Abstract
OBJECTIVE
Common variants in PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 genes have been shown to be associated with type 2 diabetes in European populations by genome-wide association studies. We have studied the association of common variants in these eight genes with type 2 diabetes and related traits in Indians by combining the data from two independent case–control studies.
RESEARCH DESIGN AND METHODS
We genotyped eight single nucleotide polymorphisms (PPARG-rs1801282, KCNJ11-rs5219, TCF7L2-rs7903146, SLC30A8-rs13266634, HHEX-rs1111875, CDKN2A-rs10811661, IGF2BP2-rs4402960, and CDKAL1-rs10946398) in 5,164 unrelated Indians of Indo-European ethnicity, including 2,486 type 2 diabetic patients and 2,678 ethnically matched control subjects.
RESULTS
We confirmed the association of all eight loci with type 2 diabetes with odds ratio (OR) ranging from 1.18 to 1.89 (P = 1.6 × 10−3 to 4.6 × 10−34). The strongest association with the highest effect size was observed for TCF7L2 (OR 1.89 [95% CI 1.71–2.09], P = 4.6 × 10−34). We also found significant association of PPARG and TCF7L2 with homeostasis model assessment of β-cell function (P = 6.9 × 10−8 and 3 × 10−4, respectively), which looked consistent with recessive and under-dominant models, respectively.
CONCLUSIONS
Our study replicates the association of well-established common variants with type 2 diabetes in Indians and shows larger effect size for most of them than those reported in Europeans.
Type 2 diabetes is a complex metabolic disorder with both genetic and environmental factors such as food habits and lifestyle contributing to its pathogenesis (1). Due to its complex etiology, the progress of discovery of genetic components for type 2 diabetes had been very slow until the advent of high throughput genome-wide association (GWA) studies (2). Until recently, only a few common variants in PPARG (3), KCNJ11 (4), and TCF7L2 (5) were shown to be associated with type 2 diabetes. With the advent of GWA studies, there are at least 20 loci identified today that are associated with the risk of type 2 diabetes (6). The first GWA study in the French population revealed SLC30A8 and HHEX as new loci for type 2 diabetes in addition to replicating the strong association with TCF7L2 (7). Further, GWA studies added several new genes including CDKAL1, CDKN2A, IGF2BP2, and FTO to the list of type 2 diabetes–associated loci and confirmed the associations for PPARG, KCNJ11, and TCF7L2 (8–12).
India harbors the maximum number of diabetic patients, which is projected to double by the year 2030 (13). Indians are diagnosed with diabetes a decade earlier and at a lower BMI than Europeans, which may be partly explained by their excess central obesity (14,15). Hence, determination of genetic risk factors predicting the risk of type 2 diabetes in the Indian population is highly desirable. Recent evidence suggests that the genetic basis of several diseases in Indians might be different from that of Europeans (16,17), which could be due to differences in the risk allele frequency and pattern of linkage disequilibrium. A report from the Indian Genome Variation Consortium also suggested that most of the populations in the Indian subcontinent are distinct from HapMap populations (18). Hence, genes associated with a disease in other populations need to be assessed for their role in the Indian population. The present study evaluated the association of eight most replicated and well-established genetic variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 with type 2 diabetes and related quantitative traits in Indians. We also performed allele dosage analysis of these variants and investigated their influence on quantitative metabolic traits related to type 2 diabetes.
RESEARCH DESIGN AND METHODS
The study involved the participation of 5,164 individuals comprising 2,486 patients with type 2 diabetes of Indo-European ethnicity and 2,678 ethnically matched control subjects recruited from two places—Delhi in northern India and Pune in western India. Subjects in the Delhi study (comprised of 1,019 patients and 1,006 control subjects) were enrolled on the basis of inclusion and exclusion criteria as described earlier (19). The consecutive subjects diagnosed as type 2 diabetic patients according to World Health Organization criteria (20) at the Endocrinology Clinic of the All India Institute of Medical Sciences were included in the study. The inclusion criteria for control subjects were ≥40 years of age, A1C ≤6.0%, fasting glucose ≤6.11 mmol/l, no history of diabetes in first- or second-degree relatives, and urban dweller of Indo-European ethnicity.
From Pune, 1,467 type 2 diabetic patients and 1,672 control subjects of Indo-European ethnicity were recruited for the study. Type 2 diabetic patients were diagnosed before 45 years of age according to World Health Organization criteria (20). The control group consisted of ethnically matched individuals recruited from different population cohorts including the Pune Maternal Nutrition Study, the Pune Children's Study, and the Coronary Risk of Insulin Sensitivity in Indian Subjects study (21).
Subjects with ketoacidosis at diagnosis, clinically judged to be insulin dependent, with exocrine pancreatic disease (fibrocalculous pancreatic diabetes), and who fulfilled the clinical criteria of maturity-onset diabetes of the young were excluded from the study. Pregnant women were also excluded from the study. Informed consent was obtained from all the participants and the study was approved by the ethics committees of the participating institutions in accordance with the principles of the Helsinki Declaration.
Clinical measurements.
All the subjects in both studies were extensively characterized for different anthropometric and quantitative metabolic traits. Anthropometric measurements, including height, weight, and waist and hip circumferences, were done per standardized protocols, and BMI and waist-to-hip ratio were calculated. Biochemical measurements including levels of A1C, fasting plasma glucose (FPG), 2-h postprandial glucose (PPG), fasting plasma insulin (FPI), total cholesterol, HDL cholesterol, and triglycerides were performed using standard laboratory assays as described earlier (19,22). Homeostasis model assessment of insulin resistance (HOMA-IR) was calculated using the formula: (FPI in mU/ml × FPG in mmol/l)/22.5 and homeostasis model assessment of β-cell function (HOMA-B) was calculated using the equation: (FPI in mU/ml × 20)/(FPG in mmol/l − 3.5) as reported previously (23).
