Abstract
In breast cancer, SRC-1 expression positively correlates with HER2 expression and poor prognosis. In MMTV-polyoma middle T (PyMT) breast cancer mouse model, SRC-1 strongly promotes mammary tumor metastasis. However, the molecular targets and mechanisms that mediate the role of SRC-1 in metastasis are unknown. In this study, SRC-1 wild type (WT) and knockout (KO) cell lines were developed from the mammary tumors of WT/PyMT and KO/PyMT mice. WT cells exhibited strong migration and invasion capabilities, reduced E-cadherin and β-catenin epithelial markers, gained N-cadherin and vimentin mesenchymal markers and formed undifferentiated invasive structures in three-dimensional (3D) culture. In contrast, KO cells showed slow migration and invasion, retained E-cadherin, had less N-cadherin and vimentin, and developed partially differentiated 3D structures. Importantly, WT cells expressed Twist, a master regulator of metastasis, at significantly higher levels versus KO cells. SRC-1 knockdown in WT cells reduced Twist expression, while SRC-1 restoration in KO cells also rescued Twist expression. Furthermore, SRC-1 was found to coactivate Twist transcription through physical interaction with the transcription factor PEA3 at the proximal Twist promoter. Accordingly, Twist knockdown in WT cells increased E-cadherin and reduced cell invasion and metastasis, and Twist expression in KO cells decreased E-cadherin and increased cell invasion. SRC-1 knockdown in human breast cancer cells also decreased Twist, cell migration and invasion. Therefore, SRC-1 promotes breast cancer invasiveness and metastasis by coactivating PEA3-mediated Twist expression. Intervention of SRC-1 function may provide new strategies to inhibit breast cancer metastasis.
Keywords: nuclear receptor, SRC-1, coactivator, Twist, breast cancer
Introduction
Breast cancer mortality is largely attributed to metastasis. Despite its clinical importance, the molecular mechanisms responsible for metastasis remain poorly understood, partially due to lack of appropriate experimental models and difficulties of identifying metastasis-specific regulators and mediators (1). Cancer metastasis consists of several steps: epithelial mesenchymal transition (EMT), local invasion, intravasation into the circulation systems, transport in the circulation, extravasation, and survival in a secondary organ site, followed by proliferation into micrometastasis and formation of overt metastatic lesions (1, 2). Conceivably, each of these steps may be promoted by a specific group of factors. During EMT, for example, breast tumor cells express one or more transcription factors including Twist, Snail, Slug and/or SIP1, which mediate the transition through repression of epithelial markers such as E-cadherin and β-catenin and induction of mesenchymal markers such as N-cadherin, fibronectin and vimentin (3, 4). In addition to the transcription factors that control EMT, many other proteins are also associated with breast cancer metastasis. These proteins include certain oncoproteins such as ras, HER2, epidermal growth factor receptor and c-myc, motility factors such as hepatocyte growth factor, extracellular matrix (ECM) degradative enzymes such as uPA and metalloproteinases (MMPs) and posttranslational modification enzymes for protein glycosylation such as Mgat5 (5–7). Proteins that enhance metastasis also include S100A4, Mta-1, Muc1 and osteopontin (6). However, none of these proteins functions exclusively in cancer metastasis as functional inactivation of each also inhibits primary tumor growth.
SRC-1 is a transcriptional coactivator in the SRC family that also contains SRC-2 (TIF2 or GRIP1) and SRC-3 (AIB1 or ACTR) (8). These coactivators promote transcription by interacting with nuclear receptors such as estrogen and progesterone receptors, as well as other transcription factors such as AP-1, NF-κB, Ets-2 and PEA3 (9–15). Studies using mutant mouse models demonstrate SRC family members to have both unique and partially redundant functions in development, somatic growth, steroid response, metabolism, reproduction, vasoprotection and inflammation (8, 16–21). In human mammary epithelial cells, SRC-1 expression is very low (14). However, SRC-1 expression in HER2-positive breast cancers is increased and correlated with disease recurrence and resistance to endocrine therapy (14, 22). SRC-1 expression inversely correlates with estrogen receptor β (ERβ) expression, a marker for better prognosis in breast cancer (23). In addition, SRC-1 interacts with Ets-2 to enhance c-Myc expression in endocrine resistant breast cancer cells (11, 12) and with ERα to promote SDF-1 expression, facilitating cell proliferation and invasion (24).
Unlike other oncogenes, SRC-1 functions specifically to promote breast cancer metastasis (25). During mammary tumorigenesis in MMTV-PyMT mice, SRC-1 is upregulated. Inactivation of SRC-1 in these mice does not alter mammary tumor initiation and growth, but drastically suppresses mammary tumor cell intravasation and lung metastasis. Although SRC-1 enhances HER2 expression, Akt activity, colony-stimulating factor-1 expression and macrophage recruitment to the tumor site, the molecular mechanisms responsible for SRC-1 to promote breast cancer metastasis are largely unknown (25).
In this study, we have developed cell lines from mammary tumors of WT/PyMT and SRC-1−/−/PyMT mice and used these cell lines to investigate the molecular mechanisms by which SRC-1 promotes metastasis. We find SRC-1 promotes EMT, mammary cancer cell migration and invasion by enhancing PEA3-mediated transcriptional activation of Twist, a master regulator of breast cancer metastasis (3).
Materials and Methods
Generation of mouse mammary tumor cell lines
Individual mammary tumors were collected from WT/PyMT and SRC-1−/−/PyMT mice. Tumor cell lines were established from WT/PyMT and SRC-1−/−/PyMT mammary tumors as described (15). The epithelial character of these cells was confirmed by immunocytochemistry for cytokeratin 8 (K8) as described (15).
