Abstract
Non-viral gene delivery by immobilization of complexes to cell-adhesive biomaterials, a process termed substrate-mediated delivery, has many in vitro research applications such as transfected cell arrays or models of tissue growth. In this report, we quantitatively investigate the efficiency of gene delivery by surface immobilization, and compare this efficiency to the more typical bolus delivery. The ability to immobilize vectors while allowing cellular internalization is impacted by the biomaterial and vector properties. Thus, to compare this efficiency between vector types and delivery methods, transfection conditions were initially identified that maximized transgene expres- sion. For surface delivery from tissue culture polystyrene, DNA complexes were immobilized to pre-adsorbed serum proteins prior to cell seeding, while for bolus delivery, complexes were added to the media above adherent cells. Mathematical modeling of vector binding, release, and cell association using a two-site model indicated that the kinetics of polyplex binding to cells was faster than for lipoplexes, yet both vectors have a half-life on the surface of approximately 17 min. For bolus and surface delivery, the majority of the DNA in the system remained in solution or on the surface, respectively. For polyplexes, the efficiency of trafficking of cell-associated polyplexes to the nucleus for surface delivery is similar or less than bolus delivery, suggesting that surface immobilization may decrease the activity of the complex. The efficiency of nuclear association for cell-associated lipoplexes is similar or greater for surface delivery relative to bolus. These studies suggest that strategies to enhance surface delivery for polyplexes should target the vector design to enhance its potency, whereas enhancing lipoplex delivery should target the material design to increase internalization.
Keywords: gene delivery, reverse transfection, surface delivery, polyethylenimine (PEI), lipoplex, substrate-mediated
Introduction
Gene delivery from surfaces has considerable potential in many in vitro research applications, such as transfected cell arrays for studies in functional genomics or patterned gene delivery for models of tissue growth (Bailey et al., 2002; Houchin-Ray et al., 2007; King et al., 2007; Ziauddin and Sabatini, 2001). For surface delivery, a process that has been termed substrate-mediated delivery (Bengali and Shea, 2005; Bengali et al., 2005; Pannier and Shea, 2004; Segura and Shea, 2002; Segura et al., 2003), reverse transfection (Ziauddin and Sabatini, 2001) or solid-phase delivery (Bielinska et al., 2000), a biomaterial immobilizes and maintains the vector at the surface, while allowing for cellular internalization. Immobilization to a cell adhesive substrate limits aggregation, places the vector directly in the cell microenvironment to reduce mass transport limitations, and localizes delivery (Bailey et al., 2002; Chang et al., 2004; Honma et al., 2001; Houchin-Ray et al., 2007; Luo and Saltzman, 2000; Schaffer and Lauffenburger, 1998; Segura and Shea, 2002; Segura et al., 2003; Shea et al., 1999; Silva et al., 2004; Vanhecke and Janitz, 2004; Webb et al., 2003; Wheeler et al., 2005; Yamauchi et al., 2004; Yoshikawa et al., 2004). DNA can be immobilized by entrapment of naked plasmid in gelatin followed by addition of the transfection reagent (Ziauddin and Sabatini, 2001) or polyelectrolyte layering (Jewell et al., 2005). Alternatively, DNA complexes can be preformed and immobilized to the substrate through specific tethers (Segura and Shea, 2002; Segura et al., 2003, 2005) or non-specific adsorption, either on an uncoated substrate (Bengali et al., 2005; Bielinska et al., 2000; Jang et al., 2006; Pannier et al., 2005; Shen et al., 2004; Yoshikawa et al., 2004) or substrates coated with serum or extracellular matrix proteins to mediate cell adhesion and complex immobilization (Bengali et al., 2007). Immobilization by non-specific adsorption can reduce the amount of DNA required for expression and increase transgene expression and the number of cells expressing the transgene relative to similar quantities delivered as a bolus (Bengali et al., 2005; Jang et al., 2006); however, the delivery efficiency is a function of both the properties of the vector and substrate (Bengali et al., 2005; Pannier et al., 2005; Segura et al., 2003).
The utility of surface delivery and the potential applications could be greatly enhanced by identifying the limiting steps in gene delivery. The barriers to successful gene delivery include extracellular stability and transport, cellular association and internalization, endosomal escape, cytoplasmic transport and stability, and nuclear localization (Wiethoff and Middaugh, 2003). Recently, quantitative studies of DNA trafficking following bolus addition of nonviral vectors to cultures have been employed to identify the rate limiting steps (Akita et al., 2004; Hama et al., 2006; Varga et al., 2005). Polyplexes were limited by intracellular trafficking, with the specific step determined by the polyplex design. Lipoplexes, conversely, were limited by cellular association and internalization (Varga et al., 2005). Although the potential to transfect cells from the surface has been demonstrated, quantitative studies for delivery of surface immobilized vectors are needed to identify the specific barriers for surface-mediated gene delivery, which can elucidate specific design parameters for the material surface and vector.
This report investigates the efficiency and principle barriers to gene delivery from a protein-coated surface relative to bolus delivery. We have previously demonstrated the potential of substrate-mediated delivery to produce significantly higher transfection than bolus delivery for the same quantity of DNA, which indicated the ability to overcome mass transport limitations and that transfection was dependent upon both the surface and vector properties (Bengali et al., 2005). In this study, transfection conditions were identified that maximized transgene expression for both surface and bolus delivery in order to compare efficiency across vectors and delivery systems. Proteincoated tissue culture polystyrene was employed for the immobilization of polyplexes and lipoplexes. Specific observations investigated include (i) protein expression and the distribution of complexes among the cell population, (ii) the relative amounts of DNA that remain in solution or on the surface, and (iii) the amount of DNA associated per cell and associated to the nucleus, which may reflect the efficacy of the complexes. Mathematical modeling was employed to characterize the kinetics of vector binding, release, and cellular association. These studies determine the relative efficacy of the complexes delivered from the substrate and identify the limiting steps, which is essential to developing systems that maximize gene transfer.
Materials and Methods
Materials
Plasmid encoding for enhanced green fluorescent protein and firefly luciferase (pEGFP-Luc) with a CMV promoter was purified from bacterial culture using Qiagen (Valencia, CA) reagents and stored in Tris–EDTA buffer (10 mM Tris, 1 mM EDTA, pH 7.4). Branched polyethylenimine (PEI, 25 kDa) was purchased from Sigma-Aldrich (St. Louis, MO). Lipofectamine 2000, SYTO 61 nucleic acid stain, and LysoTracker Yellow HCK-123 were purchased from Invitrogen (Carlsbad, CA). All other reagents were obtained from Fisher Scientific (Fairlawn, NJ) unless otherwise noted.