Genotyping.
Eight well-replicated single nucleotide polymorphisms (SNPs) from eight genes identified through GWA studies were selected for association with type 2 diabetes. These included SNPs-rs1801282 (PPARG), rs5219 (KCNJ11), rs7903146 (TCF7L2), rs13266634 (SLC30A8), rs1111875 (HHEX), rs10811661 (CDKN2A), rs4402960 (IGF2BP2), and rs10946398 (CDKAL1). In the samples from Delhi, these eight SNPs were genotyped using Illumina Golden Gate assay as a part of 1,536-plex assay and following the manufacturer's instructions (http://www.illumina.com/downloads/GOLDENGATEASSAY.pdf). Quality control criteria for the SNPs to qualify for further analysis included: genotype confidence score of >0.25, call frequency >0.9, GenTrans score >0.6, cluster separation score >0.4, minor allele frequency >0.05, and Hardy-Weinberg equilibrium (HWE) in control subjects (P > 0.0031). A total of 147 samples (7.2%) were genotyped in duplicate, and an error rate of <0.01 was estimated. Further, to validate the genotype calls, 10% of the SNPs were re-genotyped for 180 samples using Sequenom-based MassARRAY technology, and a concordance rate of >99.7% was observed.
The genotyping of all the SNPs in the subjects from Pune was performed using Sequenom-based MassARRAY technology. Genotypes for ∼10% of the samples were confirmed by sequencing on an ABI 3730 Genetic Analyzer using specific primers, and inconsistency of only 0.06% (2/3,452) was observed.
Statistical analysis.
We examined the association of the above-mentioned SNPs in eight genes with type 2 diabetes and related traits in the Delhi and Pune studies separately and also after combining the samples from both of the studies. The distributions of genotype for all the SNPs were analyzed for deviation from HWE using χ2 analysis. Cochran Q-statistics was performed to assess heterogeneity between the two groups. Logistic regression analysis assuming log additive model was performed to determine the association between SNPs and the risk for type 2 diabetes. The associations were adjusted for sex, age, BMI, and geographical region, as appropriate. The odds ratios (ORs) with 95% CIs were presented with respect to the risk allele as observed in the initial studies on Europeans (9). A P value of < 0.0062 (α = 0.05/8) was considered significant for association with type 2 diabetes in the combined samples after correcting for multiple testing. The association of each SNP with continuous traits was performed by pooling the data of control subjects from both of the studies using the Kruskal-Wallis test. Initially a priori power was calculated based on the risk allele frequency (RAF) and effect sizes reported in the Europeans. Subsequently, posterior power calculation was performed based on the RAF and effect sizes from our study, assuming log additive model using Quanto software (http://hydra.usc.edu/gxe/) at α = 0.05 and α = 0.0062 (corrected P value), and assuming 10% prevalence of type 2 diabetes. For rs7903146 (TCF7L2), we combined the published data of the Pune study (22) with that of the Delhi study data.
The combined effect of the eight SNPs on the risk of type 2 diabetes was determined through allele dosage analysis by categorizing the subjects based on the number of “effective” risk alleles. The analysis included only those individuals in whom genotypes at all eight SNPs were available. Since the effect sizes (defined by the SNP-specific ORs) were not uniform, the allele dosage score of an individual was computed by the weighted mean of the proportion of risk alleles at the eight SNPs (i.e., one for two risk alleles, 0.5 for one risk allele, and 0 for no risk allele) with weights as the relative log ORs of different SNPs. The “effective” number of risk alleles was obtained as the allele dosage score multiplied by 16 (maximum number of risk alleles corresponding to eight SNPs). Considering subjects with six or fewer number of “effective” risk alleles as the reference group, ORs and P values for every unit increase in the number of “effective” risk alleles were calculated after adjusting for sex, age, BMI, and geographical region. All statistical analyses were performed using PLINK version 1.05 (http://pngu.mgh.harvard.edu/_purcell/plink) (24) and SPSS version 17.0 (SPSS, Chicago, IL) unless specified otherwise.
RESULTS
The clinical characteristics of the subjects in both of the studies are presented in Table 1. The genotype distributions at all the SNPs were in HWE (P > 0.0031 in control subjects).
TABLE 1.
