This work identifies a group of WRKY transcription factors that function as negative abscisic acid (ABA) signaling regulators acting as transcription repressors directly downstream of a previously identified ABA receptor ABAR, describing an ABA signaling pathway from primary events to downstream gene expression.
Abstract
The phytohormone abscisic acid (ABA) plays a vital role in plant development and response to environmental challenges, but the complex networks of ABA signaling pathways are poorly understood. We previously reported that a chloroplast protein, the magnesium-protoporphyrin IX chelatase H subunit (CHLH/ABAR), functions as a receptor for ABA in Arabidopsis thaliana. Here, we report that ABAR spans the chloroplast envelope and that the cytosolic C terminus of ABAR interacts with a group of WRKY transcription factors (WRKY40, WRKY18, and WRKY60) that function as negative regulators of ABA signaling in seed germination and postgermination growth. WRKY40, a central negative regulator, inhibits expression of ABA-responsive genes, such as ABI5. In response to a high level of ABA signal that recruits WRKY40 from the nucleus to the cytosol and promotes ABAR–WRKY40 interaction, ABAR relieves the ABI5 gene of inhibition by repressing WRKY40 expression. These findings describe a unique ABA signaling pathway from the early signaling events to downstream gene expression.
INTRODUCTION
The phytohormone abscisic acid (ABA) regulates many aspects of plant development, such as seed maturation, germination, and seedling growth, and plays a central role in plant adaptation to environmental challenges (Finkelstein et al., 2002; Adie et al., 2007). ABA functions through a highly complex network of signaling pathways, and during the past decades, numerous signaling components have been identified. These involve diverse regulators, such as membrane-associated proteins, phospholipases C/D, G proteins, and receptor-like kinases, various protein kinases, and phosphatases including SNF1-related protein kinases (SnRKs), calcineurin B-like protein kinases, calcium-dependent protein kinases, mitogen-activated protein kinases, and type-2C/A protein phosphatases (PP2C/A). Also involved are E3 ligases involved in degradation of ABA signaling proteins as well as various classes of transcription factors (for reviews, see Finkelstein et al., 2002; Wang, 2002; Himmelbach et al., 2003; Shinozaki et al., 2003; Fan et al., 2004; Hirayama and Shinozaki, 2007; Seki et al., 2007). The considerable progress in characterization of the cellular components of ABA signaling deepens our understanding of the underlying mechanisms of ABA functions.
ABA receptors are the most upstream components in ABA signaling, and research in this field has attracted much attention. ABA signal perception by ABA receptors is considered to be the primary event that triggers downstream signaling cascades to induce the final physiological responses. It has been believed that the ABA signal is perceived by multiple receptors, including plasma membrane and intracellular receptors (Assmann, 1994; Finkelstein et al., 2002; Verslues and Zhu, 2007). Two plasma membrane ABA receptors, an unconventional G protein–coupled receptor (GCR2) and a novel class of G protein–coupled receptor (GTG1 and GTG2), have been reported (Liu et al., 2007a, 2007b; Johnston et al., 2007; Pandey et al., 2009), though it is controversial whether GCR2 functions in ABA signaling (Gao et al., 2007). GTGs interact with the sole Arabidopsis thaliana G protein α-subunit GPA1 to regulate ABA signaling (Pandey et al., 2009). Most recently, PYR/PYL/RCAR proteins, the members of a START domain superfamily, were reported to function as cytosolic ABA receptors by inhibiting directly type 2C protein phosphatases (Ma et al., 2009; Park et al., 2009). The ABA receptor identity of the START proteins was confirmed by recent studies of structural biology (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009; Yin et al., 2009). A PYL/PYR/RCAR-mediated ABA signaling pathway from ABA perception to downstream gene expression has been reconstituted in vitro (Fujii et al., 2009). However, it is widely believed that the networks of ABA signaling pathways are highly complex, and additional pathways of ABA signaling from early events to downstream gene expression remain to be elucidated.
We previously reported that the magnesium-protoporphyrin IX chelatase large subunit (Mg-chelatase H subunit [CHLH]/putative ABA receptor [ABAR]) binds ABA and functions in ABA signaling, thus meeting the essential criteria of a receptor for ABA in Arabidopsis (Shen et al., 2006). Although the receptor nature of the homolog of the Arabidopsis ABAR, XanF, was questioned in barley (Hordeum vulgare; Muller and Hansson, 2009), we provided new biochemical and genetic evidence for the ABA binding ability and ABA signaling functionality of the ABAR in Arabidopsis and further observed that the C-terminal half of ABAR plays a central role in ABA binding and signaling (Wu et al., 2009). Moreover, the function of the Arabidopsis ABAR in ABA signaling has been verified by an independent group (Legnaioli et al., 2009), who showed that ABAR is a key component connecting the circadian clock with ABA-mediated plant responses to drought. All these data consistently support the idea that the Arabidopsis ABAR is a chloroplast-localized intracellular ABA receptor.
ABAR is a chloroplast/plastid protein and has multiple functions in plant cells. It catalyzes the introduction of magnesium to protoporphyrin IX in the chlorophyll biosynthesis pathway (Gibson et al., 1996; Guo et al., 1998; Papenbrock et al., 2000) and also plays a key role in mediating plastid-to-nucleus retrograde signaling as Genomes Uncoupled 5 (GUN5) (Mochizuki et al., 2001; Nott et al., 2006). We showed that the ABAR-mediated ABA signaling is distinct from the tetrapyrrole/chlorophyll biosynthesis pathways (Shen et al., 2006; Wu et al., 2009). Here, we report a mechanism by which ABAR relieves ABA-responsive genes of inhibition by antagonizing the negative ABA-signaling regulators WRKYs, a group of the WRKY transcription factors that negatively modulate ABA signaling in seed germination and postgermination growth. These findings have discovered a unique ABA signaling pathway from the primary signaling events to downstream gene expression. Additionally, we observed that XanF, the barley homolog of Arabidopsis ABAR, interacts with the Arabidopsis transcription repressor WRKY40 and may function to positively regulate ABA signaling in Arabidopsis.
RESULTS
ABAR Spans the Chloroplast Envelope with Its N and C Termini Exposed to Cytosol
It remains an open question how chloroplast protein ABAR transmits a signal across the chloroplast envelope to the nucleus in response to ABA signaling (Shen et al., 2006; Wu et al., 2009) as in chloroplast retrograde signaling (Mochizuki et al., 2001; Nott et al., 2006). A previous report showed that ABAR localizes in both envelope and stroma fractions in vitro depending on the Mg2+ concentrations of the medium used in chloroplast fractionation: ABAR localizes predominantly to the envelope fraction in the medium containing a relatively high Mg2+ concentration (>5 mM), while it resides predominantly in the stroma fraction with a lower concentration (1 mM) of Mg2+ in the medium (Gibson et al., 1996). We confirmed this observation in a chloroplast fractionation assay (Figure 1D, c).
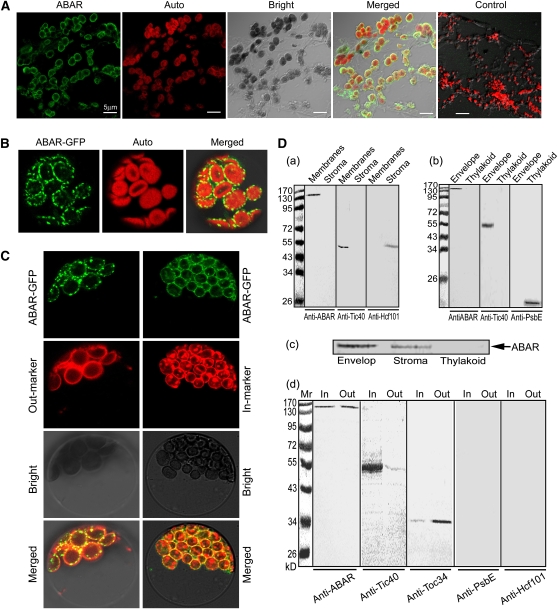
Figure 1.
ABAR Predominantly Localizes to Both Inner and Outer Envelopes of Chloroplast.
(A) Immunofluorescence assay shows that ABAR localizes predominantly to the periphery of chloroplasts. The frozen sections were prepared from Arabidopsis leaves, immunolabeled with the anti-full-length ABAR serum and goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) antibody (green fluorescence), and observed with a confocal laser scanning microscope. ABAR, Auto, Bright, and Merged indicate fluorescence of ABAR, chlorophyll autofluorescence, bright-field, and merged image of ABAR and Auto in the bright field, respectively. Control, a control section treated with rabbit preimmune serum instead of the rabbit antiserum, shows no immunosignal and thus reveals the specificity of the ABAR fluorescence localization.
(B) Transient expression of the ABAR-GFP fusion protein in Arabidopsis protoplasts, showing that ABAR-GFP localizes to the periphery of chloroplasts. Auto, chloroplast autofluorescence; Merged, merged image of ABAR-GFP and Auto.
(C) Transient expression in Arabidopsis protoplasts shows that ABAR-GFP colocalizes to the chloroplast envelope with both a chloroplast inner envelope marker fusion TIC21-RFP/mCherry (In-marker) and an outer marker fusion OEP7-RFP/mCherry (Out-marker). Bright, bright field; Merged, merged image of ABAR-GFP and In-marker or Out-marker in bright field.
(D) Immunoblotting of chloroplast fractions shows additional evidence that ABAR predominantly localizes to both inner and outer envelopes of chloroplast. (a) ABAR (detected by anti-ABAR serum) localizes to the membrane fractions including envelope and thylakoid membranes, but not to the stroma fraction. (b) ABAR (detected by anti-ABAR serum) localizes to the envelope fraction but not to the thylakoid fraction. (c) ABAR appears in both the envelope and stroma fractions when Mg2+ was used at 1 mM instead of 5 mM in the isolation and suspending buffers during preparation of the chloroplast fractions. (d) ABAR localizes to both inner and outer envelope membranes. The anti-ABAR serum recognizes ABAR protein in both inner (In) and outer (Out) envelope membranes in a similar amount. The left numbers in (a), (b), and (d) indicate molecular mass in kilodaltons.
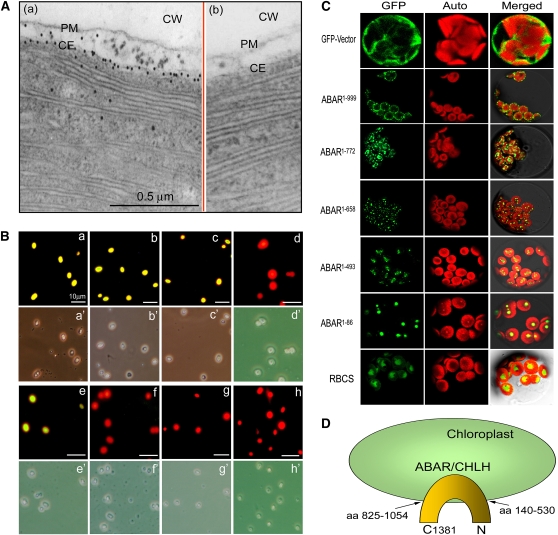
However, it is known that plant tissues contain Mg2+ at levels >5 mM, which should be enough to maintain ABAR at the chloroplast envelope. We used a combination of biochemical, cellular, and molecular approaches to localize this protein in Arabidopsis cells. An in situ immunofluorescence labeling assay in the frozen tissues of Arabidopsis leaves showed that ABAR localizes to the periphery of chloroplasts in planta (Figure 1A). An assay of transient expression in Arabidopsis protoplasts showed that ABAR predominantly resides at the chloroplast envelope (Figure 1B). We used chloroplast envelope markers to verify the envelope-associated ABAR localization, which was confirmed by overlapping of the ABAR-green fluorescent protein (GFP) signal with both an inner (TIC21) (Teng et al., 2006) and an outer (OEP7) (Lee et al., 2001) chloroplast envelope marker (Figure 1C). Two classes of biochemical assays, chloroplast fractionation combined with immunoblotting detection and in situ immunogold labeling, consistently showed chloroplast envelope localization of this protein (Figures 1D, a, b, and d, and 2A). An immunofluorescence assay with the isolated intact chloroplasts further showed that, whereas the antibodies against either the N or C terminus of ABAR recognized ABAR on the chloroplast outer surface, the antibody against a middle fragment of ABAR did not (Figure 2B). This revealed that ABAR spans the chloroplast envelope with its N and C termini exposed to the cytosol. Assays of transient expression of the truncated ABAR-GFP fusion protein in Arabidopsis protoplasts showed that the truncated ABAR1-999 (amino acid residues 1 to 999) localizes predominantly to the chloroplasts envelope, and the truncated ABAR1-772 (amino acids 1 to 772) and ABAR1-658 (amino acids 1 to 658) to both envelope and intrachloroplast compartment; and the truncated ABAR1-493 (amino acids 1 to 493) and ABAR1-86 (amino acids 1 to 86) entirely to intrachloroplast compartment (Figure 2C). These data suggested that the C-terminal transmembrane domain is situated approximately from amino acid residues 770 to 1000. This is essentially consistent with a transmembrane prediction model where several transmembrane domains may occur in the N-terminal amino acids 140 to 530 and in the C-terminal amino acids 825 to 1054 (Figure 2D; see Supplemental Figure 1 online). The cytosol-exposed C-and N termini of ABAR provide ABA signaling with a potential bridge across the chloroplast envelope.
Figure 2.
ABAR Spans the Chloroplast Envelope and Is Exposed to Cytosol with Its N and C Termini.
(A) Immunogold electronic microscopy shows that ABAR (visualized by gold particles) predominantly localizes in chloroplast envelope membranes (a). Panel (b) shows a control where the purified IgG of rabbit preimmune serum was used instead of the anti-ABAR serum in the immunolabeling, and no signal was detected. CW, cell wall; PM, plasma membrane; CE, chloroplast envelope.
(B) Immunodetection in the isolated, intact chloroplasts shows that ABAR spans the chloroplast envelope and is exposed to cytosol with its N and C terminus. The anti-ABAR (a), anti-ABAR-C terminus (b), and anti-ABAR-N terminus (c) sera recognize immunosignal (marked by yellow-green fluorescence), while the anti-ABAR middle fragment serum (d) does not. Note that the anti-Toc34 (outer envelope marker) detects immunosignal (e), but the anti-Tic40 (inner envelope marker; [f]) and anti-Hcf101 (stroma marker; [g]) sera and the purified IgG of the rabbit preimmune serum (h) do not detect signal. The corresponding bright field is displayed below each fluorescence image and indicated by the same letter marked by an apostrophe.
(C) Subcellular localization of the truncated ABARs. RBCS was used as a stroma marker. The superscript numbers indicate the numbers of amino acid residues in the order from the N terminus to the truncation site. GFP, Auto, and Merged indicate fluorescence of the truncated ABAR-GFP or RBCS-GFP fusion protein, chlorophyll autofluorescence, and merged image of GFP and Auto in the bright field, respectively.
(D) A model showing that ABAR is predominantly a trans-chloroplast membrane protein. aa 140-530 and aa 825-1052 indicate predicated transmembrane domains at N and C termini, respectively (see Supplemental Figure 1 online). C1381, the amino acid residue 1381 in the ABAR C end.