Complex Formation and Immobilization
DNA complexes were formed by the addition of cationic polymer (PEI) (Boussif et al., 1995) or cationic lipid (Lipofectamine, 2000) to plasmid resulting in self-assembled colloidal particles. PEI/DNA complexes, termed polyplexes, were formed at N/P ratios (nitrogen/phosphate ratios, i.e., charge ratios) of 10 (“low-content polyplexes”) and 25 (“high-content polyplexes”) using 6 μg DNA for polyplexes to be immobilized and 2.4 μg DNA for polyplexes to be delivered as a bolus. Lipofectamine 2000 complexes were formed at ratios of 1:0.5 μg DNA:mL lipid (“low-content lipoplexes”), with 4 μg DNA for immobilized lipoplexes and 1.6 μg DNA for lipoplexes delivered as a bolus, and 1:2 μg DNA:mL lipid (“high-content lipoplexes”), with 1.32 μg DNA for immobilized lipoplexes and 1.6 μg DNA for lipoplexes delivered as a bolus. Polyplexes were formed in tris-buffered saline (TBS, pH 7.4) and lipoplexes were formed in serum-free cell growth media (DMEM). Polyplexes to be immobilized were formed by adding PEI (500 μL) dropwise to a solution containing DNA (750 μL), vortexed and incubated for 15 min at room temperature. Lipoplexes to be immobilized were formed by adding Lipofectamine 2000 (625 μL) dropwise to solution containing DNA (625 μL), mixed gently by pipetting and incubated for 20 min at room temperature. Polyplexes and lipoplexes for bolus delivery were formed in 10% of the volume used for immobilized complexes above.
Complexes were immobilized on serum-coated tissue culture polystyrene. Tissue culture polystyrene was serum-coated by incubation with heat-inactivated fetal bovine serum (FBS, GIBCO-Invitrogen, Greenland, NY, 200 μL, 10% in PBS, pH 7.4) for 2 h, followed by two wash steps with phosphate-buffered saline (PBS). Incubation of preformed DNA complexes with the substrate for 4 h immediately followed complex formation (1,250 μL) and was followed by washing twice with TBS for polyplexes or DMEM for lipoplexes.
Cell Culture and Transfection
Transfection studies were performed with NIH/3T3 (ATCC, Manassas, VA) cells cultured at 37°C and 5% CO2 in DMEM (Invitrogen, CA) supplemented with 1% sodium pyruvate, 1% penicillin–streptomycin, 1.5 g/L NaHCO3,and10%heat-inactivated FBS to produce complete DMEM (cDMEM). Cells were seeded at a density of 62,500 cells per well in 12-well plates. For substrate-mediated delivery, cells were seeded immediately following complex immobilization and wash steps. Studies were also performed with standard bolus delivery to cells plated in 12-well dish at a density of 62,500 cells per well and cultured overnight before complexes were added. The number of cells per well in culture 24 h after delivery of DNA was determined in triplicate. Cells were harvested after transfection and counted using a hemacytometer. Transfection was analyzed 24 h after cells were exposed to complexes. Transfection was characterized through the extent of transgene expression (luciferase levels) and the number of transfected cells (GFP expression). The extent of transgene expression was quantified by measuring the luciferase activity using the Luciferase Assay System (Promega, Madison, WI). Cells were lysed with 500 μL Reporter Lysis Buffer (Promega) and assayed for enzymatic activity 24 h after exposure to DNA complexes. The luminometer was set for a 3 s delay with signal integration for 10 s. Luciferase activity was normalized to total cellular protein using the bicinchoninic acid (BCA) assay kit (Pierce, Rockford, IL). Transfected cells were visualized using an epifluorescence microscope (Leica, Bannockburn, IL) with a FITC filter and equipped with a digital camera, and transfection efficiency was determined using flow cytometry (see below). All studies were carried out in triplicate.
DNA Distribution in Culture System
Quantification of plasmid in the system utilized radiolabeled plasmid with α-32P dATP (PerkinElmer, Waltham, MA). Briefly, a nick translation kit (GE Healthcare Life Sciences, Piscataway, NJ) was used following the manufacturer's protocol with minor modifications (Segura et al., 2003). Plasmid integrity was characterized using agarose gel electrophoresis with 1% agarose in TAE. Plasmid labeling yielded super-coiled and open-circular conformations, with no small fragments that would indicate plasmid degradation (data not shown). The unbound DNA was determined by adding complex incubation solution and wash solution to scintillation cocktail (Biosafe II, 10 μL) for measurement on a scintillation counter. The amount of DNA immobilized was determined by subtracting the amount of unbound DNA from the DNA initially incubated on the substrate (Table I). The amount of cell-associated DNA was determined by harvesting cells from the substrate and adding samples to scintillation cocktail. Cultured cells were washed once with PBS (500 μL), exposed to trypsin (500 μL) for 3 min and quenched with 1 μL cDMEM. Cells were further removed from the substrate using a cell scraper. For the quantification of cell-associated DNA, cells were centrifuged at 500g for 5 min and resuspended in PBS (500 μL) for subsequent counting. DNA in the media was measured by counting the media above the cells in scintillation cocktail. DNA remaining on the substrate was determined by subtracting the cellular-associated DNA and DNA quantities in the media from the total DNA immobilized or delivered as a bolus. Quantities of DNA per cell were calculated by dividing the total amount of DNA associated with cells by the number of cells in culture 24 h after exposure to DNA complexes.
Table I.
DNA quantities applied to the substrate (μg), bound to the substrate (μg), or added as a bolus (μg/well) ± SD.
| Incubated DNA (μg) | Surface (μg) | Bolus (μg) | |
|---|---|---|---|
| PEI (N/P 10) | 6.0 | 5.4 ± 0.0 | 2.4 |
| PEI (N/P 25) | 6.0 | 4.8 ± 0.2 | 2.4 |
| Lipofectamine 2000 (1:0.5) | 4.0 | 2.2 ± 0.1 | 1.6 |
| Lipofectamine 2000 (1:2) | 1.3 | 1.0 ± 0.2 | 1.6 |
Mathematical Model
Polyplex and lipoplex desorption, cellular association, and transport in the bulk media are modeled using a series of differential equations. The substrate is modeled as having two sites for the immobilized complexes (Eqs. 1 and 2), with the density of immobilized DNA denoted as Dtot. Complex dissociation is modeled as a first order process, with the initial fractional density for sites 1 and 2 ( fs1, fs2) and their corresponding kinetic constants (ks1, ks2) dependent upon the vector. Note that the second site is modeled as irreversible binding (ks2 = 0 cm−1), with values for ks1 determined from the experimental data. Complexes are released into the bulk solution immediately adjacent to the surface. The concentration profile in solution is described by Equation (3), and allows for transport by diffusion within solution with diffusivity DDNA. The boundary condition at the substrate surface (x = 0 cm) is that the flux of DNA is equal to the amount released from the substrate. The other boundary condition is at the liquid surface, (x = 0.3 cm) which is a no-flux boundary condition. Finally,=DNA in solution adjacent to the substrate can associate with cells with a kinetic constant kassoc (Eq. 4). These equations were coded into and solved using Matlab. The values for the diffusivity were obtained from literature reports (Clamme et al., 2003; Lai and van Zanten, 2002), whereas the kinetic constants were determined using the mathematical model and minimizing the second norm. The model parameters are presented in Table II.
