Abstract
Using ultrasound-guided in utero infections of fluorescently traceable lentiviruses carrying RNAi or Cre recombinase into mouse embryos, we have demonstrated noninvasive, highly efficient selective transduction of surface epithelium, in which progenitors stably incorporate and propagate the desired genetic alterations. We achieved epidermal-specific infection using small generic promoters of existing lentiviral short hairpin RNA libraries, thus enabling rapid assessment of gene function as well as complex genetic interactions in skin morphogenesis and disease in vivo. We adapted this technology to devise a new quantitative method for ascertaining whether a gene confers a growth advantage or disadvantage in skin tumorigenesis. Using α1-catenin as a model, we uncover new insights into its role as a widely expressed tumor suppressor and reveal physiological interactions between Ctnna1 and the Hras1-Mapk3 and Trp53 gene pathways in regulating skin cell proliferation and apoptosis. Our study illustrates the strategy and its broad applicability for investigations of tissue morphogenesis, lineage specification and cancers.
Dissecting the complex cellular behaviors regulating tissue growth in embryogenesis and cancers necessitates a physiologically relevant in vivo model and a method for exploring gene function in the context of signaling pathways that govern homeostasis. In Caenorhabditis elegans and Drosophila melanogaster, studies of normal tissue balance and growth control have been aided by an array of genetic approaches including RNA interference (RNAi). In mouse models, where the relationship to human cancers is often clearer, functional analyses of genes regulating cellular growth have been limited to labor-intensive knockout technologies.
Several groups have used intra-amniotic ultrasound-guided micro-injections of viruses to deliver genes in vivo to organs and tissues of early mammalian embryos in situ1–6. Although promising, such studies have not yet achieved cell type selectivity and targeting efficiency suitable for functional analyses probing the physiological relevance of genetic interactions in mammals. Our goal was to develop a highly efficient, noninvasive, cell type–specific in vivo method that takes advantage of existing generic RNAi lentiviral libraries7,8. Exploiting skin epidermis’ use as a classical model for studies of tumorigenesis and epithelial biology, here we describe this method and document its feasibility for novel quantitative investigations of mouse tissue growth, morphogenesis, homeostasis and cancer.
RESULTS
Transduction of surface epithelium with lentiviral vectors
Endo et al.5 recently used ultrasound-guided lentiviral microinjection into the amniotic cavity of mouse embryos at 8–12 days post coitum (E8–E12) to achieve postnatal expression of an epidermal promoter-driven GFP transgene. Extending these findings, we demonstrated that lentivirus-sized fluorescent beads were internalized by E9.5–E10.5 surface epithelium comprising a single layer of non-neural ectoderm. Thereafter, bead uptake was confined to the transient periderm layer, which developed over the embryo surface (Supplementary Fig. 1). These results suggested that epidermal specificity might be achieved by the delivery mechanism itself.
To explore this possibility, we performed microinjections into the amniotic cavities of E9.5 embryos, where the ratio of amniotic volume to embryo surface area is optimal (Supplementary Fig. 2). Using pLKO.1, a generic lentiviral vector designed for RNU6-1 promoter–driven short hairpin RNA (shRNA) expression8, we replaced its PGK promoter–driven puromycin-resistance gene with the histone gene Hist2h2be fused to Gfp, Rfp, Cfp and Yfp cDNAs (yielding a vector we refer to as LV-XFP; Fig. 1a)9,10. By E18.5, this H2B-GFP fusion protein was detected throughout multilayered back skin epidermis and developing hair follicles (Fig. 1b–e). Expression was maintained in adult skin, indicating that lentiviral transductions achieved stable incorporation of DNA into the host genome (Fig. 1f), in agreement with the results of Endo et al.5. We next generated the lentiviral vector LV-Cre, harboring an nls-Cre fusion gene driven by the cytomegalovirus (CMV) promoter (Fig. 1a), and performed injections on E9.5 Rosa26-YFP Cre reporter embryos (r26yfp/+)11. As demonstrated by YFP expression, LV-Cre efficiently excised loxP-flanked (floxed) sequences in single-layered embryonic epidermis (Fig. 1g,h).
Figure 1.
