This work examines translocation of the flax rust effector protein AvrM, showing that it accumulates in the haustorial wall and extracellular matrix and is delivered into the host cytoplasm. Protein fusions expressed in the plant showed that AvrM can be secreted by the plant and subsequently internalized in the absence of pathogen.
Abstract
Translocation of pathogen effector proteins into the host cell cytoplasm is a key determinant for the pathogenicity of many bacterial and oomycete plant pathogens. A number of secreted fungal avirulence (Avr) proteins are also inferred to be delivered into host cells, based on their intracellular recognition by host resistance proteins, including those of flax rust (Melampsora lini). Here, we show by immunolocalization that the flax rust AvrM protein is secreted from haustoria during infection and accumulates in the haustorial wall. Five days after inoculation, the AvrM protein was also detected within the cytoplasm of a proportion of plant cells containing haustoria, confirming its delivery into host cells during infection. Transient expression of secreted AvrL567 and AvrM proteins fused to cerulean fluorescent protein in tobacco (Nicotiana tabacum) and flax cells resulted in intracellular accumulation of the fusion proteins. The rust Avr protein signal peptides were functional in plants and efficiently directed fused cerulean into the secretory pathway. Thus, these secreted effectors are internalized into the plant cell cytosol in the absence of the pathogen, suggesting that they do not require a pathogen-encoded transport mechanism. Uptake of these proteins is dependent on signals in their N-terminal regions, but the primary sequence features of these uptake regions are not conserved between different rust effectors.
INTRODUCTION
To establish disease, plant pathogens must overcome the natural defense mechanisms of their host plants. These include both preformed barriers to infection, as well as inducible responses to infection. For instance, recognition of pathogen-associated molecular patterns (PAMPs) by membrane-bound receptors induces a set of biochemical and transcriptional responses leading to PAMP-triggered immunity (Jones and Dangl, 2006). Bacterial pathogens have evolved mechanisms to suppress these responses through the action of effector proteins that are delivered into host cells via the specialized type three secretion system and specifically inhibit components of PAMP-triggered immunity (Chisholm et al., 2006). Eukaryotic pathogens, including biotrophic and hemibiotrophic fungi and oomycetes, also secrete effectors into host cells (Birch et al., 2008; Ellis et al., 2009; Panstruga and Dodds, 2009; Tyler, 2009). Some of these effectors have functions in suppressing defense responses, and their delivery into host cells during infection probably plays an important role in the establishment of the parasitic relationship. However, plants possess a second layer of inducible defenses governed by highly specific resistance (R) proteins that either directly or indirectly recognize pathogen effector proteins, leading to effector-triggered immunity. Effector-triggered immunity activates programmed cell death at pathogen infection sites, a process also known as the hypersensitive reaction (HR; Jones and Dangl, 2006). In this context, effector proteins that are recognized by host R proteins are termed Avirulence (Avr) proteins.
Avr proteins identified in oomycete pathogens, including Hyaloperonospora arabidopsis and Phytophthora spp, encode secreted proteins that are recognized inside plant cells (Allen et al., 2004; Shan et al., 2004; Armstrong et al., 2005; Rehmany et al., 2005). These oomycete proteins are characterized by a short sequence motif just downstream of the signal peptide that consists of the consensus amino acid sequence RxLR (where x is any amino acid), and in the case of the Phytophthora proteins, this is followed shortly afterwards (5 to 21 amino acids) by a dEER motif. The RxLR motif directs the transfer of Avr3a from haustoria of Phytophthora infestans into infected potato (Solanum tuberosum) cells but is not required for secretion from the pathogen, suggesting that oomycete Avr proteins enter the host cell in a two-step process involving signal peptide–mediated secretion followed by host cell translocation mediated by the RxLR motif (Whisson et al., 2007). Additional amino acids flanking the core conserved residues of the motif are also required for host cell uptake, suggesting that the local protein structure influences the presentation of the uptake motif (Bhattacharjee et al., 2006; Dou et al., 2008). The RxLR motif is related to the PEXEL (for Plasmodium Export Element) host-targeting signal present in secreted effectors of the human malaria parasite, Plasmodium falciparum (Hiller et al., 2004; Marti et al., 2004), and the two signals are apparently interchangeable (Bhattacharjee et al., 2006; Dou et al., 2008; Grouffaud et al., 2008). One important question is whether the transfer process depends on a pathogen-encoded translocation mechanism or on host cell transport machinery. Strong support for the second hypothesis came from a recent study of Phytophthora sojae Avr1b by Dou et al. (2008), who demonstrated that the RxLR-dEER motif mediates delivery of both Avr1b and the green fluorescent protein (GFP) into plant cells in the absence of the pathogen.
Secreted effectors of fungal pathogens are also likely to be delivered into host cells during infection. For instance, the AVR-Pita and AvrPiz-t proteins of the rice blast fungus Magnaporthe oryzae are recognized by intracellular R proteins (Jia et al., 2000; Li et al., 2009). AVR-Pita colocalizes with several other effector candidates to a distinct region of the extrainvasive hyphal space, known as the biotrophic interfacial complex, while other secreted proteins are distributed throughout the extrainvasive hyphal space (Mosquera et al., 2009). Khang et al. (2010) showed that the biotrophic interfacial complex–localized proteins are subsequently delivered into host cells, suggesting that this structure serves as an assembly area for secreted effectors prior to their transport into host cells. Likewise, the Avr2 protein of Fusarium oxysporum f. sp lycopersicon is recognized by the tomato I-2 resistance protein intracellularly (Houterman et al., 2009). Several genes for secreted proteins in the maize smut fungus, Ustilago maydis, have homology to proteins with intracellular regulatory functions, suggesting that they may act inside plant cells (Mueller et al., 2008).
The flax rust fungus Melampsora lini is an obligate biotrophic pathogen, and during infection it forms haustoria, specialized structures that penetrate host cell walls and make intimate contact with the host cell membrane. During R gene–dependent resistance to rust fungi, the HR is first observed in plant cells containing developing haustoria (Kobayashi et al., 1994; Heath, 1997), and, indeed, flax rust Avr proteins encode small secreted proteins that are expressed in haustoria (Dodds et al., 2004; Catanzariti et al., 2006; Barrett et al., 2009). Agrobacterium tumefaciens–mediated transient expression of AvrL567, AvrM, AvrP123, and AvrP4 induced cell death in plants expressing the corresponding nucleotide binding-leucine rich repeat-type R proteins. Importantly, this response was observed when the Avr proteins were expressed without their signal peptides, indicating that recognition occurs inside the plant cytoplasm. This is consistent with the location of the corresponding R proteins, and in fact both the AvrL567 and AvrM proteins directly interact with the cytoplasmic L6 and M resistance proteins, respectively (Dodds et al., 2006; Catanzariti et al., 2010). These results imply that these Avr proteins are delivered from the haustorium into the cytoplasm of infected plant cells. Kemen et al. (2005) provided direct evidence for transfer of the broad bean rust protein, RTP1, into host cells. Immunolocalization detected RTP1 in the plant cytoplasm adjacent to haustoria and within host cell nuclei.
Fungal effectors show high sequence divergence even within the same species, and there are no clearly conserved motifs that may function as uptake signals. Thus, it is not known whether fungal effectors use a similar or different uptake mechanism to oomycete effectors. Preliminary evidence suggests that the flax rust AvrM protein may enter plant cells independently of the pathogen, since transient expression of the AvrM protein with or without the signal peptide induces an M gene–specific HR, but addition of the HDEL endoplasmic reticulum (ER) retention signal prevents recognition of the secreted but not the cytoplasmic version (Catanzariti et al., 2006). This is consistent with recognition of the secreted form of AvrM by the cytoplasmic M protein after AvrM secretion and reentry into the plant cell. Similarly, AvrL567, AvrP4, and AvrP123 all induce HR when expressed as secreted proteins (Dodds et al., 2004; Catanzariti et al., 2006). In this study, we show by immunolocalization that AvrM protein of the rust pathogen M. lini is transferred from haustoria to host flax cells during infection. We also demonstrate that both AvrM and AvrL567 effectors can enter plant cells in the absence of the pathogen and that their translocation across the plant plasma membrane depends on transport signals occurring in their N-terminal domains.