Basic anthropometric and clinical characteristics of the study groups from two centers
| Characteristics | Delhi |
Pune |
||
|---|---|---|---|---|
| Type 2 diabetic patients | Control subjects | Type 2 diabetic patients | Control subjects | |
| n (men/women) | 1,019 (592/427) | 1,006 (606/400) | 1,467 (826/641) | 1,672 (884/788) |
| Age (years) | 53 (45–62) | 50 (44–60) | 46 (40–52) | 33 (29–37) |
| Age of diagnosis (years) | 45 (39–52) | — | 38.5 (33.3–42.2) | — |
| Individuals on medication for lipids (%) | 4.00 | — | 22.70 | — |
| BMI (kg/m2) | ||||
| Women | 26.70 (24.20–29.20) | 24.90 (21.10–28.60) | 26.90 (24.40–29.60) | 19.53 (17.60–22.74) |
| Men | 23.80 (22.00–26.00) | 23.20 (20.20–25.70) | 24.90 (22.80–27.70) | 21.18 (19.15–23.62) |
| Waist-to-hip ratio | ||||
| Women | 1.00 (0.97–1.03) | 0.86 (0.82–0.92) | 0.89 (0.85–0.94) | 0.76 (0.73–0.80) |
| Men | 1.00 (0.97–1.03) | 0.97 (0.92–1.00) | 0.97 (0.94–1.02) | 0.91 (0.86–0.95) |
| FPG (mmol/l) | 7.90 (6.40–10.30) | 4.90 (4.50–5.30) | 8.50 (6.90–11.30) | 5.11 (4.67–5.56) |
| 2-h PPG (mmol/l) | — | 5.60 (5.80–6.30) | — | 5.50 (4.56–6.50) |
| Total cholesterol (mmol/l) | 4.20 (3.50–5.00) | 4.40 (3.70–5.10) | 4.10 (3.50–4.80) | 3.77 (3.28–4.33) |
| Triglycerides (mmol/l) | 1.60 (1.10–2.20) | 1.30 (1.00–1.80) | 1.40 (1.00–1.20) | 0.84 (0.62–1.20) |
| HDL cholesterol (mmol/l) | 1.03 (0.88–1.22) | 1.06 (0.88–1.28) | 1.03 (0.90–1.18) | 1.03 (0.87–1.23) |
| FPI (pmol/l) | — | 32.20 (17.50–57.20) | — | 32.50 (20.84–49.18) |
| HOMA-IR | — | 1.16 (0.59–2.02) | — | 1.21 (0.76–1.89) |
| HOMA-B | — | 73.40 (40.70–138.60) | — | 69.73 (44.39–109.79) |
Data are median (interquartile range).
In the Delhi study, the association analysis with type 2 diabetes revealed that all the eight SNPs were significantly associated with type 2 diabetes at α = 0.05 and remained so after adjustment for sex, age, and BMI with ORs ranging from 1.17 to 1.67 and P values ranging from 0.02 to 1.7 × 10−13 (Table 2, and supplementary Table 1 in the online appendix, available at http://diabetes.diabetesjournals.org/cgi/content/full/db09-1386/DC1). Consistent with earlier findings, we also observed the strongest association at rs7903146 of TCF7L2 with OR 1.67 (95% CI [1.46–1.92], P = 1.7 × 10−13).
TABLE 2.
Association of SNPs with type 2 diabetes in the Delhi and Pune studies and the combined study
| SNP (gene) | Risk/non-risk allele* | Delhi |
Pune |
P (difference in allele frequencies) | Combined |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n Case/control subjects | RAF |
OR (95% CI)† | P | n Case/control subjects | RAF |
OR (95% CI)† | P | n Case/control subjects | RAF |
OR (95% CI)‡ | P | ||||||
| Case subjects | Control subjects | Case subjects | Control subjects | Case subjects | Control subjects | ||||||||||||
| rs1801282 (PPARG) | C/G | 1,019/1,004 | 0.89 | 0.86 | 1.30 (1.08–1.57) | 0.006 | 1,422/1,553 | 0.91 | 0.87 | 1.47 (1.15–1.88) | 1.8 × 10−3 | 0.21 | 2,441/2,557 | 0.90 | 0.87 | 1.37 (1.19–1.59) | 1.6 × 10−5 |
| rs5219 (KCNJ11) | T/C | 1,017/1,006 | 0.39 | 0.35 | 1.17 (1.02–1.33) | 0.02 | 1,417/1,397 | 0.40 | 0.28 | 1.84 (1.55–2.18) | 4.4 × 10−12 | 8.1 × 10−7 | 2,434/2,403 | 0.40 | 0.31 | 1.39 (1.26–1.54) | 6.7 × 10−11 |
| rs7903146 (TCF7L2) | T/C | 1,017/1,006 | 0.40 | 0.29 | 1.67 (1.46–1.92) | 1.7 × 10−13 | 1,423/1,515 | 0.37 | 0.21 | 2.10 (1.77–2.49) | 1.7 × 10−17 | 4.5 × 10−9 | 2,439/2,521 | 0.38 | 0.24 | 1.89 (1.71–2.09) | 4.6 × 10−34 |
| rs13266634 (SLC30A8) | C/T | 1,010/1,000 | 0.79 | 0.75 | 1.32 (1.13–1.53) | 0.0003 | 1,456/1,539 | 0.81 | 0.78 | 1.25 (1.04–1.51) | 0.02 | 0.03 | 2,466/2,539 | 0.80 | 0.77 | 1.34 (1.20–1.50) | 3.4 × 10−7 |
| rs1111875 (HHEX) | G/A | 1,018/1,003 | 0.47 | 0.42 | 1.29 (1.13–1.47) | 0.0001 | 1,454/1,598 | 0.47 | 0.43 | 1.20 (1.03–1.39) | 0.02 | 0.81 | 2,472/2,601 | 0.47 | 0.43 | 1.27 (1.16–1.39) | 5.7 × 10−7 |
| rs10811661 (CDKN2A) | T/C | 1,019/1,006 | 0.89 | 0.87 | 1.3 (1.07–1.59) | 0.009 | 1,431/1,086 | 0.89 | 0.86 | 1.43 (1.09–1.87) | 0.01 | 0.90 | 2,450/2,092 | 0.89 | 0.86 | 1.37 (1.18–1.59) | 5.1 × 10−5 |
| rs4402960 (IGF2BP2) | T/G | 1,019/1,006 | 0.46 | 0.42 | 1.18 (1.03–1.33) | 0.02 | 1,444/868 | 0.50 | 0.46 | 1.30 (1.08–1.56) | 5.5 × 10−3 | 0.03 | 2,463/1,874 | 0.48 | 0.44 | 1.20 (1.09–1.33) | 2.6 × 10−3 |
| rs10946398 (CDKAL1) | C/A | 1,006/990 | 0.24 | 0.21 | 1.19 (1.03–1.38) | 0.02 | 1,456/1,512 | 0.27 | 0.26 | 1.17 (0.98–1.38) | 0.08 | 3.3 × 10−4 | 2,462/2,501 | 0.26 | 0.24 | 1.18 (1.07–1.32) | 1.6 × 10−3 |
*Risk alleles as identified in earlier European studies (9).