The C Terminus of ABAR Interacts with Members of a Group of WRKY Transcription Factors: WRKY40, WRKY18, and WRKY60
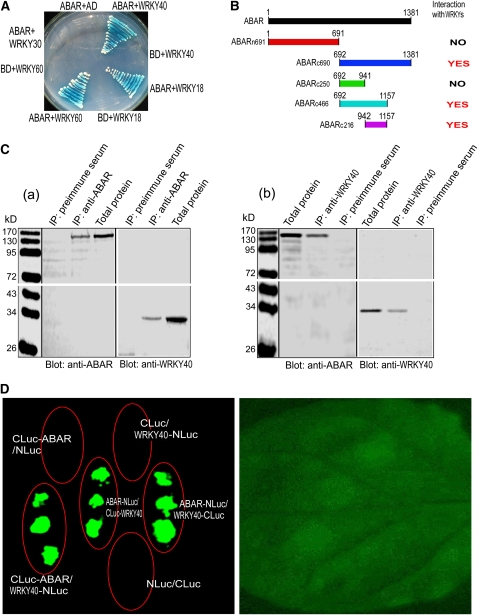
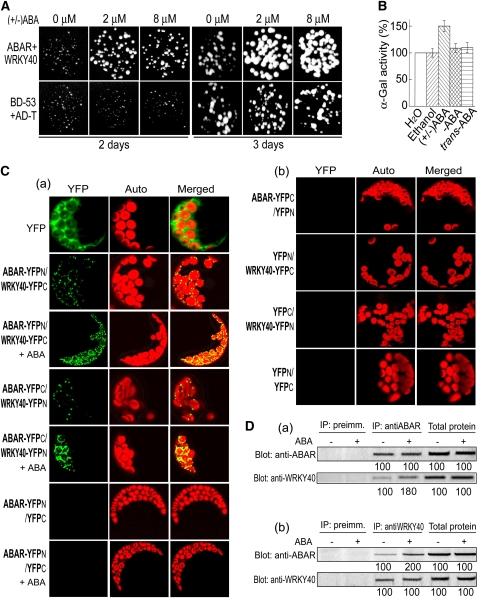
We previously showed that the C terminus of ABAR binds ABA and plays a central role in ABA signaling (Wu et al., 2009). In a yeast two-hybrid screen using the C terminus of ABAR (amino acids 692 to 1381) as a bait, we found an ABAR interaction partner that is a WRKY transcription factor (WRKY40). We confirmed this interaction using yeast two-hybrid and coimmunoprecipitation (CoIP) assays with yeast total protein (Figure 3A; see Supplemental Figure 2A online). The yeast two-hybrid assays were performed with a set of truncated proteins of the ABAR and showed that the C-terminal sequence (amino acids 942 to 1157) in ABAR is an interaction domain but that the N-terminal half (amino acids 1 to 691) of ABAR does not interact with WRKY40 (Figure 3B; see Supplemental Figure 2B online). Consistently, this interacting C-terminal domain is exposed to the cytosolic side of the chloroplast (Figure 2), indicating that the ABAR–WRKY40 interaction takes place in the C-terminal cytosolic portion of ABAR. The N-terminal half was previously shown to have no ABA binding activity but is functionally required for ABA signaling, likely through a regulatory role of the C-terminal half (Wu et al., 2009). Two closet homologs of WRKY40, viz., WRKY18 and WRKY60, were shown to interact also with ABAR (Figure 3A) but with an interaction intensity (estimated by both α-galactosidase activity and yeast growth) weaker than WRKY40 does (see Supplemental Table 1 online). We showed that the WRKY 30, a member of the WRKY family but with low identity of amino acid sequence with WRKY40 (17%), does not interact with ABAR (Figure 3A; see Supplemental Table 1 online), indicating the specificity of ABAR–WRKY40/18/60 interaction. Furthermore, we verified the ABAR–WRKY40 interaction in planta with CoIP assays using plant total protein (Figure 3C, a and b). The assay with the firefly luciferase (Luc) complementation imaging (LCI; Chen et al., 2008) confirmed the ABAR–WRKY40 interaction in vivo (Figure 3D).
Figure 3.
ABAR Interacts with Three Transcription Factors WRKY40, WRKY18, and WRKY60.
(A) Test of yeast growth in SD medium lacking Leu, Trp, His, and Ade (growth for 3 d without ABA supplementation) shows that ABAR interacts with WRKY40, -18, and -60 but not with WRKY30. BD, DNA binding domain in the bait vector; AD, activation domain in the prey vector.
(B) Summary of the interaction of the truncated ABARs with the WRKYs in the yeast two-hybrid system (without ABA supplementation). Left panel shows the truncated ABARs and right panel the interaction status, where “NO” indicates no interaction and “YES” indicates an interaction.
(C) ABAR and WRKY40 are coimmunoprecipitated from Arabidopsis total proteins. IP, immunoprecipitation; Blot, immunoblot; anti-ABAR and anti-WRKY40, antiserum specifically against ABAR and WRKY40, respectively. Immunoprecipitation with preimmune serum was taken as a control. (a) Immunoprecipitation with the anti-ABAR serum and immunoblotting with the anti-ABAR or anti-WRKY40 serum; (b) immunoprecipitation with the anti-WRKY40 serum and immunoblotting with the two antisera.
(D) Firefly Luc complementation imaging shows that ABAR interacts with WRKY40. The tobacco leaves were transformed by infiltration using a needleless syringe with construct pairs ABAR-N-terminal half of Luc (NLuc)/C-terminal half of Luc (CLuc)-WRKY40, CLuc-ABAR/WRKY40-NLuc, ABAR-NLuc/WRKY40-CLuc, CLuc-ABAR/NLuc, CLuc/WRKY40-NLuc, and NLuc/CLuc. The right panel shows the bright-field image of the treated leaf.
ABA Recruits WRKY40 from Nucleus to Cytosol
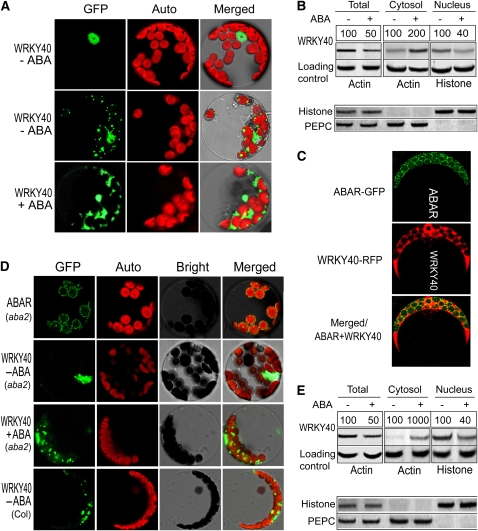
The WRKY40/18/60 group was previously shown to localize in the nucleus (Xu et al., 2006). We showed that WRKY40 localizes to the nucleus but mostly to both the nucleus and cytosol in wild-type plant cells that contain ABA at physiological concentrations by both the stable expression of the WRKY40-GFP fusion protein (Figure 4A; see Supplemental Figures 3A to 3C and 4A online) and immunoblotting of the cellular fractions (Figure 4B; see Supplemental Figures 3F online). In the protoplasts coexpressing the ABAR-GFP and WRKY40-RFP fusion proteins, we observed the colocalization of ABAR and WRKY40 on the surface of chloroplasts (Figure 4C; see Supplemental Figure 4B, a and b, online). However, in the ABA-deficient mutant aba2 cells, WRKY40 resides almost uniquely in nucleus. This was shown with the same assays as mentioned above for wild-type plants (Figures 4D and 4E; see Supplemental Figure 3F online). By contrast, ABAR localization to the chloroplast envelope was not altered in the aba2 cells (Figure 4D). Interestingly, while the exogenous ABA application downregulated WRKY40 (Figures 4B and 4E; see below for this ABA-induced downregulation of WRKY40), the ABA treatment enhanced significantly the amount of WRKY40 in the cytosol of wild-type plant cells (Figures 4A and 4B; see Supplemental Figures 3F and 4A online) and restores the cytosolic distribution of WRKY40 in the aba2 cells (Figures 4D and 4E; see Supplemental Figures 3F and 4A online). These findings reveal that ABA is important for the cytosolic distribution of WRKY40 and recruits this transcription factor from the nucleus to the cytosol.
Figure 4.
ABA Is Important for the Trafficking of WRKY40 into Cytosol.
(A) WRKY40 localizes in the nucleus (WRKY40-ABA, top panels) and also in the cytosol (WRKY40-ABA, middle panels), and ABA enhances the cytosolic distribution of WRKY40 (WRKY40 + ABA, bottom panels). The GFP-tagged WRKY40 was stably expressed in the wild-type Col plants. ABA treatment was done by spraying the transgenic plants with (+) or without (–) 100 μM ABA 6 h before the protoplasts isolated from the plants were observed. Images were taken with identical parameters to allow comparison of fluorescence intensities. GFP, fluorescence of the WRKY40-GFP fusion protein; Auto, chloroplast autofluorescence; Merged, merged image of GFP and Auto under the bright-field background.
(B) Immunoblotting of the cytosolic and nuclear fractions shows additional evidence that ABA enhances the cytosolic distribution of WRKY40. The experiments were repeated three times with the similar results. ABA treatment was done by spraying the wild-type Col plants with 100 μM (±)ABA 6 h before the cell extracts were prepared from the plants. The symbols – and + indicate ABA-free and ABA treatment, respectively. Protein amounts were evaluated by scanning the protein bands, and relative band intensities, normalized relative to the intensity with the value from the sample of the ABA-free treatment (as 100%), are indicated by numbers above the bands. For the total and cytosolic proteins, Actin was used as a loading control, and for the nuclear fraction, Histone H3 was used as a control. Bottom panel: immunoblotting assays to test the purity of the cytosolic and nuclear fractions. Histone H3 (nuclear marker) and PEPC (phosphoenolpyruvate carboxylase; cytosolic marker) were tested in the cytosolic and nuclear fractions and total proteins.
(C) Colocalization of ABAR (tagged by GFP) and WRKY40 (tagged by RFP/mCherry) in the Arabidopsis protoplasts that transiently coexpressed the constructs and were treated with 2 μM (±)ABA 2 h before observation. Images were taken with identical parameters to allow comparison of fluorescence intensities.
(D) WRKY40 is predominantly localized in nucleus in the ABA-deficient mutant aba2 cells (without ABA treatment, indicated by WRKY40 - ABA), and ABA treatment (WRKY40 + ABA) restores the cytoplasmic distribution of this protein in the mutant cells. ABAR (aba2) indicates the unchanged ABAR localization in the aba2 cells. The bottom panels show the control images in the protoplasts from the wild-type Col plants (without ABA treatment, indicated by WRKY40 - ABA). “+ ABA” indicates that the transgenic protoplasts were treated with 2 μM (±)ABA 2 h before observation. GFP, Auto, Bright, and Merged indicate fluorescence of the ABAR-GFP or WRKY40-GFP fusion protein, chlorophyll autofluorescence, bright-field, and merged image of GFP and Auto in the bright field, respectively. Images were taken with identical parameters to allow comparison of fluorescence intensities.
(E) Immunoblotting of the cytosolic and nuclear fractions, performed in the ABA-deficient mutant aba2 cells with the same procedures as used in Figure 4B for the wild-type plants, shows additional evidence that ABA is important for the cytosolic distribution of WRKY40. The experiments were repeated three times with the similar results. The symbols – and + indicate ABA-free and ABA treatment, respectively. Protein amounts were evaluated by scanning the protein bands, and relative band intensities, normalized relative to the intensity with the value from the sample of the ABA-free treatment (as 100%), are indicated by numbers above the bands. For the total and cytosolic proteins, Actin was used as a loading control, and for the nuclear fraction, Histone H3 was used as a control. Bottom panel: the same assays performed as in (B) to test the purity of the cytosolic and nuclear fractions.
ABA Stimulates ABAR–WRKY40 Interaction
In the yeast two-hybrid system, ABAR interacts with WRKY40/18/60 without supplementation with ABA (Figures 3A and 3B), suggesting that this bimolecular interaction occurs in the absence or low levels of ABA. We used a combination of yeast two-hybrid and a yellow fluorescence protein (YFP) bimolecular fluorescence complementation (BiFC) system and biochemical approaches to investigate this possible ABA-responsive phenomenon. We found that, in the yeast two-hybrid system, the exogenous ABA application specifically promoted ABAR–WRKY40 interaction as tested by yeast growth (Figure 5A) and α-galactosidase activity (Figure 5B). By contrast, two ABA inactive/less active isomers, (–)ABA and trans-ABA, had no significant effect (Figure 5B), indicating the specificity of the physiological active form, (+)ABA, for this stimulation. The BiFC assays, confirming ABAR–WRKY40 interaction in vivo occurring in the surface of chloroplasts (Figure 5C, a and b; see Supplemental Figures 4C to 4E online) showed that the exogenous ABA treatment enhanced apparently ABAR–WRKY interaction in vivo (Figure 5C, a and b). The CoIP assays showed clearly that, with the exogenous ABA treatment, the WRKY40 was more efficiently precipitated by ABAR (Figure 5D, a), and, similarly, ABAR was more efficiently precipitated with WRKY40 (Figure 5D, b). These data showed consistently that ABA stimulates ABAR–WRKY40 interaction.
Figure 5.
ABA Promotes ABAR–WRKY40 Interaction.
(A) ABA stimulates ABAR–WRKY40 interaction in a yeast two-hybrid system with the drop test assay, which is displayed by promoting growth of the yeast cells harboring ABAR plus WRKY40. Yeast cells were incubated in the SD medium lacking Leu, Trp, His, and Ade and containing 0, 2, or 8 μM and were observed 2 to 3 d after the incubation. The yeast line harboring positive control vectors BD-53 plus AD-T is taken as a control.
(B) ABA stimulates the ABAR–WRKY40 interaction in yeast two-hybrid system, which is indicated by enhancing α-gal activity of the yeast lines harboring both ABAR and WRKY40. Treatments with water or ethanol (for solubilizing ABA) are two negative controls. α-Gal activity is presented as relative units (%), normalized relative to the activity of the water control. Each value is the mean ± se of five independent biological determinations.
(C) ABA stimulates the interaction of ABAR and WRKY40 tested in vivo by BiFC. (a) Protoplasts were transformed with the construct pairs ABAR-YFPN plus WRKY40-YFPC or ABAR-YFPC plus WRKY40-YFPN or ABAR-YFPN plus YFPC (as a negative control) and treated with 0 or 2 μM (±)ABA 2 h before observation. + ABA indicates the 2 μM ABA treatment, and absence of the symbol “+ ABA” indicates the ABA-free treatment. (b) Protoplasts transformed with the construct pairs ABAR-YFPC plus YFPN or YFPN plus WRKY40-YFPC or YFPC plus WRKY40-YFPN or YFPN plus YFPC were used as additional negative controls. No fluorescence signal was detected in the transgenic control protoplasts. Images were taken with identical parameters to allow comparison of fluorescence intensities.
(D) CoIP assays show that ABA promotes ABAR–WRKY40 interaction. (±)ABA at 0.1 μM was added to the isolated total protein extract and incubated for 4 h at 4°C before the immunoprecipitated complexes (complexes of the protein A-agrose-immunoprecipitated proteins) were separated from the CoIP system. (a) Immunoprecipitation (IP) with anti-ABAR serum and immunoblotting (Blot) with both sera, and (b) IP with anti-WRKY40 serum and Blot with both sera. The symbols – and + indicate ABA-free and ABA treatment, respectively. Protein amounts were evaluated by scanning the protein bands, and relative band intensities, normalized relative to the intensity with the value from the sample of the ABA-free treatment (as 100%), are indicated by numbers below the bands. The experiments were repeated three times with the similar results.
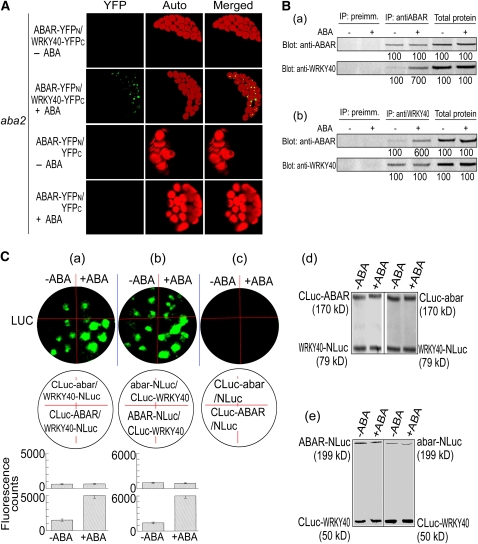
In the BiFC system, the ABAR–WRKY40 interaction was not detected in the aba2 cells, but exogenous ABA could partly restore this interaction in the mutant cells (Figure 6A; see Supplemental Figure 4 online). The CoIP assays, performed with the same procedures as mentioned above for wild-type plants, confirmed this observation (Figure 6B, a and b). These findings, together with the observations that ABA recruits WRKY40 from the nucleus to the cytosol (Figure 4), indicate that ABA is required for the WRKY40 molecule migration from the nucleus to the cytosol, where it interacts with the cytosolic portion of ABAR.
Figure 6.
ABA Is Important for ABAR–WRKY40 Interaction, and the Promoting Effect of ABA on ABAR–WRKY40 Interaction Requires Function of ABAR.
(A) Interaction of ABAR and WRKY40 in the ABA-deficient mutant aba2 cells. ABAR-YFPN/WRKY40-YFPC –ABA, BiFC in the absence of exogenous ABA treatment; ABAR-YFPN/WRKY40-YFPC +ABA, BiFC with 2 μM ABA treatment 2 h before observation. The protoplasts transformed with the construct pair ABAR-YFPN/YFPC with 0 μM (– ABA) or 2 μM ABA (+ ABA) treatment were taken as negative controls. Images were taken with identical parameters to allow comparison of fluorescence intensities.
(B) Immunoprecipitation (IP) assays in the aba2 plants show that ABA is required for the ABAR–WRKY40 interaction. (±)ABA at 0.1 μM was added into the isolated total protein for an incubation of 4 h at 4°C before the IP assays were conducted. (a) IP with anti-ABAR serum and immunoblotting (Blot) with both sera, and (b) IP with anti-WRKY40 serum and Blot with both sera. The symbols – and + indicate ABA-free and ABA treatment, respectively. Protein amounts were evaluated by scanning the protein bands, and relative band intensities, normalized relative to the intensity with the value from the sample of the ABA-free treatment (as 100%), are indicated by numbers below the bands. The experiments were repeated three times with the similar results.