RESULTS
AvrM-A Is Secreted from M. lini Haustoria and Is Translocated into Host Plant Cells
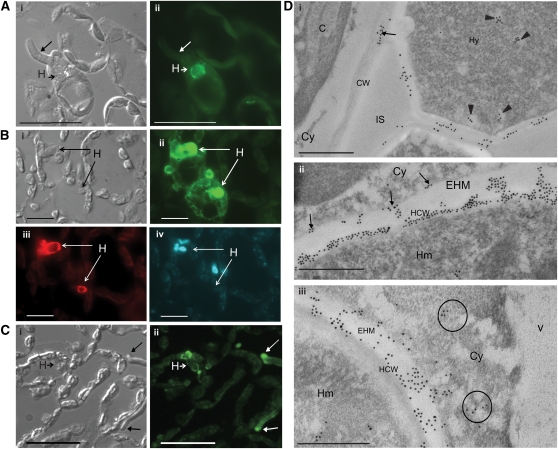
Intracellular recognition of flax rust Avr proteins provides indirect evidence for their delivery into the plant cytoplasm during infection. To directly assess AvrM delivery into host cells, we used immunolabeling to locate this protein in flax leaves infected by flax rust. His-tagged AvrM-A protein was expressed in Escherichia coli and purified by immobilized metal ion affinity chromatography. Two independent polyclonal antisera were raised in rabbits, and the antisera were purified by negative adsorption against protein extracts from uninfected flax leaves. Immunoblot analysis showed that both antisera detected a protein of ∼35 kD, the predicted size for AvrM-A, in extracts from flax leaves infected by rust strain CH5 (see Supplemental Figure 1 online). No labeling was detected in protein extracts from uninfected flax plants, confirming the specificity of the antisera. The purified antibodies were used to immunolocalize AvrM in infected flax leaves fixed at different time points from 12 to 120 h after inoculation (HAI; Figure 1, Table 1). No labeling was observed at 12 HAI, at which time no rust haustoria were visible. Haustoria were first seen at 16 HAI and at this stage were strongly labeled by the AvrM-antisera (Figure 1A). Haustoria continued to show strong labeling at subsequent time points, indicating that AvrM is present throughout haustorial development. This was further supported by colocalization studies with the haustorium-specific monoclonal antibody ML1 (Murdoch and Hardham, 1998), which showed that all haustoria detected by ML1 also showed labeling for AvrM (Figure 1B). Fluorescence labeling was also detected at the tips of fungal hyphae at 64 and 120 HAI (Figure 1C). At 120 HAI, we also detected strong AvrM-associated fluorescence in the cytoplasm of ∼13% (11 out of 87) of flax cells containing haustoria (Figure 1B). Cytoplasmic labeling was not detected in host cells lacking haustoria, including those adjacent to positively labeled hyphae (Figure 1C), indicating that the transfer of AvrM occurs predominantly through haustoria. No fluorescence was observed in sections incubated with the preimmune serum, confirming the specificity of the AvrM antisera labeling. No fluorescence was detected in host cells at earlier time points, although fewer haustoria were observed (∼15 haustoria at 16 and 24 HAI and ∼30 at 64 HAI).
Figure 1.
Immunolocalization of AvrM in Rust-Infected Leaf Material.
(A) Section of rust infected flax leaf fixed 16 HAI. Panel i shows a differential interference contrast image, while panel ii shows labeling with anti-AvrM antibody (green). Labeling was detected around this first haustorium (H, short arrow). Long arrow points to a hypha that lacks labeling. Bars = 100 μm.
(B) Section of rust infected flax leaf fixed 5 d (120 h) after inoculation. Panel i shows a differential interference contrast image, while panel ii shows labeling with anti-AvrM antibody (green). AvrM was detected in the cytoplasm of a proportion of host cells containing haustoria, but not in neighboring, uninfected cells. Haustoria were colocalized (iii) with monoclonal ML1 antihaustorial antibody (red). Nuclear material (iv) was labeled with 4',6-diamidino-2-phenylindole (blue). Bars = 50 μm.
(C) Section of rust infected flax leaf fixed 5 d after inoculation. Panel i shows a differential interference contrast image, while panel ii shows labeling with anti-AvrM antibody (green). Labeling was detected at the tips of fungal hyphae (long arrows) and around haustoria (H, short arrow). Bars = 100 μm.
(D) Electron micrographs of rust infected flax leaves 5 d after inoculation. Sections were probed with anti-AvrM antiserum and detected with goat anti-rabbit secondary antibodies conjugated to 10-nm gold particles. Panel i shows secretion of AvrM from fungal hyphae (Hy). Labeling occurs in the hyphal wall and on the surface of the adjacent plant cell wall (CW; arrow). Potential hyphal secretory vesicles are labeled (arrowheads). No labeling is observed in the intercellular space (IS). Panel ii shows a section through a fungal haustorium (Hm) with gold labeling evident in the haustorium cell wall (HCW), in the extrahaustorial matrix (EHM), and along the extrahaustorial membrane (arrows). Panel iii shows AvrM localization on the surface of the haustorium cell wall, in regions of the EHM and, to a lesser degree, in the plant cell cytoplasm (Cy) (circled). C, chloroplast; V, vacuole. Bars = 0.5 μm.
Table 1.
Localization of AvrM at 12, 16, 24, 64, and 120 HAI
| Time after Inoculation (in Hours) |
|||||
| Location | 12 h | 16 h | 24 h | 64 h | 120 h |
| Perimeter of haustoria | NA | + | + | + | + |
| Perimeter of hyphae, especially apex | − | − | − | + | + |
| Plant cytoplasm | − | − | − | − | + (in 13% of cells with haustoria) |
The labeling pattern of anti-AvrM antibody was observed in sections from at least three different leaves infected with CH5 flax rust. No haustoria were observed at 12 HAI. NA, not applicable.
AvrM localization was also examined by electron microscopy using immunogold labeling of sections of flax leaves 5 d after rust infection. Labeling was detected in the cell walls of infection hyphae and in plant cell walls adjacent to these hyphae (Figure 1Di), suggesting that AvrM is secreted from hyphae and can diffuse through the apoplast although no labeling was detected within adjacent plant cells. Strong labeling was also detected around haustoria with gold particles predominantly occurring over the haustorial cell wall (Figure 1Dii). Some labeling was evident in the extrahaustorial matrix (EHM), the space between the haustorial wall and the extrahaustorial membrane, often occurring in small clumps associated with electron-dense material of similar appearance to the haustorial cell wall. Potential secretory vesicles containing AvrM were also observed in the cytoplasm of fungal haustoria and hyphae. In most cases, immunogold labeling was not observed within the cytoplasm of plant cells containing haustoria; however, in a few cases, individual or small clusters of gold particles were seen in the plant cytoplasm near the haustoria (Figure 1Diii). Apart from an occasional gold particle over starch granules within the chloroplasts, no labeling was observed in sections incubated with the preimmune serum. Overall, the immunofluorescence and immunogold localization results indicate that during flax rust infection, the AvrM protein is secreted from rust haustoria into the extrahaustorial space and, from there, is translocated into the host cytoplasm.
Secreted AvrM-Cerulean Is Internalized into Plant Cells
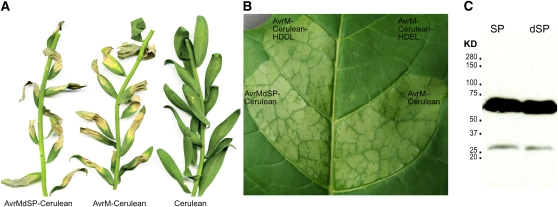
Previously, we showed that in planta expression of AvrM-A with and without the signal peptide induces an HR dependent on the cytoplasmic resistance protein M (Catanzariti et al., 2006). To further investigate the ability of AvrM-A to enter plant cells after secretion, we generated expression constructs encoding this protein (with and without the secretion signal peptide) with a C-terminal fusion to Cerulean, a variant of enhanced cyan fluorescent protein (Rizzo et al., 2004). Agrobacterium-mediated transient expression of both AvrM-Cerulean and AvrMdSP-Cerulean (i.e., a construct lacking the signal peptide) triggered HR in leaves of flax plants carrying the M resistance gene, as well as in transgenic tobacco (Nicotiana tabacum W38) expressing M (Figures 2A and 2B), confirming the functionality of the fusion proteins. No response was observed in flax or tobacco lines lacking the M gene. Recognition of the secreted AvrM-Cerulean fusion confirmed our previous observation and again suggested uptake of the secreted protein from the apoplast. Immunoblot analysis of proteins extracted from agroinfiltrated tobacco leaves with an anti-GFP antibody detected an abundant protein band of ∼66 kD corresponding to the predicted size of AvrM-Cerulean (Figure 2C). A small amount of a 27-kD protein could sometimes be detected, which probably results from cleavage of GFP in block from AvrM protein during extraction (Xu et al. 1999). This indicates that the vast majority of the fusion protein remains stable in vivo.
Figure 2.
Expression of AvrM-Cerulean Protein Fusions in Plants.
(A) Agrobacterium-mediated transient expression of AvrM-Cerulean, AvrMdSP-Cerulean (lacking the signal peptide), and cerulean alone in leaves of flax plants containing the M resistance gene. Cell death is indicative of HR induction by R-Avr recognition. Image prepared at 14 d after infiltration.
(B) Agrobacterium-mediated transient expression of AvrM-Cerulean, AvrMdSP-Cerulean (lacking the signal peptide), AvrM-Cerulean-HDEL, and AvrM-Cerulean-HDDL in leaves of transgenic tobacco plants containing the M resistance gene. Cell death is indicative of HR induction by R-Avr recognition. Image prepared at 48 HAI.
