity of RAVL1
The nuclear localization of RAVL1 was confirmed by generating transgenic rice plants harboring the following GFP fusion vectors: (1) the full-length RAVL1 cDNA (RAVL1-GFP), (2) the B3-domain deletion [RAVL1(ΔB3)-GFP], and (3) the C-terminal deletion [RAVL1(Δ3′)-GFP]. All constructs were driven by the 35S promoter (see Supplemental Figure 5 online). RAVL1-GFP plants showed the strong lamina inclination, similar to what was observed for the RAVL1 overexpressor (Figure 3C). By contrast, both RAVL1(ΔB3)-GFP and RAVL1(Δ3′)-GFP plants did not exhibit any phenotypic characteristics related to BR. RAVL1-GFP and RAVL1(ΔB3)-GFP proteins were detected in nuclei (see Supplemental Figure 5 online). However, the RAVL1(Δ3′)-GFP protein was not detected in nuclei (see Supplemental Figure 5 online). This indicates that the C-terminal region seems to contain a nuclear localization signal.
Because the B3 domains of rice RAVL1(Os RAVL1) and Arabidopsis thaliana RAV1 (At RAV1) share 87% identity at the amino acid sequence level, we next examined the binding affinity of Os RAVL1 to the target element of At RAV1 (CACCTG) (Kagaya et al., 1999). An electrophoretic mobility shift assay (EMSA) revealed that Os RAVL1 strongly bound to the At RAV1 target sequence (see Supplemental Figure 6 online). Based on the facts that the target element CACCTG is an E-box element (CANNTG) and that the promoters of many BR-induced genes contain E-box motifs (Toledo-Ortiz et al., 2003; Yin et al., 2005), we tested the possibility that RAVL1 may bind to the E-box motif. All the possible variations of this motif were tested using EMSA (see Supplemental Figure 6 online). The results of the in vitro assay showed that RAVL1 bound to all the E-box elements tested.
RAVL1 Directly Activates the Expression of BRI1
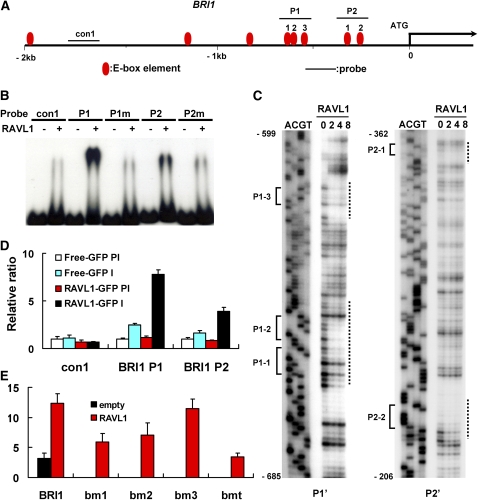
As BRI1 showed correlations in mRNA levels with RAVL1 and several E-box candidates are present within 1 kb upstream of the ATG codon of the BRI1 gene (Figure 4A), we next tested the possibility that the expression of the BRI1 gene may be directly regulated by RAVL1. First, EMSA was performed to test the direct in vitro binding of RAVL1 to the BRI1 promoter region, which contains E-box elements. Two DNA fragments (P1 and P2) located from positions −719 to −569 and from positions −380 to −220 from the ATG, respectively, were used as EMSA probes. The P1 and P2 sequences contained three and two E-box candidate elements, respectively. The same DNA fragments were mutated for all the E-box candidate elements and were used as control probes (Pm1 and Pm2). The gel shift assay revealed that RAVL1 bound to P1 and P2, but not to the two mutated probes (Pm1 and Pm2) (Figure 4B). The binding competition assay showed that P1 exhibited a stronger affinity for RAVL1 when compared with P2 (see Supplemental Figure 7 online).
Figure 4.
In Vitro and in Vivo Assays of RAVL1 Affinity to E-Boxes of the BRI1 Promoter.
(A) The schematic indicates the locations of the E-box candidate elements and of the probes used in the EMSA, DNase I footprinting, and ChIP assays. E-box elements used for the assays are numbered as P1-1, -2, -3, P2-1, and P2-2.
(B) EMSAs for the BRI1 promoter were performed with P1, P2, and their mutated forms (P1m and P2m).
(C) DNase I footprinting assays were performed with radiolabeled P1' and P2' DNA fragments slightly shorter than P1 and P2, respectively. Lanes 0, 2, 4, and 8 indicate the amount (μg) of RAVL1 proteins added to reactions. Lanes A, C, G, and T are Maxam-Gilbert chemical sequencing lanes. E-box elements and protected regions are indicated to the left and right of the autoradiographs, respectively.
(D) An antiserum against GFP (I) and a control preimmune serum (PI) were used for ChIP assay in rice calli expressing RAVL1-GFP. The same BRI1 regions used in the EMSA were amplified from immunoprecipitated DNA. The relative ratios of immunoprecipitated DNA were determined by qRT-PCR. Input DNA was used to normalize the data. Error bars are se of the means of three qPCR replicates.
(E) A transient expression assay was performed by cotransfection of pUbi:RAVL1 and each of the vectors expressing GUS from native and E-box-mutated BRI1 promoters. The bm1 construct is a BRI1 promoter in which two adjacent E-box motifs, P1-1 and -2, are mutated, while the bm2 and bm3 promoters carry mutations in the P1-3 and P2-1 E-boxes, respectively. The bmt promoter contains mutations in all six E-box motifs within 1 kb of the BRI1 promoter. Error bars are se of the means of three replicates.
[See online article for color version of this figure.]