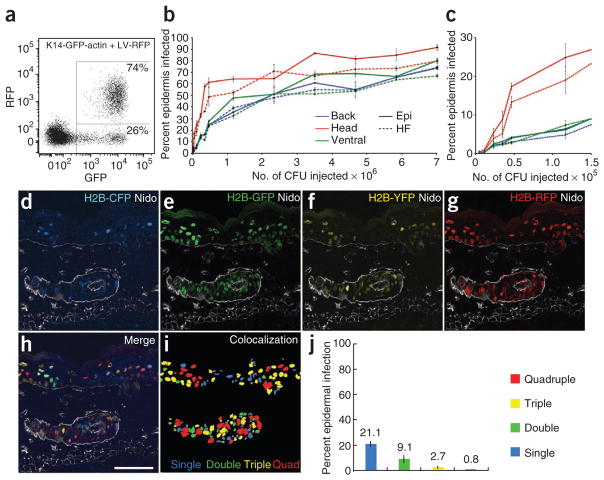
Intra-amniotic injection of lentivirus at E9.5 results in noninvasive, high-efficiency, stable and epidermally restricted transduction. (a) Lentiviral constructs used in the study. Modifications are of pLKO.1, a generic lentiviral vector for expressing human RNU6-1 promoter-driven short hairpin RNAs (shRNAs; red loop)8. LTR, long terminal repeat; ψ, retroviral packaging element; RRE, Rev response element; cPPT, central polypurine tract; PGK, phosphoglycerate kinase promoter; H2B-XFP, Hist2h2be fused to cDNA of genes encoding fluorescent proteins GFP, RFP, CFP or YFP; nls, nuclear localization signal; CMV, cytomegalovirus promoter; LV, lentivirus; Cre, bacterial Cre recombinase. (b–d) LV-GFP infection of E9.5 embryos analyzed at E15.5 (b) or E18.5 (c) relative to non-infected control (d). (e–h) Back skin sections of E9.5 LV-GFP infected control (e,f) and LV-Cre infected r26yfp/+ Cre-reporter embryos (g,h) analyzed at E18.5 (e,g) or 12 weeks (f,h). Transduced cells are YFP+ or H2B-GFP+. Nidogen (Nido) demarcates basement membrane and dermal blood vessels. (i) Newborn back skin section at high LV-Cre infection efficiency; boxed area, enlarged and shown as an inset, shows a single non-infected hair follicle with an overlying patch of YFP− epidermis. Epi, epidermis; Der, dermis; HF, hair follicle. Scale bars, 3 mm (b), 5 mm (c,d), 50 μm (e–h, inset), 100 μm (i).
Under high infection rates, large regions of skin epithelium expressed YFP (Fig. 1i), with an occasional YFP− hair follicle and overlying epidermis (Fig. 1i, inset). Conversely, at lower infectivity rates, discrete areas encompassing a uniformly YFP+ hair follicle and adjacent epidermis could be visualized (Fig. 1h). As our system provides control over the degree of labeling (Supplementary Fig. 3), it affords a means for future exploration of issues such as the arrangement of epidermis into discrete units12.
Most importantly, our transductions led to apparent surface epithelial–specific expression without the use of tissue-specific promoters. To explore this further, we injected LV-RFP into E9.5 transgenic mouse embryos expressing actin-GFP under an epidermal-specific Krt14 (also known as K14, encoding keratin 14) promoter, K14-GFP-actin13. When subjected to fluorescence activated cell sorting (FACS), H2B-RFP+ newborn back skin cells were all GFP+ (Fig. 2a), confirming that our skin transductions are epidermal specific. Moreover, FACS quantification revealed that high infection rates were consistently achieved over the entire embryo surface in both hair follicles and overlying epidermis, with highest transduction in the head region (Fig. 2b,c). High head skin infectivity at E9.5 resulted in LV-Cre achieving excision 2–3 d earlier (at E10.5) than transgenic K14-Cre (at E12.5–E13.5) (Supplementary Fig. 4). This difference could be useful for functional analyses of genes involved in early epidermal development. Notably, with higher viral titers, progressively smaller increases in overall infection levels were observed. For instance, with injections of >106 colony-forming units (CFU), a further 700% increase in viral titer elevated head skin infection by only 30%. This raised the intriguing possibility that at high infectivity, cells might be transduced with multiple viruses (Fig. 2b).
Figure 2.
Epidermal infection depends on viral titer and permits delivery of multiple viral constructs. (a) FACS analysis of K14actin-GFP embryos infected at E9.5 with LV-RFP and analyzed at E18.5. Note that only GFP+ cells are RFP+, consistent with epidermally restricted transduction. (b) Relationship between viral titer and epidermal infection efficiency as determined by FACS analysis of hair follicle and epidermal compartments of different skin regions of E18.5 embryos infected with LV-RFP. Epi, epidermis; HF, hair follicle. (c) Close-up view of lower titer part of the graph in b. Note the more efficient transduction of head skin at lower viral titers. (d–i) Representative back skin section from an E18.5 embryo simultaneously infected with LV-CFP, LV-GFP, LV-YFP and LV-RFP. Shown are single-color (d–g) and merged (h) images. Note that for this particular embryo, overall infection was ~34%, and ~66% of the cells were uninfected. (i) Colocalization of binarized single fluorescence images, color coded to mark single, double, triple and quadruple infection. (j) FACS quantification of co-infection efficiency, color coded as in i. Abbreviations: Epi, epidermis; HF, hair follicle. Nidogen (Nido) demarcates basement membrane and dermal blood vessels. Scale bar, 50 μm.