Table II.
Constants for mathematical model.
| Polyplex | Lipoplex | |
|---|---|---|
| D tot | 1.2 μg/cm2 | 0.25 μg/cm2 |
| f s1 | 0.25 | 0.43 |
| f s2 | 0.75 | 0.57 |
| k s1 | 4.0 × 10−2 cm−1 | 4.2 × 10−2 cm−1 |
| D DNA a | 3.6 × 10−6 cm2/min | 1.5 × 10−6 cm2/min |
| k assoc | 5.0 × 10−10 cm/min/cell | 5.7 × 10−11 cm/min/cell |
For polyplexes, the diffusivity was obtained from reference (Clamme et al., 2003). For lipoplexes, the diffusivity was calculated using the Stokes–Einstein equation using literature values for the radius of gyration (Lai and van Zanten, 2002).
| (1) |
| (2) |
| (3) |
| (4) |
Transfection Efficiency and Distribution of Cellular Internalization
The number of transfected cells and the number of cells that internalized DNA complexes were measured with a FACSscan flow cytometer (Becton Dickinson, San Jose, CA) equipped with a 15 mW, 488 nm air cooled argon-ion laser. A 530 nm bandpass filter was used to measure fluorescence from GFP and fluorescein labeled DNA. Approximately 10,000 cells were analyzed per sample. For all samples, a threshold was set such that the negative control had 1% positive events (i.e., 1% of the negative control was transfected or had internalized DNA). DNA was labeled with fluorescein using the Label IT Nucleic Acid Labeling Kit (Mirus, Madison, WI) following the manufacturer's protocol and complexes formed as described above. Cells were harvested as described above and resuspended in 500 μL of 0.5% BSA and 0.1% sodium azide in PBS. Fluorescence from extracellular DNA was quenched by addition of trypan blue as described (Rejman et al., 2004, 2005).
Subcellular Distribution
Quantification of internalized and nuclear-associated radiolabeled plasmids was performed as described previously with minor modifications (Moriguchi et al., 2006). For quantification of internalized DNA delivered by lipoplexes, harvested cells were suspended in 100 μL of CellScrub Buffer (Gene Therapy Systems, Inc., San Diego, CA) to remove cell- surface bound lipoplexes. The cell suspension was incubated at room temperature for 30 min, and then centrifuged at 2,000 rpm for 3 min. The cells were then resuspended in PBS for subsequent counting. DNA internalization efficiency was calculated as internalized plasmids per cell divided by cell-associated plasmids per cell. Internalization of polyplexes was not measured, as the cell scrubbing method used to remove cell-associated DNA was only designed for lipid/ DNA systems. For quantification of nuclear-associated DNA, harvested cells were suspended in a 3:1 mixture of CellScrub Buffer and cell lysis solution (2% IGEPAL CA630, 40 mM NaCl, 12 mM MgCl2 and 40 mM Tris–HCl, pH 7.4). CellScrub was present in the lysis solution to limit DNA association with the nucleus after lysis (Moriguchi et al., 2006). The suspension was centrifuged at 9,200g for 2 min, and the supernatant was removed. The pellet was resuspended in CellScrub/cell lysis solution and centrifuged three more times. The nuclear fraction was then resuspended in PBS for subsequent counting. Nuclear-trafficking efficiencies for polyplexes and lipoplexes were calculated as nuclear-associated plasmids per cell divided by cell-associated plasmids per cell.
Imaging and Quantification of Lysosomal Plasmid
Plasmids encoding for β-galactosidase were labeled with Cy3 using a Label IT nucleic acid labeling kit (Mirus) according to the manufacturer's instructions. Briefly, DNA and Label IT reagents were mixed and incubated at 37°C for 1 h. After incubation, DNA was precipitated with 70% ethanol and resuspended in 10 μL Tris-EDTA (TE) buffer.
NIH/3T3 cells were transfected in 8-well glass chamber slides (Nalge Nunc International, Rochester, NY) as described above. An inverted Leica LCS DM-IRE2 confocal microscope was used to image live cells, which were imaged through a 40× oil-immersion objective with a field of view of 375 μm × 375 μm, with Z-sections taken every 366.3 nm. The resulting image resolution was 2,048 × 2,048 pixels, and Z-sections were taken to image the entirety of cells within the X,Y field of view, producing a voxel size of 183 nm × 183 nm × 366 nm (X,Y,Z). Cy3-labeled plasmid DNA was excited by a 1 mW GreNe laser at 543 nm and fluorescence was observed at 555–600 nm. Lysosomal compartments were visualized by incubating cells with 0.5 μL Lysotracker Yellow HCK-123 for 30 min at room temperature prior to excitation by a 5 mW Ar laser at 488 nm and fluorescence was observed at 495–533 nm. Nuclear compartments were visualized by incubating cells with 0.2 μL Syto 61 nucleic acid stain for 5 min at room temperature prior to excitation by a 10 mW HeNe laser at 633 nm and fluorescence was observed at 645–725 nm. Sequential scanning was used to control for spectral overlap between fluorophores, with 2× line averaging to improve signal-to-noise ratios.
Quantification of DNA complex localization within subcellular compartments was performed using NIH ImageJ (http://rsb.info.nih.gov/ij/). Several plug-ins were used to facilitate colocalization. Adaptive3DThreshold, a part of the MBF “ImageJ for Microscopy” Collection (www. macbiophotonics.ca/imagej/) was used to identify cytoplasmic and nuclear compartments based on the differential intensity of Syto 61 (Invitrogen, Carlsbad, CA) staining of cytoplasmic mRNA and nuclear DNA. The Object Counter3D plug-in (http://rsb.info.nih.gov/ij/plugins/track/objects.html) was used to define cellular compart- ments based upon their volumes, allowing multiple cells to be simultaneously identified and characterized. ColocalizationHighlighter, also part of the MBF Collection, was used to colocalize DNA complexes to cytoplasmic or lysosomal compartments. Colocalized voxels were counted and compared to total cytoplasmic DNA for each condition. At least 17 cells were captured per image stack, and at least 3 image stacks were analyzed per condition.
Statistical Analysis
Results are represented by the mean and standard deviation of experiments with a sample size equal to three. Statistical analyses were performed using a one-way ANOVA followed by Tukey-HSD in the software package JMP 4.0.4 (SAS Institute, Cary, NC). A value of P less than 0.05 was considered significant.