To further verify the physical binding of RAVL1 to E-box sequences in the BRI1 promoter, we performed a DNase I footprinting protection assay. Two DNA fragments, P1' and P2', were used as DNA fragments for the footprinting assay. P1', from positions −685 to −599, contained three E-box motifs, while P2', from positions −362 to −206, carried two E-box motifs. The assay showed that regions protected from DNase I are coincident with the E-box sequences (Figure 4C). E-box elements present in the P1' DNA fragment were prominently protected from DNase I, and two E-box motifs in P2' were less protected than those in P1'. This observation is consistent with the EMSA assay, indicating different binding strengths of E-boxes. The footprinting assay further supports the observation that RAVL1 binds to E-box elements in the BRI1 promoter.
To test the in vivo binding ability of RAVL1 to the E-box elements, a chromatin immunoprecipitation (ChIP) assay was performed in calli derived from 35S:RAVL1-GFP and 35S:GFP transgenic plants. After immunoprecipitation with an antiserum against GFP, the same regions used in the EMSA were amplified using three sets of primers. Two of them spanned the E-box regions, while the other one set excluded these regions. The relative amounts of immunoprecipitants were measured by qRT-PCR (Figure 4D). Two primer sets generated PCR products from the RAVL1-GFP line. No DNA was detected with preimmune serum or with samples from transgenic lines expressing GFP alone (35S:GFP). Also, PCR products were not detected in any samples amplified with primer sets that did not span the E-box elements.
To verify the RAVL1-mediated activation of BRI1, we employed a transient expression assay using rice protoplasts. The assay was performed by coinfection of the pUbi:RAVL1 activator with each of the native and four E-box mutated (bm1, bm2, bm3, and bmt) BRI1 promoters, which were fused to the GUS reporter. bm1 is a BRI1 promoter in which the adjacent E-box motifs P1-1 and -2 were mutated, while bm2 and bm3 carry single mutations in P1-3 and P2-1, respectively (Figure 4A). GUS expression was increased for the native promoter when compared with the mutated promoters (Figure 4E). The bmt promoter, which was mutated at all six E-box elements within −1 kb of the BRI1 promoter, led to the weakest expression of the reporter. The bm3 promoter showed the least effects on transcription, which can be explained by the observation that the E-box element of the bm3 promoter showed the weakest binding affinity to RAVL1. This assay confirmed that RAVL1 activates the expression of BRI1 via E-box cis-elements in its promoter.
RAVL1 Directly Binds to the Promoters of BR Synthetic Genes
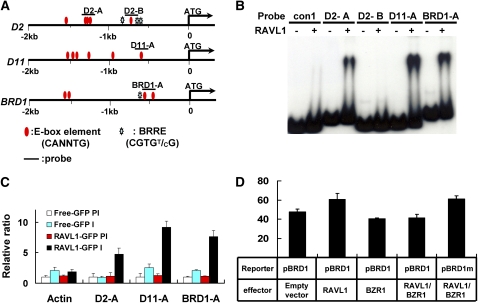
E-box elements are found in the promoters of BR biosynthetic genes D2, D11, and BRD1 (Figure 5A) whose expressions are repressed in ravl1 plants. Using EMSA and ChIP assays, we examined the possibility that RAVL1 regulates BR synthetic genes via binding to their E-boxes. For this, we selected from the promoters of D2, D11, and BRD1, two genomic DNA regions (D2-A and D2-B), one genomic DNA region (D11-A), and one genomic DNA region (BRD1-A), respectively. D2-A carried three E-box candidate elements; D2-B and D11-A contained one E-box candidate, while BRD1-A had two candidates. The EMSA assay showed that RAVL1 binds all E-boxes tested, with the exception of D2-B (Figure 5B). The binding competition assay revealed that the affinity to RAVL1 was variable among these DNA probes: BRD1-A and D11-A showed stronger affinity than D2-A (see Supplemental Figure 7 online). Seedlings and calli expressing RAVL1-GFP were used to perform the ChIP assay. Immunoprecipitants obtained using a GFP antiserum were amplified to detect the genomic DNA sequences of D2-A, D11-A, and BRD1-A. A set of primers was designed to detect an actin promoter without an E-box, which was used as a control. PCR products were obtained exclusively from reactions using primers that spanned E-box elements. The relative amounts of immunoprecipitants were measured by qRT-PCR (Figure 5C). These results strongly suggest that RAVL1 also activates BR synthetic genes by binding to their E-boxes.
Figure 5.
In Vitro and in Vivo Assay of RAVL1 Affinity to E-Boxes of the Promoters of BR Synthetic Genes (D2, D11, and BRD1).
(A) The schematic indicates the locations of the E-box and BRRE candidate elements and of the probes used in the EMSA and ChIP assays.
(B) EMSA for BR synthetic gene (D2, D11, and BRD1) promoters was performed using the D2-A, D2-B, D11-A, and BRD1-A probes, as shown in the schematic. For comparison with the data for BRI1 (Figure 4A), the con1 probe is the same as that used in EMSA with the BRI1 promoter.
(C) For ChIP assay, DNA immunoprecipitated using a GFP antiserum was amplified to detect E-box regions in the promoters of BR synthetic genes. The relative ratios of immunoprecipitated DNA to input DNA were determined by qRT-PCR. Input DNA was used to normalize the data. Error bars are se of the means of three qPCR replicates.
(D) The activity of native and BRRE-mutated promoters with different combinations of RAVL1 and BZR1. A luciferase gene driven by a 35S promoter is an internal control that was used to normalize GUS expression data. Error bars are se of the means of five replicates. BRD1 is the native promoter, and BRD1m contains a mutation of the BRRE element (−669CGTGTG−664) to CCCGGG.