Cell motility and invasion assays
Individual cell movement was traced for 18 hours in 96-well culture plate coated with blue fluorescent beads. Track areas were quantitatively analyzed using NIH image software as described (15). Cell invasion was assayed by using BioCoat Matrigel Invasion Chambers (BD Biosciences) as described (15). 25,000 cells in serum-free medium containing 0.2% BSA were loaded to the upper chambers. The lower chambers were filled with fibroblast-conditioned medium containing 5% fetal bovine serum (FBS) as chemo-attractant. After cultured for 22 hours, the invading cells were fixed, stained and counted. Real-time cell invasion assay was carried out using an RT-CES system with 16-well E-Plates equipped with a Matrigel-coated membrane with 8 μm pores (ACEA Biosciences Inc.). 50,000 cells in serum-free medium were loaded to the top chamber and medium with 5% serum was added to the bottom chamber. Cell invasion was monitored every 30min till the experimental endpoint.
SRC-1 Knockdown and adenovirus-mediated expression
To knock down SRC-1, WT/PyMT mouse cells and MDA-MB-231 human cells (5 × 105) in 6-well plates were transfected with mouse and human SRC-1 siRNA Smart Pools as described (Dharmacon) (15). Cells transfected with scrambled siRNA served as controls. The knockdown efficiency was examined by qPCR and Western blot analyses as described (15). For SRC-1 expression and its control, SRC-1−/−/PyMT cells were infected with adenoviruses carrying either an SRC-1 or a control GFP expression cassette as described (15, 26). Exogenous expression of SRC-1 in SRC-1 knockout cells was examined by western blot as described (15).
Stable shRNA knockdown and retroviral expression of Twist
Five different shRNA lentiviruses targeting non-overlapping regions of the mouse Twist mRNA (SHVRS-NM-011658) as well as a non-targeting shRNA lentivirus (SHC002V) were purchased from Sigma. WT1 mammary tumor cells were infected with either the targeting lentiviral mixture or control non-targeting lentiviral particles. Infected cells were selected in puromycin-containing medium. Surviving clones were isolated and examined by real-time RT-PCR to determine Twist knockdown efficiency. To express Twist in KO1 cells, a DNA sequence for translational initiation and Flag tag were added to the 5' human Twist coding sequence and the resulting fusion DNA was cloned into the pQCXIH retroviral vector (BD Biosciences). The parent vector was used as a control. Retroviruses were produced in pT67 packaging cells. Stable KO1 cells with human Twist expression or control vector were generated by retroviral infection followed by hygromycin selection.
Three-dimensional (3D) culture
The 3D culture system for WT/PyMT and SRC-1−/−/PyMT cells was set up as described (15). 5,000 cells in 400μl of growth medium containing 2% Matrigel were seeded on 8-well chamber slides (BD Biosciences) pre-coated with 40ul Matrigel and cultured for 8–18 days. Cells were fixed in methanol and acetone (1:1). Immunofluorescence staining was performed with primary antibodies against E-cadherin, N-cadherin (BD Biosciences) and ZO-1 (Zymed Lab.), and fluorescence-conjugated secondary antibodies. DNA was stained using propidium iodide (PI). Stained slides were mounted in 50% glycerol and examined by confocal microscopy.
Western blot
Cells in 80% confluent monolayer culture were extracted with a lysis buffer containing 0.05 M Tris-HCl, pH 7.6, 0.15 M NaCl, 1% NP-40, 0.5% Deoxycholate, 2 mM EDTA and protease inhibitor cocktail. Cell lysates with 50 μg protein were subjected to Western blot analyses using primary antibodies against SRC-1 (Abcam), E-cadherin, N-cadherin (BD Biosciences), β-catenin, vimentin, PEA3 (Santa Cruz), PEA3, Flag-Twist and β-actin (Sigma). Horseradish peroxidase-conjugated secondary antibodies and a chemiluminiscence kit from Pierce Biotechnology were used to visualize the immunochemical signals.
Real-time quantitative RT-PCR (qPCR)
Total RNA was isolated from cells using TRIZOL reagents (Invitrogen). cDNA template was produced by reverse transcription from 1 μg of RNA using Reverse Transcriptase Core kit (Eurogentec). The relative concentrations of mouse SRC-1, Twist, Snail, SIP1, E-cadherin, N-cadherin and vimentin mRNAs and human SRC-1 and Twist mRNAs were measured by qPCR using matched Universal Taqman probes and gene-specific primers (Roche). The measurement of 18s mRNA was used as an endogenous control.
Cell transfection and luciferase assays
The pGL3-mTwist-Luciferase (Luc) promoter/reporter construct was provided by Dr. Howe (27). The pGL3-hTwist-Luc promoter/reporter plasmid was constructed with a 2300-bp DNA fragment amplified by PCR from the proximal enhancer/promoter region of the human Twist gene. The primers for PCR were 5'-GTTGGTACCGCTGTGGACTTGGTTTCTCC and 5'-GATAAGCTTCTGCAGACTTGGAGGCTCTT. WT/PyMT and SRC-1−/−/PyMT cells were grown to 80% confluence in 12-well plate and transiently transfected with 200ng of pGL3-mTwist-Luc and 50ng of pSV40-gal plasmids as described (15). pSV40-gal supported constitutive expression of β-galactosidase (β-gal), which was used to normalize transfection efficiency. Hela cells were co-transfected with pSV40-gal, pCR3.1-SRC-1, pGL3-hTwist-Luc and expression plasmids for PEA3, ERM, ER81, NF-κB, c-Jun, TCF, HIF-1α or E2F1. Total DNA for each well was added to the same amount using respective parent vectors when different combinations of plasmids were applied. Cells were cultured for 48 hours and then lysed with Reporter Assay Lysis Buffer (Promega). Luciferase activity was measured using the Luciferase Assay System (Promega). β-gal activity was assayed as described (28). Relative luciferase activity was normalized to β-gal activity.
Co-immunoprecipitation (Co-IP)
Cell lysates with 500μg protein prepared from MDA-MB-231 cells were pre-cleaned with protein G/A beads and subjected to Co-IP using 2μg of PEA3 antibody. Equal amount of IgG was used as negative control. Immuno-complexes were denatured by boiling in SDS-PAGE sample buffer and separated in 6% SDS-PAGE gels for Western blot using SRC-1 and PEA3 antibodies. All Co-IP assays were repeated for 3 times.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed with MDA-MB-231 cells, 2μg of antibodies against SRC-1 or PEA3 and the protein-G/A beads as described (15). PCR was performed with specific primer pairs to amplify proximal regions of the human Twist promoter that contain PEA3-binding sites (27).