†Analysis adjusted for sex, age, and BMI.
‡Analysis adjusted for sex, age, BMI, and geographical region (Delhi and Pune). n, number of subjects genotyped; OR, odds ratio per allele; RAF, risk allele frequency.
In the Pune study, all the SNPs except for rs10946398 (CDKAL1) were significantly associated with type 2 diabetes and retained significance after adjusting for sex, age, and BMI (Table 2, supplementary Table 1). Similar to the observation in the Delhi study, the TCF7L2 variant showed the strongest association with OR 2.10 (95% CI [1.77–2.49], P = 1.7 × 10−17).
There was no significant heterogeneity of ORs for any of the variants between the Delhi and Pune studies (P > 0.05 for Cochran Q-statistics), hence we performed association analysis by combining their genotype data. All the eight SNPs were significantly associated with type 2 diabetes and adjustment for covariates like age, sex, BMI, and geographical region did not nullify the significance (Table 2, supplementary Table 1). ORs for the associations in the combined analysis ranged from 1.18 to 1.89 with P values ranging from 1.6 × 10−3 to 4.6 × 10−34. The RAF of three variants (rs5219, rs7902146, and rs10946398) differed significantly between the control subjects of the two studies (Table 2). Meta-analysis performed by combining the summary data of the two studies, both under the fixed- and random-effect models, also confirmed the association of these SNPs with type 2 diabetes, except for rs5219 under the random-effect model (supplementary Table 2). The estimated OR for rs5219 was higher under the random-effect model than the fixed effect model, which is known to yield more biased estimates. A priori power calculations showed that our study had >62% power at α = 0.05 and >32% at α = 0.0062 (supplementary Table 3) for detecting the association for the risk of type 2 diabetes for all SNPs. The effect sizes observed for all variants in our study populations were higher compared with that of Europeans (Table 3). However, the CI for two of them (IGF2BP2 and CDKAL1) included the ORs observed in Europeans.
TABLE 3.
Comparison of effect sizes of the SNPs in Indians and Europeans
| Gene | SNP | Risk/non-risk allele* | RAF in Indians | RAF in Europeans | Indians† |
Europeans‡ |
||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |||||
| PPARG | rs1801282 | C/G | 0.87 | 0.86 | 1.37 (1.19–1.59) | 1.6 × 10−5 | 1.14 (1.08–1.20) | 1.7 × 10−6 |
| KCNJ11 | rs5219 | T/C | 0.31 | 0.47 | 1.39 (1.26–1.54) | 6.7 × 10−11 | 1.14 (1.10–1.19) | 6.7 × 10−11 |
| TCF7L2 | rs7903146 | T/C | 0.24 | 0.26 | 1.89 (1.71–2.09) | 4.6 × 10−34 | 1.37 (1.31–1.43) | 1.0 × 10−48 |
| SLC30A8 | rs13266634 | C/T | 0.77 | 0.65 | 1.34 (1.20–1.50) | 3.4 × 10−7 | 1.12 (1.07–1.16) | 5.3 × 10−8 |
| HHEX | rs1111875 | G/A | 0.43 | 0.53 | 1.27 (1.16–1.39) | 5.7 × 10−7 | 1.13 (1.09–1.17) | 5.7 × 10−10 |
| CDKN2A | rs10811661 | T/C | 0.86 | 0.83 | 1.37 (1.18–1.59) | 5.1 × 10−5 | 1.20 (1.14–1.25) | 7.8 × 10−25 |
| IGF2BP2 | rs4402960 | T/G | 0.44 | 0.29 | 1.20 (1.09–1.33) | 2.6 × 10−3 | 1.14 (1.11–1.18) | 8.9 × 10−36 |
| CDKAL1 | rs10946398 | C/A | 0.24 | 0.31 | 1.18 (1.07–1.32) | 1.6 × 10−3 | 1.12 (1.08–1.16) | 4.1 × 10−11 |
*Risk alleles in Europeans, as indicated by DGI study (9).
†ORs and P values obtained for combined analysis of data from the Delhi and Pune studies.
‡ORs and P values obtained from combined analysis of the Diabetes Genetics Initiative (DGI), Finland-United States Investigation of NIDDM Genetics (FUSION), and Wellcome Trust Case Control Consortium (WTCCC) studies (9). OR, odds ratio per allele; RAF, risk allele frequency.
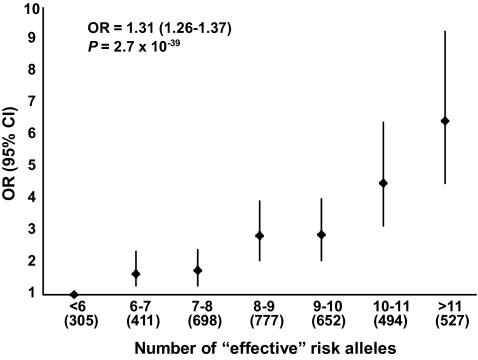
Allele dosage analysis revealed a significantly enhanced risk of type 2 diabetes by 1.31-fold with the increase in each unit of “effective” risk allele (P = 2.7 × 10−39) (Fig. 1). Individuals with 11 or more “effective” risk alleles (13.64%) carried 6.45-fold increased risk for type 2 diabetes in comparison with individuals having 6 or less “effective” risk alleles (7.89%) (P = 7 × 10−24).