[See online article for color version of this figure.]
In Arabidopsis and rice, BR synthetic genes are known to be suppressed by BZR1 (Wang et al., 2002; He et al., 2005; Bai et al., 2007). Indeed, candidate BZR1 binding motifs (BRRE elements) are found in the promoters of the BR biosynthetic genes D2 and BRD1, but not D11 (Figure 5A). To determine whether RAVL1 interacts with BZR1 in regulating the transcriptional activity of BR synthetic genes, we examined the BRD1 promoter, in which a BRRE element is located next to E-box elements that showed strong binding affinity to RAVL1 (Figure 5B). Transient expression assays with rice protoplasts were performed by coinfection of the plasmids pUbi:RAVL1 and pUbi:BZR1 with a construct bearing a native or a BRRE-mutated BRD1 promoter fused to the GUS reporter (Figure 5D). The data showed that BZR1 inhibits the transcriptional activity of BRD1 via the BRRE element and that BZR1 appears to predominate over RAVL1 in controlling BRD1 transcription. Briefly, BRD1 transcriptional activity in cells expressing both RAVL1 and BZR1 was similar to that in cells expressing only BZR1. However, when the BREE-mutated BRD1 promoter was expressed in the presence of both RAVL1 and BZR1, it was activated to a level similar to that expressed by the native BRD1 promoter in the presence of RAVL1 alone. These data demonstrate that the BR biosynthetic pathway is controlled by both negative and positive transcription factors.
BR-Specific Action of RAVL1
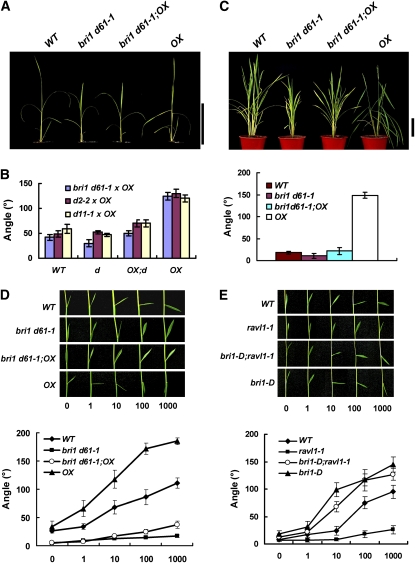
The fact that a null mutation of RAVL1 causes plants to exhibit BR-insensitive phenotypes is not sufficient to demonstrate that RAVL1 primarily acts on BR pathways. To further investigate the relationship between RAVL1 and BR signaling or BR synthesis, we examined the phenotypes developed by overexpression of RAVL1 in BR mutants. Transgenic plants expressing the RAVL1 cDNA driven by the 35S promoter showed normal plant height with strong inclination of lamina joints (Figures 6A and 6C). BR-signaling (bri1 d61-1) and synthetic mutants (d2-2 and d11-1) were semidwarf with slightly narrow (bri1 d61-1 and d11-1) or normal (d2-2) lamina angles (Figure 6B). The d2-2 mutation arose from a single amino acid substitution in the CYP90D2 gene (Hong et al., 2003). d11-1 is a nonsense mutant of CYP724B1 (Tanabe et al., 2005). Genetic crosses between these BR mutants and the RAVL1 overexpressor generated mutant plants that contained the transgene. These genotypes were selected from the F2 generation and were compared with other genotypes obtained from the same F2 population. At the seedling and mature plant stages, the phenotypes of the RAVL1 overexpressors were suppressed in these BR signaling and biosynthetic mutants (Figures 6A to 6C; see Supplemental Figure 8 online). Double genetic combinations exhibited near normal lamina angles and semidwarf phenotypes, similarly to bri1 d61-1, d2, and d11 plants (Figure 6A; see Supplemental Figure 8 online). However, it is noteworthy that the lamina angles of plants with all the double combinations (OX;bri1 d61-1, OX;d2-2, and OX;d11-1) were slightly increased (Figure 6B). As these BR mutants were weak alleles with residual activities, the partial recovery of the phenotype might be due to an increase in their mRNA levels caused by RAVL1 overexpression. The skotomorphogenesis phenotype of double combination plants was similar to that of the BR mutants (see Supplemental Figure 8 online). Etiolated seedlings of double combination genotypes exhibited short internodes as ones of BR mutants did. To confirm whether changes in lamina angles were due to differences in BR sensitivities, the lamina inclination assay was performed with OX;bri1 d61-1, OX;d2-2, and OX;d11-1 plants (Figure 6D; see Supplemental Figure 9 online). Even though OX;bri1 d61-1 plants became much less sensitive to exogenous epiBL than OX, the double combination was more sensitive to exogenous epiBL than were bri1 d61-1 plants. The sensitivities of d2-2 and d11-1 to epiBL were similar to those of wild-type siblings. However, OX;d2-2 and OX;d11-1 were as sensitive to epiBL as OX plants. In summary, RAVL1 overexpression led to partial recovery of lamina angles and epiBL sensitivities of lamina inclination. However, all plants with double combinations had the same height and skotomorphogenesis as BR mutants. Therefore, upregulation of d2-2 and d11-1 by overexpression of RAVL1 does not compensate effectively for dwarfism or aberrant skotomorphogenesis. These observations imply that different amounts of BRs may be required for the development of different phenotypes.
Figure 6.
RAVL1 Overexpression in the BR Mutants bri1 d61-1, d2-2, and d11-1 and Lamina Inclination of bri1-D, ravl1-1, and bri1-D;ravl1-1 Plants.