(C) Promoting effect of ABA on ABAR–WRKY40 interaction requires ABAR function. Tobacco leaves were transformed with the construct pairs CLuc-abar/WRKY40-NLuc and CLuc-ABAR/WRKY40-NLuc (a), abar-NLuc/CLuc-WRKY40 and ABAR-NLuc/CLuc-WRKY40 (b), or CLuc-abar/NLuc and CLuc-ABAR/NLuc ([c]; as a negative control). The leaves were observed for fluorescence imaging 6 h after the (±)ABA infiltration (0 μM, indicated by −ABA, or 80 μM, indicated by +ABA) by a needleless syringe. The term abar denotes the ABAR gene harboring the cch mutation. Top panels in (a) to (c) show fluorescent images (LUC); middle panels show corresponding locations of transformation in the leaf with the given construct pairs. The bottom panels show the corresponding quantitative data (each value is the mean ± se with five independent determinations): top and bottom columns in (a), CLuc-abar/WRKY40-NLuc and CLuc-ABAR/WRKY40-NLuc, respectively; top and bottom columns in (b), abar-NLuc/CLuc-WRKY40 and ABAR-NLuc/CLuc-WRKY40. The amounts of the expressed Luc proteins were assayed by immunoblotting with the goat anti-full-length firefly Luc antibody, which detect the N- and C-terminal firefly Luc fragments. The Luc amounts were used to assess the protein amounts of ABAR, abar, and WRKY40 in the tobacco leaves, and the data presented in (d) correspond to the assays in (A), and those presented in (e) correspond to (b). All the assays were repeated five times with similar results.
We then tested whether the ABAR–WRKY40 interaction is dependent on functional ABAR. The cch mutation in ABAR, which results in ABA insensitivity (Shen et al., 2006; Wu et al., 2009), substantially disrupted the responsiveness of the ABAR–WRKY40 interaction to ABA in the firefly LCI system (Figure 6C, a to c). The ABA treatment did not significantly affect the protein levels of the ABAR and mutated ABAR harboring the cch mutation (Figure 6C, d and e; estimated by the amounts of the truncated-Luc reporter protein), indicating that the disruption of the ABA responsiveness of the cch-mutated ABAR was not caused nonspecifically by alteration of its protein levels in the LCI system. These data indicate that ABA-stimulated ABAR–WRKY interaction requires the function of the ABAR-mediated signaling.
WRKYs Cooperate to Negatively Regulate ABA Signaling Where WRKY40 Plays a Central Role
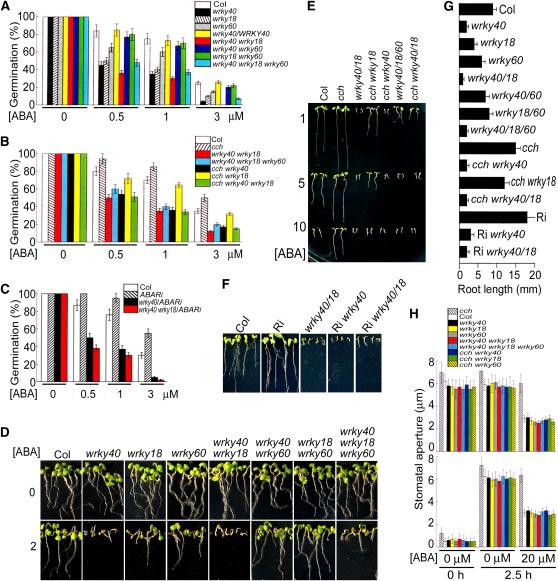
The WRKY domain proteins are a superfamily of transcription factors with up to 100 representatives in Arabidopsis, and the family members appear to be involved in the regulation of plant development and pathogen defense (Eulgem et al., 2000; Ulker and Somssich, 2004; Pandey and Somssich, 2009). Several WRKY proteins were suggested to be involved in ABA signaling in creosote bush (Larrea tridentata; Zou et al., 2004), rice (Oryza sativa; Xie et al., 2005, 2006), and barley (Xie et al., 2007). Most recently, genetic evidence was provided for the involvement of the Arabidopsis WRKY2 transcription factor in regulation of ABA signaling (Jiang and Yu, 2009). The subgroup of the WRKY40/18/60 transcription factors was previously shown to modulate cooperatively plant defense (Xu et al., 2006). In this work, we observed that all the knockout mutants wrky40, wrky18, and wrky60 showed ABA hypersensitive phenotypes in ABA-induced inhibition of seed germination and ABA-induced postgermination growth arrest, with the intensity of the phenotypes wrky40 > wrky18 > wrky60 (Figures 7A, 7D, and 7G). The introduction of the native promoter-driven cDNAs of the WRKY40, WRKY18, and WRKY60 into the wrky40, wrky18, and wrky60 mutants, respectively, rescues the ABA sensitivities of these mutants, demonstrating that these mutations are responsible for the observed ABA hypersensitive phenotypes (see Supplemental Figure 5 online).
Figure 7.
Disruption of WRKYs Results in ABA Hypersensitivity and Suppresses ABA Insensitivity of ABAR Mutants.
(A) The null mutation in WRKY40, WRKY18, or WRKY60 confers ABA hypersensitivity in ABA-induced inhibition of seed germination. White columns, wild-type Col; black, wrky40; hatched, wrky18; gray, wrky60; yellow, native promoter, WRKY40-wrky40 (a complemented line); red, wrky40 wrky18; blue, wrky40 wrky60; green, wrky18 wrky60; cyan, wrky40 wrky18 wrky60. Each value is the mean ± se of five independent biological determinations.
(B) The mutations of wrkys are epistatic to the cch mutation of ABAR gene in ABA-induced inhibition of seed germination. White columns, wild-type Col; hatched, cch; red, wrky40 wrky18; cyan, wrky40 wrky18 wrky60; black, cch wrky40; yellow, cch wrky18; green, cch wrky40 wrky18. Each value is the mean ± se of five independent biological determinations.
(C) The ABAR-RNAi lines (hatched columns) show ABA-insensitive phenotypes in ABA-induced inhibition of seed germination, but introduction of the RNAi into the wrky40 (black) or wrky40 wrky18 (red) mutants does not affect the ABA hypersensitive phenotypes of these mutants in seed germination. White columns, wild-type Col. Each value is the mean ± se of five independent biological determinations. In (A) to (C), units of ABA concentration [(±)ABA] are μM, and the germination rate was recorded 48 h after stratification.
(D) The null mutation in WRKY40, WRKY18, or WRKY60 confers ABA hypersensitivity in ABA-induced postgermination growth arrest. 0 and 2 indicate seedling growth in the ABA-free medium and 2 μM (±)ABA-containing medium, respectively.
(E) The mutations of wrkys are epistatic to the cch mutation of ABAR gene in the ABA-induced postgermination growth arrest. Col, wild-type plants; wrky40/18, wrky40 wrky 18; wrky40/18/60, wrky 40 wrky18 wrky60. The medium contained 1, 5, or 10 μM ABA as indicated to the left.
(F) The ABAR-RNAi lines (Ri) show ABA-insensitive phenotypes in postgermination growth, but introduction of the RNAi into the wrky40 (Ri wrky40) or wrky40 wrky18 (Ri wrky40/18) mutants does not affect the ABA-hypersensitive phenotypes of these mutants in postgermination growth. Col, wild-type plants. 5 μM (±)ABA was applied in the medium.
(G) Root growth in the different mutant lines. 5 μM (±)ABA was applied in the medium. Each value is the mean ± se of five independent biological determinations. In (D) to (G), the seedlings were transferred from the ABA-free medium into the ABA-containing medium 48 h after stratification, and 7 (D) or 10 d ([E] to [G]) later, the growth was recorded.
(H) Introduction of the null mutation in WRKY40, WRKY18, or WRKY60 into the cch mutant rescues ABA sensitivity in stomatal movement. Top panel, ABA-induced stomatal closure; bottom panel, ABA-inhibited stomatal opening. Hatched columns, cch; white, Col; black, wrky40; yellow, wrky18; gray, wrky60; red, wrky40 wrky18; cyan, wrky40 wrky18 wrky60; blue, cch wrky40; green, cch wrky18; yellow-hatched, cch wrky60. 0 μM ABA at 0 h, initial stomatal aperture. Stomatal aperture was recorded 2.5 h after the treatment with 0 or 20 μM ABA. Each value is the mean ± se of five independent biological determinations.
The double mutant wrky40 wrky18 showed the strongest ABA hypersensitive phenotypes, which was followed by the wrky40 single mutant and the wrky40 wrky18 wrky60 triple mutant, both of which showed comparable ABA hypersensitivity (Figures 7A, 7D, and 7G). These data indicate that WRKY40 plays a more important role than the two other WRKYs in ABA signaling, consistent with its stronger interaction with ABAR (see Supplemental Table 1 online). The wrky60 mutation, however, repressed the ABA hypersensitive phenotypes of the wrky40 and wrky18 mutations to partly restore wild-type growth when combined with either of these two mutations (Figures 7A, 7D, and 7G), indicating the complexity of the WRKY-mediated signaling processes. A previous report showed that, among the three WRKYs, WRKY18 plays the most important role in plant defense (the wrky18 mutant was most resistant to the bacterial pathogen but most susceptible to the fungal pathogen), though WRKY40 binds DNA (via the TGAC W-box) most tightly, and that WRKY60 inhibits the binding of WRKY40 to W-box, acting likely as an antagonist to WRKY18/40 in plant defense (Xu et al., 2006). Similarly, this experiment showed that WRKY60 is a regulator to balance the WRKY40/WRKY18-mediated ABA signaling. Due to the essential role of WRKY40 in the WRKY-mediated ABA signaling, we focused further detailed analysis on WRKY40.
Neither of the wrky mutations changed the concentrations of endogenous ABA, ProtoIX, magnesium-protoporphyrin IX, or chlorophyll (see Supplemental Figure 6 online), indicating that WRKY-mediated ABA signaling is distinct from the processes of ABA metabolism and chlorophyll biosynthesis. The three WRKYs are expressed ubiquitously in different organ/tissues, including stomata (see Supplemental Figure 7 online), as is ABAR (see Supplemental Figure 8 online), supporting their cooperative roles at the whole-plant level.
Disruption of WRKYs Suppresses ABA Insensitivity of abar Mutants
No ABA-related phenotype in stomatal movement was observed in any of the wrky single mutants, double mutants, and even triple mutants (Figure 7H). However, we observed that the ABA sensitivity of the guard cells of the cch mutant, which has stomata strongly insensitive to ABA in their movement (Shen et al., 2006; Wu et al., 2009), was restored by introducing any of the wrky40, wrky18, or wrky60 single mutations into the cch mutant (Figure 7H). The mechanism underlying this phenomenon needs further study. In addition, we introduced the wrky40 and wrky18 single mutations into the two other abar mutant alleles (harboring point mutations), abar-2 and abar-3 (Wu et al., 2009), and the transgenic mutant ABAR-RNAi (RNA interference) lines (see Supplemental Figure 9 online), and the wrky40 wrky18 double mutation was introduced into the three abar mutant alleles, cch, abar-2, and abar-3 mutants and the ABAR-RNAi lines (see Supplemental Figure 9 online). All these mutants combining wrky and abar alleles showed ABA hypersensitivity in ABA-induced inhibition of seed germination and ABA-induced postgermination growth arrest, resembling their respective original wrky mutants (Figures 7B, 7C, and 7E to 7G; see Supplemental Figures 10A to 10D online). This reveals that the different wrky mutations suppress the ABA-insensitive phenotypes caused by the different mutations in ABAR gene. These genetic data argue that the WRKYs mediate ABA signaling downstream of ABAR.
ABA Inhibits WRKY40 Expression through ABAR-Mediated Signaling
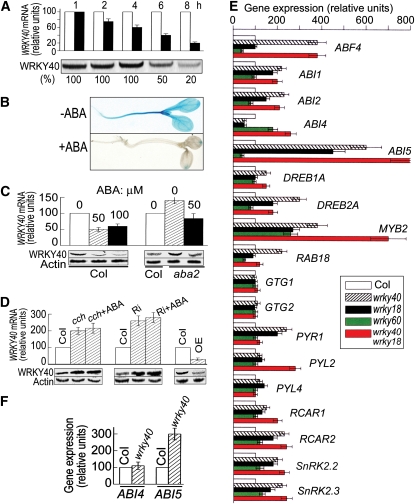
Next, we showed that the exogenous ABA treatment reduced levels of both WRKY40 mRNA (assayed by both promoter-β-glucuronidase (GUS) test and real-time PCR; Figures 8A to 8C) and protein (Figures 8A and 8C). In the ABA-deficient aba2 mutant cells, WRKY40 expression was upregulated at both mRNA and protein levels, and exogenous ABA application repressed this upregulation to restore the wild-type expression level of WRKY40 in the aba2 mutant (Figure 8C). In the ABA signaling mutant cch plants and ABAR-RNAi lines, WRKY40 expression was also upregulated as in the aba2 cells, but this upregulation could not been repressed to restore the wild-type expression level of WRKY40 by exogenous ABA application (Figure 8D). By contrast, WRKY40 expression was constitutively downregulated in the ABAR overexpressors (Figure 8D) as in the ABA-treated wild-type Columbia (Col) cells (Figures 8A to 8C). These findings indicate clearly that ABA-induced WRKY40 downregulation requires the ABAR-mediated ABA signaling.
Figure 8.
Characterization of Expression of WRKY40 and Other ABA-Responsive Genes.
(A) to (C) ABA inhibits WRKY40 expression.
(A) Time course of ABA-induced WRKY40 repression. Twenty-day-old seedlings of the wild-type Col were sprayed with 0 or 100 μM ABA and were sampled 1, 2, 4, 6, or 8 h after the ABA treatment. Top panel shows real-time PCR analysis (WRKY40 mRNA; white columns, the ABA-free treatment, and black columns, ABA treatment). The real-time PCR value obtained from the sample of the ABA-free treatment was taken as 100%, and the value from the sample of the ABA treatment was normalized relative to the ABA-free treatment value obtained at the same sampling time. Bottom panel shows immunoblotting with WRKY40. Protein amounts were evaluated by scanning the protein bands, and relative band intensities, normalized relative to the intensity with the value for the sample of the 1-h treatment (as 100%), are indicated by numbers below the bands.
(B) Three-day-old Col seedlings transformed by the WRKY40-promoter-GUS construct were transferred to medium containing 5 μM ABA (+ABA) or 0 μM ABA (−ABA) for another 3 d growth before observation.
(C) Twenty-day-old seedlings of the wild-type Col and aba2 mutant were sprayed with 0, 50, or 100 μM ABA as noted above bars and were sampled 6 h after the ABA treatment for real-time PCR analysis (top columns) and immunoblotting (bottom). Actin was used as a control.
(D) Inhibition of WRKY40 by ABA requires ABAR function. Twenty-day-old seedlings of Col, cch, ABAR-RNAi line (Ri), or an ABAR-overexpressing line (OE) were sprayed with 0 (Col, cch, Ri, and OE) or 50 μM ABA (cch+ABA and Ri+ABA) and sampled 6 h after the ABA treatment for real-time PCR analysis (top columns) and immunoblotting (bottom). Actin was used as a control. Each value is the mean ± se of five independent biological determinations.
(E) Expression of a set of ABA-responsive or signaling genes is altered in the wrky mutants (14-d-old seedlings). White columns, Col; hatched, wrky40; black, wrky18; green, wrky60; red, wrky40 wrky18. Each value is the mean ± se of five independent biological determinations.
(F) Expression of ABI4 and ABI5 in Col and wrky40 mutant 24 h after stratification in germinating seeds. Each value is the mean ± se of five independent biological determinations.