FIG. 1.
Effect of increase in the number of “effective” risk alleles on the risk of type 2 diabetes in combined samples. OR and 95% CI are plotted on the y-axis for the corresponding number of “effective” risk alleles on the x-axis. Numbers in parentheses on x-axis indicate sample size in each category.
We then examined the association of genetic variants with various quantitative traits in the control subjects from the two studies (Table 4, supplementary Table 5). We observed significant associations for PPARG and TCF7L2 with HOMA-B (P = 6.9 × 10−8 and 3 × 10−4, respectively). The association of PPARG looked consistent with a recessive model and that of TCF7L2 looked consistent with an under-dominant model (heterozygotes have lower HOMA-B than either of the homozygotes as opposed to showing a continuous trend across the genotypes). We also found nominal association of KCNJ11 with FPG (P = 0.004), HOMA-IR (P = 0.04), and waist-to-hip ratio (P = 0.01); TCF7L2 with FPG (P = 0.03) and 2-h PPG (P = 0.03); and HHEX with FPI (P = 0.04) and HOMA-IR (P = 0.02). However, none of the associations for the quantitative trait analysis, except for PPARG and TCF7L2 with HOMA-B, remained significant after correcting for multiple testing (Pcorrected = 0.000625, 0.05/80).
TABLE 4.
Association of SNPs with quantitative traits (glucose and insulin metabolism) in combined control subjects
| Gene SNP | Genotypesmajor hetero minor | n | FPG (mmol/l) |
2-h PPG (mmol/l) |
FPI (pmol/l) |
HOMA-IR |
HOMA-B |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median (IQR) | P‡ | Median (IQR) | P‡ | Median (IQR) | P‡ | Median (IQR) | P‡ | Median (IQR) | P‡ | |||
| PPARG | CC | 1,932 | 5.03 (4.61–5.43) | 5.50 (4.56–6.50) | 32.99 (20.00–51.46) | 1.21 (0.71–1.92) | 72.58 (43.10–116.82) | |||||
| rs1801282 | CG | 578 | 5.03 (4.61–5.41) | 5.61 (4.72–6.43) | 30.42 (19.69–47.14) | 1.13 (0.69–1.86) | 67.19 (43.87–117.43) | |||||
| (C/G) | GG | 47 | 4.94 (4.72–5.28) | 0.39 | 5.50 (4.61–6.26) | 0.49 | 27.80 (15.45–44.60) | 0.13 | 1.02 (0.51–1.62) | 0.33 | 65.36 (36.36–105.99) | 6.9 × 10−8* |
| KCNJ11 | CC | 1,143 | 5.00 (4.59–5.39) | 5.50 (4.56–6.50) | 32.00 (19.93–48.82) | 1.18 (0.70–1.87) | 71.68 (42.47–116.18) | |||||
| rs5219 | CT | 1,016 | 5.07 (4.71–5.44) | 5.50 (4.57–6.44) | 32.66 (20.02–51.81) | 1.21 (0.73–1.94) | 68.27 (44.32–112.37) | |||||
| (C/T) | TT | 244 | 5.06 (4.64–5.46) | 0.004* | 5.44 (4.50–6.33) | 0.63 | 32.33 (19.26–55.95) | 0.70 | 1.22 (0.65–1.96) | 0.04* | 73.89 (39.33–122.34) | 0.09 |
| TCF7L2 | CC | 1,449 | 5.00 (4.61–5.44) | 5.33 (4.50–6.28) | 32.26 (19.23–49.64) | 1.18 (0.69–1.87) | 71.83 (42.75–115.76) | |||||
| rs7903146 | CT | 919 | 5.06 (4.61–5.44) | 5.67 (4.70–6.56) | 31.39 (20.04–52.00) | 1.18 (0.71–1.96) | 68.89 (42.29–118.40) | |||||
| (C/T) | TT | 153 | 5.00 (4.66–5.38) | 0.03* | 5.64 (4.78–6.52) | 0.03* | 34.32 (19.32–53.74) | 0.65 | 1.23 (0.72–1.87) | 0.97 | 72.56 (41.30–111.24) | 3 × 10−4* |
| SLC30A8 | CC | 1,487 | 5.03 (4.64–5.44) | 5.44 (4.56–6.44) | 32.50 (19.80–50.00) | 1.19 (0.71–1.89) | 69.79 (43.88–115.71) | |||||
| rs13266634 | CT | 911 | 5.03 (4.61–5.39) | 5.61 (4.78–6.44) | 32.16 (19.86–51.00) | 1.21 (0.69–1.90) | 71.09 (41.60–118.98) | |||||
| (C/T) | TT | 141 | 4.94 (4.50–5.34) | 0.28 | 5.22 (4.34–6.47) | 0.17 | 28.25 (18.17–45.16) | 0.08 | 1.00 (0.63–1.79) | 0.05 | 74.62 (42.25–114.14) | 0.46 |
| HHEX | AA | 847 | 5.03 (4.61–5.39) | 5.44 (4.56–6.44) | 34.00 (20.00–53.86) | 1.26 (0.72–2.02) | 73.85 (45.07–119.81) | |||||
| rs1111875 | AG | 1,274 | 5.00 (4.61–5.44) | 5.44 (4.56–6.51) | 31.39 (20.34–48.18) | 1.16 (0.72–1.81) | 68.49 (43.03–114.09) | |||||
| (A/G) | GG | 480 | 5.06 (4.64–5.46) | 0.17 | 5.67 (4.67–6.39) | 0.20 | 32.93 (18.99–54.00) | 0.04* | 1.25 (0.68–2.02) | 0.02* | 72.74 (41.51–120.19) | 0.48 |
| CDKN2A | TT | 1,554 | 5.06 (4.67–5.44) | 5.39 (4.50–6.39) | 32.64 (19.74–51.12) | 1.21 (0.69–1.94) | 70.52 (42.57–114.69) | |||||
| rs10811661 | TC | 497 | 5.02 (4.57–5.44) | 5.44 (4.56–6.41) | 30.72 (19.14–53.85) | 1.15 (0.68–1.96) | 66.10 (40.97–115.21) | |||||