To monitor multiple viral deliveries, we injected E9.5 embryos with four different fluorescently tagged viruses and analyzed them at E18.5. Even with low infections (30%), ~1% of skin cells coexpressed all four fluorescently tagged histones (Fig. 2d–j). Because the backbone of each of these viruses can accommodate additional features, including multiple shRNAs and transgenes, the potential of this technique for rapid analysis of multiple gene functions far exceeds that of conventional mouse genetics.
In regard to the efficacy of the transduction system, it is noteworthy that our micromanipulations yielded high survival rates (79%) and efficient targeting to the amniotic cavity (81%). Moreover, lentiviral infection did not affect tissue proliferation, apoptosis, morphology or differentiation (Supplementary Fig. 5). Additionally, leukocyte and lymphocyte numbers were comparable and low in lentivirally infected and non-infected postnatal day (P) 0 mouse skins (Supplementary Fig. 6). This was relevant because other perturbations are known to elicit immune response during embryonic skin development and because retroviral-based vectors are known to elicit innate and/or adaptive immune responses in gene therapy trials14,15. Lastly, low levels of lentiviral transduction were detected at other sites, including the corneal, oral, nasal and otic epithelia (not shown).
Development of a quantitative cellular growth assay
The ability to co-infect epidermal cells with multiple viruses and to accurately quantify infection levels by FACS facilitated our adaptation of the system to assay whether a genetic deficiency results in a growth advantage or disadvantage in the context of tissue development or homeostasis in vivo. The general principle is outlined below, and should be particularly useful in the field of cancer, where skin carcinogenesis is often the model of choice (Fig. 3a–d).
Figure 3.
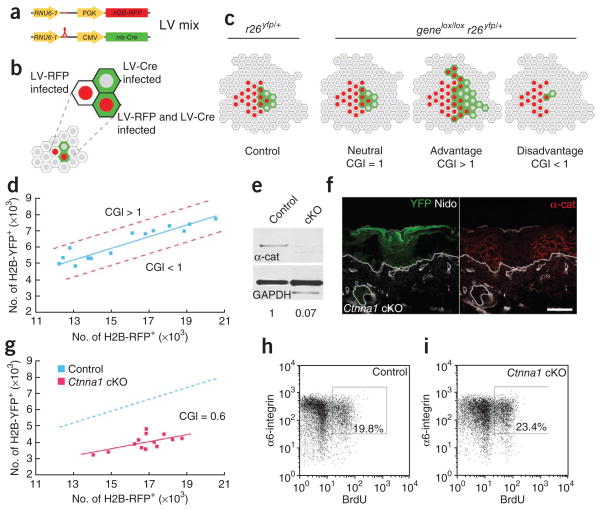
Rapid assay for measuring an epidermal growth advantage or disadvantage conferred by a gene mutation reveals an unexpected growth disadvantage following α1-catenin loss despite hyperproliferation. (a–c) Schematic of the cellular growth index (CGI) assay. E9.5 Cre reporter embryos are infected with a mix of LV-Cre and LV-RFP, resulting in epidermal cells that have been transduced and that express H2B-RFP, H2B-YFP or both (a,b). At E18.5, the relative ratios of H2B-RFP+ to YFP+ cells in control (r26yfp/+) and gene knockout (genelox/loxr26yfp/+) mice are compared (c). Phenotypes are scored as either being neutral or having a growth advantage or disadvantage depending on this CGI value. (d) Graph of FACS-quantified numbers of H2B-RFP+ cells relative to YFP+ cells in control mice at E18.5. Genes whose depletion results in a growth advantage or disadvantage would shift the curve toward the upper or lower dashed red lines, respectively. (e) Quantified anti–α1-catenin (α-cat; test) and glyceraldehyde phosphate dehydrogenase (GAPDH; control) immunoblots of protein lysates from cells FACS-sorted from LV-Cre–infected control and Ctnna1-floxed (cKO) embryos. (f) Back skin sections of LV-Cre Ctnna1lox/lox r26yfp/+ (Ctnna1 cKO) embryos immunolabeled with anti–α1-catenin. Transduced cells are indicated by their YFP expression. (g) Graph of numbers of H2B-RFP+ cells relative to YFP+ cells in control (as in d) and Ctnna1 cKO mice at E18.5. Note the reduced CGI (0.6; P < 0.001) in the Ctnna1 cKO clones. (h,i) FACS plots and quantification of % basal (α6-integrin+) cells that incorporated BrdU after a 6-h labeling of E18.5 embryos. Note elevated BrdU incorporation despite the growth disadvantage in Ctnna1 cKO skin. Nidogen (Nido) marks the epidermal-dermal boundary as well as dermal blood vessels. Scale bar, 50 μm.