Results
Polyplex and Lipoplex Mediated Gene Expression
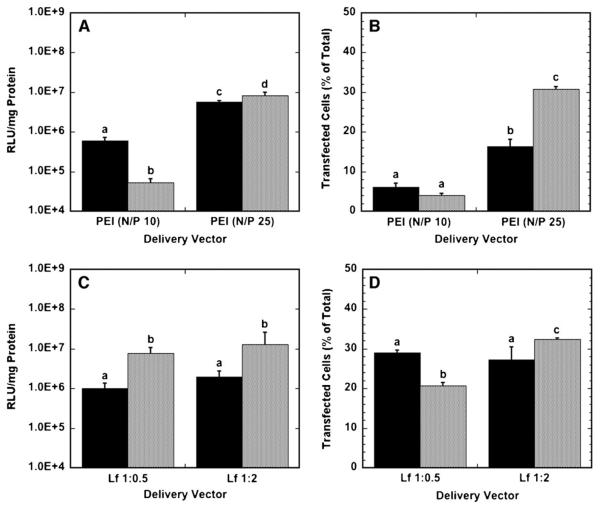
To compare efficiency between vector types and delivery method, transfection conditions, that is, the amount of DNA and transfection reagent to be used, were initially identified that maximized transgene expression (Table I). Transgene expression and number of transfected cells were determined to be a function of the vector properties in substrate-mediated and bolus delivery (Fig. 1). Substrate-mediated delivery of N/P 10 polyplexes produced greater levels of transgene expression than with bolus delivery, yet yielded similar quantities of transfected cells. However, at an N/P ratio of 25, bolus delivery yielded greater expression and percentage of transfected cells than surface delivery. Polyplexes formed at an N/P ratio of 25 resulted in greater expression for both surface and bolus delivery compared to polyplexes formed at an N/P ratio of 10.
Figure 1.
Transgene expression by polyplex and lipoplex delivery from a substrate (black bars) and as a bolus (gray bars) to NIH/3T3 cells. A: Luciferase expression levels for polyplexes delivered at N/P ratios of 10 and 25. B: The number of cells expressing GFP after transfection with PEI polyplexes at N/P ratios of 10 and 25. C: Luciferase expression levels for lipoplexes delivered at DNA:lipid (μg:μL) ratios of 1:0.5 and 1:2. D: The number of cells expressing GFP after transfection with lipoplexes delivered at DNA:lipid (μg:μL) ratios of 1:0.5 and 1:2. Data are presented as an average of triplicate measurements ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters.
Bolus delivery yielded higher protein expression compared to surface delivery with lipoplexes. Interestingly, the lipid content of the lipoplexes did not influence transgene expression for substrate-mediated or bolus delivery. The number of transfected cells was not different for different lipid amounts for surface delivery, but the number of transfected cells differed for different lipid amounts for bolus delivery, with high-content lipoplexes resulting in more transfected cells than low-content lipoplexes.
DNA Localization to Culture System Components
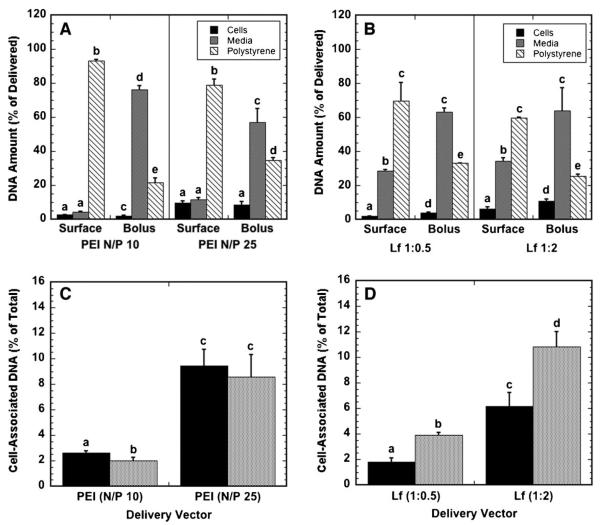
We examined the efficiency of surface and bolus delivery by determining the fraction of delivered DNA associated with the cells, the culture surface, or the culture media (Fig. 2). The relative distribution of DNA between the cellular compartment, culture media, and surface depended primarily upon the delivery mechanism and the N/P ratio; for substrate-mediated delivery, the majority of polyplex-delivered DNA, 79% or greater, remained immobilized to the surface, while polyplexes delivered as a bolus primarily remained in the media (Fig. 2A). Similar to polyplexes, lipoplex delivery had most of the DNA on the substrate for substrate-mediated delivery, and in the media for bolus delivery (Fig. 2B). For bolus delivery, high lipid-content lipoplexes had an increased efficiency of cellular association and a reduced percentage on the substrate relative to low lipid content lipoplexes, while the percentage in the media remained unchanged. A greater percentage of the immobilized DNA was found in the media above the cells when compared to polyplexes, indicating that lipoplexes are more readily released from the substrate compared to polyplexes.
Figure 2.
Localization of DNA following polyplex and lipoplex delivery. The percentage of DNA delivered that is associated with cells, in growth media and on the substrate for polyplexes (A) and lipoplexes (B), and the percentage of DNA delivered that associates with cells for polyplexes (C) and lipoplexes (D) delivered from a substrate (black bars) and as a bolus (gray bars) to NIH/3T3 cells are shown. Data are presented as an average of triplicate measurements ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters, with comparisons made only within the same N/P ratios.
The percentage of DNA that associated with cells was determined as the amount of cell-associated DNA divided by the cumulative amount of DNA in the media, with the cells, and on the surface. For all conditions, less than 12% of DNA was associated with cells after 24 h (Fig. 2C and D). The efficiency by which polyplexes associated with cells was comparable for both substrate-mediated delivery and bolus delivery (Fig. 2C). The efficiency of association significantly increased as the N/P ratio increased from 10 to 25. In contrast to polyplexes, bolus delivery was more efficient at associating the lipoplexes with cells than with surface delivery for both low and high lipid contents (Fig. 2D). The high-lipid content lipoplexes were more efficient at associating with cells than the low-lipid lipoplexes for both surface and bolus delivery.
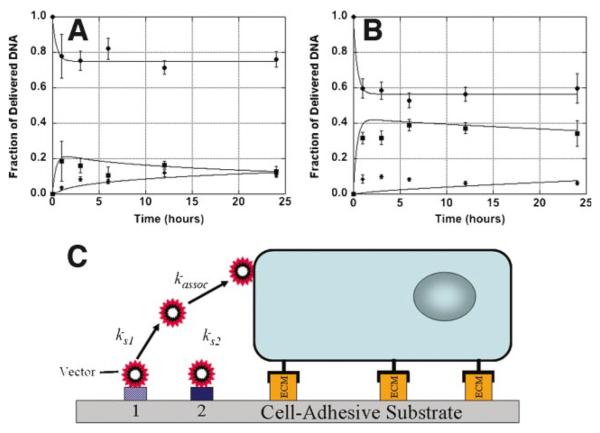
The distribution of polyplexes and lipoplexes in the culture system over time was subsequently investigated for delivery from serum-coated substrates (Fig. 3). Both polyplexes and lipoplexes had a majority of the release occur within 1 h. Polyplexes had a release of approximately 20% from the substrate (Fig. 3A), whereas lipoplexes had a release of approximately 40% (Fig. 3B). For polyplexes, a majority of the released DNA initially enters the media, with a gradual accumulation of DNA with the cells. Lipoplexes similarly have a majority of the released DNA entering the bulk media; however, the cellular associated quantities are maximal within 1 h and persist for the remainder of the study.