(A) Three-week-old plants are shown from the wild type (WT), bri1 d61-1, OX;bri1 d61-1, and OX. OX indicates RAVL1 overexpressor. Double combinations of the d2-2 and d11-1 mutations with the RAVL1 OX construct are presented in Supplemental Figure 8 online. Bar = 20 cm.
(B) The graph shows the angle of fourth-leaf lamina inclination. Plants were grown for 3 weeks in the greenhouse. Among the 100 to 200 F2 plants examined for each genetic cross, specimens carrying each of the four different genotypes were selected. Mean values are measurements of 5 to 10 plants. Error bars are se of the means.
(C) In the top panel, 2-month-old plants grown in the field are shown: the wild type, bri1 d61-1, OX;bri1 d61-1, and OX. In the bottom panel, the graph shows the lamina angles of the leaf just below the flag (topmost) leaf. Bar = 20 cm.
(D) The lamina inclination assay was performed with wild-type, bri1 d61-1, OX;bri1 d61-1, and OX plants.
(E) The lamina inclination assay was performed with the wild type, ravl1-1, bri1-D, and bri1-D;ravl1-1.
For the lamina inclination assay, the indicated amounts of epiBL in 1 μL of ethanol were spotted on the tip of the second lamina of 6-d-old seedlings (top panels). The graph shows measurements of lamina inclinations in response to epiBL. Mean values are derived from measurements of 10 plants (bottom panels). Error bars are se of the means.
To further verify that the BR-insensitive phenotype of ravl1 mutants is caused by the reduced expression of BRI1, a dominant bri1-D overexpression line was crossed with ravl1-1. bri1-D arose from an insertion of an activation tagging T-DNA into the 0.3-kb region downstream of the BRI1 gene (Jeong et al., 2002). To examine whether BRI1 overexpression suppresses the BR-insensitive phenotype of ravl1 mutants, the lamina inclination assay was performed with wild type, ravl1-1, bri1-D, and bri1-D;ravl1-1 of the F2 generation (Figure 6E). qRT-PCR analysis confirmed that the levels of BRI1 mRNA were greatly enhanced in both bri1-D;ravl1-1 and bri1-D plants (see Supplemental Figure 9 online). The assay showed that bri1-D;ravl1-1 plants were as hypersensitive to exogenous epiBL as bri1-D plants (Figure 6E). Taken together, this study confirmed that RAVL1 exerts its biological action mainly by mediating the expression of BRI1 and of several BR synthetic genes.
RAVL1 Is Necessary for Maintenance of BR Homeostasis
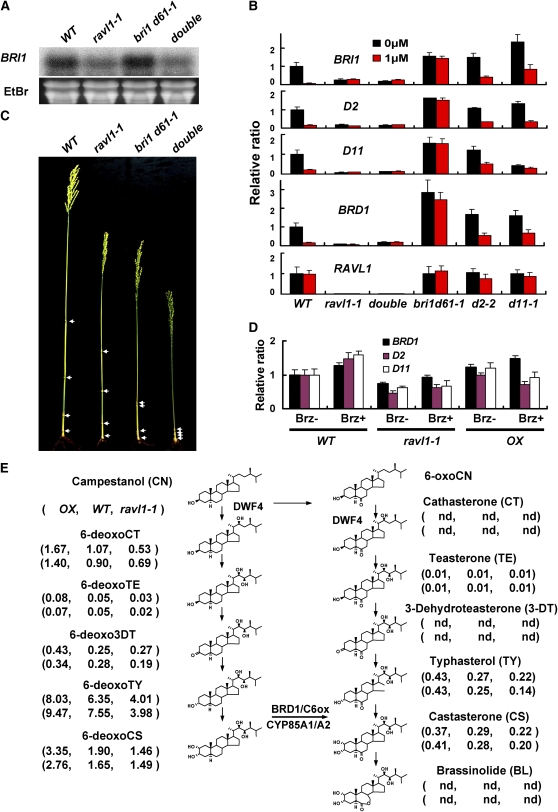
Because RAVL1 activates both BRI1 and biosynthetic genes, we tested the possibility that RAVL1 is also necessary for the BR feedback response of these genes. The following two experiments were performed: first, exogenous BRs were applied to plants. Second, the expression of BR-dependent genes was examined in double mutants for the bri1 d61-1 and ravl1-1. Exposure of wild-type plants and BR-synthetic mutants to exogenous epiBL led to the repression of the expression of both BRI1 and the BR-synthetic genes (Figure 7B). By contrast, in bri1 d61-1 mutants, these genes were no longer subject to feedback inhibition by exogenous epiBL treatment (Figure 7B). It should be noted that RAVL1 expression is not altered by exogenous epiBL treatment. Therefore, suppression of BRI1 and the BR-synthetic genes by epiBL treatment appears to be independent of RAVL1 action. To clarify whether RAVL1 influences the feedback relationship between BRI1 and the BR biosynthetic pathway, we examined double mutants of bri1 d61-1 and ravl1-1 for the expression of signaling and biosynthetic genes (Figure 7B). RNA gel blot analysis showed that the expression level of BRI1 was decreased in the double mutants and was as low as one detected in ravl1-1 mutant plants (Figure 7A). In addition, BR treatment did not alter BRI1, D2, D11, or BRD1 expression in either the double mutant or ravl1-1 mutant plants (Figure 7B). Therefore, the expression of RAVL1 seems to be necessary for the establishment of the feedback relationship. Phenotypically, double mutant (ravl1-1; bri1 d61-1) specimens showed more severe dwarfism than either of their ravl1-1 or bri1 d61-1 F2 siblings (Figure 7C). Since bri1 d61-1 is a leaky allele (Yamamuro et al., 2000), the decrease in the mRNA level represents an additive phenotypic effect.