(C) Immunoblot of AvrM-Cerulean (SP) and AvrMdSP-Cerulean (dSP) fusion proteins expressed in tobacco. Proteins extracted from tobacco leaves 3 d after infiltration were separated by SDS-PAGE and the fusion proteins detected with anti-GFP antibodies. Positions and sizes (kD) of protein molecular mass markers are shown.
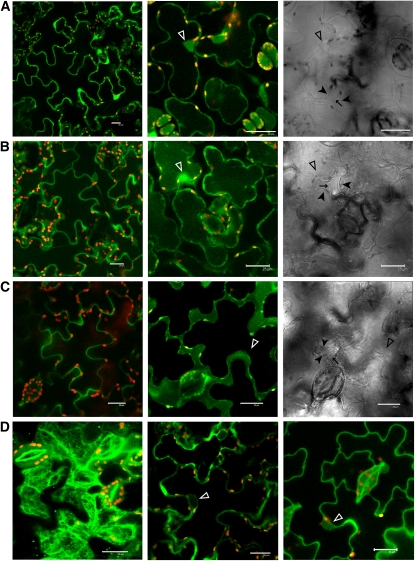
Confocal fluorescence microscopy was used to examine the subcellular localization of AvrM-Cerulean in intact cells of W38 tobacco leaves transiently expressing this protein. Cells transformed with a control construct expressing Cerulean alone showed fluorescence in the nuclei and cytoplasm (Figure 3A). AvrM-Cerulean exhibited a similar pattern of fluorescence to that of Cerulean alone (Figure 3B), suggesting an intracellular location for this fusion protein despite the presence of the secretion signal peptide. Plasmolysis of plant cells by treatment with 0.8 M mannitol confirmed that AvrM-Cerulean accumulated in the cytoplasm. However, when Cerulean was fused to just the signal peptide of AvrM-A, fluorescence was observed exclusively in the apoplastic space after plasmolysis (Figure 3C). This confirms that the leader peptide of AvrM-A functions correctly to direct efficient secretion of the fusion protein from plant cells. Nevertheless, it remained possible that the full-length AvrM-Cerulean was somehow blocked from entering the secretory pathway. To test this possibility, the four amino acid HDEL tag, which functions as an ER retention signal (Denecke et al., 1992), was added to the C terminus of the AvrM-Cerulean fusion. Expression of AvrM-Cerulean-HDEL resulted in fluorescence accumulating in the ER, Golgi, and the periphery of the nucleus (Figure 3D). This fusion protein also failed to induce the HR on M tobacco (Figure 2B), suggesting that the protein failed to be internalized into the cytoplasm. However, addition of the chemically similar HDDL tetrapeptide tag, which does not function in ER retention, resulted in intracellular accumulation of fluorescence (Figure 3D) and did not affect HR induction (Figure 2B). This indicates that the full-length AvrM-Cerulean fusion is efficiently delivered into the secretory pathway and that retention in the ER prevents subsequent internalization into the cytoplasm. Thus, the observed cytoplasmic accumulation in transformed cells is most likely the result of uptake of AvrM-Cerulean into the cytoplasm across the plant plasma membrane after passing through the secretory pathway. Importantly, this uptake can occur in the absence of flax rust haustoria, suggesting that it does not rely on pathogen-encoded transport machinery.
Figure 3.
Subcellular Localization of Transiently Expressed Avr-Cerulean Fusions in Tobacco.
(A) to (C) Confocal images of wild-type W38 tobacco cells expressing Cerulean alone (A), AvrM-Cerulean (B), or the AvrM signal peptide only fused to Cerulean (C). The left panel shows fluorescence of untreated cells, the middle panel shows fluorescence of cells plasmolysed with 0.8 M mannitol, and the right panel shows corresponding transmission images of the plasmolysed cells. Closed arrowheads indicate the plant plasma membrane where it is pulled away from the cell wall (arrows), and open arrowheads indicate nuclei. Cerulean fluorescence and chloroplast autofluorescence (red) were imaged using the 405- and 543-nm laser excitation light sources, respectively, with appropriate emission filters. Bars = 25 μm.
(D) Confocal images of tobacco cells expressing AvrM-Cerulean with HDEL (left and middle panels) or HDDL (right panel) C-terminal tags. The left-hand panel shows a z-stack of five optical sections showing reticulate distribution of AvrM-Cerulean-HDEL indicative of localization within the ER. The middle panel is a single optical section showing perinuclear labeling of AvrM-Cerulean-HDEL (open arrowhead). The right-hand panel shows AvrM-Cerulean-HDDL localization within the plant cell cytoplasm and nucleus (open arrowhead). Bars = 25 μm.
An N-Terminal Region of AvrM Determines Uptake by Plant Cells
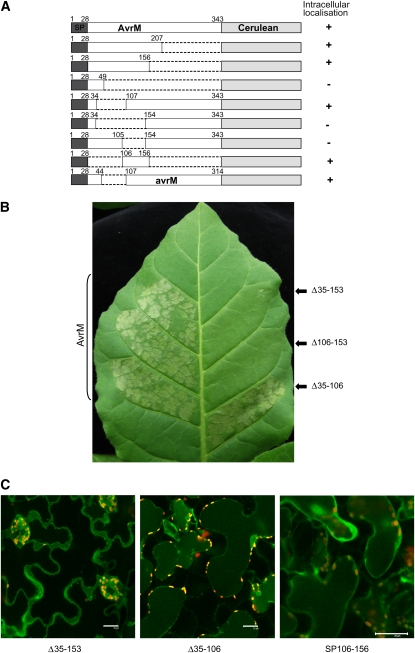
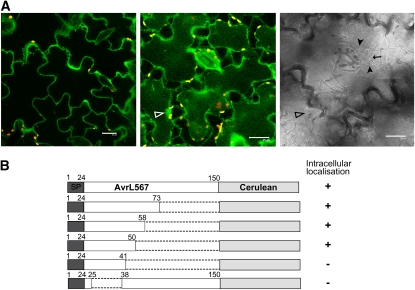
Uptake of oomycete effector proteins depends on the RxLR transport motif (Whisson et al., 2007; Dou et al., 2008). To determine whether AvrM-A contains a functionally equivalent uptake signal, we constructed a series of Cerulean fusions to C-terminal truncations of AvrM-A, which include the signal peptide along with various regions of the predicted mature protein (Figure 4A). The localization of these proteins was examined by confocal microscopy of transiently transformed tobacco cells after plasmolysis. The smallest N-terminal fragment that allowed internalization of the Cerulean fusion contained 156 amino acids, while a 49–amino acid N-terminal fragment was extracellular, indicating that sequences between positions 49 and 156 are required for internalization. The virulence allele of this gene, avrM, lacks a 62–amino acid region between positions 45 and 108 of AvrM-A. We therefore tested an avrM-Cerulean fusion protein and found that this was also localized to the cytoplasm. This suggests that the virulence allele does indeed encode for a functionally translocated effector and that amino acids in the deleted region are not required for uptake. We further tested several internal deletions of AvrM-A for their effect on Cerulean localization (Figure 4A). Deletion of the region between amino acids 35 and 106 had no effect on accumulation of intracellular fluorescence (Figure 4). However, deletion of amino acids 35 to 153 or 106 to 153 led to the accumulation of Cerulean fluorescence solely in the apoplastic space. Because the C-terminal region of AvrM-A after amino acid 196 is sufficient for recognition by the M resistance protein (Catanzariti et al., 2010), we also tested the effect of these truncations on recognition of the secreted proteins in M transgenic tobacco (Figure 4B). Expression of the 35-106 deletion protein still induced an HR in M tobacco, while the 35-153 and 106-153 deletions abrogated the M-dependent HR, consistent with the extracellular localization of these proteins. Finally, we generated a Cerulean fusion containing just the signal peptide and the 106-156 region of AvrM (SP106-156). Transient expression of this construct gave rise to intracellular fluorescence (Figures 4A and 4C). Immunoblot analysis confirmed that the internally deleted protein fusions accumulated to levels equivalent to the wild-type protein (see Supplemental Figure 2E online). Taken together, these results indicate that residues in the region between amino acids 106 and 156 are both necessary and sufficient for internalization of secreted AvrM.
Figure 4.
Deletion Analysis of AvrM.
(A) A series of C-terminal and internal truncations of AvrM fused to Cerulean were compared with wild-type AvrM-Cerulean in transient expression assays in tobacco cells. The fusion proteins included the 28–amino acid signal peptide (SP, dark gray), various regions of AvrM protein (unshaded), and a C-terminal cerulean domain (light gray). One fusion included the full-length virulence allele of AvrM (avrM), which contains an internal deletion of 62 amino acids. Localization of Cerulean fluorescence was assessed by confocal microscopy of infiltrated tissues after plasmolysis. Protein fusions showing intracellular fluorescence are indicated by +, with – indicating those showing solely extracellular localization.
(B) Fusions of Cerulean to either wild-type AvrM (left leaf panels) or truncated AvrM proteins with internal deletion of amino acids 35 to 106, 35 to 153, or 106 to 153 (Δ35-106 etc, right leaf panels) were expressed in W38:M tobacco by agroinfiltration. Images prepared 48 h after infiltration.