Results
SRC-1 Enhances Mammary Tumor Cell Migration and Invasion
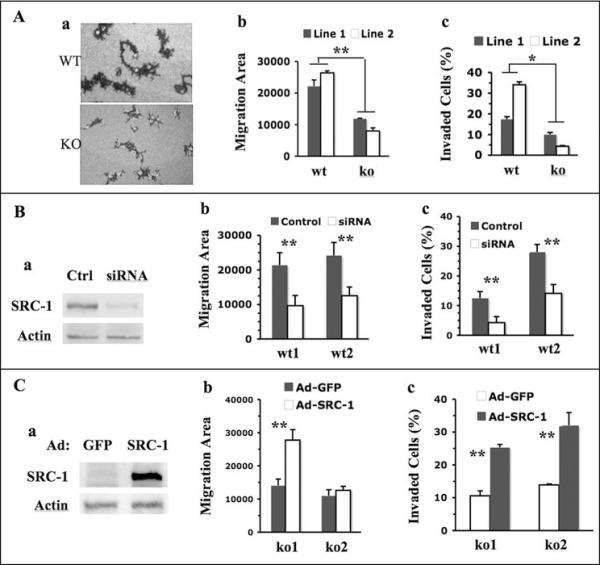
To investigate the role of SRC-1 in regulation of cell behaviors and molecular pathways relevant to breast cancer metastasis, we developed WT and SRC-1 KO mammary tumor cell lines. Two independent WT cell lines, WT1 and WT2, were derived from mammary tumors of WT/PyMT mice, and two independent KO cell lines, KO1 and KO2, were derived from mammary tumors of SRC-1−/−/PyMT mice. After culturing under selective conditions for epithelial cell growth, these established cell lines were positive to K8 immunostaining (Supplementary Fig. S1) and thus to be of mammary epithelial origin. Since metastatic potential correlates with the ability of cancer cells to migrate out of tumor mass and invade tissues, we subjected cell lines to migration and invasion assays. We traced the migration of individual cells on culture plates by pre-coated fluorescence beads. Within 17 hours, most WT1 and WT2 cells had migrated extensively and left much broader and longer trails than SRC-1-deficient KO1 and KO2 cells did (Fig. 1A, a). The average migration area swept by individual WT1 or WT2 cells was significantly larger compared with that swept by individual KO1 or KO2 cells (Fig. 1A, b). We also cultured WT and KO cells in chambers containing Matrigel barrier that simulates ECM and measured their invasion capabilities. We found 17% of WT1 and 38% of WT2 cells invaded through the Matrigel barrier, versus only 10% of KO1 and 5% of KO2 cells (Fig. 1A, c). The different invasion capabilities between WT1 and WT2 cells and between KO1 and KO2 cells might reflect variable genetic and epi-genetic changes in different tumors. Overall, these results suggest that SRC-1 deficiency reduces migration and invasion capabilities of the mammary tumor cells.
Fig. 1. SRC-1 promotes mammary tumor cell migration and invasion.
A. SRC-1 deficiency decreases tumor cell migration and invasion. The trails of WT and SRC-1 KO cell migration were traced using fluorescence beads (a). The migration areas of at least 40 cells were individually measured by pixels. The average migration areas per cell for WT1, WT2, KO1 and KO2 cell lines are presented (b). Cell migration data for these cell lines are presented as percentages of invaded cells to total cells in the invasion assay chambers (c). B. SRC-1 knockdown in WT cells decreases cell migration and invasion. Western blot analysis for SRC-1 and β-actin (loading control) in WT1 cells transfected with either scrambled double strand RNA (Ctrl) or SRC-1 siRNA (a). Cell migration and invasion data for WT1 and WT2 cells treated with control or SRC-1 siRNA are presented (b and c). C. SRC-1 restoration in SRC-1 KO cells rescues their migration and invasion. Western blot analysis confirmed SRC-1 expression in KO1 cells infected with SRC-1 adenoviruses. KO1 cells infected with GFP adenoviruses served as a control (a). The cell migration and invasion data for these cells are presented in panels b and c. *, p<0.05 and **, p<0.01 by unpaired t test.
To confirm the role of SRC-1 in promoting cell migration and invasion in WT1 and WT2 cells, we knocked down SRC-1 by transfecting these cells with SRC-1 siRNA. Scrambled siRNA served as a control (Fig. 1B, a). Upon SRC-1 knockdown, both WT1 and WT2 cell lines showed more than 50% reduction in their migration velocity and invasion capability (Fig. 1B, b and c). We also expressed SRC-1 in KO1 and KO2 cells by adenoviral infection to evaluate the role of SRC-1 in cell migration and invasion (Fig. 1C, a). SRC-1 restoration in KO1 cells resulted in an increase in both cell migration and invasion versus GFP control. SRC-1 restoration in KO2 cells caused a similar increase in cell invasion, but did not increase cell migration (Fig. 1C, b and c). Taken together, these results demonstrate that SRC-1 plays an important role in promotion of mammary tumor cell migration and invasion.