Disruption of WRKYs Alters Expression of a Set of ABA-Responsive Genes
Real-time PCR analysis showed that the expression of a set of ABA-responsive genes was altered in the wrky single and double mutant seedlings (Figure 8E). These genes include ABF4 (Kang et al., 2002), ABI1 (Gosti et al., 1999), ABI2 (Leung et al., 1997), ABI4 (Finkelstein et al., 1998), ABI5 (Finkelstein and Lynch, 2000), DREB1A (Liu et al., 1998), DREB2A (Liu et al., 1998), MYB2 (Abe et al., 2003), PYL2/RCAR13 (Park et al., 2009), PYR1/RCAR11 (Park et al., 2009), RAB18 (Lang and Palva, 1992), RCAR1/PYL9 (Ma et al., 2009), RCAR2/PYL7 (Ma et al., 2009), SnRK2.2 (Fujii and Zhu, 2009), and SnRK2.3 (Fujii and Zhu, 2009). However, the expression of the genes encoding the plasma membrane ABA receptors GTG1/GTG2 (Pandey et al., 2009) and PYL4/RCAR10 (a member of the ABA receptor PYR/PYL family; Park et al., 2009) was not changed in these wrky mutants (Figure 8E). The expression-altered genes were upregulated in the wrky mutants in most cases, especially in the wrky40 single mutant and wrky40 wrky18 double mutant seedlings (Figure 8E). It is noteworthy that ABI5 and MYB2 were most remarkably upregulated in the wrky40 wrky18 double mutant (Figure 8E). It is also notable that ABI4 expression was downregulated significantly in the wrky40 and wrky18 mutant seedlings, but upregulated in the wrky60 single mutant and wrky40 wrky18 double mutant (Figure 8E). All these genes are ABA positively responsive genes or positive ABA-signaling regulator genes except for ABI1 and ABI2, which encode two negative ABA signaling regulators (Gosti et al., 1999; Leung et al., 1997). However, the expression of ABI1 and ABI2 was also upregulated in the wrky40 and wrky18 single mutant and wrky40 wrky18 double mutant like the most positive ABA signaling regulator genes (Figure 8E).
It is interesting to observe that the expression of the genes involved in the PYR/PYL/RCAR ABA receptor–mediated signaling cascade, PYR1/RCAR11, PYL2/RCAR13, PYL9/RCAR1, PYL7/RCAR2, ABI1, ABI2, SnRK2.2, and SnRK2.3 (Fujii et al., 2009), was upregulated in some of the wrky mutants (Figure 8E). However, a recent report showed that ABA downregulates expression of PYR1/RCAR11, slightly upregulates expression of PYL2/RCAR13 and PYL9/RCAR1, and does not significantly alter expression of PYL7/RCAR2 (Szostkiewicz et al., 2010). Given that ABA represses expression of WRKY40 and, thus, disruption of the WRKY40 gene should mimic the effects of ABA, the expression of PYR1/RCAR11 and PYL7/RCAR2 should have been downregulated or not altered in these wrky mutants. These discrepancies implicate complexity of the ABA signaling network, and further studies will be necessary to assess if crosstalk occurs between the PYR/PYL/RCAR-mediated signaling and ABAR-mediated signaling. Also, the results suggest that a complex mechanism involving forward and reverse feedback effects may function, and particularly a tightly regulated cooperation among the three WRKYs should be important in this ABAR-WRKY–mediated signaling pathway, which is consistent with the above genetic findings.
We further assayed ABI4 and ABI5 expression in the germinating seeds of the wrky40 mutant because the two genes are expressed at low level in seedlings after germination (Finkelstein et al., 1998, 2002; Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001, 2002). The expression of ABI4 was not altered, but ABI5 was upregulated in wrky40 mutant (Figure 8F), suggesting that the regulation of ABI4 expression by WRKY40 is developmental stage dependent.
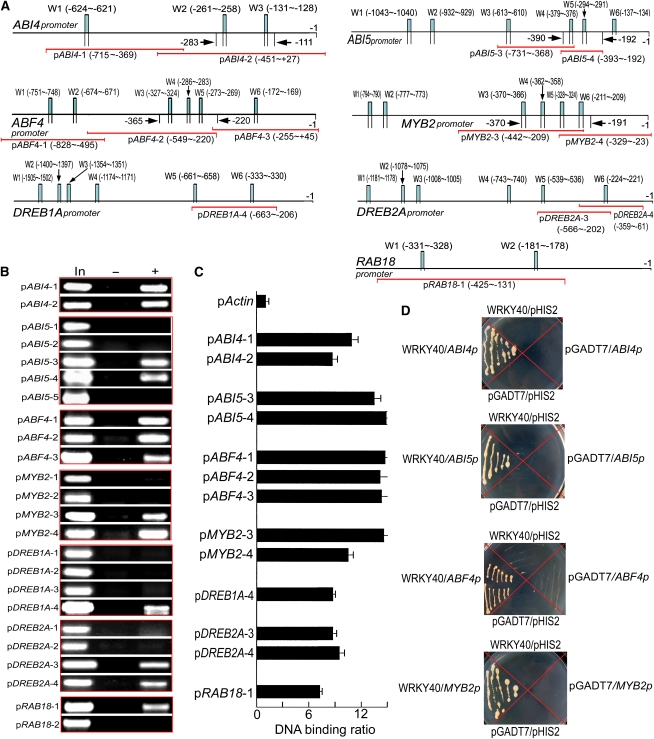
WRKY40 Binds W-Box of the Promoters of Several Important ABA-Responsive Genes
A search of the Arabidopsis genomic sequence showed that several ABA signaling genes have, in their promoter regions, a W-box sequence, the core of a cis-element to which the WRKY transcription factors bind. Among these ABA signaling regulators, ABF4 and ABI5 both belong to a class of basic leucine zipper transcription factors (Finkelstein and Lynch, 2000; Kang et al., 2002) and ABI4, DREB1A, and DREB2A to a class of Apetala-2 domain transcription factors (Finkelstein et al., 1998; Liu et al., 1998), MYB2 is a MYB-related transcription factor (Abe et al., 2003), and RAB18 is a rab-related protein (Lang and Palva, 1992). The three classes of transcription factors are considered to be some members of the most important ABA regulators in the ABA signaling framework (Finkelstein et al., 2002; Zhang et al., 2004). With chromatin coimmunoprecipitation (ChIP) analysis combined with PCR and quantitative real-time PCR, we showed that WRKY40 binds the promoters of all these genes via the core W-box sequence TGAC (Figures 9A to 9C; see Supplemental Table 2 online). We further confirmed these WRKY40-promoter interactions for the more important ABA-responsive transcription factors, ABF4, ABI4, ABI5, and MYB2, with both yeast one-hybrid system (Figure 9D) and gel shift assays (Figures 10A to 10D). Together with the observations that the expression of these transcription factor-encoding genes was upregulated in the wrky knockout mutants as mentioned above (Figures 8E and 8F), these data indicate that WRKY40, binding to the W-box of the promoters, represses expression of many of the most important ABA signaling regulator genes. It is noteworthy, however, that, as mentioned above, the wrky40 and wrky18 single mutations did not upregulate the ABI4 gene expression, but the expression levels of the ABI4 gene expression were enhanced in the wrky60 single mutant and wrky40 wrky18 double mutant (Figure 8E). These observations indicate that the regulation of the ABI4 gene expression by WRKYs involves a complex mechanism involving cooperation among the three transcription factors that functions to repress ABI4 expression.
Figure 9.
Downstream Target Genes of WRKY40: ChIP and Yeast One-Hybrid Assays.
(A) The promoter structure of ABI4, ABI5, ABF4, MYB2, DREB1A, DREB2A, and RAB18 genes. W1, W2…denote each W-box numbered from left to right with sequence sites relative to the star code. Red lines indicate the sequences detected by ChIP assays described in (B). Arrows indicate the sequence fragments used in the gel shift assays described in Figure 10.
(B) WRKY40 interacts with the promoters of a set of ABA-responsive or signaling genes (ABI4, ABI5, ABF4, MYB2, DREB1A, DREB2A, and RAB18): PCR data from ChIP assay with the antibody against WRKY40N. In the promoter fragment names, prefix “p” indicates promoter. The sequences for each promoter fragment (indicated by the suffix number) are indicated in (A) and listed in detail in Supplemental Table 2 online. In, PCR product from the chromatin DNA; –, PCR product from ChIP with preimmune serum (as a negative control); +, PCR product from ChIP with the antibody against WRKY40N.
(C) WRKY40 interacts with the promoters of a set of ABA-responsive or signaling genes (ABI4, ABI5, ABF4, MYB2, DREB1A, DREB2A, and RAB18): real-time PCR data from ChIP assay with the antibody against WRKY40N with the Actin promoter (pActin) as a negative control. All the symbols for promoters present the same significances as described in (B).
(D) WRKY40 interacts with the promoters of ABA-responsive/signaling genes ABI4, ABI5, ABF4, and MYB2 in the yeast one-hybrid assay. The prey vector harboring WRKY40 (pGADT7-WRKY40, indicated by WRKY40) and the bait vector pHIS2 harboring different promoters (indicated by ABI4p, ABI5p, ABF4p, and MYB2p) were used to transform yeast cells. The transformation with empty vectors pGADT7 and pHIS2 was taken as negative controls. The experiments were repeated three times with the same results.
Figure 10.
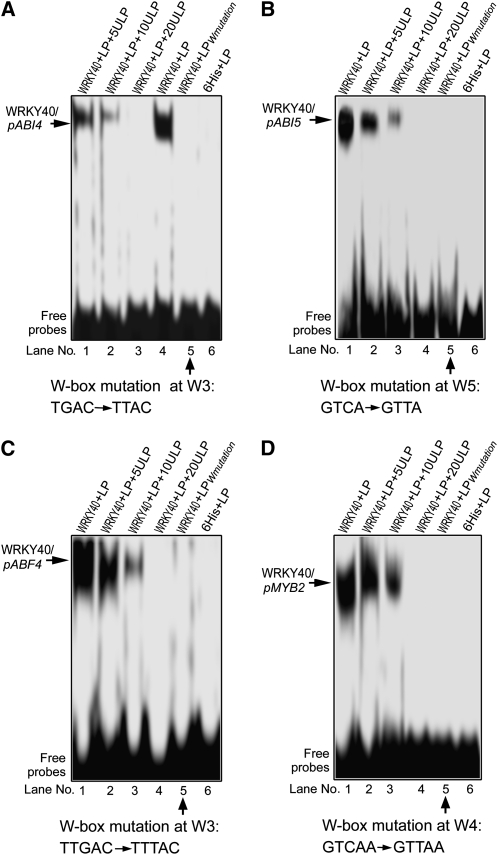
Analysis of Downstream Target Genes of WRKY40 by Gel Shift Assay.
Gel shift assay shows that WRKY40 binds the promoters of the ABA-responsive/signaling transcription factor genes ABI4 (pABI4) (A), ABI5 (pABI5) (B), ABF4 (pABF4) (C), and MYB2 (pMYB2) (D). LP, labeled probe; LPWmutation, labeled probe with the mutated W-box as indicated below the panels, which was used as a negative control; 6His, 6His tag peptide fused to WRKY40 protein, which was used as another negative control. W3 ([A] and [C]), W4 (D), and W5 (B) indicate the positions of the W-box as described in Figure 9A. ULP, unlabeled probe. 5ULP, 10ULP, and 20ULP indicate 5-, 10-, and 20-fold unlabeled probe addition, respectively. The probe sequences are listed in detail in Supplemental Table 2 online. The experiments were repeated five times with the same results.
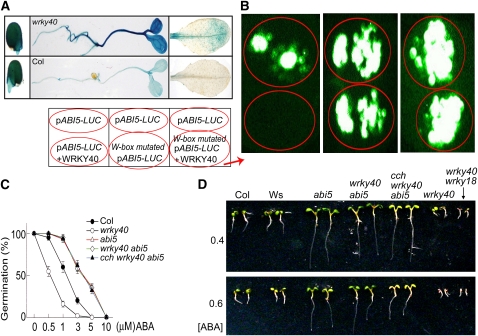
WRKY40 Directly Represses ABI5 Expression
Next, we focused the analysis on ABI5 because, among the other three transcription factors, ABI4 appears to function in a more complicated manner in the WRKY-mediated signaling according to the gene expression analysis (Figure 8). Also, disruption of ABF4 or MYB2 has no ABA-related phenotypes due to functional redundancy. ABI5 is one of the most important and genetically well-characterized ABA-signaling regulators that control seed germination and postgermination growth (Finkelstein and Lynch, 2000; Finkelstein et al., 2002). We thus analyzed the function of the WRKY40-promoter interaction in vivo. First, we used transformation of the ABI5 promoter-driven GUS to confirm the observation by real-time PCR analysis (Figure 8). Consistently, the wrky40 mutation was shown to enhance remarkably the expression level of ABI5 (Figure 11A). Next, we investigated whether WRKY40 directly represses ABI5 expression by coexpression of WRKY40 and ABI5 promoter in a heterologous system. In the tobacco (Nicotiana tabacum) leaves cotransformed with both WRKY40 and ABI5 native-promoter-Luc constructs, WRKY40 specifically inhibited expression of ABI5 (Figure 11B). Introduction of abi5 mutation into the wrky40 and cch wrky40 mutants resulted in the ABA-insensitive phenotypes in seed germination and postgermination growth, thus suppressing the ABA hypersensitivity of the wrky40 and cch wrky40 mutants (Figures 11C and 11D). Taken together, our findings consistently show that WRKY40 binds directly to the ABI5 promoter to repress ABI5 gene expression; thus, ABI5 functions directly downstream of WRKY40 in the ABAR-WRKY40–mediated ABA signaling.
Figure 11.
Identification of ABI5 as a Direct Target of WRKY40.
(A) ABI5-promoter-driven GUS expression in germinating seeds (left), 3-d-old seedlings (middle), and mature leaves (right) in the wrky40 mutant (top) and wild-type Col (bottom), showing that the wrky40 mutation significantly enhances ABI5 expression.
(B) WRKY40 inhibits the promoter activity of ABI5 in vivo. The tobacco leaves were transformed with pABI5-LUC alone and pABI5-LUC plus WRKY40 (left panel) or with pABI5-LUC alone and W-box-mutated pABI5-LUC alone (middle panel) or with pABI5-LUC alone and W-box-mutated pABI5-LUC plus WRKY40 (right panel). Prefix “p” indicates promoter. Note that WRKY40 abolishes pABI5-LUC expression.
(C) abi5 mutation is epistatic to wrky40 mutation in ABA-induced inhibition of seed germination. The germination rates were recorded 72 h after stratification. Each value is the mean ± se of five independent biological determinations.
(D) abi5 is epistatic to wrky40 in ABA-induced postgermination growth arrest. Two wild-types, Col and Wassilewskija (the background of the abi5-1mutant) were used as controls. Seeds were directly planted in medium containing either 0.4 or 0.6 μM ABA and photographs were taken 7 d after stratification.
DISCUSSION
ABAR Is a Transmembrane Protein Spanning the Chloroplast Envelope
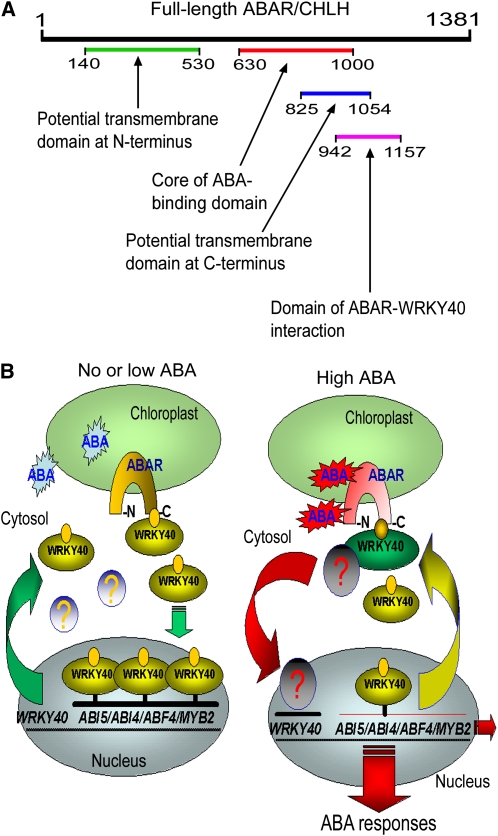
Using molecular and biochemical combined approaches, we showed that ABAR is a transmembrane protein that spans the chloroplast envelope, exposing its N and C termini to the cytosolic side in cells (Figures 1 and 2). The transient expression assays indicate the occurrence of C-terminal transmembrane domains between amino acid residues 770 and 1000 of ABAR (Figure 2C), essentially consistent with a transmembrane prediction model suggesting both N-terminal (amino acids 140 to 530) and C-terminal (amino acids 825 to 1054) transmembrane domains (Figure 2D; see Supplemental Figure 1 online). However, the C-terminal deletion with an intact N-terminal half (amino acids 1 to 658) results predominantly in the localization of ABAR (Figure 2C) to the stroma, indicating that the C-terminal half may cooperate with the N-terminal half to determine the intracellular localization of this protein. It is noteworthy that a portion of the ABAR protein may reside in the stroma when the concentration of Mg++ is lower (1 mM, for example; Figure 1D). The mechanism underlying this movement from envelope membranes to stroma remains unknown. It might be caused by a endocytosis-like mechanism involving chloroplast membrane trafficking within cells in response to low Mg2+ stress. Nevertheless, the transmembrane location of ABAR from chloroplast to cytoplasm provides the possibility of ABA signaling across the chloroplast envelope. A model for the functional domains in the ABAR molecule is proposed in Figure 12A, which shows, in a clearer manner, that key functional domains reside in the C-terminal half of the protein exposed to cytosolic side.