Briefly, we infected E9.5 mouse embryos with two lentiviruses: (i) LV-Cre, which, depending on the genetic background, marks control (r26yfp/+) or mutant (genelox/lox r26yfp/+) cells and (ii) LV-RFP, which labels a corresponding group of cells to serve as an internal control for overall infection levels (Fig. 3a,b). By FACS-quantifying H2B-RFP+ and YFP+ cells in control and mutant E18.5 embryos, we obtained a cellular growth index (CGI), defined as the ratio between YFP+ cells observed in the test condition (genelox/lox r26yfp/+) and YFP+ cells in the control (r26yfp/+) at an equivalent infection level (H2B-RFP+ cells). A CGI of 1 reflects no effect of gene deficiency on cellular growth, whereas a higher or lower value indicates an advantage or disadvantage, respectively (Fig. 3c). As depicted by regression analysis, the ratio of H2B-RFP+ to YFP+ cells in a control tissue remained linear across a range of infection levels (Fig. 3d).
To illustrate the power of the approach, we focused on α1-catenin (encoded by Ctnna1), an essential actin-binding component of adherens junctions16, which shows reduced expression in most mouse and human carcinomas17–20. To establish the fidelity of the r26yfp/+ Cre reporter as an indicator of Ctnna1 knockout clones, we used LV-Cre to infect r26yfp/+ E9.5 embryos that were either wild-type or homozygous for the floxed Ctnna1 allele16. At E18.5, YFP+ back skin epidermal cells were isolated by FACS and analyzed by immunoblotting. α1-catenin protein levels were ~7% of the control, suggesting that LV-Cre–mediated excision was efficient (Fig. 3e). Moreover, by immunofluorescence, clonal patches of YFP+ epidermis were always negative for α1-catenin and vice versa (Fig. 3f).
When we compared the Ctnna1lox/loxr26yfp/+ mice to the r26yfp/+ controls using our CGI assay, the ratio of H2B-RFP+ cells and Ctnna1 mutant (YFP+) cells was independent of overall infection levels, as observed for the controls. However, the calculated CGI was 0.6, indicating a significant (P < 0.001) 67% reduction in YFP+ cells observed in Ctnna1 mutants relative to wild-type mice (Fig. 3g). Consistent with the results of the CGI assay, a progressive loss of YFP+ Ctnna1 mutant but not control cell clones occurred during postnatal development. Thus, following loss of α1-catenin, epidermal cells are at a growth disadvantage.
The growth disadvantage of α1-catenin–null clones seemed at odds with the elevation in proliferating nuclear antigen Ki67 reported previously16. To verify that α1-catenin deficiency indeed leads to hyper-proliferation in our LV-Cre–infected embryos, we administered BrdU to r26yfp/+ and Ctnna1lox/loxr26yfp/+ E18.5 embryos that had been infected at E9.5. Six hours later, basal epidermal cells were FACS-sorted for α6-integrin+ ± YFP+. For control LV-Cre–infected r26yfp/+ embryos, the animal-matched ratio of BrdU+YFP+ versus BrdU+YFP− cells remained constant as expected. By contrast, a significant increase (~20%, P < 0.001) was seen in BrdU+YFP+ versus BrdU+YFP− cells in LV-Cre–infected Ctnna1lox/loxr26yfp/+ animals (Fig. 3h,i; see additional details below).
Although revealing the power of our strategy, these results unveiled an unexpected conundrum: how does α1-catenin loss result in a cellular growth disadvantage and yet promote proliferation and tumorigenesis16,19,20? To dissect the cellular mechanisms responsible, we first needed to demonstrate the utility of our system for conducting rapid functional and genetic interaction analyses in vivo.
Efficient gene knockdown using lentiviral RNAi in vivo
Utility of our system for rapid RNAi-mediated loss-of-function studies requires efficient gene knockdown and faithful recapitulation of the knockout phenotype (Fig. 4). The RNAi Consortium (TRC) mouse lentiviral library9 carried three Ctnna1 shRNA constructs (Fig. 4a). When introduced into cultured wild-type epidermal keratinocytes, they reduced Ctnna1 mRNA levels to ~70% (shCtnna1-186), 30% (shCtnna1-1764) and 9% (shCtnna1-912), respectively, of those seen with a control scrambled shRNA (shScram; Fig. 4c).
Figure 4.