SRC-1 Reduces Mammary Tumor Cell Differentiation and Promotes EMT
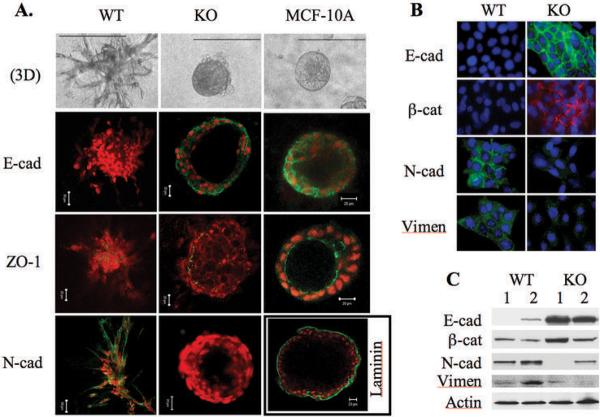
To evaluate the degree of epithelial differentiation of mammary tumor cells with and without SRC-1, we performed 3D culture and examined the expression and distribution patterns of E-cadherin, ZO-1, laminin and N-cadherin. In the 3D culture system, the MCF-10A human mammary epithelial cells formed highly differentiated hollow spheres consisting of a single layer of polarized epithelial cells (Fig. 2A). E-cadherin was highly expressed and localized primarily at the lateral cell membrane for cell-cell adhesion. The tight junction protein ZO-1 was located at the apilateral membrane of polarized epithelial cells, and the basement membrane protein laminin was located around the sphere (Fig. 2A). N-cadherin, a marker of mesenchymal cells, was undetectable in MCF-10A cells (data not shown). In contrast, WT1 and WT2 cells were unable to develop hallow spheres and they only formed an undifferentiated dendritic architecture. In these invasive cellular architectures, E-cadherin, ZO-1 and laminin were undetectable but N-cadherin was detected, suggesting that WT1 and WT2 cells lost epithelial differentiation and polarity, and gained certain mesenchymal features (Fig. 2A and data not shown). Interestingly, cultured KO1 and KO2 cells formed 3D structures with a partially differentiated phenotype. These cells formed mammary epithelial spheres, although the shells of some spheres consisted of multiple layers of cells. E-cadherin was detected between most cells and ZO-1 was detected in about 50% of cells in the outer cell layer of the spheres (Fig. 2A and data not shown). Laminin and N-cadherin were undetectable. Overall, these results demonstrate that the SRC-1-deficient KO1 and KO2 mammary tumor cells were more differentiated than SRC-1-positive WT1 and WT2 cells, but less differentiated than normal MCF-10A cells.
Fig. 2.
SRC-1 deficiency helps to retain epithelial differentiation of mammary tumor cells. A. 3D structures formed from WT1, KO1 and MCF-10A cells. Immunofluorescence staining was performed for E-cadherin (E-cad), ZO-1, N-cadherin (N-cad) and Laminin. N-cad was not detected in the 3D structures of MCF-10A cells (data not shown). Laminin was not detected in the 3D structure of WT and KO mammary tumor cells (data not shown). B. Immunofluorescence staining for E-cad, β-catenin (β-cat), N-cad and vimentin (Vimen) in WT and KO mammary tumor cells cultured as monolayer. C. Western blot analyses of E-cad, β-cat, N-cad and Vimen in WT1, WT2, KO1 and KO2 mammary tumor cells. The β-actin served as a loading control.
In WT1 and WT2 cells cultured as monolayer, immunofluorescent labeling did not detect epithelial markers E-cadherin and β-catenin but instead detected mesenchymal markers N-cadherin and vimentin. In contrast, parallel assays in KO1 and KO2 cells detected E-cadherin and β-catenin but not N-cadherin and vimentin (Fig. 2B and data not shown). Western blot analyses confirmed that E-cadherin and β-catenin levels were significantly higher in KO1 and KO2 cells versus in WT1 and WT2 cells, while N-cadherin and vimentin levels were higher in WT1 and WT2 cells versus in KO1 and KO2 cells (Fig. 2C). These results suggest SRC-1 plays an important role in promoting EMT of mammary tumor cells.
SRC-1 Partially Inhibits E-cadherin Expression
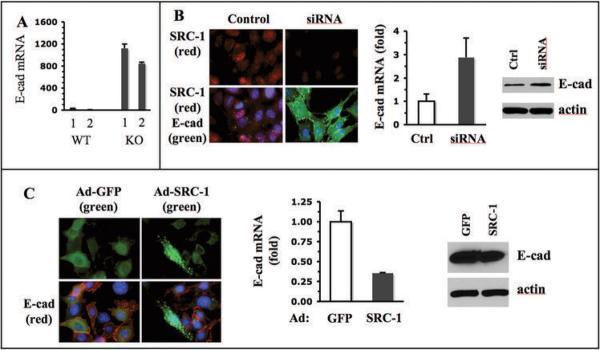
Since loss of E-cadherin is an essential event in EMT during breast cancer progression (4), we focused our attention on addressing how SRC-1 regulates E-cadherin expression. E-cadherin mRNA was detected at high levels in KO1 and KO2 cells but at very low levels in WT1 and WT2 cells (Fig. 3A), which was consistent with the results from immunochemical analyses in Fig. 2. Southern blot analysis revealed that the genomic DNA of E-cadherin gene was present in both WT and KO cell lines, indicating that the loss of E-cadherin mRNA expression in WT cells was not due to a loss of the E-cadherin gene (Supplementary Fig. S2). In order to confirm the impact of SRC-1 on E-cadherin expression, we knocked down SRC-1 in WT1 and WT2 cells using siRNA, restored SRC-1 in KO1 and KO2 cells by adenovirus-mediated expression, and examined E-cadherin expression. E-cadherin protein and mRNA expression showed no change in WT cells treated with a short scrambled double strand RNA as a control. However, when WT cells were treated with SRC-1 siRNA, E-cadherin protein and mRNA expression were induced in these cells (Fig. 3B). On the other hand, when KO cells were infected with SRC-1-expressing adenoviruses, E-cadherin protein became undetectable by immunostaining only in some cells with high SRC-1 expression; E-cadherin mRNA levels assayed by qPCR were significantly reduced; and E-cadherin protein levels assayed by immunoblotting were partially reduced (Fig. 3C). Although it was unclear why short-term SRC-1 expression had a more prominent effect on E-cadherin mRNA than protein, these results demonstrate that SRC-1 levels inversely correlate with E-cadherin levels in these mammary tumor cells.
Fig. 3.
SRC-1 inhibits E-cadherin expression. A. qPCR analysis of E-cadherin mRNA in WT1, WT2, KO1 and KO2 mammary tumor cells. The relative E-cadherin mRNA levels were normalized to the 18 S RNA. B. SRC-1 knockdown and Immunofluorescence staining of SRC-1 and E-cadherin in WT1 cells transfected with control or SRC-1 siRNA. The E-cadherin mRNA and protein levels in these cells were measured by qPCR and Western blot, respectively. C. SRC-1 expression and Immunostaining of SRC-1 and E-cadherin in KO1 mammary tumor cells with adenovirus-mediated GFP or SRC-1 expression. GFP signal and E-cadherin immunoreactive signal were imaged by fluorescence microscopy. The E-cadherin mRNA and protein levels in these cells were measured by qPCR and Western blot, respectively.