Figure 3.
Dynamics of DNA complex distribution. Polyplex (A) and lipoplex (B) quantities associated with the substrate (●), media (■), and cells (◆) following delivery from serum coated substrates are shown. Experimental measurements are shown as the data points, and predictions from the model are represented by lines that pass through the data points. A schematic of the model for release of immobilized DNA and cellular association is shown (C). Data are presented as an average of triplicate measurements ± standard deviation of the mean. [Color figure can be seen in the online version of this article, available at www.interscience.wiley.com.]
A mathematical model with two distinct DNA binding sites, a reversible and an irreversible site, was utilized to describe the dynamics of DNA release and cellular association (Fig. 3C, Table II). The kinetic constants for desorption were approximately 0.04 min−1 for both polyplexes and lipoplexes, which correspond to a half-life of approximately 17 min. The kinetic constant for cellular association for polyplexes was 5.0 × 10−10 cm/min/cell, which is approximately an order of magnitude greater than that determined for lipoplexes.
Distribution of Cells With Internalized Complexes
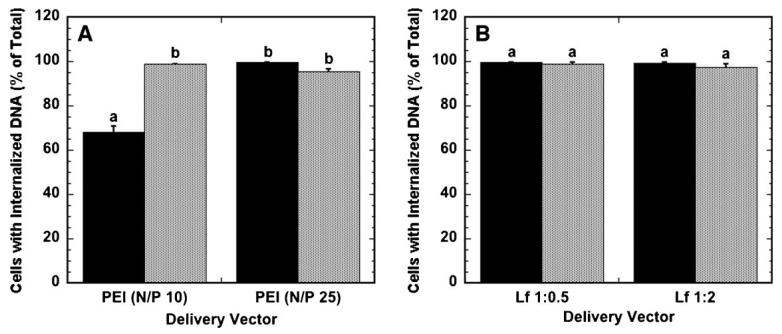
The efficiency of polyplexes and lipoplexes was further investigated by examining the percentage of cells that have internalized complexes. Nearly all cells (greater than 95%) exposed to polyplexes or lipoplexes as a bolus had internalized DNA after 24 h (Fig. 4A and B). Similarly, nearly all cells (greater than 99%) on surfaces with immobilized complexes had internalized DNA, except for N/P 10 polyplexes. Importantly, the percentage of cells with internalized DNA does not correspond to the percentage of cells expressing protein for both surface and bolus delivery, suggesting that internalization of DNA does not necessarily result in protein expression. The amount of DNA internalized and nuclear-associated per cell likely determines whether a cell expresses the delivered transgene, and subsequent studies quantify the cell-associated, internalized and nuclear-associated DNA.
Figure 4.
Percentage of cells with internalized DNA. The percentage of cells that have internalized DNA when delivered as polyplexes (A) and lipoplexes (B) from the substrate (black bars) and as a bolus (gray bars) is shown. Data are presented as an average of triplicate measurements ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters.
Subcellular Distribution of DNA
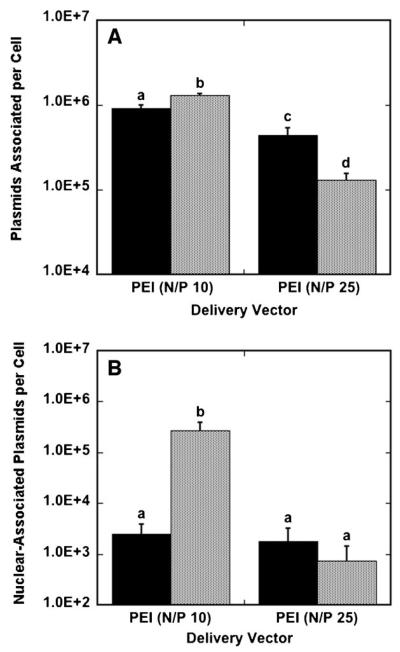
The amount of cell-associated, internalized and nuclear-associated DNA at the maximal transfection conditions was subsequently investigated for polyplexes and lipoplexes. Polyplex delivery from the surface provided lower levels of DNA per cell compared to traditional bolus delivery at N/P 10, while polyplex delivery from the surface provided greater levels of DNA per cell compared to traditional bolus delivery at N/P 25 (Fig. 5A). Low PEI-content polyplexes yielded higher amounts of DNA per cell than high PEI-content polyplexes for both bolus and surface delivery. The highest polyplex association was achieved with N/P 10 polyplexes delivered as a bolus, with 1.3 × 106 plasmids associated per cell. Intracellular DNA was also quantified for polyplex delivery (data not shown); however, these numbers matched the quantity of cell-associated DNA, likely due to the inability of CellScrub buffer to dissociate polyplexes from cell membranes. Bolus delivery resulted in more nuclear-associated DNA for low-content polyplexes compared to surface delivery, while nuclear-associated DNA was similar for both bolus and surface delivery with high-content polyplexes (Fig. 5B). This reduced amount of nuclear association is particularly noted with polyplexes formed at an N/P ratio of 10, which resulted in higher nuclear-association of DNA with bolus delivery, but significantly lower protein expression compared to substrate-mediated gene delivery. Nuclear-trafficking efficiencies for polyplexes, which is calculated as nuclear-associated plasmids divided by cell-associated plasmids, were comparable for surface and bolus delivery with high-content polyplexes, with bolus delivery resulting in a nuclear-trafficking efficiency of 0.56% and surface delivery resulting in a 0.40% nuclear-trafficking efficiency. Nuclear-trafficking efficiencies for low-content polyplexes were 20.8% for bolus delivery and 0.27% for surface delivery.
Figure 5.
Plasmids associated per cell and nuclear-associated plasmids per cell for polyplexes. Cells were exposed to DNA for 24 h in culture and harvested. The amount of DNA associated per cell (A) and nuclear-associated per cell (B) with polyplexes delivered from a substrate (black bars) and as a bolus (gray bars) to NIH/3T3 cells are shown. Data are presented as an average of triplicate measurements ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters.