Efficient epidermal-specific lentivirus RNAi-mediated knockdown of Ctnna1 faithfully recapitulates phenotypic abnormalities shown by K14-Cre conditional and LV-Cre induced knockout counterparts. (a) TRC RNAi library shRNA constructs (arrowheads) corresponding to Ctnna1. Numbers correspond to TRC nomenclature. (b) Anti–α1-catenin and GAPDH immunoblots of protein lysates of cells FACS-sorted from embryos infected with LV-GFP harboring Ctnna1-specific shRNAs (shCtnna1) and control scrambled shRNA (shScram). (c) Quantification of α1-catenin levels from blot in b. (d–i) Comparative analyses of representative P0 skin sections of conditional Ctnna1 knockout (Ctnna1 cKO), shCtnna1-912 knockdown (Ctnna1 RNAi) and shScram control (Scram RNAi) embryos. (d) α1-Catenin immunolabeling reveals efficient knockdown in all shCtnna1-912 infected (H2B-GFP+) but not uninfected cells. (e–g) Morphological and adherens junction defects, not found in shScram RNAi–infected skin, are similar between cells infected with Ctnna1 cKO (f) and Ctnna1 RNAi (g). Boxed areas are shown in insets. Arrowheads denote hair follicles derived from infected epidermis. Note that asymmetric E-cadherin localization seen in shScram RNAi–infected (e) or uninfected areas (f) is consistently lost in Ctnna1 RNAi–infected skin (g), indicative of a planar cell polarity defect. (h,i) Suprabasal keratin 6 (K6), often reflective of enhanced basal cell proliferation, is detected in Ctnna1 cKO (h) and Ctnna1 RNAi (i) cell clones. Transduced cells are identified by their YFP or H2B-GFP expression. Nidogen (Nido) marks the basement membrane and dermal blood vessels. Epidermal adherens junctions are marked by antibody to E-cadherin (Ecad). Primary antibodies are noted on each frame, with color coding according to secondary antibodies used. Scale bars, 50 μm.
After cloning these shRNAs into our LV-GFP backbone, we performed amniotic injections on E9.5 embryos. At E18.5, H2B-GFP+ back skin cells were isolated by FACS and used for immunoblot analyses. In agreement with the transcript reductions observed in vitro, α1-catenin protein expression in vivo was reduced to ~70% (shCtnna1-186), 45% (shCtnna1-1764) and 18% (shCtnna1-912) of control levels, respectively (Fig. 4b,c). Immunofluorescence analyses corroborated these results, revealing the strongest reduction in α1-catenin in H2B-GFP+ epidermal patches from embryos transduced with shCtnna1-912 (Fig. 4d).
Previous Ctnna1 gene targeting by transgenic K14-Cre expression resulted in defects in intercellular adhesion and actin dynamics, as well as disorganized epidermal stratification, MAPK-mediated hyperproliferation and precancerous epithelial invaginations16,19,21. To verify that in vivo RNAi-mediated gene knockdown and LV-Cre–mediated knockout can phenocopy these known loss-of-function consequences, we compared LV-GFP shCtnna1-912 knockdown and LV-Cre Ctnna1 knockout with conditional K14-Cre Ctnna1 knockout. For gene targeting, we used Ctnna1lox/loxr26yfp/+ mice so that levels of LV-Cre and K14-Cre–mediated knockout cells could be quantified by measuring the proportion of YFP+, α6-integrin+ basal epidermal cells. Similarly, α6-integrin and H2B-GFP+ were used to score the proportion of knockdown cells.
Ctnna1 knockdown, LV-Cre knockout and conditional K14-Cre knockout embryos shared several phenotypic characteristics: eyelid closure failure; curled tails; shortened limbs; fused digits; and skins that were shiny, taut and fragile16 (Supplementary Fig. 7). Only Ctnna1 knockdown and LV-Cre–mediated Ctnna1 knockout embryos showed a paucity of skin in the head region (Supplementary Fig. 7c,d). This increased severity in head skin phenotype defects in the lentiviral versus K14-transgenic Cre phenotype was consistent with the high infectivity at this site (Fig. 2b,c) as well as with the 2–3-d difference in the timing of the excision achieved by LV-Cre (at E10.5) as compared to K14-Cre (at E12.5–E13.5; Supplementary Fig. 4).
In addition to recapitulating the gross defects caused by α1-catenin loss of function, knockdown also generated tissue defects characteristic of those observed in Ctnna1 knockouts. These included perturbations in epidermal architecture and stratification, accompanied by induced suprabasal keratin 616 (Fig. 4e–i). In addition, cultured Ctnna1 knockout and knockdown keratinocytes showed expected defects in their actin cytoskeletons and failed to establish and maintain cell-cell adhesion junctions21 (Supplementary Fig. 8). Altogether, the striking phenotypic parallels between mice with Ctnna1 RNAi-mediated knockdown and those with loss-of-function mutations, but not those treated by control scrambled RNAi, made off-target effects unlikely and underscored the efficiency of this strategy for dissecting physiological mechanisms. In subsequent experiments, we guarded against possible off-target effects by using shScram RNAi controls and multiple shRNA hairpins against each gene transcript.
Using RNAi in vivo to dissect a genetic network
The ability to conduct knockdowns for functional studies provided the means to probe deeper into why Ctnna1 mutant cells have a growth disadvantage despite being hyperproliferative (Fig. 5). We first tested whether Ras-MAPK activity, previously found to be elevated in cultured Ctnna1-null cells16, might be responsible for the elevated proliferation in our embryos. We selected shRNAs corresponding to (i) Hras1, encoding the most abundant Ras family member and predominant target of oncogenic mutations in skin22,23, and (ii) Mapk3, encoding Mapk3, the most downstream component of the MAPK signaling cascade governing epidermal proliferation24. In keratinocytes in vitro, shHras1-267 and shMapk3-357 resulted in 86% Hras1 and 93% Mapk3 transcript reductions, respectively (Fig. 5a).