Figure 7.
Expression Relationship among RAVL1, BRI1, and Synthetic Genes and Measurement of BR Intermediates.
(A) RNA gel blot analysis of the expression levels of BRI1 in double mutants and single mutants of ravl1-1 and bri1 d61-1. WT, wild type.
(B) The effects of epiBL treatment on the expression of BRI1 and BR synthetic genes (D2, D11, and BRD1) were examined in ravl1-1, bri1 d61-1, and BR-deficient mutants. The relative ratios of mutants to the wild type without BL treatment were measured by qRT-PCR. 25S rRNA was used control to normalize the expression data. Error bars are se of the means of three qPCR replicates.
(C) Double mutants (ravl1-1 and bri1 d61-1) showed more severe dwarfism than ravl1-1 or bri1 d61-1 single mutant plants.
(D) The effects of brassinazole treatment on the expression of BR synthetic genes (BRD1, D2, and D11) were examined in the wild type, ravl1-1, and RAVL1 OX. Brz+ or Brz− indicate with or without treatment of brassinazole, respectively. The relative ratios of mutants and OX to the wild type without Brz treatment were measured by qRT-PCR. 25S rRNA was used control to normalize the expression data. Error bars are se of the means of three qPCR replicates.
(E) The concentration of various BRs was measured in shoots. The amounts of BL and its precursors are shown in the following order: OX, wild type, and ravl1-1. OX indicates RAVL1 overexpressor. BR levels were measured in two independent experiments. Units are ng/g of fresh weight of samples. **, Not detected.
To further support a role of RAVL1 in regulating BR homeostasis, BR synthetic genes (BRD1, D2, and D11) were examined in the seedlings of ravl1-1 and RAVL1 overexpressor lines treated with brassinazole (Figure 7D). In both the wild type and ravl1-1, brassinazole application led to a slight enhancement of BRD1, D2, and D11. However, mutants accumulated BRD1, D2, and D11 mRNAs at much lower levels than the wild type under the same treatment conditions. In the RAVL1 overexpressor, the overall expression patterns of these genes were not significantly altered under the same treatment conditions.
Endogenous accumulation of BRs was measured in the ravl1-1 mutant and in the RAVL1 overexpressor (Figure 7E). With the exception of teasterone, which accumulated the least, all BRs tested were uniformly decreased in the ravl1-1 mutants and were uniformly increased in the RAVL1 overexpressor. 6-deoxocathasterone (6-deoxoCT), 6-deoxotyphasterol (6-deoxoTY), and 6-deoxocastasterone (6-deoxoCS) are the most abundant BRs of rice. The 6-deoxoCT, 6-deoxoTY, and 6-deoxoCS compounds were reduced to 62, 57, and 83%, respectively, in ravl1-1 mutants and were increased to 156, 126, and 172%, respectively, in RAVL1 overexpressors when compared with wild-type plants. This is in contrast with what is observed in BR biosynthetic mutants, in which the blockage of a specific reaction leads to the significant accumulation of intermediate species prior to the reaction step (Hong et al., 2002, 2003; Tanabe et al., 2005). Previous studies showed that BR signaling mutants, which included bri1 d61, accumulate high levels of all the BR intermediates (Noguchi et al., 1999; Nomura et al., 1999; Yamamuro et al., 2000); however, endogenous BR levels were lower in ravl1-1 mutants when compared with wild-type plants, even if the expression of BRI1 was severely repressed. This may be a consequence of the inactivation of BR biosynthetic genes in ravl1-1 plants. These data strongly suggest that RAVL1 directly controls the level of endogenous BRs. Together, our results demonstrate that RAVL1 is not only an activator of BRI1 and of BR synthetic genes, but is also necessary for the maintenance of BR homeostasis in plant cells.
DISCUSSION
The perception of BRs by plant cells is initiated by the activation of the receptor kinase gene BRI1. Recent work has focused on the elucidation of the mechanisms of activation of BRI1 and of triggering of downstream actions. In spite of the demonstrated significant role of BRI1, the mechanism underlying the regulation of the expression of the BRI1 gene remains unknown. This study shows that the RAVL1 gene coordinately regulates the expression of the BR receptor and of the biosynthetic genes (D2, D11, and BRD1) that are subjected to the BR negative feedback relationship. Phenotypically, ravl1 mutants exhibit alterations in BR response pertaining to root growth and lamina inclination and show defective skotomorphogenesis. In overexpressors, higher levels of RAVL1 mRNA correlated with greater growth stunting. This observation indicates that excessive activation of BR signaling might exert inhibitory effects on plant growth, which is consistent with the observation that roots became shorter with increases in exogenously applied epiBL levels (see Supplemental Figure 3 online).
To further solidify the genetic evidence that the action of RAVL1 is exerted via BRI1, two strategies have been employed: one is to analyze ravl1 phenotypes during the constitutive expression of BRI1, and the other is to examine bri1 d61-1 plants that constitutively express RAVL1. Our findings showed not only that the phenotypes of RAVL1-overexpressing plants are suppressed in the bri1 d61-1 and BR synthetic mutants but that the dominant overexpression of bri1-D suppresses the BR insensitivity of ravl1. Evidently, the primary action of RAVL1 takes place by mediating the expression of genes involved in BR signaling and biosynthesis. Also, RAVL1 is necessary for the response of BRI1 and biosynthetic genes to changes in cellular BR homeostasis. In addition, this study showed that RAVL1 interacts with BZR1 that is the best known factor of the BR negative feedback relationship. RAVL1 thus belongs to a regulatory circuit controlling the homeostasis of BRs.