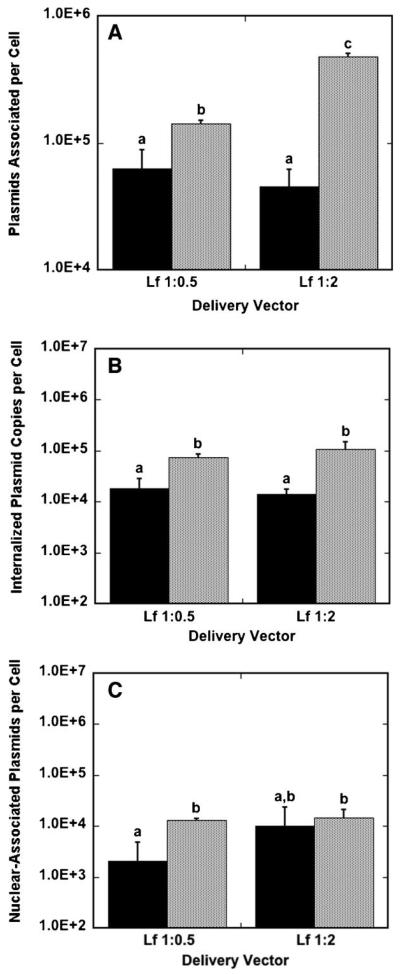
Lipoplexes had greater quantities of cell-associated DNA for bolus delivery compared to surface delivery for both low-and high-content lipoplexes (Fig. 6A). The highest lipoplex association was achieved with high-content lipoplexes delivered as a bolus, with 4.8 × 105 plasmids per cell. For lipoplexes at both high and low lipid content, bolus delivery resulted in more internalized DNA relative to surface delivery (Fig. 6B). Low-content lipoplexes had internalization efficiencies of approximately 30%, with no significant difference between surface and bolus conditions, while high-content lipoplexes achieved 52.2% and 21.9% internalization efficiency for surface and bolus delivery, respectively. In addition, bolus delivery resulted in more nuclear-associated DNA for low-content lipoplexes compared to surface delivery, while nuclear-associated DNA was similar for both bolus and surface delivery with high-content lipoplexes (Fig. 6C). Nuclear-trafficking efficiencies for low-content lipoplexes were of the same order of magnitude for surface and bolus delivery, with bolus delivery resulting in a nuclear-trafficking efficiency of 3.3% and surface delivery resulting in a 9.4% nuclear-trafficking efficiency. Nuclear-trafficking efficiency for high-content lipoplexes delivered from the surface was an order of magnitude greater than for bolus delivery, with surface delivery resulting in a nuclear-trafficking efficiency of 22.0% and bolus delivery resulting in only a 3.1% nuclear-trafficking efficiency. Interestingly, surface delivery with lipoplexes resulted in nuclear-trafficking efficiencies that are one to two orders of magnitude greater than polyplex surface delivery, suggesting that lipoplexes are not deactivated by immobilization to a surface compared to polyplexes.
Figure 6.
Plasmids associated per cell, internalized plasmids per cell, and nuclear-associated plasmids per cell for lipoplexes. Cells were exposed to DNA for 24 h in culture and harvested. The amount of DNA associated per cell (A), internalized per cell (B) and nuclear-associated per cell (C) with lipoplexes delivered from a substrate (black bars) and as a bolus (gray bars) to NIH/3T3 cells are shown. Data are presented as an average of triplicate measurements ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters.
We subsequently characterized the quantity of DNA complexes within lysosomes as a percentage of cytoplasmic totals. DNA complexes and lysosomes were imaged by confocal microscopy, and the colocalization of DNA in lysosomes was quantified using NIH ImageJ (Fig. 7A). For bolus delivery, the amount of DNA within the lysosomes did not significantly vary between polyplexes and lipoplexes (Fig. 7B). Conversely, for surface delivery, the mean percentages of lysosomal DNA by delivery of lipoplexes was increased approximately fourfold relative to delivery of polyplexes (Fig. 7C). Significant differences were not observed between the different polyplex formulations (N/P 10 vs. 25) or the different lipoplex formations (high vs. low). These differences in the quantity of DNA in the lysosomes suggest that the internalization pathway or cellular trafficking may vary for lipoplexes relative to polyplexes delivered from the surface.
Figure 7.
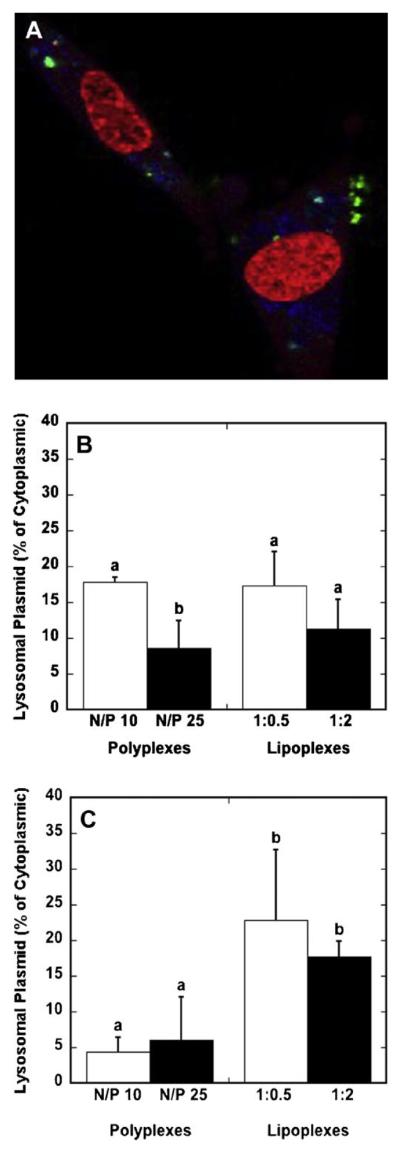
Lysosomal DNA levels. Cells were exposed to DNA complexes for 24 h prior to imaging. A representative confocal microscopy image depicting lysosomes (blue), Cy3-labeled plasmid (green) and nucleic acid staining (red) is shown (A). The quantity of lysosomal DNA as a percentage of cytosplasmic DNA was quantified by confocal microscopy for polyplexes and lipoplexes for delivery as a bolus (B) and from a substrate (C). Data are presented as the mean ± standard deviation of the mean. A statistical significance with P < 0.05 is denoted for values with different letters. [Color figure can be seen in the online version of this article, available at www. interscience.wiley.com.]
Discussion
This report investigates the efficacy and limiting steps of gene delivery from surfaces relative to that observed with bolus delivery, and is examined as a function of the vector properties and delivery method. Both bolus and substrate-mediated delivery produced substantial transgene expression and numbers of transfected cells. The percentage of transfected cells did not always correlate to the amount of protein expressed, as individual cells may produce different amounts of protein (Bengali et al., 2005). For lipoplexes, bolus delivery resulted in higher protein expression than substrate-mediated gene delivery. For polyplexes, protein expression depended on the N/P ratio as well as the delivery method, with bolus delivery of N/P 25 polyplexes resulting in the highest protein expression. Inherent inefficiencies of both bolus and surface delivery were investigated by measuring the amount of DNA with cells, in the media, and on the substrate. The majority of the DNA exposed to cells from substrates remained on the culture dish for both polyplexes and lipoplexes, while the majority of DNA delivered as a bolus remained in media. In addition, polyplexes delivered from the surface traffic to the nucleus with similar or worse efficiency than polyplexes delivered as a bolus. Lipoplexes delivered from the surface, in contrast, traffic to the nucleus with similar or greater efficiency than lipoplexes delivered as a bolus. Finally, lipoplexes delivered from the surface result in higher amounts of DNA in the lysosomes compared to polyplexes, suggesting that the internalization pathway or cellular trafficking may vary for lipoplexes relative to polyplexes delivered from the surface.