SRC-1 Potentiates PEA3-mediated Twist Expression
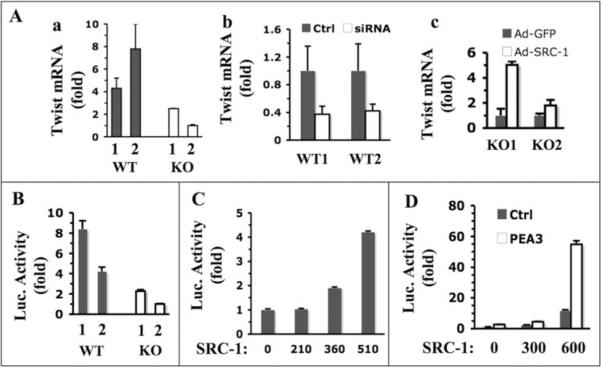
Several transcription factors including Snail, SIP1 and Twist can suppress E-cadherin promoter and promote EMT and cancer metastasis (3, 29). Since SRC-1, as a transcriptional coactivator, might enhance EMT and promote metastasis through influencing the expression levels of these master regulators, we measured the mRNA levels of Snail, SIP1 and Twist in WT and KO mammary tumor cells. Snail mRNA levels in KO1 and KO2 cells were similar, while it was lower in WT1 cells but slightly higher in WT2 cells. There was no statistical difference in average expression levels of Snail mRNA in WT and KO cells (Supplementary Fig. S3A). SIP1 mRNA levels were also similar in all four cell lines (Supplementary Fig. S3B). Twist mRNA levels, however, were markedly higher in WT1 and WT2 cells than in KO1 and KO2 cells (Fig. 4A, a). To confirm the role of SRC-1 in Twist expression, we also measured Twist mRNA levels in WT cells treated with SRC-1 siRNA and KO cells infected with SRC-1 expression adenovirus. Our measurements revealed that SRC-1 knockdown in WT cells decreased Twist expression more than 50%, while SRC-1 restoration increased Twist expression more than 5 fold in KO1 cells and 2 fold in KO2 cells (Fig. 4A, b and c). These results indicate that SRC-1 plays an important role in upregulating Twist expression in mammary tumor cells.
Fig. 4.
SRC-1 enhances Twist expression. A. qPCR analyses of relative Twist mRNA levels in the following groups of cells: WT1, WT2, KO1 and KO2 cells (a); WT1 and WT2 cells transfected with control or SRC-1 siRNA (b); and KO1 and KO2 cells with adenovirus-mediated GFP or SRC-1 expression (c). B. Transfection assays of Twist-Luc promoter/reporter construct in WT1, WT2, KO1 and KO2 cells. C. SRC-1 enhances Twist promoter activity in a dosage-dependent manner. Hela cells were co-transfected with Twist-Luc and different amounts (ng) of SRC-1 expression plasmids as indicated. D. SRC-1 and PEA3 synergistically enhance Twist promoter activity. Hela cells were transfected with Twist-Luc plasmids, variable amounts of SRC-1 expression plasmids as indicated, and with PEA3 expression plasmids or its parent vector. In all of the above transfection assays, the reporter luciferase activities were normalized to β-galactosidase activity from a co-transfected constitutively active expression vector.
To understand how SRC-1 regulates Twist promoter activity, we performed transient transfection assays using a Twist promoter-driven luciferase reporter (Twist-Luc). The reporter showed much higher activities in WT1 and WT2 cells compared with KO1 and KO2 cells (Fig. 4B). Furthermore, co-transfection of SRC-1 and Twist-Luc into Hela cells activated Twist promoter in a SRC-1 dose-dependent manner (Fig. 4C). These results indicate that SRC-1 is required for Twist promoter activation.
The 2.8-kb Twist promoter region used in the Twist-Luc reporter contains binding elements for NF-κB, c-Jun, TCF, HIF1α, E2F1 and PEA3, and some of these transcription factors have been shown to regulate Twist promoter (27, 30). To search for transcription factor candidates working with SRC-1, we co-transfected Hela cells with Twist-Luc reporter, one of the above transcription factors and SRC-1 expression plasmids. Our assays showed that expression of NF-κB, c-Jun, TCF or PEA3 could slightly enhance Twist promoter activity, while expression of E2F1 or HIF-1α had no effect or an inhibitory effect on the Twist promoter (Fig. 4D and Supplementary Fig. S4). Co-expression of SRC-1 with NF-κB, c-Jun, TCF, HIF-1α or E2F1 did not further enhance Twist promoter activity (Supplementary Fig. S4), suggesting that SRC-1 is not required for these transcription factors to activate Twist promoter. On the contrary, co-expression of SRC-1 with PEA3 synergistically activated Twist promoter activity, suggesting that SRC-1 may serve as a coactivator for PEA3 to potentiate Twist promoter activity (Fig. 4D). In addition, co-expression of SRC-1 with other Ets family members ERM and ER81 showed no increase in Twist promoter activity. Thus, the role of SRC-1 in promoting PEA3-mediated Twist promoter activation is quite specific.