(C) Confocal images of W38 tobacco cells expressing AvrM-Cerulean fusion proteins with internal deletions of amino acids 35 to 153, 35 to 106, or SP106-156 after plasmolysis. The Δ35-106 fusion protein is intracellular, while Δ35-153 is extracellular. Bars = 25 μm.
Mutational Analysis of AvrM Uptake Motifs
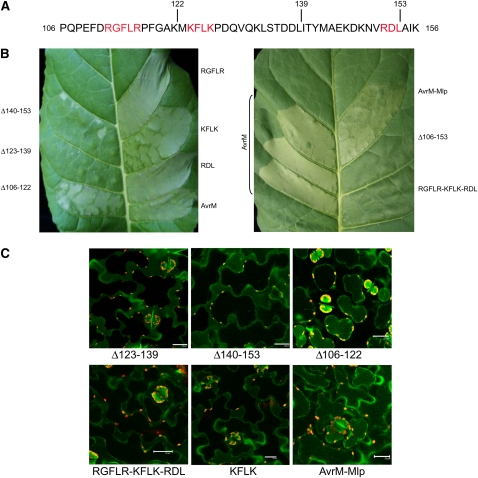
Although the 106-156 region of AvrM does not contain a clearly conserved RxLR motif such as found in the uptake region of the oomycete effectors, there are three short sequences, RGFLR, KFLK, and RDL, that contain positively charged and hydrophobic residues in a similar arrangement (Figure 5A). Unlike the RxLR signal, these sequences do not occur within the N-terminal 60 amino acids, and there is no downstream dEER acidic motif. We generated Ala substitutions in these sequences and assayed their effects on AvrM uptake. The RDL-AAA and RGFLR-AAAAA substitutions had no effect on Cerulean localization or HR induction; however, the KFLK-AAAA mutation resulted in a slight but consistent delay in HR induction on M tobacco (Figure 5B). This was evident as a weaker HR phenotype at 48 h, but by 3 d, the phenotype was similar to the wild-type AvrM-Cerulean fusion. Consistent with this observation, Cerulean fluorescence was observed largely in the extracellular space at 24 to 48 h but was detected intracellularly by 3 d after infiltration (Figure 5C). This suggested that this mutation partially compromised, but did not completely abolish, the uptake process. To test for possible functional redundancy of these motifs, we generated a mutant in which all three motifs were converted to Ala (triple mutant). This mutation abolished uptake (Figure 5B), suggesting these motifs may contribute independently to translocation. We therefore tested three further deletions within this region to determine what other features may be involved in the uptake signal. Deletion of the region between 106 and 122, which includes the RFGLR sequence, had no effect on either HR induction or Cerulean internalization (Figures 5B and 5C). However, deletion of amino acids 123 to 139 (containing KFLK) or 140 to 153 (containing RDL) abolished HR induction and led to extracellular accumulation of Cerulean, indicating that amino acids in these regions are required for uptake. In this case, both regions are required and do not seem to act independently.
Figure 5.
An Uptake Signal in the AvrM N-Terminal Region.
(A) Amino acid sequence of AvrM from positions 106 to 156. Three short motifs with some similarity to the oomycete RxLR motifs are shown in red, and the sites of three internal deletions, 106-122, 123-139, and 140-153 are indicated.
(B) The effects of agroinfiltration and expression in W38:M tobacco of fusions of Cerulean to either wild-type AvrM or mutant AvrM with the RGFLR, KFLK, or RDL motifs substituted with Ala residues, AvrM with the 123 to 147 region replaced by equivalent sequence in Mlp-37970 (AvrM-Mlp), or AvrM with internal deletions of amino acids 106-122, 123-139, 140-153, or 106-153 (Δ106-122, etc.). Images were prepared 48 HAI.
(C) Confocal images of plasmolysed tobacco cells expressing AvrM-Cerulean fusions containing the 106-122, 123-139, and 140-153 deletions, the triple mutation RGFLR-KFLK-RDL-AAAAA-AAAA-AAA, the KFLK-AAAA mutation, and the Mlp-37970 substitution. Fluorescence occurs in the cytoplasm for Δ106-122 fusion protein but occurs in the apoplast for the other three fusion proteins. Images were prepared 48 HAI. Bars = 25 μm.
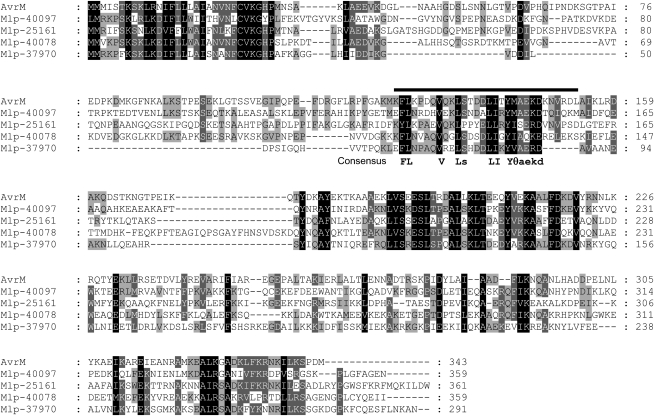
To examine the conservation of the predicted AvrM uptake region, we compared AvrM to homologous sequences present in the recently released genome sequence of poplar rust, Melampsora larici-populina (Mlp), which belongs to the same genus as flax rust. Figure 6 shows an alignment of AvrM to the four most similar predicted proteins in poplar rust. The amino acid sequences of these proteins show significant similarity to AvrM in the C-terminal region corresponding to the recognition domain of AvrM and in the N-terminal signal peptide. However, the intervening regions are highly divergent, except for an ∼23–amino acid sequence within the 123-153 region of AvrM that is required for uptake. This suggests that this sequence shares a conserved function in these related proteins. Most of the conserved positions within this sequence contain hydrophobic residues. In the KFLK motif, whose mutation partially compromised AvrM uptake, the F and L positions are highly conserved, while the two Lys residues are substituted by other hydrophilic residues (E or N). We confirmed that the conserved region of Mlp-37970 contains a functional uptake signal using this sequence to replace the equivalent region of AvrM (amino acids 124 to 147). The AvrM-Mlp-Cerulean fusion protein induced HR similar to wild-type AvrM on M transgenic tobacco and showed intracellular fluorescence (Figure 5B).
Figure 6.
AvrM Homologs in Poplar Rust (M. larici-populina).
Alignment of AvrM to four related proteins predicted in the M. larici-populina genome sequence. The putative uptake region of AvrM between amino acids 123 and 153 in AvrM is indicated by the bar, and the consensus sequence of this strongly conserved region is shown below the alignment. A text file of this alignment is provided as Supplemental Data Set 1 online.
An N-Terminal Region of AvrL567 Determines Uptake by Plant Cells
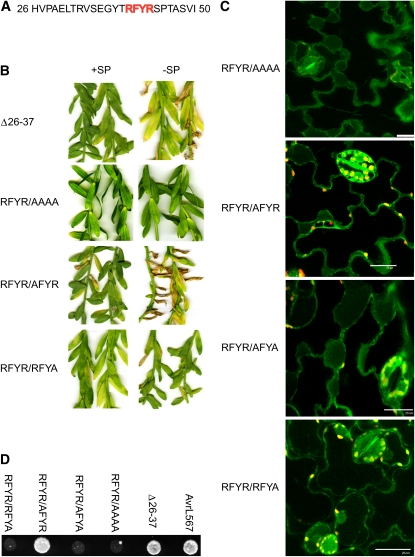
We also tested the ability of the flax rust effector AvrL567-A to enter plant cells after secretion. Similarly to AvrM, we found that Agrobacterium-mediated transient expression of AvrL567-Cerulean with and without the signal peptide triggered HR in leaves of flax plants carrying the L6 resistance gene (see Supplemental Figure 2A online), again suggesting that the AvrL567-Cerulean protein was internalized after secretion. Immunoblot analysis detected the expected 44-kD protein corresponding to intact AvrL567-Cerulean fusion (see Supplemental Figure 2B online), confirming the stability of the fusion protein in vivo. Confocal microscopy showed that AvrL567-Cerulean accumulated in the plant cytoplasm (Figure 7A), while fusion of the signal peptide alone to Cerulean resulted in extracellular fluorescence (data not shown). Analysis of C-terminal truncations of AvrL567 revealed that amino acids 1 to 50 were sufficient to direct intracellular localization of Cerulean, while a smaller fragment consisting of amino acids 1 to 41 allowed only apoplastic accumulation of Cerulean (Figure 7B). Thus, amino acids between position 41 and 50 are required for the uptake of this protein. We also tested an internal deletion lacking amino acids 26 to 37, which abrogated both HR induction and accumulation of intracellular Cerulean. This deletion did not affect recognition of a nonsecreted AvrL567 protein (lacking the signal peptide) in flax and also had no effect on interaction with the L6 protein in a yeast two-hybrid (y2h) assay (Figures 8B and 8D).