Figure 12.
Proposition of an ABAR-Mediated Signaling Model.
(A) A model of functional domains in ABAR molecule. Numbers of the amino acid residues are shown in the molecule. See text for the model.
(B) A model of ABAR-mediated signaling pathway. The symbol <?> indicates an unknown factor or signaling cascade that may repress the WRKY40 gene expression. Note that ABAR interacts with WRKY40 in the absence or low levels of ABA, and ABA at high levels promotes this bimolecular interaction and triggers downstream signaling cascade. See text for this model in detail.
ABAR Antagonizes Negative ABA Signaling Regulators WRKYs to Derepress ABA-Responsive Genes
Using a yeast two-hybrid assay, we identified a group of the WRKY transcription factors, WRKY40, WRKY18, and WRKY60, as interaction partners of ABAR. We also characterized the interaction of ABAR with WRKY40 using a combination of yeast two-hybrid system, CoIP in yeast and in planta, LCI, and YFP BiFC (Figures 3, 5, and 6). Furthermore, we demonstrate that an ABA-ABAR-WRKY40-ABI5–linked signaling cascade from the primary signaling event to downstream gene expression operates in plant cells essentially from the following evidence: first, we showed, by the same approaches as mentioned above, that the ABAR–WRKY40 interaction requires stimulation by ABA, and the bimolecular interaction depends on function of the ABAR-mediated signaling (Figures 5 and 6); second, the movement of the transcription factor WRKY40 from the nucleus to the cytoplasm occurs and is essential for the ABAR–WRKY40 interaction, and ABA is required for this molecular trafficking (Figure 4); third, we showed that ABA represses the WRKY40 expression, and this transcription repression depends on the function of ABAR-mediated signaling (Figure 8); fourth, we provided genetic evidence that the three WRKY transcription factors, WRKY40, WRKY18, and WRKY60, cooperate to negatively regulate ABA signaling downstream ABAR (Figure 7; see Supplemental Figure 10 online); and fifth, using the combined approaches of ChIP, yeast one-hybrid assay, gel shift assay, coexpression of the transcription factor and the potential target promoter, and genetic analysis, we identified an important ABA signaling regulator, ABI5, as a direct downstream signaling component of WRKY40 (Figures 8 to 11).
Thus, we establish a model to reveal a missing link in the ABAR-mediated ABA signaling pathway between the primary signaling events to downstream gene expression. In this model, ABAR spans the chloroplast envelope, exposing its N and C termini to the cytosol. The cytosolic C terminus of ABAR interacts with a group of WRKY transcription factors, WRKY40, WRKY18, and WRKY60, that negatively regulate ABA signaling. WRKY40, a central regulator in the WRKYs-mediated ABA signaling, inhibits expression of ABA-responsive genes, such as ABI5. In response to a high level of ABA that recruits WRKY40 from the nucleus to the cytosol and promotes ABAR–WRKY40 interaction, ABAR relieves ABI5 gene of inhibition by downregulating WRKY40 expression to induce physiological responses (Figure 12B). In this model, however, an unknown factor or signaling cascade may be involved in the repression of WRKY40 expression in response to ABA after the interaction between ABAR and WRKY40 occurs (Figure 12B). This needs further studies to assess the identity of this (these) important signaling component(s).
ABAR functions as the most upstream component in this signaling pathway, consistent with the idea that ABAR acts as an ABA receptor to sense the ABA signal. How ABAR perceives the ABA signal remains unknown. ABAR was initially isolated by an ABA affinity chromatography column to which ABA binds through its carboxylic group (Zhang et al., 2002); later, this affinity column was used as an alternative technique to detect ABA binding activity to ABAR (Wu et al., 2009). This suggests that ABA could bind to ABAR with its carboxyl fixed to the chromatography column. However, it has been known that the carboxyl of ABA is important to its binding to the PYR/PYL//RCAR receptor (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009; Yin et al., 2009). It remains mysterious what role the carboxyl of ABA plays in the ABA binding to ABAR. Based on our previous studies, we consider that ABAR may bind ABA but probably with a decreased affinity when its carboxyl is partly inactive (i.e., fixed to a chromatography column), and so we do not exclude the possibility that the ABA molecule having a free carboxyl binds ABAR more tightly. The progress in the ABAR structure studies will answer these open questions.
Is the ABAR-WRKY–Coupled Signaling Pathway Conserved in the Monocotyledonous Plants?
Interestingly, using both the yeast two-hybrid assay and LCI in vivo system in tobacco leaves, we showed that the homolog of the Arabidopsis ABAR in barley, XanF, also interacts with the Arabidopsis WRKY40, WRKY18, and WRKY60 (see Supplemental Figures 11A and 11B and Supplemental Methods online). The interaction of the barley XanF with WRKY40 was shown to be promoted by ABA treatment in both yeast cells and tobacco leaf tissues (see Supplemental Figures 11C to 11E and Supplemental Methods online). As a positive control, ABA treatment stimulated significantly the interaction of PYL1 (a member of the PYR/PYL ABA receptors) with its interaction partner ABI2 (Ma et al., 2009; Park et al., 2009) in the LCI system (see Supplemental Figure 11D and Supplemental Methods online), demonstrating the reliability of this LCI system. Most importantly, we observed that the expression of barley XanF in wild-type Arabidopsis plants confers ABA hypersensitivity in seed germination, postgermination growth, and stomatal movement (see Supplemental Figures 12A to 12E, 12G, and 12H and Supplemental Methods online), and the XanF expression in the cch mutant plants rescues the ABA sensitivity of this mutant in all the major ABA responses (see Supplemental Figures 12A to 12D, 12F to 12H, and Supplemental Methods online). Taken together with our previous observations that the barley Xan-F binds ABA (Wu et al., 2009), all the data indicate that the barley XanF may function in ABA signaling in Arabidopsis, and further studies are needed to assess if an ABA signaling pathway similar to the Arabidopsis ABAR-WRKY–coupled signaling pathway functions in the monocotyledonous plants, such as rice and barley.
Significance of Cytosolic Exposure of the C and N Termini of Chloroplast ABAR in Plant Cell Signaling
Exposure of its C and N termini to the cytosol enables the chloroplast ABAR to interact with cytosolic-nucleus proteins, WRKYs, to transmit ABA signal to the nucleus. Consistent with this model, we previously observed that the C-terminal fragments of ABAR function in the cytosol to induce ABA hypersensitivity in wild-type plants and to restore ABA sensitivity in the cch mutant without involvement in chlorophyll biosynthesis (Wu et al., 2009), suggesting that the C terminus of ABAR may sense the ABA signal in cytosol at least partly independently of chloroplast. A truncated C-terminal half of CHLH/ABAR exists in the monocotyledonous plants, such as rice, which has two copies of the CHLH/ABAR gene, one coding for full-length protein and another for the C-terminal half. This C-terminal half may function in ABA signaling, though is likely expressed in cytosol because of lack of transit peptide (Wu et al., 2009). The previously reported null ABA-related phenotypes of the knockdown mutants of the full-length Xan-F gene in barley (Muller and Hansson, 2009) may most likely be due to the functional redundancy: a truncated C-terminal half of XanF encoded by a small XanF gene may occur in cytosolic space like in rice (Wu et al., 2009), and this cytosolic, small XanF may function in ABA signaling but not in chlorophyll biosynthesis. A negative result of DNA gel blot analysis for the second copy of the XanF gene (Muller and Hansson, 2009) could not exclude the possibility of occurrence of a C-terminal truncated copy of XanF in barley likely due to technical limitation because, for example, the second copy of the rice CHLH was not detected by DNA gel blot in a previous study (Jung et al., 2003), but this small CHLH copy does exist in the rice genome as determined by genomic sequence analysis.
It remains mysterious why plants have evolved a chloroplast protein for an intracellular ABA receptor. The chloroplast ABAR may function as an efficient signaling mechanism to sense ABA signal both from chloroplast that is an intracellular ABA pool and from cytosol. Additionally, the cytosolic C and N termini of ABAR may be of significance in chloroplast-to-nucleus retrograde signaling. It remains unclear how the chloroplast retrograde signal traverses chloroplast envelope (Nott et al., 2006). As a key player in this chloroplast signaling process, may ABAR/GUN5 transmit chloroplast retrograde signal with its C and N termini across chloroplast envelope to the nucleus?
Functions of a Group of WRKY Transcription Factors as Upstream ABA Signaling Regulators
We provide biochemical, cellular, and genetic evidence to identify a group of WRKY transcription factors (WRKY40, WRKY18, and WRKY60), which function as negative regulators of ABA signaling directly downstream of the ABA receptor ABAR. These WRKY transcription factors directly regulate a set of ABA-responsive transcription factors, such as ABI4, ABI5, ABF4, and MYB2, that, in turn, regulate many key genes involved in ABA-induced physiological responses. This indicates that these WRKYs functions upstream of ABA signaling, consistent with their identity of the direct ABA receptor-interaction partners involved in the primary events of ABA signaling. The WRKY transcription factors are encoded by a superfamily of genes (Eulgem et al., 2000), and other members, besides the three WRKYs, may interact with ABAR to coordinate complex ABA signaling. It will be of interest to identify other WRKY proteins involved in ABA signaling to reveal the mechanism of the ABAR-mediated ABA signaling coordination. Currently, ABA also is believed to be an essential signal to regulate plant defenses against pathogens (Adie et al., 2007), and WRKY transcription factors, including WRKY40/18/60, have been shown to be a class of important regulators of plant defense signaling (Ulker and Somssich, 2004; Xu et al., 2006; Pandey and Somssich, 2009). It will be of interest to assess if and how the ABA-ABAR-WRKY40 pathway functions in the crosstalk between ABA and defense signaling in plant cells.
Finally, it is noteworthy that, in this ABA-ABAR-WRKY40-ABI5 model, the underlying mechanisms of several important steps remain open questions. Where is the ABA signal perceived by ABAR, within the chloroplast, in the cytosolic side, or in both sides? By what mechanisms does ABA recruit WRKY40 from the nucleus to the cytosol, and by what mechanism is the WRKY40 expression downregulated? Further research to answer these questions will shed new light on ABAR-mediated ABA signal transduction.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used to generate transgenic plants. An ABAR gene (At5g13630) fragment, encoding a truncated ABAR with 1 to 1303 amino acid residues, was introduced into Col plants as a GFP fusion protein. Because overexpression of the full-length ABAR generally induced cosuppression of ABAR gene, and this truncated ABAR-GFP fusion protein functions similarly to full-length ABAR in plants when it is overexpressed, leading to ABA hypersensitivity in all the three ABA major responses (Wu et al., 2009), we used the truncated ABAR overexpression lines as ABAR overexpressors instead of overexpression of full-length ABAR. The cDNA isolation and transgenic manipulation were as previously described (Wu et al., 2009). The ABAR-RNAi lines of ABAR gene were generated with the same procedures as described previously (Shen et al., 2006) using Col ecotype. Briefly, a gene-specific 653-bp fragment, amplified by PCR, located downstream 2363 to 3015 bp of the start codon was used as the sense arm, and a 596-bp fragment, located downstream 2420 to 3015 bp of the start codon, was used as the antisense arm. The PCR fragments were ligated into pBI121 vector (13.0 kb; Clontech) under the control of the cauliflower mosaic virus (CaMV) 35S promoter, which was used to transform wild-type Col plants. The homologous T3 generation seeds or plants were used for analysis. At least 10 transgenic lines were obtained for each construct, and all the lines had similar ABA-related phenotypes. The results from one representative line were presented here. The cch mutant was a generous gift from J. Chory (The Salk Institute, La Jolla, CA). The seeds of the abar-2 (CS89100) and abar-3 (CS92346) mutants in the ABAR gene were obtained from the ABRC via the Arabidopsis TILLING (Targeting Induced Local Lesions in Genomes) project (Henikoff et al., 2004) and were characterized as previously described (Wu et al., 2009). The abar-2 allele was identified as an ABA-insensitive mutant in both seed germination and postgermination growth. The abar-3 mutant is hypersensitive to ABA in seed germination but insensitive to ABA in postgermination growth. Both alleles have no ABA-related phenotypes in stomatal movement. The cch, abar-2, and abar-3 mutants were isolated from the ecotype Col. The seeds of the abi5 (CS8105: abi5-1, with Wassilewskija ecotype as background) and aba2 (CS156: aba2-1, with Col ecotype as background) mutants were also obtained from ABRC.
The wrky40-1 (Stock number: ET5883, with Landsberg erecta ecotype as background) was obtained from Cold Spring Harbor Laboratory gene and enhancer trap lines and contains a Ds transposon inserted within the second exon of WRKY40 (Arabidopsis genomic locus tag: At1g80840). wrky18-1 (SALK_093916) and wrky60-1 (SALK_120706) are T-DNA insertion knockout mutants with a T-DNA insertion within the first exon, respectively, in WRKY18 (At4g31800) and WRKY60 (At2g25000) genes. Both mutants were isolated from Col ecotype. All the three mutants were previously identified as null alleles in their respective genes (Xu et al., 2006) and were confirmed in our laboratory by PCR genotyping. The wrky40-1 mutation was transferred from its Ler ecotype background into Col ecotype by backcrossing as previously described (Xu et al., 2006). The ABA-related phenotypes of the wrky40-1, wrky18-1, and wrky60-1 mutants were complemented by introducing into the mutant plants the WRKY40, WRKY18, and WRKY60 cDNAs, respectively, driven by their corresponding native promoters (∼1.1 kb for WRKY40, ∼1.3 kb for WRKY18, and ∼1.35 kb for WRKY60) that were amplified by PCR. Double and triple mutants were generated by genetic crosses and identified by PCR genotyping.
The ABAR-RNAi construct was used to transform directly the wrky40 and wrky40 wrky18 mutants to downregulate ABAR expression in these mutants because we observed that the ABAR-RNAi construct in background wild-type Col (or gl1; see Shen et al., 2006) was generally lost in the T4 RNAi lines, so we could not transfer the ABAR-RNAi construct from transgenic wild-type plants into the mutants by crossing. At least 10 RNAi lines for each mutant background were obtained. For comparing the RNAi lines of the mutant background with those of the wild-type Col background, the ABAR levels were checked by real-time PCR and immunoblotting to ensure that the RNAi lines of different backgrounds have similarly low levels of ABAR mRNA and protein product. The homologous T3 generation seeds or plants were used for analysis.
Plants were grown in a growth chamber at 19 to 20°C on Murashige and Skoog (MS) medium (Sigma-Aldrich) at ∼80 μmol photons m−2 s−1 or in compost soil at ∼120 μmol photons m−2 s−1 over a 16-h photoperiod.
Protein Production of ABAR, Truncated ABARs, WRKY40, and Truncated WRKY40 in Escherichia coli
We produced proteins of the full-length ABAR, several truncated ABARs (ABARN corresponding to N-terminal 258 amino acids from 53 to 310, ABARC corresponding to C-terminal 421 amino acids from 800 to 1220, and ABARM corresponding to a middle fragment 201 amino acids from 505 to 705), full-length WRKY40, and a truncated WRKY40 (WRKY40N corresponding to N-terminal 110 amino acids from 21 to 130) in E. coli essentially as described previously (Wu et al., 2009). The cDNAs encoding these proteins were amplified by PCR (see Supplemental Table 3 online for the primers). For the full-length open reading frame (ORF) of ABAR, ABARN, ABARC, ABARM, and WRKY40, the forward primers introduced an EcoRI restriction site and the reverse primers introduced a SalI restriction sites into the fragments, and the PCR products were digested and cloned into pET48b(+) (Novagen) (for the full-length ABAR, ABARM, and WRKY40) or pGEX-4T-1 (Novagen) (for ABARN and ABARC) between EcoRI and SalI sites. For WRKY40N, the forward primers introduced an EcoRI restriction site and the reverse primers introduced an XhoI restriction site into the fragment, and the PCR products were digested and cloned into pGEX-4T-1between EcoRI and XhoI sites. The fragments in the plasmids were sequenced to check for errors. The recombinant cDNAs were expressed in E. coli BL21 (DE3) (Novagen) strains as 6×His-tagged fusion proteins (for pET48b vector-harbored constructs) or glutathione S-transferase–tagged fusion proteins (for pGEX-4T-1 vector-harbored constructs). The E. coli strains containing the expression plasmids were grown at 37°C in 1 liter of Luria-Bertani medium containing 50 μg/mL kanamycin until the OD600 of the cultures was 0.6 to 0.8. Protein expression was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mM in a condition at 16°C with 150 rotations per minute. After 16 h, the cells were lysed and proteins purified on a Ni2+-chelating column (for proteins expressed by pET48b vector-harbored constructs) or Sepharose 4B (for proteins expressed by pGEX-4T-1 vector-harbored constructs) column as described in the manufacture's system manual.