The percentage of DNA that is delivered to cells is similar for the bolus and substrate-mediated delivery for polyplexes, though both approaches have significant quantities of DNA that are either in solution or on the culture surface. Bolus delivery left approximately 60% of the complexes in solution, with 30% associated with the surface. Conversely, for substrate-mediated delivery, approximately 60-80% of the DNA was associated with the substrate while only a small percentage was released into the media, consistent with previous observations (Bengali et al., 2005). For surface delivery, an increase in release from the surface would decrease the amount of DNA required for expression. A challenge in interpreting these data is that the mechanism of internalization is unknown, particularly for surface delivery. Complexes may directly internalized from the surface, or they may be released into the media and subsequently associate with the plasma membrane. The varied levels of DNA within the lysosomal compartment for surface delivery of lipoplexes or polyplexes may result from internalization through different pathways. Different pathways for internalization could influence whether complexes are internalized through clathrin-coated pits, caveolae, or by macropinocytosis, which influence subsequent intracellular trafficking (Rejman et al., 2004, 2005). Reversible adsorption to the substrate is a potential mechanism for maintaining the DNA in the cellular microenvironment for both surface and bolus delivery. Designing surfaces that reversibly bind complexes to allow for cellular internalization yet retain potency is a fundamental challenge in surface delivery.
For polyplexes, the results suggest that the best path to enhancing gene transfer is to maintain or enhance the activity of the complexes. Bolus delivery has been reported to deliver 105–106 copies of plasmid to a cell, with 102–105 copies reaching the nucleus for expression (James and Giorgio, 2000; Tachibana et al., 2002). For surface delivery, polyplexes had threefold more plasmid associated with the cell at N/P 25, yet the quantity of nuclear-associated plasmid was similar to bolus delivery, suggesting that polyplexes internalized from the surface did not traffic to the nucleus as efficiently. At N/P 10, the efficiency of nuclear trafficking was reduced for substrate-mediated delivery by two orders of magnitude compared to bolus delivery. The efficacy of the polyplexes could be reduced by interaction with some of the serum components, which are known to modulate gene transfer (Dash et al., 1999). Alternatively, surface association could alter the activity of the immobilized polyplex (Pannier et al., 2008), such that the complex is less efficacious than bolus-delivered complexes for internalization, trafficking, and nuclear association. The differences in expression and efficacy between low and high N/P polyplexes have been previously reported for bolus delivery (Boussif et al., 1995; Fischer et al., 2003), and thus should be expected for surface delivery. Increasing the N/P ratio increases the charge on polyplexes (Bengali et al., 2005), which could influence substrate immobilization, and free PEI has been reported in solution that may also influence substrate immobilization. Taken together, for polyplex delivery, the challenge lies in developing either surfaces or vectors to maintain the vector activity, especially for high-content polyplexes, with which surface delivery provides efficient cellular and nuclear association of polyplexes.
For lipoplexes, the challenge to enhancing gene transfer lies with designing surfaces or vectors that allow DNA to be more efficiently delivered to cells for internalization. For the high-lipid lipoplexes, the amount of plasmid associated with cells was threefold less with surface delivery, yet the nuclear levels were similar, indicating that the trafficking of lipoplexes was enhanced for surface delivery. For low-content lipoplexes, the efficiency of nuclear association relative to cell-association was similar for surface and bolus delivery, though surface delivery had lower quantities of plasmid in both compartments. The results herein suggest that increasing internalization of DNA delivered by lipoplexes may increase transfection. Additionally, with the percentage of plasmid in lysosomes increased with surface delivery relative to polyplexes, strategies to enhance endosomal escape may also be beneficial. For lipoplexes delivered as a bolus, altering lipoplex properties can increase transfection through increased cellular association (Rea et al., 2008). Surface delivery would also benefit from increasing cellular association and internalization, and the material properties could potentially be designed to promote release and cellular association of the lipoplexes.
The quantities of DNA associated with cells represent a fraction of the DNA released from the surface. Release is relatively rapid (half-life ≈17 min), yet is sufficient to maintain the DNA at the surface until cells deposit on the surface. Two mechanisms for cellular association by a substrate-mediated approach have been proposed: the direct internalization of complexes from the substrate or the release of complexes from the substrate followed by cell association (Bengali and Shea, 2005). Internalization following release is supported by previous observations that incubation of substrates with cell-conditioned medium led to significantly greater release than incubation with PBS (Bengali et al., 2005). Additionally, we have observed that washing surfaces by pre-incubation with conditioned media for 24 h followed by cell seeding did not produce significant transfection (unpublished observations). For polyplexes, the amount of DNA that associated with cells increased with time, and corresponds with a decreasing amount of DNA in solution and stable quantities on the surface, which again suggests internalization from the media. Polyplexes that associate with cells several hours after cell seeding may be deactivated by aggregation or disassembly of the polyplexes, which may lead to the observed reduction in efficacy. Lipoplexes, conversely, demonstrated a maximal association after 3 h indicating differences in the dynamics of cellular association between lipoplexes and polyplexes. The rapid cell association of lipoplexes and stable levels with increasing time may be described more effectively by a model that includes saturation of cell binding sites. The relatively rapid release of complexes from a surface is sufficient for transfection in vitro, as the cells are present immediately after seeding for cellular association; however, a longer half-life of vector release from the surface may be beneficial for in vivo applications to provide time for cell infiltration into the biomaterial.
Although surface delivery does not generally increase transfection relative to bolus delivery, the delivery of vectors by surface immobilization provides a versatile tool to localize or pattern gene delivery for applications such as transfected cell arrays or modeling tissue formation (Houchin-Ray et al., 2007; Pannier et al., 2007; Ziauddin and Sabatini, 2001). For delivery from tissue culture polystyrene, surface delivery produces protein expression that is within an order of magnitude of protein expression with bolus delivery. These levels of expression are sufficient for use in the traditional in vitro applications of gene delivery, such as cell screening systems (Pannier et al., 2007; Ziauddin and Sabatini, 2001). Importantly, the studies reported herein used standard transfection reagents and culture surfaces. These results demonstrate that surface and bolus delivery may employ different cellular pathways, and thus transfection reagents that maximize gene transfer by bolus delivery may not necessarily maximize transfection with surface delivery. Although protein expression levels are a function of many factors (e.g., vector unpacking, transcriptional availability of DNA, etc.), these results suggest that appropriately modifying the vector or the material for binding and release of complexes offers the potential to increase transfection. Based on the results reported herein, modifying polyplexes to achieve increased polyplex potency or modifying lipoplexes to achieve increased internalization and nuclear-association are rational strategies for increasing substrate-mediated transfection efficiency.
In conclusion, substrate-mediated gene delivery can provide localized and efficient expression in vitro for numerous research applications. These applications require systems that deliver DNA locally with high efficiency. Lipoplexes and polyplexes can be delivered from the surface, but their immobilization and cellular association can differ relative to bolus delivery. Immobilized polyplexes are readily delivered to cells, and strategies that maintain the activity of the complex could enhance transgene expression. For lipoplexes, immobilization does not appear to reduce the activity of the complexes, and the primary challenge is thus to increase the amount of DNA that is internalized and nuclear-associated. Developing biomaterial substrates and non-viral vectors specifically for substrate-mediated gene delivery approaches can increase transfection efficiency, which may lead to more utility in in vitro and in vivo applications.