| (T/C) | CC | 41 | 4.94 (4.73–5.61) | 0.27 | 5.20 (4.35–6.26) | 0.74 | 32.71 (21.74–59.87) | 0.58 | 1.27 (0.77–2.03) | 0.90 | 62.75 (40.79–103.38) | 0.31 |
| IGF2BP2 | GG | 591 | 5.00 (4.65–5.38) | 5.36 (4.56–6.39) | 33.02 (20.28–51.86) | 1.24 (0.73–1.95) | 74.87 (44.54–122.09) | |||||
| rs4402960 | GT | 919 | 5.03 (4.61–5.44) | 5.39 (4.50–6.26) | 30.54 (18.54–50.77) | 1.13 (0.65–1.88) | 65.27 (40.79–111.48) | |||||
| (G/T) | TT | 364 | 5.06 (4.72–5.38) | 0.69 | 5.50 (4.50–6.44) | 0.64 | 31.43 (18.85–49.22) | 0.23 | 1.15 (0.69–1.81) | 0.31 | 67.48 (42.35–109.40) | 0.18 |
| CDKAL1 | AA | 1,466 | 5.05 (4.61–5.44) | 5.52 (4.67–6.50) | 32.16 (19.00–51.81) | 1.17 (0.68–1.93) | 68.88 (40.87–116.96) | |||||
| rs10946398 | AC | 875 | 5.06 (4.68–5.44) | 5.56 (4.50–6.56) | 32.50 (20.44–49.18) | 1.23 (0.74–1.90) | 72.10 (45.05–113.93) | |||||
| (A/C) | CC | 160 | 5.08 (4.61–5.39) | 0.78 | 5.00 (4.39–6.11) | 0.14 | 34.40 (21.60–55.58) | 0.05 | 1.28 (0.79–2.14) | 0.06 | 78.54 (48.10–121.61) | 0.05 |
Data are median (interquartile range).
*Significant P value.
‡Analysis adjusted for sex, age, BMI, and geographical region. IQR, interquartile range.
DISCUSSION
Genome-wide association studies have resulted in the identification of a number of loci associated with type 2 diabetes—from just 3 confirmed loci in 2006 to 20 in 2009 (6). In the present study, we investigated the association of eight common and well-established common genetic variants with type 2 diabetes in 5,148 Indians. We combined the data from two independent studies of Indo-European individuals to increase the power of the study. To our knowledge, this is the largest study reported for the association of type 2 diabetes in Indians.
Among all the loci, TCF7L2 so far has shown the strongest association with the largest effect size for type 2 diabetes in Europeans (5,7–12), Amish (25), and Indians (22,26,27), but not in Chinese (28) and Japanese (29) subjects. The present study confirms the association of TCF7L2 with type 2 diabetes with the largest effect size. The TCF7L2 gene product has been implicated in blood glucose homeostasis (5,30), and the variant rs7903146 is reported to be associated with measures of glucose metabolism (25). Consistent with these observations, we also found a strong association of TCF7L2 with HOMA-B and a nominal association with FPG and 2-h PPG, confirming the physiological role of TCF7L2 in glucose homeostasis.
Studies evaluating the association of the Pro12Ala variant of the PPARG gene with type 2 diabetes have reported contradictory results. The minor allele (Ala) at this locus has been shown to confer protection against type 2 diabetes by many studies (31–34) while in several other studies it has been found to predict susceptibility for type 2 diabetes (35,36). Moreover, association analysis of Pro12Ala in Asian Indians has also shown inconsistent findings (27,37) that might be attributed to the differences in the ethnicity of the subjects in these studies, or the studies may have been underpowered for detecting the direction of the association. We established the strong protective effect of the Ala allele against the development of type 2 diabetes in Indians.
Variant E23K (rs5219) in KCNJ11 has been shown to be associated with type 2 diabetes in Europeans and Japanese (4,38) subjects, and our study replicates this significant association. The polymorphisms in SLC30A8 and HHEX were identified by GWA studies in several European populations and subsequently was replicated in Asians (38–40). However, the association could not be validated in the Indian Sikh population, which was probably due to the small sample size and the low power of the study (27). With a larger sample size and higher power, we confirmed the strong association of SLC30A8 and HHEX SNPs with type 2 diabetes in our combined data. The variant rs13266634 in SLC30A8 has been shown to be associated with FPG and BMI (39,40), but we did not find any association with either of these parameters.