Figure 5.
Use of RNAi knockdown in vivo to functionally dissect why loss of α1-catenin results in hyperproliferation but a growth disadvantage to the epidermis. (a–b) Efficiency of Hras1 and Mapk3 RNAi knockdowns in vitro and in vivo as determined in embryos and cultured keratinocytes subjected to lentivirus-mediated RNAi knockdown with shHras1-267, shMapk-357 or control scrambled shRNA (shScram). (a) Real-time PCR quantification of Hras1 and Mapk3 transcripts from keratinocytes. (b) Immunoblot analysis of Hras1 and Mapk3 protein levels in embryos (data normalized to control values of 100%). (c) Effects of Hras1 and Mapk3 RNAi on BrdU incorporation in control and LV-Cre Ctnna1 cKO embryos following a 6-h pulse of BrdU. Data were quantified by FACS. (d–i) Trp53-dependent apoptosis occurs in Ctnna1 cKO skin in vivo and is directly responsible for the reduced CGI. (d) Apoptotic cells (marked by active caspase 3) in clonal patches of Ctnna1 cKO skin, marked by YFP expression. Quantifications are shown in g. Nidogen (Nido) marks basement membrane and dermal blood vessels. DAPI (blue) labels the nuclei. (e) Fold changes in in vivo transcript levels, normalized to LV-Cre–infected control embryos (red dashed line), of TRP53 signature target genes in cells FACS-sorted from Ctnna1 cKO embryos alone or Ctnna1 cKO embryos infected with shTrp53-1223 (Ctnna1 cKO + Trp53 KD). (f) Levels of Trp53 transcripts in vitro and in vivo after lentivirus-mediated RNAi knockdown in keratinocytes and embryos. Two different Trp53 shRNAs are tested. (g) Percentage of active caspase 3+ cells in control and Ctnna1 cKO cells in the presence and absence of Trp53 RNAi in vivo. (h) Graph of numbers of RFP+ cells relative to YFP+ cells at E18.5 in control and Ctnna1 KO mice infected with shTrp53-1223. CGI = 0.8 (P < 0.001). (i) Comparison of CGI for control (wt), Ctnna1 knockout (cKO) and Ctnna1 knockout with Trp53 knockdown (cKO + Trp53 KD) embryos. * denotes CGI values that show significant differences (P < 0.001). Scale bar, 50 μm.
After modifying our LV-Cre vector to express these shRNAs, we infected r26yfp/+ embryos with these and control viruses. Quantitative immunoblot analyses of lysates from FACS-purified infected (YFP+) cells revealed marked reductions of Hras1 (79%) and Mapk3 (86%, Fig. 5b). Newborn mice with strongly reduced Hras1 and Mapk3 were viable and had normal skin, consistent with these proteins’ non-essential function in skin embryogenesis25,26.
To investigate the effects of Hras1 and Mapk3 knockdowns on the hyperproliferative behavior of Ctnna1 mutant cells, we again analyzed BrdU incorporation, this time in r26yfp/+ and Ctnna1lox/loxr26yfp/+ E18.5 embryos that had been infected with shRNA-modified LV-Cre at E9.5 (Fig. 5c). For each animal, BrdU incorporation in YFP+ cells was normalized to that of animal-matched YFP− cells. In Hras1 knockdown control animals, the proportion of BrdU+ cells in YFP+ and YFP− populations was constant, indicating that Hras1 reduction alone did not affect proliferation. By contrast, equivalent Hras1 knockdown in Ctnna1 mutant cells abolished the increase in BrdU incorporation. Mapk3 knockdown also restored normal proliferation to Ctnna1 mutant cells, while showing no effect on control cells. These data imply that the hyperproliferation following α1-catenin loss in vivo is dependent upon downstream Ras-MAPK activity. These experiments further illustrate the strength of our system, in which a combination of RNAi-mediated knockdown and Cre-mediated knockout can be used for rapid assessment of physiologically important genetic interactions.
Although our findings established a pathway whereby α1-catenin deficiency leads to enhanced Ras-MAPK signaling and hyperproliferation in vivo, this mechanism acted counter to the decreased CGI. To understand why, we first checked for senescence following α1-catenin loss. When the senescence-associated β-galactosidase protocol27 revealed no signs of enhanced senescence (Supplementary Fig. 9), we addressed whether the hyperproliferation might be counterbalanced by enhanced apoptosis. Interestingly, a 960% increase in active caspase 3– and TdT-mediated dUTP nick end-labeling (TUNEL)–positive cells was found in Ctnna1 mutant relative to control tissues (Fig. 5d,g; data not shown).