Acknowledgments
Contract grant sponsor: National Institutes of Health (NIH)
Contract grant numbers: EB003806; GM066830
We would like to thank Chris Ramsborg and Antonia McKinney for technical assistance and Angela Pannier and Tiffany Houchin-Ray for their conversations and their help with the manuscript.
References
- Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9(3):443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov Today. 2002;7(18 Suppl):S113–S118. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]
- Bengali Z, Shea LD. Gene delivery by immobilization to cell-adhesive substrates. MRS Bull. 2005;30(9):659–662. doi: 10.1557/mrs2005.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotechnol Bioeng. 2005;90(3):290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengali Z, Rea JC, Shea LD. Gene expression and internalization following vector adsorption to immobilized proteins: Dependence on protein identity and density. J Gene Med. 2007;9(8):668–678. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinska AU, Yen A, Wu HL, Zahos KM, Sun R, Weiner ND, Baker JR, Jr., Roessler BJ. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21(9):877–887. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FH, Lee CH, Chen MT, Kuo CC, Chiang YL, Hang CY, Roffler S. Surfection: A new platform for transfected cell arrays. Nucleic Acids Res. 2004;32(3):e33. doi: 10.1093/nar/gnh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamme JP, Azoulay J, Mely Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84(3):1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PR, Read ML, Barrett LB, Wolfert MA, Seymour LW. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999;6(4):643–650. doi: 10.1038/sj.gt.3300843. [DOI] [PubMed] [Google Scholar]
- Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol Ther. 2006;13(4):786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Honma K, Ochiya T, Nagahara S, Sano A, Yamamoto H, Hirai K, Aso Y, Terada M. Atelocollagen-based gene transfer in cells allows high-throughput screening of gene functions. Biochem Biophys Res Commun. 2001;289(5):1075–1081. doi: 10.1006/bbrc.2001.6133. [DOI] [PubMed] [Google Scholar]
- Houchin-Ray T, Swift LA, Jang JH, Shea LD. Patterned PLG substrates for localized DNA delivery and directed neurite extension. Biomaterials. 2007;28(16):2603–2611. doi: 10.1016/j.biomaterials.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol Ther. 2000;1(4):339–346. doi: 10.1006/mthe.2000.0054. [DOI] [PubMed] [Google Scholar]
- Jang JH, Bengali Z, Houchin TL, Shea LD. Surface adsorption of DNA to tissue engineering scaffolds for efficient gene delivery. J Biomed Mater Res A. 2006;77(1):50–58. doi: 10.1002/jbm.a.30643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell CM, Zhang J, Fredin NJ, Lynn DM. Multilayered polyelectrolyte films promote the direct and localized delivery of DNA to cells. J Control Release. 2005;106(1–2):214–223. doi: 10.1016/j.jconrel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- King K, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush M. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7(1):77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, van Zanten JH. Real time monitoring of lipoplex molar mass, size and density. J Control Release. 2002;82(1):149–158. doi: 10.1016/s0168-3659(02)00104-9. [DOI] [PubMed] [Google Scholar]
- Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18(8):893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- Moriguchi R, Kogure K, Iwasa A, Akita H, Harashima H. Non-linear pharmacodynamics in a non-viral gene delivery system: Positive non-linear relationship between dose and transfection efficiency. J Control Release. 2006;110(3):605–609. doi: 10.1016/j.jconrel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol Ther. 2004;10(1):19–26. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Pannier AK, Anderson BC, Shea LD. Substrate-mediated delivery from self-assembled monolayers: Effect of surface ionization, hydrophilicity, and patterning. Acta Biomaterialia. 2005;1(5):511–522. doi: 10.1016/j.actbio.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier AK, Ariazi EA, Bellis AD, Bengali Z, Jordan VC, Shea LD. Bioluminescence imaging for assessment and normalization in transfected cell arrays. Biotechnol Bioeng. 2007;98(2):486–497. doi: 10.1002/bit.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannier AK, Wieland JA, Shea LD. Surface polyethylene glycol enhances substrate-mediated gene delivery by nonspecifically immobilized complexes. Acta Biomaterialia. 2008;4(1):26–39. doi: 10.1016/j.actbio.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea JC, Barron AE, Shea LD. Peptide-mediated lipofection is governed by lipoplex physical properties and the density of surface-displayed amines. J Pharm Sci. 2008;97(11):4794–4806. doi: 10.1002/jps.21338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377(Pt 1):159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12(3):468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Schaffer DV, Lauffenburger DA. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J Biol Chem. 1998;273(43):28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13(3):621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- Segura T, Volk MJ, Shea LD. Substrate-mediated DNA delivery: Role of the cationic polymer structure and extent of modification. J Control Release. 2003;93(1):69–84. doi: 10.1016/j.jconrel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid-collagen hydrogels via a substrate-mediated approach. Biomaterials. 2005;26(13):1575–1584. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17(6):551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- Shen H, Tan J, Saltzman WM. Surface-mediated gene transfer from nanocomposites of controlled texture. Nat Mater. 2004;3(8):569–574. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: High-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci USA. 2004;101(17):6548–6552. doi: 10.1073/pnas.0400165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana R, Harashima H, Ide N, Ukitsu S, Ohta Y, Suzuki N, Kikuchi H, Shinohara Y, Kiwada H. Quantitative analysis of correlation between number of nuclear plasmids and gene expression activity after transfection with cationic liposomes. Pharm Res. 2002;19(4):377–381. doi: 10.1023/a:1015162722295. [DOI] [PubMed] [Google Scholar]
- Vanhecke D, Janitz M. High-throughput gene silencing using cell arrays. Oncogene. 2004;23(51):8353–8358. doi: 10.1038/sj.onc.1208027. [DOI] [PubMed] [Google Scholar]
- Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12(13):1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- Webb BL, Diaz B, Martin GS, Lai F. A reporter system for reverse transfection cell arrays. J Biomol Screen. 2003;8(6):620–623. doi: 10.1177/1087057103259324. [DOI] [PubMed] [Google Scholar]
- Wheeler DB, Carpenter AE, Sabatini DM. Cell microarrays and RNA interference chip away at gene function. Nat Genet. 2005;37:S25–S30. doi: 10.1038/ng1560. [DOI] [PubMed] [Google Scholar]
- Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- Yamauchi F, Kato K, Iwata H. Micropatterned, self-assembled monolayers for fabrication of transfected cell microarrays. Biochim Biophys Acta. 2004;1672(3):138–147. doi: 10.1016/j.bbagen.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Uchimura E, Kishi M, Funeriu DP, Miyake M, Miyake J. Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J Control Release. 2004;96(2):227–232. doi: 10.1016/j.jconrel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]