Antiserum Production, Protein Extraction, and Immunobloting
The antisera against ABAR, ABARN, ABARC, ABARM, and WRKY40N were produced and tested for specificity as described previously (Wu et al., 2009). The extraction of the Arabidopsis protoplasts was performed essentially according to procedures described by Walter et al. (2004). The extraction of the Arabidopsis total protein from leaves or whole plants, SDS-PAGE, and immunoblotting were done essentially according to previously described procedures (Shen et al., 2006; Wu et al., 2009).
Immunohistochemical Detection of ABAR in Leaf Tissues
The Arabidopsis leaves were prepared as frozen sections (8 μm thick) and immediately fixed with 4% (w/v) paraformaldehyde at 4°C for 10 min. The tissues were then rinsed with PBS1 solution (0.12 M KH2PO4 and 0.017 M K2HPO4, pH 7.2) three times for 10 min each. After the sections were incubated in blocking buffer (PBS2 solution containing 6 mM Na2HPO4, 3.5 mM KH2PO4, and 2.6 mM KCl, pH 7.0, and supplemented with 0.1% [v/v] Tween 20, 1.5% [w/v] glycine, and 5% [w/v] BSA) overnight at 4°C, they were labeled with the anti-full-length ABAR antiserum diluted 200-fold in a PBS2 solution supplemented with 0.8% BSA for 2 h at 37°C. After extensive rinsing with PBS2, the samples were incubated in goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) antibody diluted 100-fold in a PBS2 solution supplemented with 0.8% BSA for 2 h at 37°C. The specimens were rinsed in PBS2 and mounted with 80% glycerol in PBS2 and observed under a confocal laser scanning microscope (Zeiss LSM 510 META). The fluorescence of FITC was pseudocolored in green and the autofluorescence of chloroplasts in red.
The specificity and reliability of the immunohistochemical assays were tested. The first control was to omit antiserum to test possible unspecific labeling of the goat anti-rabbit IgG-FITC antibody. The second control was to use rabbit preimmune serum instead of the rabbit antiserum to test the specificity of the antiserum. No substantial FITC signal was observed in either of these negative controls (Figure 1A), showing that the immunochemical detection was specific to ABAR. More than three repetitions of the control experiments were conducted for each sample.
Transient Expression in Arabidopsis Protoplasts and Stable Expression in Plants for Assaying Subcellular Localization
Transient expression in the Arabidopsis protoplasts was performed essentially with the procedures described by Walter et al. (2004). For the transient expression, ABAR and WRKY40 were tagged by GFP, and WRKY40 was also tagged by mCherry (a red fluorescence protein [RFP]). The two chloroplast inner (TIC21; Teng et al., 2006) and outer (OEP7; Lee et al., 2001) envelope markers were tagged by mCherry and used for observation of precise localization of ABAR. The corresponding cDNAs were amplified by PCR (see Supplemental Table 3 online for the primers). The cDNAs of ABAR, the ABAR fragments, WRKY40 (for WRKY40-mCherry fusion), and RBSC were driven by the CaMV 35S promoter and downstream tagged by GFP. The cDNAs of the two chloroplast envelope markers, TIC21 and OEP7, were also driven by CaMV 35S promoter but downstream tagged by mCherry (an RFP). Each of the 35S promoter-driven and GFP- or mCherry-tagged cDNAs was fused to the pMD 19-T vector (Takara) at the SphI (5′-end) and EcoRI (3′-end) sites. The cDNA of WRKY40 (for WRKY40-GFP fusion) was also linked to its genomic native promoter to be transiently expressed in protoplasts, and in this case, the WRKY40 native promoter was isolated using the forward primer 5′-AACTGCAGAGCCGTGTGGGCTTGACTTT-3′ and reverse primer 5′-GCTCTAGACGGTGGATCTTCTTC-3′ and cloned into the PstI (5′-end) and XbaI (3′-end) sites upstream of GFP in the pMD 19-T vector (Takara) in which the 35S promoter was replaced by the genomic native promoter of WRKY40. Protoplasts were isolated from the leaves of 3- to 4-week old plants of Arabidopsis (ecotype Col) or from leaves of the ABA biosynthesis mutant aba2 plants (for assaying ABAR and WRKY40 localization) and transiently transformed using polyethylene glycol essentially according to Sheen's protocol (http://genetics.mgh.harvard.edu/sheenweb/). Fluorescence of GFP or RFP was observed by a confocal laser scanning microscope (Zeiss LSM 510 META) after incubation at 23°C for 16 h. For assaying the effects of ABA treatment on the ABAR or WRKY40 distribution in cells, (±)ABA at 2 μM concentration was used to incubate the transformed protoplasts 2 h before the observation under the confocal laser scanning microscope.
For stable expression of the GFP-tagged ABAR and WRKY40 in Arabidopsis plants, the cDNA encoding ABAR and WRKY40, respectively, was cloned using the same primers as described above for transient expression in protoplasts. The cDNAs were cloned into the binary vector pCAMBIA1300 (Cambia) that contains the CaMV 35S promoter and a C-terminal GFP flag. These constructs were introduced into the GV3101 strain Agrobacterium tumefaciens and transformed into Arabidopsis Col plants by floral infiltration. The protoplasts were prepared from the 3- to 4-week-old homozygous T3 plants for observation under a confocal laser scanning microscope. For observation of WRKY40 in roots, the young roots of the 8- to 10-d-old seedlings were directly observed under the confocal laser scanning microscope. For assaying the effects of ABA treatment on the ABAR or WRKY40 distribution in cells, (±)ABA solution at 100 μM concentration was used to spray the 3- to 4-week-old homozygous T3 plants, and the leaves were sampled 6 h after the spraying. The protoplasts were isolated from the ABA-treated and nontreated plants for observation under the confocal laser scanning microscope.
Isolation of Intact Chloroplasts
Arabidopsis intact chloroplasts were isolated from ∼3-week-old healthy plants, essentially as described previously (Aronsson and Jarvis, 2002) with modifications. All the procedures were performed at 4°C. Briefly, leaves were homogenized in the isolation buffer consisting of 330 mM sorbitol, 5 mM MgCl2, 2 mM EDTA, 1 mM MnCl2, and 50 mM HEPES/KOH, pH 7.8. The homogenate was filtered and centrifuged, and the pellet was resuspended in 100 μL suspending buffer composed of 330 mM sorbitol, 5 mM MgCl2, 50 mM HEPES/KOH, pH 7.8, and complete protease inhibitor cocktail. The resuspended chloroplasts were loaded onto a two-step Percoll gradient and were centrifuged in a swinging-bucket rotor at 1500g for 10 min. The band that appeared between the two phases contained intact chloroplasts and was recovered. The intact chloroplasts were washed with the suspending buffer at a rate of chloroplast to buffer 1/10 (v/v) by inverting the tubes carefully. The chloroplasts were centrifuged in a swinging-bucket rotor at 1000g for 3 min, and the pellet was recovered and resuspended in the suspending buffer. The intactness of the chloroplasts was verified by phase contrast microscopy (Nikon TE 2000U).
Chloroplast Fractionation Combined with Immunoblotting
Chloroplast membrane and soluble/stroma fractions were prepared essentially as described previously (Olsson et al., 2003) with modifications. Briefly, the suspended intact chloroplasts obtained as described above were pelleted by centrifugation. The pellets were thus suspended in the sorbitol-free suspending buffer and ruptured by homogenization. Chloroplast membranes were pelleted by ultracentrifugation in a swinging-bucket rotor at 125,000g for 30 min, and the supernatant was collected as the chloroplast stroma extract. The stroma fraction was concentrated by ultrafiltration. The recovered-membrane pellet was solubilized in the suspending buffer supplemented with 2% Triton X-100 (v/v) for 1 h of extraction, which gave the chloroplast membrane protein fractions for SDS-PAGE and immunoblotting.
The envelope and thylakoid fractions were separated as described previously (Douce and Joyard, 1982) with modifications, from the chloroplast membrane fractions obtained as described above but without the Triton X-100 extraction procedure. Discontinuous sucrose gradients were prepared in the chloroplast suspending buffer as a 0.6 M sucrose (12 mL) layer superposed on a 0.93 M sucrose (12 mL) layer. The chloroplast membrane fractions were loaded on the discontinuous sucrose gradients and centrifuged in a swinging-bucket rotor at 75,000g for 60 min. The chloroplast membranes were separated into the thylakoid fraction that appeared as a dark-green pellet at the bottom of the tube, and the envelope fraction that appeared as a yellow band at the interface of the two sucrose layers. The thylakoid fraction was collected and solubilized in the suspending buffer supplemented with 2% Triton X-100 for 1 h of extraction. The envelope fraction was collected and diluted in the chloroplast suspending buffer to give 0.2 to 0.3 M sucrose and centrifuged in a swing-out rotor at 113,000g for 40 min. The pellet was recovered and solubilized in the suspending buffer supplemented with 2% Triton X-100 for 1 h of extraction. The Triton X-100–treated thylakoid and envelope fractions were used for SDS-PAGE and immunoblotting.
The chloroplast inner and outer envelope membranes were separated as described previously (Keegstra and Yousif, 1986) with modifications. The suspended intact chloroplasts obtained as described above were pelleted by centrifugation at 350g for 3 min. The pellet was suspended in the chloroplast suspending buffer supplemented with 0.6 M sucrose for a hypertonic treatment for 10 min. The chloroplasts were then subjected to a series of procedures as described above for final isolation of the envelope fraction. The envelope membrane fraction obtained as such (without treatment by Triton X-100) was used to separate the inner and outer envelope membranes. Discontinuous sucrose density gradients were prepared in the chloroplast suspending buffer as 1.0 to 0.8 to 0.46 M (in order of the bottom to the top layer) three sucrose layers. The chloroplast membrane fractions were loaded on the discontinuous sucrose gradients and centrifuged in a swinging-bucket rotor at 180,000g for 2 h. The chloroplast envelope membranes were divided into the inner membrane fraction that appeared between 1.0 and 0.8 M sucrose layers and the outer membrane fraction that appeared at the interface of the 0.8 and 0.46 M sucrose layers. Both membrane fractions were recovered separately and diluted in the chloroplast suspending buffer to give 0.2 to 0.3 M sucrose and centrifuged at 125,000g for 90 min. The pellet was recovered and solubilized in the suspending buffer supplemented with 2% Triton X-100 for 1 h of extraction. The Triton X-100–treated fractions were used for SDS-PAGE and immunoblotting. All the procedures described above were performed at 4°C.
Chloroplast outer envelope marker Toc 34 (Jelic et al., 2002), inner marker Tic 40 (Chou et al., 2003; Li and Schnell, 2006), thylakoid marker PsbE (Garcia-Cerdan et al., 2009), and stroma marker Hcf101 (Stockel and Oelmuller, 2004) were used to characterize purity of the chloroplast fractions. The commercial antibodies against these proteins were purchased from AgriSera. For testing the purity of chloroplast membrane fraction, the inner envelope marker Tic-40 (detected by anti-Tic40 serum) was shown to be enriched in this membrane fraction, while the stroma marker Hcf101 (assayed by anti-Hcf101) was not detected in this membrane fraction, indicating that the membrane fractions were not contaminated by the stroma fraction (Figure 1D). For the purity of chloroplast envelope fraction, the inner envelope marker Tic-40 (detected by anti-Tic40 serum) was enriched in the envelope fraction, while the thylakoid marker PsbE (assayed by anti-PsbE) was not detected in the envelope fractions, indicating that the envelope fraction was not contaminated by thylakoid fraction (Figure 1D). For the purity of chloroplast inner and outer envelope fractions, the inner envelope marker Tic-40 (Chou et al., 2003), detected by anti-Tic40 serum, was enriched in the inner envelope fraction, and the outer envelope marker Toc34 (Jelic et al., 2002), detected by anti-Toc34 serum, was enriched in the outer envelope fraction, while the inner envelope contamination was low in the outer envelope fraction, and the outer envelope contamination was low in the inner envelope fraction (Figure 1D). Neither the thylakoid marker PsbE (Garcia-Cerdan et al., 2009) (assayed by anti-PsbE) nor stroma marker Hcf101 (Stockel and Oelmuller, 2004) (assayed by anti-Hcf101) was detected in the inner or outer envelope fraction, showing that the chloroplast envelope fractions were not substantially contaminated by these two fractions (Figure 1D).
Immunofluorescence Detection of ABAR on Outside of Intact Chloroplasts
The intact chloroplasts were prepared and tested for intactness as described above. The assays for detecting ABAR on the outside of intact chloroplasts were performed essentially according to the previously described procedures (Joyard et al., 1983) with modifications. Briefly, the intact chloroplasts were suspended in an incubation buffer consisting of 0.3 M sucrose and 10 mM HEPES/KOH, pH8.0, in a final volume of 100 μL containing 0.1 mg chloroplasts per mL. The suspended chloroplasts were centrifuged at 200g for 2 min, and the pellet was resuspended, for 30 min at 4°C, in the blocking buffer (100 μL) composed of 3% BSA (w/v) dissolved in the incubation buffer. After a centrifugation at 200g for 2 min, the pellet was collected and resuspended in appropriate, affinity-purified, rabbit antibody solution (diluted 1:200 in the incubation buffer) for 30 min at 4°C. The chloroplasts were pelleted by a centrifugation at 200g for 2 min and then washed two times (5 min each) at 4°C with 400 μL incubation buffer for each time. After a centrifugation at 200g for 2 min, the pellet was resuspended in the solution containing FITC-labeled goat-against-rabbit antibodies (diluted in the incubation buffer at 1:200) for an incubation at 4°C for 30 min. The pellet, recovered after a centrifugation at 200g for 2 min, was washed again two times (5 min each) at 4°C with 400 μL incubation buffer for each time. After the last centrifugation at 200g for 2 min, the pellet was resuspended in the incubation buffer (50 μL) and observed under a fluorescence phase contrast microscope (Nikon TE 2000U).
To test the specificity and reliability of the fluorescence labeling, two negative controls were performed. In the first one, the antibody was omitted to test possible unspecific labeling of the FITC-labeled gaot-against-rabbit antibodies. In the second one, purified IgG from rabbit preimmune serum was used instead of the rabbit antibody to test the specificity of the antibody. No substantial FITC fluorescence was observed in either of these negative controls (Figure 2B), showing that the fluorescence labeling was specific. More than three repetitions of the control experiments were conducted for each sample.
As for characterization of the chloroplast fractions, commercial antibodies (AgriSera) against chloroplast outer envelope marker Toc 34, inner marker Tic 40, thylakoid marker PsbE, and stroma marker Hcf101 were used as controls to verify the chloroplast intactness.
Immunogold Labeling of ABAR
Subcellular immunogold labeling of ABAR was done essentially as described previously (Fan et al., 2009) using the rabbit anti-full-length ABAR polyclonal antibody.
Analysis of Protein Interaction by Yeast Two-Hybrid System and CoIP in Yeast and in Planta
Interaction between proteins was assayed by a yeast Gal4-based two-hybrid system as described by the manufacturer (Clontech). The primers used for cloning the related cDNAs are listed in Supplemental Table 3 online. The cDNAs encoding the truncated ABARs were inserted into the pGBKT7 plasmid by the EcoRI (5′-end) and SalI (3′-end) sites to generate bait plasmids, and the cDNA encoding WRKYs were cloned into pGADT7 plasmid by the EcoRI (5′-end) and XhoI (3′-end) sites to generate prey plasmids. The assays, including measurement of α-galactosidase activity, were performed according to the manufacturer's protocols using as substrate p-nitrophenyl-α-d-galactoside, which is hydrolyzed to p-nitrophenol and d-galactose.
To test the effects of ABA on the ABAR–WRKY40 interaction in the yeast two-hybrid system, we used drop test to assay yeast growth. Yeast cells expressing varies constructs were grown on the SD medium lacking Leu, Trp, His, and Ade overnight and then transferred to the fresh, liquid, Leu-Trp-His-Ade–deficient medium to OD600 0.2. After a further incubation of 5 h, the OD values were measured. The cells were diluted in sterile water, and 8 μL was spotted at OD600 = 0.001 on the Leu-Trp-His-Ade–deficient medium supplemented with various concentrations of ABA (as indicated). The yeast cells were further grown at 30°C for 2 or 3 d for observations. The control yeast cells (supplied in the kit) were grown in the medium lacking Leu, Trp, His, and Ade and supplemented with the same concentrations of ABA as for the ABAR-WRKY40-transgenic yeast lines (as indicated).