Loss of transformation-related protein 53 (Trp53) rescues the apoptotic defects in Cdh1(encoding cadherin 1) mutant mammary gland cells in vivo28, and a variety of human epithelial cancers show reduced E-cadherin and α1-catenin levels along with Trp53 activation17,18. To test whether Trp53 is activated following α1-catenin loss in skin, we isolated mRNA from FACS-purified YFP+ cells of LV-Cre infected r26yfp/+ and Ctnna1lox/loxr26yfp/+ E18.5 embryos and profiled them for known Trp53 targets. The highest transcript increases in Ctnna1 mutant embryos were Bbc3 (1,130%) and Pmaip1 (530%) (encoding, respectively, PUMA and Noxa, the primary mediators of Trp53-dependent cell death)29 (Fig. 5e, black bars). We also detected smaller increases in the Trp53 targets Cdkn1a, Gadd45a and Apaf1.
To test for functional interactions between α1-catenin and Trp53-mediated apoptosis, we measured the effect of Trp53 knockdown on apoptosis in Ctnna1 mutant cells. First, we selected shTrp53-1223 and shTrp53-9132, which showed ~50–60% reduction in Trp53 transcripts in keratinocytes and embryonic skin (Fig. 5f). Next, we infected E9.5 Ctnna1lox/loxr26yfp/+ embryos with LV-Cre harboring shTrp53 and analyzed them at E18.5. In Ctnna1 mutant cells, Trp53 reduction in vivo reduced increases in Bbc3 and Pmaip1 transcripts observed following loss of Ctnna1 alone by 190% and 350%, respectively (Fig. 5e, green bars). Furthermore, whereas Trp53 knockdown in control clones showed no measurable effect on apoptosis, comparable Trp53 knockdowns in Ctnna1 mutant cells resulted in a 300% decrease in the elevation of active caspase 3–positive cells seen following Ctnna1 loss (Fig. 5g). This phenomenon was not attributable to off-target effects, as knockdown with two different Trp53 shRNAs gave similar results (Fig. 5g). Both transcript and apoptosis reductions refer to changes in values observed in Ctnna1 mutant cells that are substantially increased as compared to the wild type.
Next we tested if the observed reduction in CGI could be reversed on Trp53 knockdown in Ctnna1 mutant animals. Indeed, when the mix of LV-RFP– and LV-Cre–expressing shTrp53-1223 was injected into Ctnna1lox/loxr26yfp/+ test and r26yfp/+ control embryos at E9.5 and analyzed at E18.5, the calculated CGI was 0.8. Although this was <1, the difference from control was not statistically significant at P < 0.05 (Fig. 5h,i). This result differed significantly from the CGI following loss of α1-catenin alone (Fig. 3g; P < 0.001). Together, these findings provide compelling evidence that Trp53 activation is responsible for the growth disadvantage following loss of α1-catenin.
DISCUSSION
Although primary keratinocyte cultures have been instrumental for elucidating many features of epidermal biology, they undergo significant morphological and biochemical changes that limit their value for studying normal tissue physiology. This is true even for organotypic cultures and in vivo engraftment approaches, which generate wound-like perturbations in tissue integrity. Although in vivo xenotransplantation makes it possible to test relationships between human and mouse biology and hence represents an important approach to skin cancer research30, engraftment procedures require immunocompromised mice, and thus can not recapitulate the full complement of cellular behaviors likely to play a role in cancers.
Our strategy for conducting comprehensive functional analyses couples the accessibility of epidermis with the utility and expediency of RNAi and commercially available shRNA libraries, and thus greatly expands the molecular toolbox for dissecting complex genetic pathways in mammalian tissue biology. In its simplest form as a single-gene functional analysis, our method necessitates only a few weeks between target selection and phenotypic analysis. As such, it offers a distinct advantage over classical mouse genetics, the conventional methods currently used to study embryonic development and tissue homeostasis in an unperturbed physiological setting.
The ability to selectively target and label epidermal progenitors allowed us to develop the CGI assay as a quantitative tool for dissecting pathways that regulate cell growth. Given that the epidermis has long served as a major model in cancer studies, this new strategy becomes particularly powerful. In addition, the combination of RNAi-mediated knockdown and lentiviral Cre-mediated knockout allows for a rapid assessment of genetic epistasis. Moreover, at least four different viruses can be used for simultaneous tissue infection, expanding the utility of this system, for example, for eliminating functional redundancies or conducting knockdown and/or replacement studies.
It is noteworthy that epidermal transduction with LV-Cre provides temporal and spatial benefits over the existing epidermis-specific Cre lines. The earlier and more uniform generation of epidermal- specific gene knockouts with LV-Cre permits future exploration of early developmental functions such as stratification, planar cell polarity and epithelial-mesenchymal interactions. The ability to control infection levels by varying lentiviral titers offers (i) an ideal source of animal-matched internal control cells, (ii) a means of analyzing gene function in adult skin, which is often precluded by newborn lethality, and (iii) the ability to distinguish between cell-autonomous and non-cell-autonomous protein roles in vivo.