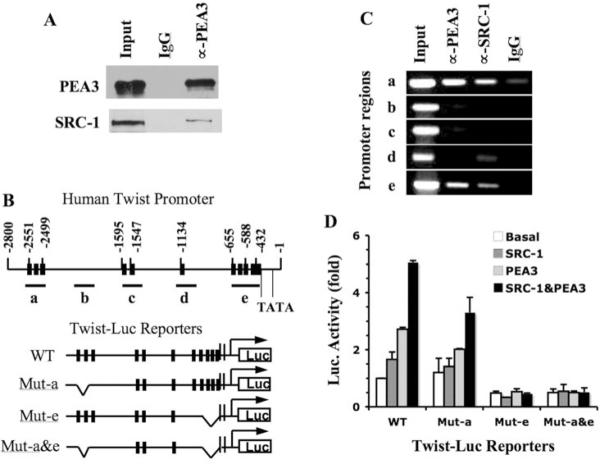
SRC-1 Is Associated with PEA3 and Twist Promoter for Transcriptional Activation
We also investigated the molecular mechanisms responsible for SRC-1 and PEA3 to regulate Twist promoter. From the total cell extract prepared from the metastatic MDA-MB-231 breast cancer cells, SRC-1 was specifically co-immunoprecipitated with PEA3 (Fig. 5A), which was consistent with a previous study showing physical interactions of SRC family members with Ets family transcription factors (13). Next, we employed ChIP assays to address whether PEA3 and SRC-1 were recruited to Twist promoter. The 2.8-kb Twist promoter region contains 11 potential PEA3 binding sites. Five primer pairs at locations depicted in Fig. 5B were designed to amplify DNA fragments termed a – e that cover Twist promoter. Regions a, c, d, and e contained one or multiple PEA3-binding sites, and region b without PEA3-binding site served as a control (Fig. 5B). ChIP assays, performed in MDA-MB-231 cells, revealed both PEA3 and SRC-1 specifically associated with regions a and e, but not with regions b, c and d (Fig. 5C). These results suggest that both PEA3 and SRC-1 are recruited to regions a and e of Twist promoter for transcriptional activation.
Fig. 5.
SRC-1 directly regulates PEA3-mediated Twist expression. A. Co-IP analysis. MDA-MB-231 cell lysate was subjected to Co-IP with PEA3 antibody. None immunized IgG was used as a negative control. Input represents 5% of lysate used for Co-IP. Western blot analyses were performed with PEA3 and SRC-1 antibodies. B. PEA3 binding sites in the Twist promoter/enhancer region and Twist-Luc promoter/reporter constructs. The twist promoter region (bp −1 – −2800) contains 11 PEA3-binding elements (black bars). Fragments a–e were chosen for PCR amplification in ChIP assays. The positions of two TATA boxes were indicated. Four Twist-Luc promoter/reporter constructs in the pGL3 vector are depicted, which are WT, Mut-a (with deletions of the three PEA3-binding sites in region a), Mut-e (with deletions of the five PEA3 binding sites in region e) and Mut-a&e (with deletions of the PEA3 binding sites in both regions a and e). C. ChIP assays using MDA-MB-231 cells. Input represents 3% of material used for ChIP analysis. ChIP analyses were performed using PEA3 and SRC-1 antibodies. None immunized IgG was used as a negative control. PCR reactions were performed with specific primer pairs to amplify fragments a–e depicted in Panel B. D. Transfection assays for wild type and mutant Twist promoter activities. Hela cells were transfected with wild type or mutant Twist-Luc promoter/reporter constructs depicted in panel B. For each construct, cells also were co-transfected with parent vector (basal), SRC-1 and/or PEA3 expression plasmids as indicated. Data from three repeat assays are presented as mean ± SD.
To determine whether region a, region e, or both regions were responsible for PEA3 and SRC-1-mediated Twist promoter activation, we generated three luciferase reporter constructs with deletions of region a (termed Mut-a-Luc), region e (termed Mut-e-Luc) or both regions a and e (termed Mut-a&e-Luc) (Fig. 5B), and performed transfection assays with SRC-1 expression vector, PEA3 expression vector or both SRC-1 and PEA3 expression vectors. Deletion of region a slightly reduced the Twist promoter activity only when PEA3 and SRC-1 were co-expressed. However, deletion of region e or of both regions a and e abolished PEA3/SRC-1-mediated Twist promoter activity (Fig. 5D). These results indicate that SRC-1 promotes PEA3-medicated Twist transcription mainly via the region e of Twist promoter. Region e (bp −655 to −432) is adjacent to the first TATA box and contains 5 PEA3-binding sites.
Twist Mediates the Role of SRC-1 in Inhibition of E-cadherin Expression and Promotion of Breast Cancer Cell Migration and Invasion
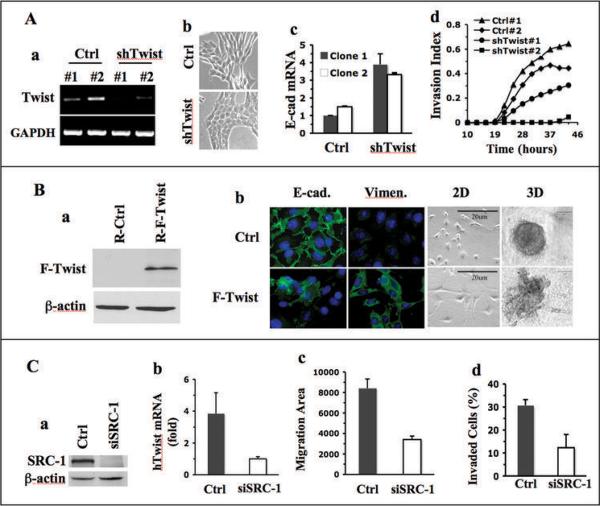
The preceding results suggest that SRC-1 may be through upregulation of Twist in breast cancer cells to suppress E-cadherin expression and promote EMT, cell motility, invasion and metastasis. If Twist mediates SRC-1 function during breast cancer progression, Twist knockdown in WT mammary tumor cells with SRC-1 would block SRC-1-dependent inhibition of E-cadherin expression and tumor cell invasion. Conversely, expression of Twist in KO mammary tumor cells lacking SRC-1 would decrease E-cadherin expression and epithelial differentiation, but increase EMT marker expression and cell invasiveness. Indeed, when Twist was stably knocked down in WT1 cells by shRNA (Fig. 6A, a), the morphology of some cells changed from spindle shape with loose cell-cell contact to cubic shape with tight cell-cell contact (Fig. 6A, b). E-cadherin expression in clones with Twist knockdown was markedly increased compared with clones with non-targeting control shRNA as assayed by qPCR (Fig. 6A, c). Accordingly, real-time cell invasion assays revealed that Twist knockdown in WT1 cells significantly reduced their invasion through a Matrigel layer. Invasive behavior in one of the two examined Twist knockdown clones (shTwist#2) was almost completely blocked (Fig. 6A, d). Furthermore, NU/NU recipient mice with xenograft mammary tumors formed from WT/PyMT cells with stable Twist knockdown possessed a significantly lower frequency of tumor cells in their blood and metastatic tumors in their lungs compared with mice with tumors from control WT/PyMT cells (Supplementary Fig. S5). On the other hand, when flag-tagged Twist was expressed in KO1 cells by adenoviral infection (Fig. 6B, a), E-cadherin expression was reduced in about 50% of cells, while vimentin was induced in a similar promotion of cells (Fig. 6B, b). Expression of Twist in KO1 cells also changed their epithelial morphology into fibroblast-like morphology and disrupted their capability to form partially differentiated 3D structure in the 3D culture system. These KO1 cells with exogenous Twist expression formed only undifferentiated and invasive structures (Fig. 6B, b), resembling structures formed by WT cells (Fig. 2A). These results demonstrate that Twist is a down-stream mediator for SRC-1 in mammary tumor cells to regulate E-cadherin expression and promote EMT and malignancy.