E-box motifs are among the most common elements found in many plant genes. The mechanism underlying the specificity of the recognition of E-box elements by transcriptional factors is largely unknown. The BES/BIM and Myc/basic helix-loop-helix transcription factors present in the BR signaling pathway also bind to E-box elements. However, in Arabidopsis, BIM/BES regulates BR-related growth or development, but not BR synthesis. To date, there is no evidence implying that they control BR biosynthetic pathway. The specificity of the selective RAVL1 binding could be controlled at many different levels. E-boxes, which are present in the promoters of BRI1, D2, D11, and BRD1, showed variation in their binding affinity to RAVL1 in our in vitro assay; therefore, the specificity of the binding could arise from the microenvironment surrounding the target sites, which include adjacent sequences and other transcriptional apparatus, which might differentiate between BIM/BES and RAVL1.
However, it should be noted that the phenotype of the ravl1 mutant was less dramatic than those associated with most null alleles of BR genes both in signal transduction and in biosynthetic pathways. Many BR signaling and synthetic mutants have compact stature and show strong dwarfism; by contrast, both ravl1 alleles yielded mild phenotypic effects. Rather, weak alleles of BRI1, D2, and D11 (bri1 d61-1, d2-2, and d11-1, respectively) develop plant statures similar to those of ravl1. One possible explanation for this discrepancy is the nature of the biological function of RAVL1, which supports the expression of genes that are subjected to the negative feedback relationship. Considering this, it may be difficult to develop severe phenotypic effects. Another possibility is that other transcriptional mechanisms support the expression of these genes, even in the absence of RAVL1 expression. This could explain the observation that loss of RAVL1 function in plants does not lead to complete blockage of the signaling or biosynthetic pathways.
Many biosynthetic pathways of plant hormones are regulated by end-product feedback. In the case of ABA, a positive feedback mechanism is proposed where stress-induced ABA further stimulates the accumulation of the hormone (Xiong and Zhu, 2003). The activity and expression levels of the 1-amino-cyclopropane-1-carboxylic acid synthase gene, which participates in ethylene biosynthesis, are controlled by negative feedback regulation (Thain et al., 2004; Wang et al., 2005b). Recent works demonstrate that key regulatory genes in signaling pathways, which interact directly or indirectly with hormones, eventually regulate the transcriptional or enzymatic activities of biosynthetic genes. In the auxin stimulation pathway, early genes induced by the ubiquitinylation activity of the SCFTIR1 complex eventually lead to the decrease of active IAA levels (Staswick et al., 2005). Moreover, gibberellin homeostasis is maintained by the processes of feedback and feedforward regulation of GA metabolism via the DELLA protein-dependent signaling pathway (Hedden and Phillips, 2000). This study elucidated a mechanism whereby participants of the BR signaling pathway are coordinately regulated with BR biosynthetic genes in an antagonistic relationship. This transcriptional relationship ensures a basal activity in both the signaling and biosynthetic pathways (see Supplemental Figure 10 online).
METHODS
Plant Materials and Growth Conditions
Wild-type, mutant, and transgenic plants were of Oryza sativa ssp japonica cv Dong Jin background. The d2-2 and d11-1 mutants were obtained from Seoul University (Korea), and the bri1 d61-1 mutant was obtained from Makoto Matsuoka's laboratory in Japan. The d2-2, d11-1, osbri1-D, and bri1 d61-1 plants were of O. sativa japonica background. The daily high and low temperatures at the growing sites (during the summer) and in the greenhouse were typically 35 and 20°C, respectively.
Constructs and Plant Transformation
The pSB11 plasmid, in which the hygromycinR gene (hygromycin phosphotransferase, HPT) was inserted in a T-DNA vector, was used to construct the pDE35S:cDNA (Os RAVL1). A cDNA encoding a full-length RAVL1 was cloned by PCR with primers 5′-ATGGAGCAGGAGCAGGATGAAG-3′ and 5′-TCACAGATCGAGATCCAACGAGG-3′. The cDNA was ligated to a double enhanced 35S promoter. The binary vector pCAMBIA-1302 (Hajdukiewicz et al., 1994) was used to construct the 35S:RAVL1-GFP, RAVL1(ΔB3)-GFP, and RAVL1(Δ3′)-GFP vectors. For RAVL1(ΔB3), the region encoding a B3 domain was deleted by PCR. For RAVL1(Δ3′), 498 nucleotides of the 3′ end were deleted from the full-length cDNA. GFP was fused to the 3′ ends of these cDNAs. T-DNA vectors were transformed into LBA4404 cells. A published method of tissue culture (Hiei et al., 1994) was followed, with slight modifications. Instead of NB medium, Murashige and Skoog–based media were used for the selection of HPT-resistant calli.
Lamina Inclination Assay
Rice plants were grown for 5 or 6 d in a chamber kept at 25 to 26°C with a light/dark cycle of 16/8 h. One microliter of ethanol containing 0, 1, 10, 100, or 1000 ng of epiBL (Sigma-Aldrich) was spotted on the tip of the lamina of 6-d-old seedlings. Treated seedlings were grown for three additional days, and the inclination of the lamina joint of the second leaf was measured.
RNA Manipulation and qRT-PCR Analysis
For the preparation of RNA, whole seedlings were harvested and immediately ground in liquid nitrogen. For the treatment of epiBL and brassinazole, 2-week-old seedlings were submerged for 6 h in distilled water containing 1 μM epiBL or 10 μM brassinazole or none. Total cellular RNA was prepared using the easy-BLUE reagents (iNtRON Biotechnology) or the RNeasy Kit (Qiagen). RNA electrophoresis, blotting, and hybridization were performed based on standard methods.