Variants of the CDKAL1 gene have been linked to glucotoxicity (8) and have been shown to be associated with insulin secretion (8), A1C, and HOMA-B (41). We found significant association of the CDKAL1 variant with type 2 diabetes but with none of the quantitative traits. We also found significant association for the variant rs10811661, which lies near the CDKN2A gene, with the risk of type 2 diabetes. This SNP was not associated with type 2 diabetes in a recent study of Indian Sikhs (27), stressing on the importance of adequate sample size with sufficient power. Consistent with previous studies, we also provided evidence for the association of rs4402960 in IGF2BP2 with type 2 diabetes (27,38–40).
While all the eight loci independently predicted the risk for type 2 diabetes, this increased significantly if all the alleles were present together. A 1.31 times enhanced risk with the increase in each unit of “effective” risk allele and 6.45-fold risk among individuals carrying 11 or more “effective” risk alleles compared with individuals with 6 or less “effective” risk alleles underlines the additive influence of these genetic variants on risk for type 2 diabetes and their likely role in predictive genetic testing. Furthermore, several variants were found to have higher effect sizes in our study population in comparison with Europeans. This might be an indication of higher penetration of these variants for the risk of type 2 diabetes in Indians. However, the CIs for two of them (IGF2BP2 and CDKAL1) include the OR estimated in the Europeans suggesting a similar effect size in both. It is worth mentioning that other six loci (PPARG, TCF7L2, KCNJ11, SLC30A8, CDKN2A, and HHEX), whose effect sizes are higher compared with those of Europeans, also show higher effect sizes when compared with East Asians (28,29,32–34, 38,39,40). Thus, the risk alleles at a few type 2 diabetes–associated loci have higher penetration in Indians compared with Europeans and East Asians subjects.
Population stratification is a major confounder for any association analysis. To address this issue, we have recruited case and control subjects from the same area for both the Delhi and Pune studies to minimize the chance for population stratification in the Indian context (18). However, given the diversity of Indian population, this possibility cannot be completely eliminated in the present study.
In conclusion, we replicated the association of eight major common variants of PPARG, KCNJ11, TCF7L2, CDKAL1, HHEX, SLC30A8, CDKN2A, and IGF2BP2 with type 2 diabetes in Indians of Indo-European origin with an increased effect size compared with Europeans. Although our study is the largest and is an adequately powered study that has investigated the genetic basis of type 2 diabetes in Indians, a larger study of different ethnic groups including Dravidians will aid in better understanding their penetration, the likely effect of these genes on various quantitative traits, and the potential use as diagnostic markers for the susceptibility to type 2 diabetes in the diabetic capital of the world.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by “Diabetes-new drug discovery R&D, molecular mechanisms and genetic & epidemiological factors” (NWP0032-8), which is funded by the Council of Scientific and Industrial Research (CSIR), Government of India.
No potential conflicts of interest relevant to this article were reported.
We thank all the participants of the study. We thank A.K. Sharma for his help in sample collection. We also thank Dr. S.D. Kale and Dr. Jayant Deshpande for their help in sample collection and phenotype information.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.McCarthy MI, Froguel P: Genetic approaches to the molecular understanding of type 2 diabetes. Am J Physiol Endocrinol Metab 2002;283:217–225 [DOI] [PubMed] [Google Scholar]
- 2.Frayling TM: Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet 2007;8:657–662 [DOI] [PubMed] [Google Scholar]
- 3.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES: The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 2000;26:76–80 [DOI] [PubMed] [Google Scholar]
- 4.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, Walker M, Levy JC, Sampson M, Halford S, McCarthy MI, Hattersley AT, Frayling TM: Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 2003;52:568–572 [DOI] [PubMed] [Google Scholar]
- 5.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K: Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 6.McCarthy MI: Casting a wider net for diabetes susceptibility genes. Nat Genet 2008;40:1039–1040 [DOI] [PubMed] [Google Scholar]
- 7.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 8.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K: A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 2007;39:770–775 [DOI] [PubMed] [Google Scholar]
- 9.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 10.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, Wellcome Trust Case Control Consortium (WTCCC) McCarthy MI, Hattersley AT: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wild S, Roglic G, Green A, Sicree R, King H: Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 2004;27:1047–1053 [DOI] [PubMed] [Google Scholar]
- 14.Ramachandran A, Snehalatha C, Viswanathan V, Viswanathan M, Haffner SM: Risk of noninsulin dependent diabetes mellitus conferred by obesity and central adiposity in different ethnic groups: a comparative analysis between Asian Indians, Mexican Americans and Whites. Diabetes Res Clin Pract 1997;36:121–125 [DOI] [PubMed] [Google Scholar]
- 15.Shelgikar KM, Hockaday TD, Yajnik CS: Central rather than generalized obesity is related to hyperglycaemia in Asian Indian subjects. Diabet Med 1991;8:712–717 [DOI] [PubMed] [Google Scholar]