Figure 7.
Deletion Analysis of AvrL567.
(A) Confocal images of tobacco cells expressing AvrL567-Cerulean. The left panel shows fluorescence of untreated cells, the middle panel shows cells plasmolysed with 0.8 M mannitol, and the right panel shows the transmission image of the plasmolysed cells. Closed arrowheads indicate the plant plasma membrane where it is pulled away from the cell wall (arrow) and open arrowheads indicate nuclei. Cerulean fluorescence and chloroplast autofluorescence were imaged using the 405- and 543-nm laser excitation light sources, respectively, with appropriate emission filters. Bars = 25 μm.
(B) A series of C-terminal and internal truncations of AvrL567 fused to Cerulean were generated and assayed by Agrobacterium-mediated transient expression in tobacco cells. The fusion proteins included the 24–amino acid signal peptide (SP, dark gray), various regions of AvrL567 (unshaded), and a C-terminal cerulean domain (light gray). Localization of Cerulean fluorescence was assessed by confocal microscopy of infiltrated tissues after plasmolysis of the plant cells.
Figure 8.
An Uptake Signal in the AvrL567 N-Terminal Region.
(A) Amino acid sequence of AvrL567 from positions 27 to 50. A short motif (RFYR) similar to the oomycete RxLR motifs is shown in red.
(B) AvrL567-Cerulean fusion constructs either with (+SP) or without (−SP) the signal peptide were expressed in the flax line Birio (which contains the L6 resistance gene). AvrL567-Cerulean products contained the amino acid 26-37 internal deletion (Δ26-37) or the RFYR to AAAA, AFYR, and RFYA mutations. Images prepared 14 d after infiltration.
(C) Confocal images of plasmolysed tobacco cells expressing AvrL567-Cerulean with various substitutions in the RFYR sequence. Bars = 25 μm.
(D) The y2h analysis of the interaction between L6 and AvrL567 mutant proteins. Growth of strain HF7c on media lacking His. These strains express the GAL4 DNA binding domain fused to the L6 protein along with the GAL4 activation domain fused to AvrL567-A (wild type), the amino acid 26-37 internal deletion (Δ26-37), and the RFYR to AAAA, AFYA, AFYR, and RFYA mutants.
Thus, amino acid sequence features in the AvrL567 N-terminal region from amino acid 26 to 50 are implicated in protein translocation (Figure 8A). This region does not show any similarity to the uptake region of AvrM. However, it contains a sequence, RFYR, which is similar to the RxLR transport motif of oomycete effectors. We therefore tested several mutations of this sequence for their effect on protein uptake. Mutation of RFYR to AAAA was found to prevent both intracellular accumulation of AvrL567-CRL and HR induction (Figures 8B and 8C). However, this mutation also prevented HR induction when introduced into a nonsecreted version of AvrL567, as well as knocking out the interaction with L6 in a y2h assay (Figures 8B and 8D), indicating that it has an additional effect on recognition separate from any effect on uptake. Indeed, in the crystal structure of AvrL567, F41 is deeply buried in the protein and is probably critical for overall folding. The second R residue (R43) is also largely buried, with part of the charged amine group exposed on the opposite side of the protein to the rest of the amino acid side chains in this sequence (see Supplemental Figure 3 online). We therefore tested single and double Ala substitutions for the two Arg residues, R40 and R43, for their effect on protein uptake and recognition (Figures 8B to 8D). Mutation of both Arg residues together (RFYR/AFYA) abolished both HR induction and intracellular localization of the secreted AvrL567-Cerulean but also prevented recognition of the nonsecreted version in planta as well as in y2h assays. However, mutation of R40 alone (RFYR/AFYR) had no effect on recognition of the nonsecreted protein in planta or in y2h assays but did prevent HR induction and intracellular localization of the secreted protein. On the other hand, R43 was required for AvrL567 recognition both in planta and in y2h assays. However, the secreted AvrL567-Cerulean containing this mutation (RFYR/RFYA) was intracellular, indicating that R43 is not important for uptake. All of the mutant protein variants were stably expressed in both yeast and in planta (see Supplemental Figures 2C and 2D online).
DISCUSSION
Translocation of pathogen effector proteins into the host cell cytosol is a key determinant for the pathogenicity of many bacterial and oomycete plant pathogens (Chisholm et al., 2006; Birch et al., 2008; Tyler, 2009). A number of fungal Avr proteins are also inferred to be delivered into host cells, based on their intracellular recognition by R proteins (Jia et al., 2000; Dodds et al., 2004; Catanzariti et al., 2006; Ridout et al., 2006; Houterman et al., 2009; Li et al., 2009). For instance, flax rust Avr proteins have been shown to trigger HR when transiently expressed as intracellular proteins in flax leaves expressing corresponding R genes (Dodds et al., 2004; Catanzariti et al., 2006), and recognition of both AvrM and AvrL567 is determined by direct interaction with cytoplasmic resistance proteins (Dodds et al., 2006; Catanzariti et al., 2010). Here, we show by immunolocalization that the flax rust AvrM secreted protein is localized within the cytoplasm of plant cells during infection, confirming its delivery from rust haustoria. Furthermore, uptake of secreted AvrL567 and AvrM proteins into plant cells can occur in the absence of the pathogen, suggesting that they do not require a pathogen-encoded transport mechanism. Uptake of these proteins is dependent on signals in their N-terminal regions, but the primary sequence features of these uptake regions are not related to each other and also differ from the conserved RxLR uptake motif that has been defined in oomycete secreted effector proteins (Rehmany et al., 2005; Birch et al., 2006; Jiang et al., 2008).
AvrM Is Delivered into Host Cells during Infection
The AvrM protein is secreted by both intercellular hyphae and haustoria of flax rust, and we observed accumulation of AvrM protein in some host cells containing haustoria (Figure 1). Translocation of AvrM into plant cells is likely to occur mostly from haustoria, as only haustoria-infected cells showed positive labeling with AvrM antibodies and no signal could be detected in cells neighboring hyphal tips. This localization is similar to that of the bean rust RTP1p protein, which was previously found to be secreted from Uromyces fabae haustoria and transferred into infected host cells (Kemen et al., 2005). The delivery of AvrM into host cells seemed to be temporally regulated. At 5 d after inoculation, we observed cytoplasmic labeling in ∼13% of host cells containing an haustorium. However, at earlier time points, AvrM was only detected in the haustorial wall or EHM, but not in host cells (Table 1). It is possible that low levels of AvrM protein were present in the host cytoplasm at earlier time points but were below the detection limit with this antibody. Nevertheless, it was clear that at all time points, the majority of AvrM signal was detected surrounding the haustorium with only low levels observed in the host cytoplasm. This may reflect a concentration dependent process, whereby significant AvrM translocation does not occur until a high concentration is reached within the EHM. Alternatively, it may reflect a developmental regulation of the transport process. The observation that AvrM was mainly accumulated in the haustorial cell wall suggests that it is not freely diffusible in the EHM. It is possible that AvrM is initially sequestered in the haustorial wall and is subsequently released from the wall allowing transport into the host (Figure 1D). Alternatively, AvrM may have a specific function in the fungal cell wall since it is also present in the hyphal wall (Figure 1Di). Another possibility is that translocated AvrM protein is rapidly degraded in the host cell. Similarly, RTP1 was first detected only in the EHM of early stage haustoria and accumulated in the host cytosol at later stages of haustorium development (Kemen et al., 2005).
Uptake of Avr Proteins Can Occur Independently of the Rust Pathogen
One important question is whether the entry of rust effectors into plant cells depends on a translocation mechanism provided by the pathogen, as is the case of Type III secreted effectors of gram-negative bacterial pathogens (Chisholm et al., 2006), or whether it occurs via a pathogen-independent process. Our data support the second hypothesis since expression of the secreted AvrM and AvrL567 Cerulean fusion proteins in plant cells resulted in their intracellular accumulation (Figure 3). The signal peptides of these proteins are functional in plants, as they directed a fused Cerulean to the apoplastic space, and addition of an HDEL tag resulted in retention of the secreted proteins in the ER, confirming that the fusion proteins do enter the secretion pathway (Figures 3 and 4). Thus, the AvrM-Cerulean and AvrL567-Cerulean fusion proteins accumulate within the plant cell cytoplasm after passing into the secretory pathway, which implies that the secreted proteins must cross a plant membrane. Since ER retention blocks cytosolic localization, membrane penetration must occur from either the late stage secretory vesicles or across the plasma membrane after secretion into the apoplast. Importantly, this occurs in the absence of the pathogen and therefore does not require a pathogen-derived translocation mechanism. These effectors may either contain autonomous membrane crossing properties or may hijack a plant-derived membrane transport system. Although transport can occur in the absence of the rust, it is nevertheless possible that the pathogen imposes a level of regulation over transport, which may explain why AvrM protein was only detected in the host cytosol late in the infection process. Similarly, ToxA, a host selective toxin delivered from Pyrenophora tritici-repentis, has been shown to translocate into wheat (Triticum aestivum) cells (Manning and Ciuffetti, 2005).