Fig. 6.
Twist mediates the role of SRC-1 in breast cancer cells. A. Effects of Twist knockdown in WT1 cells. RT-PCR analysis revealed that Twist mRNA in WT1 cells was efficiently knocked down by lentivirus-mediated expression of two different shRNAs (#1 and #2) against mouse Twist mRNA. Lentivirus expressing non-targeting shRNAs served as a control (Ctrl) (a). The images of WT1 cells expressing control or Twist shRNA were taken under a phase-contrast microscope (b). Relative E-cadherin mRNA levels in WT1 cells expressing control or Twist shRNAs were measured by qPCR (c). Cell invasion indices of these cells were monitored in real time as described in Materials and Methods (d). B. Effects of Twist restoration in KO1 cells. Flag-tagged human Twist (F-Twist) was expressed in KO1 cells by retrovirus-mediated expression. Empty retrovirus served as a control. The F-Twist protein was detected by Flag antibody (a). Immunofluorescent staining of E-cadherin and vimentin in control and F-Twist-expressing KO1 cells was carried out using E-cadherin and vimentin antibodies. Cell images of 2D cultures and morphologies of 3D structures formed from control and F-Twist-expressing cells were taken under a phase-contrast microscope. C. Effects of SRC-1 knockdown in MDA-MB-231 cells. Cells were transfected with control or SRC-1 siRNAs. SRC-1 was analyzed by Western blot (a). Relative human Twist (hTwist) mRNA in these cells was measured by qPCR (b). Cell migration was traced with fluorescence beads. Average area (pixels) swept by each cell were obtained from measurements of at least 40 cells in each group (c). Cell invasion assay chambers with a Matrigel layer were used to determine the percentages of invaded cells to total cells (d). The assays were done in triplicates.
Finally, we validated the findings obtained in mouse mammary tumor cells by recapitulating SRC-1 knockdown experiments in MDA-MB-231 metastatic human breast cancer cells. MDA-MB-231 cells express SRC-1 protein. SRC-1 knockdown in these cells by siRNA decreased Twist expression to 25% and cell migration and invasion to about 45% (Fig. 6C). These results indicate that SRC-1 also plays an important role to potentiate Twist expression, cell motility and invasiveness in human breast cancer cells.
Discussion
In the SRC family, the role of AIB1 in breast cancer has been intensively investigated. AIB1 is overexpressed in a subgroup of breast cancers (31). In mice, AIB1 overexpression causes mammary tumorigenesis, while its deficiency suppresses both mammary tumor formation and lung metastasis (15, 32, 33). These studies not only proved a crucial role of AIB1 in breast cancer but also raised the questions regarding possible roles for other SRC family members in breast cancer. To investigate the in vivo role of SRC-1 in breast cancer, we have recently generated WT/PyMT and SRC-1−/−/PyMT mice and compared mammary tumor initiation, growth and metastasis in these mice (25). Surprisingly, we found that inactivation of SRC-1 in mice does not affect primary mammary tumor initiation and growth but does drastically inhibit mammary tumor metastasis to the lung (25). Therefore, AIB1 promotes both primary mammary tumor formation and metastasis, while SRC-1 specifically promotes mammary tumor metastasis, even though their differences in molecular mechanisms are unknown.
The present study is to address how SRC-1 promotes breast cancer metastasis at cellular and molecular levels by developing and using SRC-1 WT and KO tumor cell lines. We found SRC-1 plays important roles promoting of mammary tumor cell migration, invasion, epithelial depolarization and EMT. These results were validated by analysis of SRC-1 knockdown in WT tumor cells, SRC-1 restoration in KO tumor cells, cellular morphology of WT and KO cells in 3D cultures, and epithelial/mesenchymal marker profile of each cell line. These findings, obtained from cultured cells, strongly correlate with and substantiate our former in vivo observations showing that SRC-1 is required for enhancing mammary tumor metastasis to the lung in mice (25).
EMT is an important event during cancer progression. In the EMT process, tumor cells lose polarity and epithelial differentiation and acquire certain mesenchymal features, which enable tumor cells to invade and survive in the stromal tissues, namely the first step of metastasis (34). Tumor cell EMT is associated with increased cell migration, invasion and metastasis. The well-established hallmark of EMT is the loss of E-cadherin, which disrupts the stable cell-cell adhesion between epithelial cells (34). Our data demonstrate that SRC-1 expression inversely correlates with E-cadherin expression, suggesting that SRC-1 may promote EMT through direct or indirect inhibition of E-cadherin expression.
Several transcription factors including Twist, Snail and SIP1 are known to suppress E-cadherin expression and induce tumor cell EMT (3, 4). Among these transcription factors examined in SRC-1 WT and KO tumor cell lines, Twist is significantly upregulated in WT cells compared with KO cells. Furthermore, SRC-1 knockdown in WT tumor cells decreases Twist expression while SRC-1 re-expression in KO tumor cells increases Twist expression. The Twist promoter activity is higher in WT tumor cells than in KO tumor cells, and it can be potentiated by SRC-1 in another type of cells in a dosage-dependent manner. Taken together, these results demonstrate, for the first time, that SRC-1 regulates Twist expression.