CoIP assays were performed using extracts of both yeast cells and Arabidopsis plants. Yeast strains were grown on SD medium deficient in Leu, Trp, His, and Ade to OD600 1.0 at 30°C. Total proteins were prepared from yeast cells with an extraction buffer (2 mL/g cells) containing 50 mM HEPES, pH 7.4, 10 mM EDTA,0.1% (v/v) Triton X-100, 1 mM PMSF, and 1 μg/mL each of aprotinin, leupeptin, and pepstatin A (Sigma-Aldrich). The antibodies used were mouse antibody (Santa Cruz Biotechnology) specific to MYC-tagged truncated ABAR protein and rabbit antibody specific to HA-tagged (hemagglutinin peptide epitope) (Sigma-Aldrich) WRKY40 protein. Immunoprecipitation experiments were performed with protein A/G Plus-agarose beads (Santa Cruz Biotechnology) following the manufacturer's protocol. In brief, cell lysates were precleared with the protein A/G Plus-agarose beads and incubated with the antibodies and the protein A/G Plus-agarose beads at 4°C overnight in the extraction buffer. The beads were washed twice extensively with buffer A (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% [v/v] Triton X-100) and buffer B (50 mM Tris, pH 8.0, and 0.1% [v/v] Triton X-100) and then resuspended in SDS-PAGE sample buffer. The immunoprecipitates were separated on a 12% SDS-PAGE and analyzed by immunoblotting with anti-MYC or anti-HA serum.
For immunoprecipitation in Arabidopsis extracts, the total proteins (6 mg) were resuspended in the extraction buffer (2 mL) containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% (v/v) Triton X-100, 10% (v/v) glycerol, 1 mM PMSF, and 1 μg/mL each of aprotinin, leupeptin, and pepstatin A. The immunoprecipitation was done with the same procedures as described above except that the anti-ABAR or anti-WRKY40N serum was used instead of the anti-MYC or anti-HA serum, and the beads were washed with the extraction buffer instead of the buffer A and buffer B.
Isolation of Cytosolic and Nuclear Fractions
For observation of WRKY40 trafficking between nucleus and cytoplasm, the cytosolic and nuclear fractions were isolated for immunoblotting of WRKY40. The cytosolic fraction was isolated essentially according to the procedures described previously (Zhang et al., 2001) with modifications. The Arabidopsis leaves were ground to fine powder using liquid nitrogen and prechilled mortars and pestles. Cytosolic protein isolation buffer is composed of 10 mM HEPES, pH 8.0, 250 mM sucrose, 0.5% (v/v) Triton X-100, 1 mM EDTA, 5 mM MgCl2, 50 mM NaCl, 1 mM PMSF, and 1× Roche Cocktail (protease inhibitor cocktail). The cytosolic protein isolation buffer was added at 1 mL/g to powder to generate the homogenate. After centrifuging at 10,000g for 15 min, the supernatant was mixed with 2× SDS sample buffer and denatured for 10 min in boiling water. The isolated cytosolic fraction was examined by immunodetecting the presence of the nuclear marker Histone H3 with anti-Histone H3 antibody (Sigma-Aldrich), which showed absence of the Histone H3 in the prepared cytosolic fraction, verifying that the cytosolic fraction was not contaminated by the nuclear fraction (Figure 4). The nuclear fraction was isolated according to the protocol of Cold Spring Harbor Laboratory as described at its website. The isolated nuclear fraction was examined by immunodetecting the presence of the cytosolic marker PEPC (phosphoenolpyruvate carboxylase) with anti-PEPC antibody (Agrisera), which showed absence of PEPC in the prepared nuclear fraction and so the nuclear fraction was not contaminated by cytosolic fraction (Figure 4).
Test of Protein–Protein Interaction by BiFC of YFP and LCI
We performed BiFC assays in vivo as described (Walter et al., 2004). The ORFs of ABAR and WRKY40 were amplified by PCR with the following primers: for ABAR, forward primer 5′-GACTAGTATGGCTTCGCTTGTGTA-3′, and reverse primer, 5′-ACGCGTCGACTCGATCGATCCCT-3′, with the SpeI (5′-end) and SalI (3′-end) sites; for WRKY40, forward primer, 5′-CGCGGATCCATGGATCAGTACTCATC-3′, and reverse primer, 5′-CCGCTCGAGTTTCTCGGTATGATTCTG-3′, with the XhoI (5′-end) and BamHI (3′-end) sites. The ORFs were cloned into the plasmid pUC-SPYNE-35S/pUC-SPYNE (Walter et al., 2004) to form ABAR-YFPN (N-terminal half of YFP) or WRKY40-YFPN fusion protein and into pUC-SPYCE-35S/pUC-SPYCE to form ABAR-YFPC (C-terminal half of YFP) or WRKY40-YFPC fusion protein. The protoplasts isolated from Arabidopsis Col wild-type and aba2 mutant plants were transiently transformed with these constructs, and the fluorescence of YFP was observed with the confocal laser scanning microscope as described above. To test the effects of ABA treatment on the interaction between ABAR and WRKY40, (±)ABA at 2 μM concentration was used to incubate the transformed protoplasts 2 h before the observation under the confocal laser scanning microscope.
To further confirm the results of BiFC, we used a luciferase complementation imaging assay according to previously described procedures (Chen et al., 2008) in which the firefly Luc enzyme is divided into the N- (NLuc) and C-terminal (CLuc) halves that do not spontaneously reassemble and function. Luc activity occurs only when the two fused proteins interact, resulting in reconstituted Luc enzyme. The primers used for cloning the related cDNAs are listed in Supplemental Table 3 online. The constructs were cloned into pCAMBIA-NLuc and pCAMBIA-CLuc at the KpnI and SalI sites. The constructs were mobilized into A. tumefaciens strain GV3101. Bacterial suspensions were infiltrated into young but fully expanded leaves of the 7-week-old Nicotiana benthamiana plants using a needleless syringe. It is noteworthy that the amounts of the constructs were the same among treatments and controls for each group of assay. After infiltration, plants were grown first under dark for 12 h and then with 16 h light/dark for 60 h at room temperature, and the Luc activity was observed with a CCD imaging apparatus (Andor iXon). The goat anti-full-length firefly Luc antibody (Promega) was used to immunodetect Luc fusion protein in transgenic tissues.
Quantitative Real-Time PCR
Real-time PCR for mRNA expression of various genes (see Supplemental Table 4 online for the gene-specific primers) was performed as previously described (Wu et al., 2009) essentially according to the instructions provided for the Bio-Rad Real-Time System CFX96TM C1000 thermal cycler. Total RNA was isolated from leaves of 3-week-old seedlings with the RNasy plant mini kit (Qiagen) supplemented with an on-column DNA digestion (Qiagen RNase-Free DNase set) according to the manufacturer's instructions, and then the RNA sample was reverse transcribed with the Superscript II RT kit (Invitrogen) in 25 mL volume at 42°C for 1 h. Amplification of ACTIN2/8 genes was used as an internal control. The suitability of the oligonucleotide sequences in terms of efficiency of annealing was evaluated in advance using the Primer 5.0 program. The cDNA was amplified using SYBR Premix Ex Taq (TaKaRa) using a DNA Engine Opticon 2 thermal cycler in a 10 μL volume with the following program: one cycle of 95°C for 10 s and 40 cycles of 94°C for 5 s and 60°C for 30 s. The amplification of the target genes was monitored every cycle by SYBR-green fluorescence. The Ct (threshold cycle), defined as the PCR cycle at which a statistically significant increase of reporter fluorescence was first detected, was used as a measure for the starting copy numbers of the target gene. Relative quantitation of the target gene expression level was performed using the comparative Ct method. Three technical replicates were performed for each experiment. For all the quantitative real-time PCR analysis, the assays were repeated three times along with at least three independent repetitions of the biological experiments, and the means of the three biological experiments were calculated for estimating gene expression.
Analysis of Gene Expression by Promoter-GUS Transformation
The promoter fragments of Arabidopsis genes At5g13630 (ABAR), At2g36270 (ABI5), and At1g80840 (WRKY40) were amplified by PCR (see Supplemental Table 3 online for the primers). The DNA fragments were cloned into pCAMBIA1391 (for ABAR and ABI5) or pCAMBIA1300-221 (for WRKY40) vector and introduced into the GV3101 strain A. tumefaciens and transformed into Arabidopsis (Col) plants by floral infiltration. T3 generation homologous plants were used for the analysis of GUS activity. GUS staining was performed essentially according to Jefferson et al. (1987). Whole plants or tissues were immersed in 1 mM 5-bromo-4-chloro-3-indolyl-β-GlcUA solution in 100 mM sodium phosphate, pH 7.0, 2 mM EDTA, 0.05 mM ferricyanide, 0.05 mM ferrocyanide, and 0.1% (v/v) Triton X-100 for 5 to 6 h at 37°C. Chlorophyll was cleared from the tissues with a mixture of 30% acetic acid and 70% ethanol.
ChIP Assays
ChIP assay was performed essentially according to the previously described protocols by Saleh et al. (2008). Two-week-old seedlings were immersed in cross-linking buffer composed of 0.4 M sucrose, 10 mM Tris-HCl, pH 8, 1 mM PMSF, 1 mM EDTA, and 1% formaldehyde under vacuum for 10 min followed by additional 10-min incubation with 0.1 M glycine. Seedlings were ground in liquid nitrogen and resuspended in nuclei isolation buffer consisting of 0.25 M sucrose, 15 mM PIPES, pH 6.8, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 0.9% Triton X-100, 1 mM PMSF, 2 μg/mL pepstatin A, and 2 μg/mL aprotinin. Nuclei were then collected by centrifugation at 11,000g for 20 min at 4°C, resuspended in nuclei lysis buffer composed of 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1% SDS (w/v), 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM PMSF, 1 μg/mL pepstain A, and 1 μg/mL aprotinin, and sonicated to ∼200- to 1000-bp fragments. After centrifugation at 13,800g for 10 min at 4°C, the supernatants were incubated in ChIP dilution buffer (the same as the nuclei lysis buffer) with addition of 60 μL protein A agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C with gentle rotation to preclear the diluted sonicated chromatin and then centrifuged at 3800g for 2 min at 4°C. The supernatants were recovered and incubated with the antibody against WRKY40N (with the preimmune serum instead of the antibody as a negative control) overnight at 4°C. Protein A agarose (60 μL) was added into the mixture for a further incubation for 2 h at 4°C and centrifuged at 3800g for 2 min at 4°C to collect the agarose beads and the chromatin. The agarose beads were washed for 10 min each time (except for TE buffer wash for 5 min each time) with gentle rotation at 4°C with 1 mL of each of the following buffers and centrifuged at 3800g for 2 min at 4°C: two times with low salt wash buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8, 0.2% [w/v] SDS, 0.5% Triton X-100, and 2 mM EDTA), two times with high salt wash buffer (low salt wash buffer but containing 500 mM NaCl), two times with LiCl wash buffer (0.25 M LiCl, 1% sodium deoxycholate, 10 mM Tris-HCl, pH 8, 1% Nonidet P-40, and 1 mM EDTA), and three times with TE buffer (1 mM EDTA and 10 mM Tris-HCl, pH 8). The immunocomplexes were eluted from the agarose beads with 500 μL of the elution buffer (two times for 250 μL each) composed of 1% SDS (w/v) and 0.1 M NaHCO3 by incubating at room temperature for 30 min (two times for 15 min each) with gentle rotation. Cross-links were reversed by incubation at 65°C overnight followed by proteinase K treatment for 2 h at 45°C, phenol/chloroform/isoamyl alcohol extraction, and ethanol precipitation. Pellets were washed with 70% (v/v) ethanol and resuspended in double distilled water. The primers used for PCR amplification for different promoters are listed in Supplemental Table 2 online. PCR amplification was performed using 35 cycles and 56°C for ABI5 promoter fragments and 49.5°C for ABF4-, ABI4-, MYB2-, DREB1A-, DREB2A-, and RAB18-promoter fragments. Aliquots of the PCR reactions were resolved by electrophoresis on a 1% agarose gel. Images of the ethidium bromide–stained gels were captured by the Molecular Imager System (Gel Doc XR; Bio-Rad) with ImageQuant software (Molecular Dynamics). The results presented here come from at least five independent experiments.
To determine quantitatively the WRKY40-DNA (target promoters) binding, real-time PCR analysis was performed according to the procedure described previously with the Actin2 3′ untranslated region sequence as the endogenous control (Mukhopadhyay et al., 2008). The relative quantity value calculated by the 2−ddCt method is reported as DNA binding ratio (differential site occupancy) (Figure 9C). The same primers as for the above-mentioned PCR analysis were used for the real-time PCR except for ABI4-1, ABI4-2, ABI5-4, and MYB2-4 fragments for which the primers used were as follows: forward primer 5′-GCCTATCTTTTGCTATGTTT-3′ and reverse primer 5′-GAGAAATAAGAAACGAATAATC-3′ for ABI4-1, forward primer 5′-CCAATGTGTAACAAGTAAC-3′ and reverse primer 5′-CTGAAGAGTGTTTGAATGTC-3′ for ABI4-2, forward primer 5′-CGTTTGTCGCTGTCACGATGTG-3′ and reverse primer 5′-GTCCCTTATTCAACTATCACG-3′ for ABI5-4, and forward primer 5′-AACAATTGACCAAATGGAGA-3′ and reverse primer 5′-GACTGGAACACGGCATAAGT-3′ for MYB2-4. A fragment of the Actin2 promoter was used as a negative control, and the primers used were as follows: forward primer 5′-CGTTTCGCTTTCCT-3′ and reverse primer 5′-AACGACTAACGAGCAG-3′.
WRKY40–Promoter Interaction Tested with Yeast One-Hybrid Assay
Yeast one-hybrid assays were performed with the kit provided by Clontech (Matchmaker One-Hybrid Library Construction and Screening kit) using the AH109 yeast strain according to the manufacturer's instructions. The primers used for cloning the related cDNAs or promoter DNAs are listed in Supplemental Table 3 online. The promoter DNA fragment was subcloned into the SmaI/MluI sites of pHIS2 vector. The one-hybrid assays were performed using the AH109 yeast strain according to the manufacturer's instructions. Yeast cells were cotransformed with pHIS2 bait vector harboring promoter of target genes and pGADT7 prey vector harboring ORF of WRKY40. As negative controls, the yeast cells were cotransformed with the combination of pGADT7-WRKY40 and empty pHIS2 vector, or empty pGADT7 vector and pHIS2 harboring the corresponding promoter, or two empty vectors pGADT7 and pHIS2. Transformed yeast cells were first grown in SD-Trp-Leu medium to ensure that the yeast cells were successfully cotransformed, and then the yeast cells were grown on SD-Trp-Leu-His medium plates supplemented with 3-AT (Sigma-Aldrich) at 25 mM (for WRKY40-ABI5 promoter interaction), 5 mM (for WRKY40-ABI4 or ABF4 promoter interactions), or 3 mM (for WRKY40-MYB2 promoter interaction). The plates were then incubated for 3 d at 30°C.