The RxLR-dEER motif of oomycete effectors controls effector delivery during infection (Whisson et al., 2007; Grouffaud et al., 2008), and Dou et al. (2008) showed that this motif also controls delivery of P. sojae Avr1b into plant cells in the absence of the pathogen. This motif was required for recognition of Avr1b transiently expressed in soybean (Glycine max) cells as a secreted protein but not when it was expressed intracellularly. In addition, fusion to the Avr1b RxLR-dEER domain allowed purified GFP to enter soybean root cells from solution. These results suggest that uptake of RxLR effectors into host cells is not dependent on a pathogen-derived transport mechanism. This contrasts with recent findings for malaria effectors that contain the related PEXEL motif (RxLxE/Q). These effectors are subjected to proteolytic cleavage immediately after the central Leu residue, and an N-acetyl group is added to the mature protein (Chang et al., 2008; Boddey et al., 2009). PEXEL cleavage and modification occurs within the parasite endomembrane system, meaning that the native motif is no longer present when it encounters the host membrane. de Koning-Ward et al. (2009) recently identified a protein complex present in the parasitophorous vacuole that includes an ATP-driven HSP101 chaperone and a potential membrane pore protein and could therefore constitute a pathogen-encoded translocation machine. However, these proteins are apparently unique to Plasmodium species, suggesting that despite the similarity in the uptake motifs, the translocation mechanism may not be conserved between oomycetes and plasmodium.
Uptake of Avr Proteins Depends on N-Terminal Signals
For both AvrM and AvrL567, intracellular accumulation was dependent on sequence features in the N-terminal region (Figures 4 and 7). These regions are distinct from the recognition domains; hence, these Avr proteins have a bipartite structure, with an N-terminal region directing transport of the protein first into the rust secretory pathway, and then across the host membrane, while the C-terminal regions are likely to represent the functional effector domains. These C-terminal regions are sufficient for recognition by the corresponding R proteins (Figure 8D; Catanzariti et al., 2010), but there is not yet any direct evidence for a pathogenicity function of AvrL567 and AvrM. Oomycete effectors also appear to have a bipartite structure, with distinct N-terminal uptake (RxLR) and C-terminal effector domains (Bos et al., 2006; Jiang et al., 2008). For AvrM, uptake was dependent on the region from amino acids 106 to 156 (Figure 5), which includes a 23–amino acid stretch that is strongly conserved in AvrM homologs from poplar rust, despite a high level of diversity in the rest of the N-terminal region (Figure 6). The equivalent region from Mlp-37970 was functional when incorporated into AvrM (Figure 5B). Although this region of AvrM contains some sequences loosely related to the RxLR motif (RGFLR, KFLK, and RDLA), it is not clear whether these are the critical residues for uptake. Mutation of one of these motifs (KFLK) slightly delayed uptake, while the other two had no effect. However, mutation of all three motifs together abolished uptake, suggesting that they could act redundantly. However, this is not consistent with the deletion results, which suggest that the regions 123-139 and 140-153 are both required. It is possible that either the triple mutant, which has 12 Ala substitutions, or the deletion constructs, or both may disrupt important structural elements of the uptake signal. The conserved region 125-148 shows a pattern of conserved hydrophobic residues, which may form an amphipathic structure, followed by three charged residues (EKD; Figure 6). Interestingly, the KFLK-AAAA mutation, which partially compromises uptake, disrupts two of the conserved hydrophobic residues (FL). On the other hand, the equivalent sequence in Mlp-37970 is EFLN, which is fully functional when substituted into AvrM, indicating that the positively charged Lys residues are not required at these positions.
In contrast with AvrM, the AvrL567 uptake region occurs immediately after the SP and depends on amino acids between positions 26 and 50. This region includes an RxLR-like sequence (RFYR). However, mutation analyses indicated that the first Arg (R40) is required for uptake but not recognition of AvrL567 protein, while the second Arg (R43) has a role in recognition but not uptake (Figure 8). Thus, the similarity of this sequence to the RxLR motif may be coincidental rather than reflecting a conserved function. In addition to R40, the sequence between amino acids 26 and 37 is also required for uptake. This amino acid region has no fixed structure (Wang et al., 2007) and constitutes a flexible tail that extends from the globular AvrL567 protein. R40 is located on the surface of the protein close to this flexible tail and therefore may form part of an exposed uptake signal.
These results suggest that pathogen-independent intracellular delivery of AvrM and AvrL567 depends on N-terminal uptake signals. While some sequence features are similar to RxLR in terms of positively charged residues, they do not conform to all known RxLR characteristics. For instance, Whisson et al. (2007) showed that Lys residues could not substitute for Arg residues in the RxLR motif of Avr3a. The uptake regions of AvrM and AvrL567 are also not similar to each other or to sequences present in other known effectors from rust or other fungi. The apparent divergence between the transport signals of oomycete and fungal effectors is striking, especially given the strong conservation within the oomycetes and the lack of conservation even between rust effectors. Most oomycete RxLR effectors are members of a large superfamily of related sequences that may have expanded from a single ancestral sequence (Jiang et al., 2008). Thus, the conservation of the RxLR motif may reflect their relatively recent divergence. Other non-RxLR effectors also occur in oomycetes, such as the crinkler family (Kamoun, 2006). Although the divergence between these uptake signals may suggest that different transport mechanisms are involved, it is possible that convergent evolution has led to targeting of similar host transport pathways by these effectors without leaving a clearly recognizable sequence relationship. Detailed mutagenesis of AvrM and AvrL567 uptake regions will be required to determine precise sequence requirements for translocation.
The structure formed by the defined uptake peptides can help elucidate the transport mechanism of these proteins. Internalization of P. tritici-repentis ToxA into wheat cells depends on a solvent-exposed loop of the ToxA protein, which contains the cell attachment motif RGD (Manning et al., 2008). Mutation of individual residues within the RGD loop impaired internalization, suggesting the importance of the RGD loop structure in the internalization process, possibly through recognition by a receptor at the plant cell plasma membrane.
Rust effector protein uptake signals may be considered as a class of cell penetrating peptides (CPPs), which have been identified in numerous proteins from distant phylogenetic groups, including the Tat protein of the human immunodeficiency virus type1 (Frankel and Pabo, 1988) and the Antennapedia protein of Drosophila melanogaster (Derossi et al., 1994), and facilitate protein trafficking across membranes. CPPs have diverse sequences, and while some are rich in positively charged amino acids, others consist of amphipathic structures (Eiriksdottir et al., 2010). CPPs can engage in different uptake pathways, including endocytosis, macropinocytosis, and direct plasma membrane penetration, sometimes simultaneously (Duchardt et al., 2007). Several CPPs defined in mammalian systems have also been shown to function in plants (Chugh and Eudes, 2008), indicating that they can engage appropriate uptake mechanisms in plant cells. However, these mechanisms are not yet understood; likewise, we still know little of the uptake pathway of rust effector proteins. Ultrastructure studies have detected tubular extensions of the extrahaustorial membrane with associated budding vesicles that reach into the host cell cytoplasm and can form close contact with host ER and dictyosomes (Mims et al., 2002). Although this suggests that endocytosis may be a key process in the uptake of effector proteins, it remains to be determined what uptake pathways are involved in rust Avr protein delivery.
METHODS
Rust and Plant Materials
The flax rust (Melampsora lini) strain CH5 is the F1 hybrid of a cross between parental strains C and H, while CH5-120 and CH5-132 are two individuals from the F2 family derived from selfing rust CH5 and are homozygous for the avirulence and virulence alleles of AvrM, respectively (Lawrence et al., 1981). The flax lines Dakota and Birio contain the M or L6 resistance genes, respectively (Lawrence et al., 1981). A transgenic W38 tobacco (Nicotiana tabacum) line expressing the M gene was described by Catanzariti et al. (2006). Rust inoculations were performed as described by Lawrence et al. (1981).
Expression of His-Tagged AvrM, Antibody Generation, and Affinity Purification of Antibodies
AvrM-A (lacking the signal peptide) was expressed with an N-terminal hexahistidine tag in BL21 (DE3) pLysS Escherichia coli from the vector pET15b. Luria-Bertani broth cultures were grown to midexponential phase and induced with 1 mM isopropyl-thio-β-d-galactoside for 16 h at 16°C. Cells were harvested by centrifugation and resuspended in 1/20 of the original culture volume of column buffer (20 mM HEPES, pH 8.0, and 300 mM NaCl) with 10 mM imidazole and 1× Ni-column compatible protease inhibitor cocktail (Sigma-Aldrich). Cells were lysed by three freeze-thaw cycles in the presence of lysozyme, and the cell lysate was loaded onto a 1 mL Ni-sepharose 6 Fast Flow column (GE Healthcare) preequilibrated with 10 column volumes of buffer. Then, the column was washed with 25 volumes of column buffer with 65 mM imidazole before elution of AvrM-A in column buffer with 250 mM imidazole. The protein was then buffer exchanged into (2.7 mM KCl, 138 mM NaCl, 1.5 mM KH2PO4, and 20.4 mM NaH2PO4, pH 7) and concentrated using a 30 kD MWCO Amicon Ultracentrifugation device (Millipore), snap frozen in liquid nitrogen, and stored at ×80°C.