- 16.Chandak GR, Idris MM, Reddy DN, Bhaskar S, Sriram PV, Singh L: Mutations in the pancreatic secretory trypsin inhibitor gene (PSTI/SPINK1) rather than the cationic trypsinogen gene (PRSS1) are significantly associated with tropical calcific pancreatitis. J Med Genet 2002;39:347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabassum R, Chavali S, Mahajan A, Ghosh S, Madhu SV, Tandon N, Bharadwaj D: Association analysis of TNFRSF1B polymorphisms with type 2 diabetes and its related traits in North India. Genomic Medicine 2008;2:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Indian Genome Variation Consortium Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet 2008;87:3–20 [DOI] [PubMed] [Google Scholar]
- 19.Tabassum R, Chavali S, Dwivedi OP, Tandon N, Bharadwaj D: Genetic variants of FOXA2: risk of type 2 diabetes and effect on metabolic traits in North Indians. J Hum Genet 2008;53:957–965 [DOI] [PubMed] [Google Scholar]
- 20.The Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2003;26(Suppl. 1):S5–S20 [DOI] [PubMed] [Google Scholar]
- 21.Yajnik CS, Janipalli CS, Bhaskar S, Kulkarni SR, Freathy RM, Prakash S, Mani KR, Weedon MN, Kale SD, Deshpande J, Krishnaveni GV, Veena SR, Fall CH, McCarthy MI, Frayling TM, Hattersley AT, Chandak GR: FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia 2009;52:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandak GR, Janipalli CS, Bhaskar S, Kulkarni SR, Mohankrishna P, Hattersley AT, Frayling TM, Yajnik CS: Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia 2007;50:63–67 [DOI] [PubMed] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC: Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damcott CM, Pollin TI, Reinhart LJ, Ott SH, Shen H, Silver KD, Mitchell BD, Shuldiner AR: Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes 2006;55:2654–2659 [DOI] [PubMed] [Google Scholar]
- 26.Bodhini D, Radha V, Dhar M, Narayani N, Mohan V: The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism 2007;56:1174–1178 [DOI] [PubMed] [Google Scholar]
- 27.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, Kamboh MI: Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet 2008;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YC, Chang TJ, Jiang YD, Kuo SS, Lee KC, Chiu KC, Chuang LM: Association study of the genetic polymorphisms of the transcription factor 7-like 2 (TCF7L2) gene and type 2 diabetes in the Chinese population. Diabetes 2007;56:2631–2637 [DOI] [PubMed] [Google Scholar]
- 29.Horikoshi M, Hara K, Ito C, Nagai R, Froguel P, Kadowaki T: A genetic variation of the transcription factor 7-like 2 gene is associated with risk of type 2 diabetes in the Japanese population. Diabetologia 2007;50:747–751 [DOI] [PubMed] [Google Scholar]
- 30.Yi F, Brubaker PL, Jin T: TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3 beta. J Bio Chem 2005;280:1457–1464 [DOI] [PubMed] [Google Scholar]
- 31.Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J: A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 1998;20:284–287 [DOI] [PubMed] [Google Scholar]
- 32.Hara K, Okada T, Tobe K, Yasuda K, Mori Y, Kadowaki H, Hagura R, Akanuma Y, Kimura S, Ito C, Kadowaki T: The Pro12Ala polymorphism in PPAR gamma2 may confer resistance to type 2 diabetes. Biochem Biophys Res Commun 2000;271:212–216 [DOI] [PubMed] [Google Scholar]
- 33.Mori H, Ikegami H, Kawaguchi Y, Seino S, Yokoi N, Takeda J, Inoue I, Seino Y, Yasuda K, Hanafusa T, Yamagata K, Awata T, Kadowaki T, Hara K, Yamada N, Gotoda T, Iwasaki N, Iwamoto Y, Sanke T, Nanjo K, Oka Y, Matsutani A, Maeda E, Kasuga M: The Pro12 → Ala substitution in PPAR-γ is associated with resistance to development of diabetes in the general population: possible involvement in impairment of insulin secretion in individuals with type 2 diabetes. Diabetes 2001;50:891–894 [DOI] [PubMed] [Google Scholar]
- 34.Douglas JA, Erdos MR, Watanabe RM, Braun A, Johnston CL, Oeth P, Mohlke KL, Valle TT, Ehnholm C, Buchanan TA, Bergman RN, Collins FS, Boehnke M, Tuomilehto J: The peroxisome proliferator-activated receptor-gamma2 Pro12A1a variant: association with type 2 diabetes and trait differences. Diabetes 2001;50:886–890 [DOI] [PubMed] [Google Scholar]
- 35.Hegele RA, Cao H, Harris SB, Zinman B, Hanley AJ, Anderson CM: Peroxisome proliferator-activated receptor-gamma2 P12A and type 2 diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab 2000;85:2014–2019 [DOI] [PubMed] [Google Scholar]
- 36.Ristow M, Müller-Wieland D, Pfeiffer A, Krone W, Kahn CR: Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N Engl J Med 1998;339:953–959 [DOI] [PubMed] [Google Scholar]
- 37.Radha V, Vimaleswaran KS, Babu HN, Abate N, Chandalia M, Satija P, Grundy SM, Ghosh S, Majumder PP, Deepa R, Rao SM, Mohan V: Role of genetic polymorphism peroxisome proliferator-activated receptor-gamma2 Pro12Ala on ethnic susceptibility to diabetes in South-Asian and Caucasian subjects: evidence for heterogeneity. Diabetes Care 2006;29:1046–1051 [DOI] [PubMed] [Google Scholar]
- 38.Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, Kawamori R, Nakamura Y, Maeda S: Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes 2008;57:791–795 [DOI] [PubMed] [Google Scholar]
- 39.Wu Y, Li H, Loos RJ, Yu Z, Ye X, Chen L, Pan A, Hu FB, Lin X: Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes 2008;57:2834–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, Lam VK, Ma RC, So WY, Cho YS, Kim HL, Lee HK, Chan JC, Cho NH: Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes 2008;57:2226–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chimienti F, Devergnas S, Pattou F, Schuit F, Garcia-Cuenca R, Vandewalle B, Kerr-Conte J, Van Lommel L, Grunwald D, Favier A, Seve M: In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci 2006;119:4199–4206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.