For qRT-PCR, RNA samples were treated with DNase at 37°C for 30 min. The first-strand cDNA was synthesized using M-MuLV reverse transcriptase (New England Biolabs) in 20 μL of reaction mixture containing 1 μg total RNA, according to the manufacturer's instructions. Both an oligo dT12-18 primer and random primers for reverse transcription produced the same results in the subsequent qRT-PCR (data not shown). RT-PCRs were performed in triplicate with a Bio-Rad Opticon 2. Since 25S rRNA shows more stable expression than Ubiquitin (UBQ) under hormone treatment (Jain et al., 2006), 25S rRNA was used control to normalize the expression data. Data were analyzed with Bio-Rad Chromo4 Real-time PCR Detection System (2−ΔΔCt method). The primers for qRT-PCR are listed in Supplemental Table 1 online.
ChIP Assay
Ten grams of calli from 35S:RAVL1-GFP and 35S:GFP transgenic plants were prepared for the ChIP assay, performed following a published method (Ito et al., 1997) with slight modifications. Preabsorption with preimmune serum was performed before immunoprecipitation with a GFP monoclonal antibody (Clontech). Immunoprecipitants were analyzed by qRT-PCR as described above. Each input DNA level was used control to normalize immunoprecipitant ratio. The primers used are listed in Supplemental Table 2 online.
EMSA and DNase I Footprinting Assays
For EMSA, a standard binding reaction was performed in a total volume of 20 μL by incubation of an appropriate amount of purified protein with 40k cpm of 32P-labeled probe DNA and 1 μg of poly(dI-dC) in B buffer [25 mM HEPES-KOH, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 10% (v/v) glycerol, 1 mM DTT] at room temperature for 30 min. The binding reaction products were resolved on an 8% polyacrylamide gel run in 0.5× TBE buffer. The primers described above for the ChIP assay were used to generate the probes for this assay. For mutated forms of E-box elements P1m and P2m, P1 (agcacatgccatgtgac and tacagctgtt) and P2 (tacaaatgat and ttcatttgag) of the BRI1 promoter were changed to agagccgtctcggctac, tattataatt, tataaactat, and ttacggctag, respectively.
For the DNase I footprinting assay, appropriate amounts of purified protein and a 32P-end-labeled probe DNA of 105 cpm were incubated along with 1 unit DNase I in a total volume of 50 μL at room temperature for 1 min. The binding buffer was the same as that used for EMSA. Reaction products and DNA sequencing products were resolved on a 6% polyacrylamide, 8 M urea gel run in 0.5× TBE buffer. Maxam-Gilbert chemical sequencing was performed based on the protocol for the Sequenase Version 2.0 DNA sequencing kit (USB Corporation).
Transient Expression Assay
Effectors (pUbi:RAVL1 and pUbi:BZR1), reporters (pBRI1:GUS, pBRD1:GUS, and mutated promoter-GUS fusions), and the internal control (p35S:LUC) were constructed in pUC19. The promoter lengths of BRI1 and BRD1 were ∼2 kb. RAVL1 and BZR1 cDNA were cloned into a plasmid vector harboring a maize (Zea mays) Ubi promoter (pUbi:PAT2). BRI1 and BRD1 promoters were cloned from genomic DNA by PCR. The promoter DNA was cloned into pBI221 carrying a GUS coding region. The primers used are listed in Supplemental Table 3 online. For internal control vector (p35S:LUC), 1.7-kb DNA encoding a luciferase gene was fused to a 35S promoter. The effector, reporter, and internal control vectors were cotransformed into rice Oc protoplasts (Wong et al., 2004). A luciferase gene (LUC) was used to normalize GUS expression data. Electroporation and activity assays were followed as published previously (Wong et al., 2004). For the bm1 promoter, both the P1-1 and -2 E-box sequences (agcacatgccatgtgac) were changed to agagccgtctcggcact. The bm2 promoter carrying P1-3 was mutated from tacagctgtt to tattataatt. The bm3 promoter has a mutation (tataaactat) in the P2-1 E-box (tacaaatgat). The bmt promoter has all the P1-1 to P2-2 mutations and an additional E-box mutation between −841 and −836 that was changed from ggcacgtgac to ggtacgtaac. The relative expression ratio of the control reporter 35S:GUS was 189 ± 3.8.
Quantification of Endogenous BRs
Wild-type, ravl1-1 mutant, and RAVL1 overexpressor plants were grown in a greenhouse. Shoots of 3-week-old plants were harvested and lyophilized immediately. Lyophilized shoots (equivalent to 10 g of fresh weight) were extracted twice with 250 mL of methanol:CHCl3 (4:1, v:v) and deuterium-labeled internal standards (1 ng/g of fresh weight) were added. Purification and gas chromatography–mass spectrometry analyses were performed as described by Fujioka et al. (2002).
Accession Numbers
Accession numbers for the major genes discussed in this article can be found in the GenBank/EMBL databases as follows: Os BRI1, Os01g0718300; D2, Os01g0197100; D11, Os04g0469800; BRD1, Os03g0602300; Os BZR1, Os07g0580500; and Os RAVL1, HM450152.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Patterns and Phenotypes of RAVL1 Plants.
Supplemental Figure 2. Growth Responses of Seminal Roots to ACC, ABA, GA, and NAA Treatment.
Supplemental Figure 3. Growth and Root Curling Responses of Seminal Roots to epiBL.