Gel Shift Assay
Gel shift assay was performed using recombinant His-WRKY40 protein purified from E. coli as described above. The promoter fragments used for the gel shift assay were amplified by PCR using the following primer pairs: forward primer 5′-CCAATGTGTAACAAGTAAC-3′and reverse primer 5′-CTGAAGAGTGTTTGAATGTC-3′ for ABI4 promoter (pABI4-2); forward primer 5′-CGTTTGTCGCTGTCACGATGTG-3′ and reverse primer 5′-GTCCCTTATTCAACTATCACG-3′ for ABI5 promoter (pABI5-4); forward primer 5′-CTCTGTATCTGGTGTGAATTCG-3′ and reverse primer 5′-CTGGACAAACCACATAAATGCG-3′ for ABF4 promoter (pABF4-2); and forward primer 5′-GTCAAAGGGTCAAACCTTAG-3′and reverse primer 5′-GAGTAGAATGTTGAAGAGTG-3′ for MYB4 promoter (pMYB2-3). The suffix numbers corresponds to the fragment numbers presented in Supplemental Table 2 online and Figure 9 for ChIP assay. The sequences amplified by these primer pairs are listed in Supplemental Table 2 online. The site-specific mutations from GTCA to GTTA in the core sequence of W-box of the ABI5 and MYB2 promoters or from TGAC to TTAC in the ABI4 and ABF4 promoters were introduced into the above promoters by two independent PCR with the following primers (with mutated W-box underlined) in addition to the above-mentioned primers for each promoter: 5′-CCATTCACAACGATTACATTCAAACACTCTTCAG-39 and 5′-CTGAAGAGTGTTTGAATGTAATCGTTGTGAATGG-3′ for mutated pABI4-2; 5′-GTTATTTCAATATTTTTTACGTTATTTAAACTCCACTTTACC-3′ and 5′-GGTAAAGTGGAGTTTAAATAACGTAAAAAATATTGAAATAAC-3′ for mutated pABI5-4; 5′-GACCTTTATTGATTTACTTTACTGTGCTTTTAC-3′ and 5′-GTAAAAGCACAGTAAAGTAAATCAATAAAGGT-3′ for mutated pABF4-2; and 5′-CATCTGAACAAGTTAAAGGGTTAAACCTTAGTATTTTAAAATTAC-3′ and 5′-GTAATTTTAAAATACTAAGGTTTAACCCTTTAACTTGTTCAGATG-3′ for mutated pMYB2-3. Reconstitution was done using equimolar quantities of the two fragments from the initial PCRs for each promoter, which were used as template of a third PCR. The mutations were verified by sequence analysis. Each of the promoter fragments was labeled in the base T with digoxigenin-dUTP (Roche) according to the manufacturer's instructions. Binding reactions were performed in 20 μL of binding buffer composed of 10 mM Tris (2-amino-2-(hydroxymethyl)-1,3-propanediol)-HCl, pH 8.0, 20 mM NaCl, 0.4 mM MgCl2, 0.5 mM ZnSO4, 0.25 mM EDTA, 0.25 mM DTT, and 10% glycerol and in the presence of 0.04 mg/mL of poly(deoxyinosinic-deoxycytidylic) sodium salt (Sigma-Aldrich). Binding reactions were done using 50 ng of His-WRKY40 fusion protein and 26 ng for each of the digoxigenin-labeled promoter fragments at room temperature for 30 min. Samples were separated on a 5% polyacrylamide gel (19:1 acrylamide:bisacrylamide) in 0.5× Tris-borate-EDTA at 4°C, transferred into nylon filter (Hybond-N+; GE Amersham), exposed under UV light to cross-link the samples to the filter, washed with 2× standard saline citrate buffer two times for 15 min each, blocked with blocking reagent (provided in the kit) for 2 h, incubated with the Anti-Digoxigenin-AP (alkaline phosphatase) antibody for 1 h before adding the AP substrate into the mixture and analyzed by autoradiography. Competition experiments were performed using from 5 to 20 molar excess of unlabeled fragments.
Trans-Inhibition of ABI5 Promoter Activity by WRKY40 in Tobacco Leaves
WRKY40 was used for the effector construct. The cDNA of WRKY40 was PCR amplified using the forward primer 5′-CGCGGATCCATGGATCAGTACTCAT-3′ and reverse primer 5′-CCGCTCGAGCTATTTCTCGGTATGA-3′, and the PCR product was fused to pBI121 vector downstream of the CaMV 35S promoter at the BamHI/XhoI sites. Reporter constructs were composed of the ABI5-promoter linked to LUC. The ABI5 promoters were isolated using the forward primer 5′-GGGGTACCCAGCCGAACGGATTCT-3′ and reverse primer 5′-TCCCCCGGGCAACTGCATCATATACAC-3′ (1232 bp). The site-specific mutations from GTCA to GTTA in the core sequence of W-box were introduced into the ABI5 promoter by two independent PCR with the following primers (with mutated W-box underlined) in addition to the above-mentioned primer pairs for cloning ABI5 promoter: 5′-GTTATTTCAATATTTTTTACGTTATTTAAACTCCACTTTACC-3′ and 5′-GGTAAAGTGGAGTTTAAATAACGTAAAAAATATTGAAATAAC-3′. Reconstitution was performed using equimolar quantities of the two fragments from the initial PCRs, which were used as template of a third PCR. The mutations were verified by sequence analysis. The LUC cDNA was PCR amplified using the forward primer 5′-TCCCCCGGGATGGAAGACGCCAAAAAC-3′and reverse primer 5′-CGGGATCCTTACACGGCGATCTTTCCGC-3′ from pGL3-Basic Vector harboring the LUC cDNA. The DNA sequences of ABI5 promoter and mutated ABI5 promoter were separately fused to the KpnI/SmaI sites of pCAMBIA1300 vector from which the CaMV 35S promoter was deleted, with the LUC cDNA fused to the SmaI/BamHI sites downstream of the ABI5 promoter or mutated ABI5 promoter. The constructs were mobilized into A. tumefaciens strain GV3101. Bacterial suspensions were infiltrated into young but fully expanded leaves of the 7-week-old N. benthamiana plants using a needleless syringe. It is noteworthy that the amounts of the constructs were the same among treatments and controls for each group of assay. After infiltration, plants were grown first under dark for 12 h and then with 16 h light/dark for 60 h at room temperature, and the LUC activity was observed with a CCD imaging apparatus (Andor iXon). The experiments were repeated independently at least five times with the similar results.
Phenotypic Analysis
Phenotypic analysis was done essentially as previously described (Shen et al., 2006; Wu et al., 2009). To assay germination, ∼100 seeds were sterilized and planted in triplicate on MS medium (Sigma-Aldrich; full-strength MS). The medium contained 3% sucrose and 0.8% agar, pH 5.9, and was supplemented with or without different concentrations of (±)-ABA. The seeds were incubated at 4°C for 3 d before being placed at 20°C under light conditions, and germination (emergence of radicals) was scored at the indicated times. For seedling growth experiments, seeds were germinated after stratification on common MS medium and transferred to MS medium supplemented with different concentrations of (±)-ABA in the vertical position. The time for transfer was ∼48 h after stratification. Seedling growth was investigated at the indicated times after the transfer, and the length of primary roots was measured using a ruler. Seedling growth was also assessed by directly planting the seeds in ABA-containing MS medium to investigate the response of seedling growth to ABA after germination. For stomatal aperture assays, 3-week-old leaves were used. To observe ABA-induced stomatal closure, leaves were floated in the buffer containing 50 mM KCl and 10 mM MES-Tris, pH 6.15, under a halogen cold light source (Colo-Parmer) at 200 μmol m−2 s−1 for 2.5 h followed by addition of different concentrations of (±)-ABA. Apertures were recorded on epidermal strips after 2.5 h of further incubation to estimate ABA-induced closure. To study ABA-inhibited stomatal opening, leaves were floated on the same buffer in the dark for 2.5 h before they were transferred to the cold light for 2.5 h in the presence of ABA, and then apertures were determined.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g13630 (ABAR), At1g80840 (WRKY40), At4g31800 (WRKY18), At2g25000 (WRKY60), At3g19290 (ABF4), At2g40220 (ABI4), At2g36270 (ABI5), At4g25480 (DREB1A), At5g05410 (DREB2A), At2g47190 (MYB2), and At5g66400 (RAB18). Germplasm identification numbers for mutant lines and SALK lines are as follows: aba2 (CS156: aba2-1), the abar-2 (CS89100), abar-3 (CS92346), abi5 (CS8105: abi5-1), wrky40-1 (stock number: ET5883, Cold Spring Harbor Laboratory gene and enhancer trap lines), wrky18-1 (SALK_093916), and wrky60-1 (SALK_120706).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Prediction of the Potential Transmembrane Domains in ABAR Protein.
Supplemental Figure 2. Test of Interactions of ABAR and Truncated ABARs with WRKY40 in Yeast Two-Hybrid System.
Supplemental Figure 3. Subcellular Localization of WRKY40 in Roots and Immunoblotting Test of WRKY40 in Cytosol and Nucleus in Response to ABA.
Supplemental Figure 4. Immunodetection of Expression of the Fluorescent Proteins and Fusion Proteins in the Transgenic Plants/Protoplasts.
Supplemental Figure 5. Expression of the Native Promoter-Driven WRKY40, WRKY18, and WRKY60 Rescues ABA Sensitivity of the Mutants wrky40, wrky18, and wrky60, Respectively.
Supplemental Figure 6. Concentrations of Endogenous ABA, Protoporphyrin (Proto), Mg-Protoporphyrin (Mg-Proto), and Chlorophyll in Leaves of the Different wrky Mutants.
Supplemental Figure 7. WRKYs Are Expressed Ubiquitously in Different Tissues/Organs.
Supplemental Figure 8. ABAR Is Expressed Ubiquitously in Different Tissues/Organs Except for Dry Seeds.
Supplemental Figure 9. Analysis of the ABAR Expression in the Different Transgenic Lines Used in Figures 7 and 8.
Supplemental Figure 10. Disruption of WRKYs Suppresses ABA Insensitivity of Two Mutant Alleles of the ABAR Gene, abar-2, and abar-3.
Supplemental Figure 11. Barley XanF Interacts with Arabidopsis WRKY40/WRK18/WRKY60 and ABA Stimulates This Interaction.
Supplemental Figure 12. Expression of the Barley XanF Confers ABA Hypersensitivity in Wild-Type Plants and Rescues ABA Sensitivity of the cch Mutant in Arabidopsis.
Supplemental Table 1. Test of Interaction of ABAR with WRKY Transcription Factors in the Yeast Two-Hybrid System.
Supplemental Table 2. Information for Detecting the WRKY40 Binding Promoter Sequences by PCR in the Chromatin Immunoprecipitation Assay with the Antibody against the Truncated WRKY40N and by Gel Shift Assay.
Supplemental Table 3. Primers Used for Cloning Promoter DNAs or cDNAs.
Supplemental Table 4. Gene-Specific Primers for Real-Time PCR Analysis.
Supplemental Methods. Generation of the XanF-Expressing Transgenic Lines, Identification of the cch Mutation in the XanF-Transgenic Lines of the cch Background, Analysis of the XanF-WRKY40 Interaction, and Chlorophyll and Porphyrin Measurements.
Supplemental References.
Acknowledgments
The cch seeds were a gift from J. Chory (The Salk Institute, La Jolla). The wrky40-1 seeds were provided by the Cold Spring Harbor Laboratory. Other mutant seeds were provided by the ABRC. We thank G.-Q. Liu, D.-T. Ren, D.-W. Li, D. Ye, and Z.-Z. Gong (China Agricultural University, Beijing, China); G.-Y. Wang (Chinese Academy of Agricultural Sciences, Beijing, China); and D. Liu, J.M. Pan, Y.L. Liu, and D.X. Xie (Tsinghua University, Beijing, China) for advice and help on materials. This work was supported by grants from the National Natural Science Foundation of China (Grants 90817104 and 30700053) and the Ministry of Agriculture of China (Grant 2008ZX08009-003).
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcription activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie B.A.T., Perez-Perez J., Perez-Perez M.M., Godoy M., Sanchez-Serrano J.J., Schmelz E.A., Solanoa R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Assmann S.M. (1994). Ins and outs of guard cell ABA receptors. Plant Cell 6: 1187–1190 [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R., Joyard J. (1982). Purification of the chloroplast envelope. Methods Enzymol. 16: 239–256 [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 1360–1385 [DOI] [PubMed] [Google Scholar]
- Fan L.M., Zhao Z., Assmann S.M. (2004). Guard cells: A dynamic signaling model. Curr. Opin. Plant Biol. 7: 537–546 [DOI] [PubMed] [Google Scholar]
- Fan R.C., Peng C.C., Xu Y.H., Wang X.F., Li Y., Shang Y., Du S.Y., Zhao R., Zhang X.Y., Zhang L.Y., Zhang D.P. (2009). Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 150: 1880–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S., Rock C. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Wang M.L., Lynch T.J., Rao S., Goodman H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signaling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zeng Q., Guo J., Cheng J., Ellis B.E., Chen J.G. (2007). Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant J. 52: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Garcia-Cerdan J.G., Sveshnikov D., Dewez D., Jansson S., Funk C., Schroder W.P. (2009). Antisense inhibition of the PsbX protein affects PSII integrity in the higher plant Arabdopsis thaliana. Plant Cell Physiol. 50: 191–202 [DOI] [PubMed] [Google Scholar]
- Gibson L.C.D., Marrison J., Leech R.M., Jensen P.E., Bassham D.C., Cibson M., Hunter C.N. (1996). A putative Mg-chelatase subunit from Arabidopsis thaliana cv C24. Plant Physiol. 111: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A.A.R., Vartanian N., Giraudata J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Luo M., Weinstein J.D. (1998). Magnesium-chelatase from developing pea leaves. Plant Physiol. 116: 605–615 [Google Scholar]
- Henikoff S., Till B.J., Comai L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135: 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Yang Y., Grill E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2007). Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12: 343–350 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 20: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelic M., Sveshnikova N., Motzkus M., Hörth P., Soll J., Schleiff E. (2002). The chloroplast import receptor Toc34 functions as preprotein-regulated GTPase. Biol. Chem. 383: 1875–1883 [DOI] [PubMed] [Google Scholar]
- Jiang W., Yu D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and post-germination arrest of development by abscisic acid. BMC Plant Biol. 9: 96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C.A., Temple B.R., Chen J.G., Gao Y., Moriyama E.N., Jones A.M., Siderovski D.P., Willard F.S. (2007). Comment on a G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 318: 914. [DOI] [PubMed] [Google Scholar]
- Joyard J., Billecocqll A., Bartlett S.G., Block M.A., Chua N.H., Douce R. (1983). Localization of polypeptides to the cytosolic side of the outer envelope membrane of spinach chloroplasts. J. Biol. Chem. 258: 10000–10006 [PubMed] [Google Scholar]
- Jung K.H., Hur J., Ryu C.H., Choi Y., Chung Y.Y., Miyao A., Hirochika H., An G. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 44: 463–472 [DOI] [PubMed] [Google Scholar]
- Kang J., Choi H., Im M., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K., Yousif A.E. (1986). Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118: 316–325 [Google Scholar]
- Lang V., Palva E.T. (1992). The expression of a tab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T., Cuevas J., Mas P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACD-INSENSITIVE2 (ABI2) and ABI1 encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Schnell D.J. (2006). Reconstitution of protein targeting to the inner envelope membrane of chloroplasts. J. Cell Biol. 175: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA-binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yue Y., Li B., Nie Y., Li W., Wu W.H., Ma L.G. (2007a). A G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Liu X., Yue Y., Li W., Ma L. (2007b). Response to comment on a G protein coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 318: 914. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christman A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Melcher K., et al. (2009). A gate–latch–lock mechanism for hormone signalling by abscisic acid receptor. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., et al. (2009). Structural basis of abscisic acid signaling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.A., Larkin R., Nagatani N., Chory J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A.J.M., Tissenbaum H.A. (2008). Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3: 698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A.H., Hansson M. (2009). The barley magnesium chelatase 150-kD subunit is not an abscisic acid receptor. Plant Physiol. 150: 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., Schroeder J.I., Getzoff E.D. (2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Olsson T., Thelander M., Ronne H. (2003). A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J. Biol. Chem. 278: 44439–44444 [DOI] [PubMed] [Google Scholar]
- Pandey S., Nelson D.C., Assmann S.M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J., Peter-Mock H.P., Tanaka R., Kruse E., Grimm B. (2000). Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., Cutler S.R., Rodriguez P.L., Marquez J.A. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Shen Y.Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Stockel J., Oelmuller R. (2004). A novel protein for photosystem I biogenesis. J. Biol. Chem. 279: 10243–10251 [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I., Richter K., Kepka M., Demmel S., Ma Y., Korte A., Assaad A.F., Christmann A., Grill E. (2010). Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Teng Y.S., Su Y., Chen L.J., Lee Y.J., Hwang I., Lib H. (2006). Tic21 is an essential translocon component for protein translocation across the chloroplast Inner envelope membrane. Plant Cell 18: 2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker B., Somssich I.E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7: 491–498 [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Zhu J.K. (2007). New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 10: 447–452 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang X. (2002). Phospholipase D in hormonal and stress signaling. Curr. Opin. Plant Biol. 5: 408–414 [DOI] [PubMed] [Google Scholar]
- Wu F.Q., et al. (2009). The Mg-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: New evidence in Arabidopsis. Plant Physiol. 150: 1940–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Zhang Z.L., Hanzlik S., Cook E., Shen Q.J. (2007). Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 64: 293–303 [DOI] [PubMed] [Google Scholar]
- Xie Z., Zhang Z.L., Zou X., Huang J., Ruas P., Thompson D., Shen Q.J. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Zhang Z.L., Zou X., Yang G., Komatsu S., Shen Q.J. (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46: 231–242 [DOI] [PubMed] [Google Scholar]
- Xu X., Chen C., Fan B., Chen Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Zhang D.P., Chen S.W., Peng Y.B., Shen Y.Y. (2001). Abscisic acid-specific binding sites in the flesh of developing apple fruits. J. Exp. Bot. 52: 2097–2103 [DOI] [PubMed] [Google Scholar]
- Zhang D.P., Wu Z.Y., Li X.Y., Zhao Z.X. (2002). Purification and identification of a 42-kilodalton abscisic acid-specific-binding protein from epidermis of broad bean leaves. Plant Physiol. 128: 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Creelman R.A., Zhu J.K. (2004). From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Seemann J.R., Neuman D., Shen Q.J. (2004). A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J. Biol. Chem. 279: 55770–55779 [DOI] [PubMed] [Google Scholar]