Antibodies were raised in rabbits inoculated four times with 400 μg of purified fusion protein with Freund's adjuvant. Sera were purified by negative adsorption against protein from uninfected flax plants bound to HybondC nitrocellulose membranes (Amersham) in a protocol modified from Robinson et al. (1988). Briefly, proteins extracted from uninfected flax plants were spotted onto a nitrocellulose membrane, stained with Ponceau S, and rinsed three times for 5 min in TBS (20 mM Tris-Cl, pH 7.4, and 500 mM NaCl). Then, poorly bound protein was removed by washing with 200 mM glycine, pH 2.5. This was followed by rinsing the membranes three times for 5 min with TBST (20 mM Tris-Cl, pH 7.4, 500 mM NaCl, and 0.05% [v/v] Tween 20) and blocking with 1% BSA and 2% nonfat milk for 2 h at room temperature. The membranes were then cut to 5 × 5 mm and incubated overnight at 4°C with diluted sera. Sera were then used for immunolocalization studies.
Immunoblot Analysis
Tissue from infected flax leaves or agroinfiltrated tobacco leaves was frozen in liquid nitrogen and ground in either 50 mM Tris-Cl, pH 7.5, 5 mM EDTA, pH 8.0, 10% glycerol, or 150 mM NaCl, 20 mM Tris-Cl, pH 6.5, 1 mM EDTA, 1% Triton X-100, 0.1% β-mercaptoethanol, and 1 mM PMSF. In both cases, extraction buffers were supplemented with Roche Complete protease inhibitor cocktail (Roche Diagnostics). The samples were then centrifuged to remove cellular debris before boiling for 5 min in the presence of Laemmli buffer with β-mercaptoethanol. Samples containing Cerulean fusion proteins were not boiled as this treatment was found to cause significant cleavage of the 27-kD Cerulean fragment from the fusion protein. Samples were separated by SDS-PAGE and electroblotted to Hybond C-extra nitrocellulose membranes (Amersham Biosciences). AvrM protein was detected using anti-AvrM primary antibodies and anti-rabbit secondary antibodies conjugated to alkaline phosphatase (Chemicon International) with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as the color substrate. Cerulean and Cerulean fusion proteins were detected by the SuperSignal West Pico chemiluminescence kit (Pierce) using an anti-GFP primary antibody (Roche Diagnostics) and anti-mouse secondary antibodies conjugated to horseradish peroxidase (Bio-Rad).
Immunolocalization of AvrM
Leaves from Hoshangabad flax plants were sampled at 12, 16, 24, 64, and 120 HAI with CH5 flax rust (compatible interaction). The lower epidermis was peeled off and leaves were cut into 3 × 3-mm pieces and fixed under vacuum in 4% (v/v) formaldehyde with 0.2% (v/v) glutaraldehyde in 50 mM PIPES buffer, pH 7.0, and 0.02% Triton X-100, dehydrated, and embedded in butyl-methyl methacrylate as described by Kobayashi et al. (1994). Infected leaves were sectioned using an Ultracut microtome (Reichert-Jung, Cambridge Instruments). After removing embedding resin in acetone, sections were blocked for 1 h in 1% (w/v) BSA and 0.1% (v/v) fish scale gelatin in PBS. Sections were then incubated in primary anti-AvrM antibodies for 2 h at 37°C, rinsed three times, and then incubated in secondary goat anti-rabbit antibodies conjugated to Alexa A488 (Molecular Probes). For double-labeling studies, sections were coincubated with rabbit anti-AvrM antibodies and mouse monoclonal antihaustorial antibody ML1 (Murdoch and Hardham, 1998) followed by coincubation with sheep anti-mouse antibodies conjugated to rhodamine (Silenus Laboratories) and goat anti-rabbit antibodies conjugated to Alexa A488 for 2 h at 37°C. Following incubation with secondary antibodies, slides were rinsed, stained with 1 μg/mL 4',6-diamidino-2-phenylindole (Sigma-Aldrich), and mounted in a glycerol-based mounting medium containing Mowiol 4-88 (Hoechst Australia) containing the antifade agent p-phenylenediamine (Sigma-Aldrich). Sections were viewed using an Axioplan microscope (Zeiss) and photographed with a digital camera (Princeton Instruments).
For immunogold labeling, the lower epidermis was removed from infected leaves, and leaves were fixed in a vacuum for 2 h in 2% glutaraldehyde in 500 mM PIPES buffer, pH 6.9, and embedded in Lowicryl K4M (Lowi) as previously described (Hardham, 2001). Lowicryl ultrathin sections were blocked with 1% (w/v) BSA plus 0.1% (v/v) fish scale gelatin in PBS for 20 min and then incubated with anti-AvrM antibody diluted in 1% (w/v) BSA plus 0.1% (v/v) fish scale gelatin for 1 h at room temperature. Sections were rinsed three times in PBT (PBS with 0.2% Tween 20) and incubated with goat anti-rabbit antibody conjugated to 10-nm gold (GE Healthcare) for 1 h at room temperature. Sections were counterstained with uranyl acetate and lead citrate and examined with a Hitachi H7100FA transmission electron microscope equipped with a digital camera. Image processing was performed using Adobe Photoshop CS2 (Adobe Systems) and Adobe Illustrator CS2 (Adobe Systems).
Transient in Planta Expression Assays
Gene expression constructs contained the AvrM and AvrL567 coding sequences fused to the Cerulean version of cyan fluorescent protein were generated in the vector pCAMBIA3301, driven by the nopaline synthase promoter and terminator sequences (De Block et al., 1984; Horsch et al., 1984). The AvrM-Cerulean and AvrL567-Cerulean cDNA expression constructs were prepared by inserting a cloned PCR product as a SpeI-PvuI fragment, using primers AvrM-5′, 5′-CGCACTAGTATGATGATGATTTCTACAAAGTCG-3′; AvrM-3′, 5′-GCTCACCATAGCTGCTGCAGCCGCTGCAGCCATGTCTGGAGATTTC-3′; AvrL567-5′, 5′-CGCACTAGTATGAAGATTAATCTATCATGG-3′; AvrL567-3′, 5′-GCTCACCATAGCTGCTGCAGCCGCTGCAGCGGTCTTTGGAACCAACTTGA-3′; Cerulean-5′, 5′-GCTGCAGCGGCTGCAGCAGCTATGGTGAGCAAGGGCGAGGAGCTGTT-3′; and Cerulean-3′, 5′-CTCGATCGTTACTTGTACAGCTCGTCCATGC-3′. Constructs encoding the truncated AvrM and AvrL567 proteins without the signal peptides start from the internal Met residues at positions 31 and 24, respectively. The stop codons of both AvrM and AvrL567 proteins were removed to create in-frame fusions to Cerulean. Truncation and Ala mutations within AvrM and AvrL567 coding sequences were generated by overlapping PCR. C-terminally truncated fragments were fused directly to Cerulean at an in-frame XbaI site, while the other constructs contain a seven Ala linker. AvrM-Cerulean constructs encoding C-terminal HDEL or HDDL tetrapeptides tags contained 12 additional bases (CATGATGAGCTC or CATGATGACCTC, respectively) inserted immediately prior to the stop codon of Cerulean. All constructs were fully sequenced to confirm their integrity.
Agrobacterium cultures of strain GV3101 pMP90 containing the binary vector expression constructs were prepared at an OD600 of 0.1 or 0.2 for AvrM and 0.5 for AvrL567 constructs, in Murashige and Skoog (1962) basal medium supplemented with 200 μM acetosyringone and infiltrated into flax or tobacco leaves using a 1-mL syringe.
Confocal Microscopy
Plant tissue samples were harvested from infiltrated tobacco or flax leaves at either 2 or 3 d after infiltration, and tissue was either directly imaged or infiltrated with 0.8 M mannitol solution, for plasmolysis, 15 to 20 min prior to imaging. Live tissue imaging was performed on a Leica microscope with a Leica SP2 confocal system using ×40 and ×63 C-Apochromat water immersion objectives. The 405- and 458-nm laser lines with appropriate emission filters were used to image Cerulean and chloroplast autofluorescence, respectively.
Yeast Two-Hybrid Analysis
GAL4 DNA binding domain and transcriptional activation domain fusions to AvrL567 protein variants were prepared in the pGBT9 and pGADT7 vectors, respectively (Clontech) as described (Dodds et al., 2006). L5 and L6 GBT9 and GADT7 clones were described previously (Dodds et al., 2006). Yeast transformation and His growth assays were performed as described in the Yeast Protocols Handbook (Clontech). Yeast proteins were extracted by the trichloroacetic acid method, and immunoblotting was performed as described above with detection by anti-HA (Roche Molecular Systems) monoclonal antibodies followed by goat anti-mouse/horseradish peroxidase (Pierce). Labeling was detected with the SuperSignal West Pico chemiluminescence kit (Pierce).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers DQ279864.1 (AvrM) and AY510102.1 (AvrL567).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunoblot with Affinity-Purified anti-AvrM Antibodies.