Supplemental Figure 4. Phenotypes of RAVL1 Overexpressor.
Supplemental Figure 5. Nuclear Localization of RAVL1-GFP in Transgenic Plant Roots.
Supplemental Figure 6. EMSA to Test Binding of RAVL1 to Target Sequences.
Supplemental Figure 7. Binding Competition Assays of RAVL1 Target Sequences.
Supplemental Figure 8. Phenotypes of RAVL1 Overexpression in BR Mutants.
Supplemental Figure 9. Lamina Inclination in OX;d2-2 and OX;d11-1 Plants Exposed to epiBL, and Expression of BRI1 in bri1-D and bri1-D;ravl1-1 Plants.
Supplemental Figure 10. Functional Model of RAVL1 in BR Homeostasis.
Supplemental Table 1. List of Primers Used for Real-Time PCR.
Supplemental Table 2. List of Primers Used for ChiP and EMSA.
Supplemental Table 3. List of Primers Used for the Vector Constructs in Transient Assays.
Acknowledgments
This work was supported by the Crop Functional Genomics Center of the 21st Century Frontier Research Program (CG3313) and BioGreen 21 Program (Rural Development Administration; Grant 20070401034001). B.I.J., S.J.P., H.L.P., and Y.H.X. were supported by a scholarship from the BK21 program, the Ministry of Education, Science, and Technology, Korea. J.H. was also supported by the Korea Research Foundation Grant (F00026) funded by the Korean Government (Ministry of Education and Human Resource Development). This work was also supported in part by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to S.F. (Grant 19380069). We thank Suguru Takatsuto (Joetsu University of Education) for supplying deuterium-labeled internal standards. The comments of the anonymous reviewers and the editor were greatly appreciated.
References
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A., Tretyn A. (2003). The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62: 1027–1046 [DOI] [PubMed] [Google Scholar]
- Bancos S., Nomura T., Sato T., Molnar G., Bishop G.J., Koncz C., Yokota T., Nagy F., Szekeres M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonan N. (1967). Studies on the photosynthetic tissue in the leaves of cereal crops. 4. Effect of shading on the mesophyll structure of the rice leaves. Jpn. J. Crop. Sci. 36: 297–301 [Google Scholar]
- Clouse S.D. (1996). Molecular genetics studies confirm the role of brassinosteroids in plant growth and development. Plant J. 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Davies P.J. (2004). Regulatory factors in hormone action: Level, location and signal transduction. Plant Hormones: Physiology, Davies P.J., (Dordrecht, The Netherlands: Kluwer Academic Press; ), pp. 16–35 [Google Scholar]
- Fujioka S., Noguchi T., Takatsuto S., Yoshida S. (1998). Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49: 1841–1848 [Google Scholar]
- Fujioka S., Takatsuto S., Yoshida S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove M.D., Spencer G.F., Rohwedder W.K., Mandava N., Worley J.F., Warthen J.D., Stelffens G.L., Flippen-Anderson J.L., Carter Cook J. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hong Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Takahashi N., Shimura Y., Okada K. (1997). A Serine/Threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGMOUS. Plant Cell Physiol. 38: 248–258 [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S., Kang H.G., Moon S., Han J.J., Park S., Lee H.S., An K., An G. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K., Hattori T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.M., et al. (2004). Rapid, large-scale generation of Ds transposant lines and analysis of Ds insertion sites in rice. Plant J. 39: 252–263 [DOI] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1999). Brassinosteroid actions in plants. J. Exp. Bot. 50: 332–340 [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li L., Xu J., Xu Z.H., Xue H.W. (2005). Brasssinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumo S., Hattori H., Nanoyma Y., Munakata M. (1968). The presence of novel plant growth regulators in leaves of Distylium racemosum Sieb et Zucc. Agric. Biol. Chem. 32: 528–529 [Google Scholar]
- Mitchell J.W., Mandava N., Worley J.F., Plimmer J.R., Smith M.V. (1970). Brassins: A new family of plant hormones from rape pollen. Nature 225: 1065–1066 [DOI] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Satoh T., Tanaka S., Mochizuki N., Yokota T., Nagatani A. (2005). Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J. Exp. Bot. 56: 833–840 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Nguyen S.M., Malancharuvil E.J., Fujioka S., Noguchi T., Seto H., Tsubuki M., Honda T., Takatsuto S., Yoshida S., Chory J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Kitasaka Y., Takatsuto S., Reid J.B., Fukami M., Yokota T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 119: 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G.L., Vaistij F.E., Hiranuma S., Seto H., Takatsuto S., Adam G., Yoshida S., Bowles D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rouleau M., Marsolais F., Richard M., Nicolle L., Voigt B., Adam G., Varin L. (1999). Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J. Biol. Chem. 274: 20925–20930 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Shimada Y., Goda H., Nakamura A., Takatsuto S., Fujioka S., Yoshida S. (2003). Organ-specific expression of brassinosteroid biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G.M., Ross J.J., Jager C.E., Reid J.B. (2008). Brassinosteroid transport. J. Exp. Bot. 59: 17–24 [DOI] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., Kato H., Iwasaki Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain S.C., Vandenbussche F., Laarhoven L.J., Dowson-Day M.J., Wang Z.Y., Tobin E.M., Harren F.J., Millar A.J., Van Der Straeten D. (2004). Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 136: 3751–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.N., Shih M.C., Li N. (2005b). The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 56: 909–920 [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005a). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wong H.L., Sakamoto T., Kawasaki T., Umemura K., Shimamoto K. (2004). Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.Y., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Zhu J.K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]