Finally, using Cnnta1 as the archetype, we have shown how our technology can be used to uncover new insights into the genetic interplay between intercellular adhesion and growth control. Our analyses demonstrated a measurable effect of Ras-MAPK–dependent cell proliferation and Trp53-dependent cell death on Ctnna1 loss-of-function phenotypes during skin morphogenesis. It is tempting to speculate that the genetic interactions we uncovered between these two opposing pathways allow the epidermis to suppress neoplastic growth and sustain homeostasis following loss of α1-catenin. A similar phenomenon has been observed following loss of TGF-β signaling in the skin31. Given our new findings, we posit that this genetic interplay between opposing pathways may be a common feature of tumor suppressors in skin epithelium. In this scenario, tipping the balance toward cell survival through pro-survival signals or alterations in Trp53 pro-apoptotic function could subsequently lead to development of epidermal tumors, reinforcing why the frequent occurrence of Trp53-null mutations following chronic UVB exposure contributes so greatly to skin cancers.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
We thank M. Takeichi (RIKEN CDB) for antibodies and reagents; N. Stokes and Rockefeller Comparative Bioscience Center staff for expert care of mice; M. Schober and M. Perez-Moreno for helpful discussions; A. North and Rockefeller Bioimaging Resource Center staff for assistance with image acquisition and analysis; and S. Mazel and Rockefeller Flow Cytometry Resource Center staff for assistance with FACS. S.B. is supported by the International Human Frontier Science Program Organization. S.W. is an American Cancer Society Postdoctoral Fellow. This work was supported by a grant from the US National Institutes of Health (R01-AR27883). E.F. is an investigator with the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
S.B., G.L. and S.W. designed and performed the experiments and analyzed the raw data. S.B. and E.F. wrote the manuscript. E.F. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Published online at http://www.nature.com/naturemedicine/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary information is available on the Nature Medicine website.
References
- 1.Holzinger A, Trapnell BC, Weaver TE, Whitsett JA, Iwamoto HS. Intraamniotic administration of an adenoviral vector for gene transfer to fetal sheep and mouse tissues. Pediatr Res. 1995;38:844–850. doi: 10.1203/00006450-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Lu B, Federoff HJ, Wang Y, Goldsmith LA, Scott G. Topical application of viral vectors for epidermal gene transfer. J Invest Dermatol. 1997;108:803–808. doi: 10.1111/1523-1747.ep12292254. [DOI] [PubMed] [Google Scholar]
- 3.Liu A, Joyner AL, Turnbull DH. Alteration of limb and brain patterning in early mouse embryos by ultrasound-guided injection of Shh-expressing cells. Mech Dev. 1998;75:107–115. doi: 10.1016/s0925-4773(98)00090-2. [DOI] [PubMed] [Google Scholar]
- 4.Slevin JC, et al. High resolution ultrasound-guided microinjection for interventional studies of early embryonic and placental development in vivo in mice. BMC Dev Biol. 2006;6:10. doi: 10.1186/1471-213X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endo M, et al. Efficient in vivo targeting of epidermal stem cells by early gestational intraamniotic injection of lentiviral vector driven by the keratin 5 promoter. Mol Ther. 2008;16:131–137. doi: 10.1038/sj.mt.6300332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Punzo C, Cepko CL. Ultrasound-guided in utero injections allow studies of the development and function of the eye. Dev Dyn. 2008;237:1034–1042. doi: 10.1002/dvdy.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 8.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 9.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 10.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 13.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 14.Follenzi A, Santambrogio L, Annoni A. Immune responses to lentiviral vectors. Curr Gene Ther. 2007;7:306–315. doi: 10.2174/156652307782151515. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Moreno M, et al. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 17.Xiangming C, et al. Cooccurrence of reduced expression of alpha-catenin and overexpression of p53 is a predictor of lymph node metastasis in early gastric cancer. Oncology. 1999;57:131–137. doi: 10.1159/000012020. [DOI] [PubMed] [Google Scholar]
- 18.Nozawa N, et al. Immunohistochemical alpha- and beta-catenin and E-cadherin expression and their clinicopathological significance in human lung adenocarcinoma. Pathol Res Pract. 2006;202:639–650. doi: 10.1016/j.prp.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc Natl Acad Sci USA. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamin JM, Nelson WJ. Bench to bedside and back again: molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin Cancer Biol. 2008;18:53–64. doi: 10.1016/j.semcancer.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 22.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 23.Leon J, Guerrero I, Pellicer A. Differential expression of the ras gene family in mice. Mol Cell Biol. 1987;7:1535–1540. doi: 10.1128/mcb.7.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khavari TA, Rinn J. Ras/Erk MAPK signaling in epidermal homeostasis and neoplasia. Cell Cycle. 2007;6:2928–2931. doi: 10.4161/cc.6.23.4998. [DOI] [PubMed] [Google Scholar]
- 25.Pagès G, et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 26.Ise K, et al. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–2956. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 27.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derksen PW, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Villunger A, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 30.Reuter JA, et al. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell. 2009;15:477–488. doi: 10.1016/j.ccr.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guasch G, et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.