Supplemental Figure 2. Expression of AvrL567-Cerulean and AvrM-Cerulean Protein Fusions in Plants.
Supplemental Figure 3. Location of RFYR Motif in the AvrL567 Protein Structure.
Supplemental Data Set 1. Text File of AvrM Homolog Alignment.
Acknowledgments
We thank Kim Newell and Patricia Moore for excellent technical assistance. Funding for this work was provided by the Australian Research Council (DP0771374), the Australian Grains Research and Development Corporation (CSP00099), and the U.S. National Institutes of Health (GM074265-01A2). We thank D. Piston for supplying the Cerulean plasmid.
References
- Allen R.L., Bittner-Eddy P.D., Grenvitte-Briggs L.J., Meitz J.C., Rehmany A.P., Rose L.E., Beynon J.L. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Armstrong M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102: 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett L.G., Thrall P.H., Dodds P.N., van der Merwe M., Linde C.C., Burdon J.J. (2009). Diversifying selection and geopraphic structure drive adaptive evolution of avirulence loci in a wild host-pathogen interaction. Mol. Biol. Evol. 26: 2499–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Hiller N.L., Liolios K., Win J., Kanneganti T.D., Young C., Kamoun S., Haldar K. (2006). The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch P.R.J., Boevink P.C., Gilroy E.M., Hein I., Pritchard L., Whisson S.C. (2008). Oomycete RXLR effectors: Delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11: 373–379 [DOI] [PubMed] [Google Scholar]
- Birch P.R.J., Rehmany A.P., Pritchard L., Kamoun S., Beynon J.L. (2006). Trafficking arms: Oomycete effectors enter host plant cells. Trends Microbiol. 14: 8–11 [DOI] [PubMed] [Google Scholar]
- Boddey J.A., Moritz R.L., Simpson R.J., Cowman A.F. (2009). Role of the plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I.B., Kanneganti T.D., Young C., Cakir C., Huitema E., Win J., Armstrong M.R., Birch P.R., Kamoun S. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48: 165–176 [DOI] [PubMed] [Google Scholar]
- Catanzariti A.M., Dodds P.N., Lawrence G.J., Ayliffe M.A., Ellis J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.M., Dodds P.N., Ve T., Kobe B., Ellis J.G., Staskawicz B.J. (2010). The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol. Plant Microbe Interact. 23: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.H., Falick A.M., Carlton P.M., Sedat J.W., DeRisi J.L., Marletta M.A. (2008). N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160: 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chugh A., Eudes F. (2008). Cellular uptake of cell-penetrating peptides pVEC and transportan in plants. J. Pept. Sci. 14: 477–481 [DOI] [PubMed] [Google Scholar]
- De Block M., Herrera-Estrella L., Van Montagu M., Schell J., Zambryski P. (1984). Expression of foreign genes in regenerated plants and in their progeny. EMBO J. 3: 1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward T.F., Gilson P.R., Boddey J.A., Rug M., Smith B.J., Papenfuss A.T., Sanders P.R., Lundie R.J., Maier A.G., Cowman A.F., Crabb B.S. (2009). A newly discovered protein export machine in malaria parasites. Nature 459: 945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J., De Rycke R., Botterman J. (1992). Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 11: 2345–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derossi D., Joliot A.H., Chassaing G., Prochiantz A. (1994). The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 269: 10444–10450 [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Ayliffe M.A., Ellis J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Teh T., Wang C.I., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., Kale S.D., Wang X., Jiang R.H., Bruce N.A., Arredondo F.D., Zhang X., Tyler B.M. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20: 1930–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchardt F., Fotin-Mleczek M., Schwarz H., Fischer R., Brock R. (2007). A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic 8: 848–866 [DOI] [PubMed] [Google Scholar]
- Ellis J.G., Rafiqi M., Gan P., Chakrabarti A., Dodds P.N. (2009). Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12: 399–405 [DOI] [PubMed] [Google Scholar]
- Eiríksdóttir E., Konate K., Langel U., Divita G., Deshayes S. (2010). Secondary structure of cell-penetrating peptides controls membrane interaction and insertion. Biochim. Biophys. Acta. 1798: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Frankel A.D., Pabo C.O. (1988). Cellular uptake of the Tat protein from human immunodeficiency virus. Cell 55: 1189–1193 [DOI] [PubMed] [Google Scholar]
- Grouffaud S., van West P., Avrova A.O., Birch P.R.J., Whisson S.C. (2008). Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology 154: 3743–3751 [DOI] [PubMed] [Google Scholar]
- Hardham A.R. (2001). Investigations of oomycete cell biology. Molecular and Cell Biology of Filamentous Fungi: A Practical Approach, Talbot N., (Oxford: Oxford University Press; ), pp. 127–155 [Google Scholar]
- Heath M.C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. (Lond.) 80: 713–720 [Google Scholar]
- Hiller N.L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estraño C., Haldar K. (2004). A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306: 1934–1937 [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Fraley R.T., Rogers S.G., Sanders P.R., Lloyd A., Hoffmann N. (1984). Inheritance of functional foreign genes in plants. Science 223: 496–498 [DOI] [PubMed] [Google Scholar]
- Houterman P.M., Ma L., van Ooijen G., de Vroomen M.J., Cornelissen B.J.C., Takken F.L.W., Rep M. (2009). The effector protein Avr2 of the xylem colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58: 970–978 [DOI] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.H.Y., Tripathy S., Govers F., Tyler B.M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving super-family with more than 700 members. Proc. Natl. Acad. Sci. USA 105: 4874–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44: 41–60 [DOI] [PubMed] [Google Scholar]
- Kemen E., Kemen A.C., Rafiqi M., Hempel U., Mendgen K., Hahn M., Voegele R.T. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant Microbe Interact. 18: 1130–1139 [DOI] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporther oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I., Kobayashi Y., Hardham A.R. (1994). Dynamic reorganisation of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195: 237–247 [Google Scholar]
- Lawrence G.J., Mayo G.M.E., Shepherd K.W. (1981). Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology 71: 12–19 [Google Scholar]
- Li W., et al. (2009). The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant Microbe Interact. 22: 411–420 [DOI] [PubMed] [Google Scholar]
- Manning V.A., Ciuffetti L.M. (2005). Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 17: 3203–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning V.A., Hamilton S.M., Karplus P.A., Ciuffetti L.M. (2008). The Arg-Gly-Asp-containing, solvent-exposed loop of Ptr ToxA is required for internalization. Mol. Plant Microbe Interact. 21: 315–325 [DOI] [PubMed] [Google Scholar]
- Marti M., Good R.T., Rug M., Knuepfer E., Cowman A.F. (2004). Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306: 1930–1933 [DOI] [PubMed] [Google Scholar]
- Mims C.W., Rodriguez-Lother C., Richardson E.A. (2002). Ultrastructure of the host-pathogen interface in daylily leaves infected by the rust fungus Puccinia hemerocallidis. Protoplasma 219: 221–226 [DOI] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O., Kahmann R., Aguilar G., Trejo-Aguilar B., Wu A., de Vries R.P. (2008). The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45 (Suppl. 1): S63–S70 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15: 473–497 [Google Scholar]
- Murdoch L.J., Hardham A.R. (1998). Components in the haustorial wall of the flax rust fungus,Melampsora lini, are labelled by three anti-calmodulin monoclonal antibodies. Protoplasma 201: 180–193 [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany A.P., Gordon A., Rose L.E., Allen R.L., Armstrong M.R., Whisson S.C., Kamoun S., Tyler B.M., Birch P.R.J., Beynon J.L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout C.J., Skamnioti P., Porritt O., Sacristan S., Jones J.D.G., Brown J.K.M. (2006). Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18: 2402–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo M.A., Springer G.H., Granada B., Piston D.W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22: 445–449 [DOI] [PubMed] [Google Scholar]
- Robinson P.A., Anderton B.H., Loviny T.L. (1988). Nitrocellulose-bound antigen repeatedly used for the affinity purification of specific polyclonal antibodies for screening DNA expression libraries. J. Immunol. Methods 108: 115–122 [DOI] [PubMed] [Google Scholar]
- Shan W., Cao M., Leung D., Tyler B.M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17: 394–403 [DOI] [PubMed] [Google Scholar]
- Tyler B.M. (2009). Entering and breaking: Virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 11: 13–20 [DOI] [PubMed] [Google Scholar]
- Wang C.I., et al. (2007). Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19: 2898–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118 [DOI] [PubMed] [Google Scholar]
- Xu Q., Buckley D., Guan C., Guo H.C. (1999). Structural insights into the mechanism of intramolecular proteolysis. Cell 98: 651–661 [DOI] [PubMed] [Google Scholar]