Our data indicate that SRC-1 directly enhances Twist expression by serving as a coactivator to potentiate PEA3-mediated Twist mRNA transcription. First, SRC-1 synergistically potentiates PEA3-mediated Twist promoter activation in transfection assays; second, SRC-1 directly associates with PEA3 in human breast cancer cells; third, both PEA3 and SRC-1 are recruited to Twist promoter regions a and e; and fourth, deletion of region e that contains multiple PEA3-binding sites diminishes PEA3 and SRC-1 activities in activation of Twist promoter. Conceivably, SRC-1 may also enhance Twist expression through other transcription factors and future studies may lead to identification of these yet unknown transcription factors.
Finally, this study addressed if Twist, as a SRC-1 target, is responsible for the effect of SRC-1 on the mammary tumor cells. Our data demonstrate that Twist knockdown in WT tumor cells increases E-cadherin expression and decreases cell invasion capability, while expression of Twist in SRC-1 KO tumor cells reduces E-cadherin expression and increases cell invasiveness. Since Twist expression or knockdown recapitulates the phenotypes of SRC-1 expression or knockout, SRC-1-regulated Twist expression should be responsible, at least in part, for SRC-1-controlled mammary tumor cell morphology and invasiveness. In addition, SRC-1 depletion in MDA-MB-231 human breast cancer cells also decreases Twist expression and cell migration and invasion, which validates the regulatory relationship and functional significance between SRC-1 and Twist in breast cancer.
Our data also showed that short-term expression of either SRC-1 or Twist at medium levels was not sufficient to induce complete EMT of mammary tumor cells in culture. This could be due to the lack of tumor progression environment, insufficient time for completing EMT, and/or limited contributions from other EMT-promoting factors that can be induced only under in vivo conditions. Furthermore, in addition to SRC-1, several other coactivators also have been implicated in breast cancer metastasis. For example, AIB1 can coactivate PEA3-mediated MMP2 and MMP9 expression to promote mammary tumor cell invasion and metastasis (15). The metastasis tumor antigen 1 (MTA1) can serve as a coactivator for HIF1α to enhance breast cancer metastasis (35). The functional and molecular regulatory relationships among these coactivators in metastasis will continue being an interesting research topic.
The SRC-1-linked regulatory relationship between PEA3 and Twist has important implications in breast cancer. Although controversial reports exist, recent studies suggest that PEA3 expression in breast cancer associates with HER2 expression, metastasis and poor prognosis (14, 15, 36, 37). Twist is an E-box-binding transcription factor that serves as one of the master regulators for EMT and breast cancer metastasis (3, 27). Since SRC-1 is overexpressed in many HER2-positive breast cancers (14) and it mediates PEA-3-dependent Twist expression in these cancers, inhibition of SRC-1 function may provide a useful intervening point to control the deleterious roles of both PEA3 and Twist in HER2-positive breast cancer metastasis.
Supplementary Material
Acknowledgements
Grant support: NIH grants CA112403, CA119689 and DK58242; American Cancer Society Scholar Award RSG-05-082-01-TBE; and a Susan Komen Postdoctoral Fellowship to L.Q.
We thank Y. Yuan, Q. Li, J. Hong, J. Fu, B. York and X. Chen for experimental assistance and materials, and J. Tien for assisting manuscript preparation. We thank Dr. L.R. Howe for providing the pGL3-mTwist-Luc plasmid.
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2:408–416. doi: 10.1186/bcr87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169:1122–1133. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 5.Moustafa AS, Nicolson GL. Breast cancer metastasis-associated genes: prognostic significance and therapeutic implications. Oncol Res. 1997;9:505–525. [PubMed] [Google Scholar]
- 6.Schwirzke M, Schiemann S, Gnirke AU, Weidle UH. New genes potentially involved in breast cancer metastasis. Anticancer Res. 1999;19:1801–1814. [PubMed] [Google Scholar]
- 7.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nature Medicine. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 9.Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273:16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 10.Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 11.Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S, Hill AD, Young LS. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene. 2008;27:3021–3031. doi: 10.1038/sj.onc.1210964. [DOI] [PubMed] [Google Scholar]
- 12.Myers E, Hill AD, Kelly G, McDermott EW, O'Higgins NJ, Buggy Y, Young LS. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2005;11:2111–2122. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 13.Goel A, Janknecht R. Concerted activation of ETS protein ER81 by p160 coactivators, the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J Biol Chem. 2004;279:14909–14916. doi: 10.1074/jbc.M400036200. [DOI] [PubMed] [Google Scholar]
- 14.Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O'Higgins NJ, Hill AD, Young LS. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004;57:1069–1074. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O'Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 18.Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol. 2002;22:5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao L, Chen X, Wang S, Parlow AF, Xu J. Steroid receptor coactivator 3 maintains circulating insulin-like growth factor I (IGF-I) by controlling IGF-binding protein 3 expression. Mol Cell Biol. 2008;28:2460–2469. doi: 10.1128/MCB.01163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Y, Xu J. Loss-of-function deletion of the steroid receptor coactivator-1 gene in mice reduces estrogen effect on the vascular injury response. Arterioscler Thromb Vasc Biol. 2007;27:1521–1527. doi: 10.1161/ATVBAHA.107.144477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming FJ, Hill AD, McDermott EW, O'Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004;89:375–383. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- 23.Myers E, Fleming FJ, Crotty TB, Kelly G, McDermott EW, O'Higgins NJ, Hill AD, Young LS. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer. 2004;91:1687–1693. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26:1706–1715. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Yuan Y, Liao L, Kuang S-Q, Tien J, O'Malley BW, Xu J. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci U S A. 2008;106:151–156. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–3823. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- 28.Norton PA, Coffin JM. Bacterial beta-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol Cell Biol. 1985;5:281–290. doi: 10.1128/mcb.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 31.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 32.Torres-Arzayus MI, De Mora JF, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 33.Kuang SQ, Liao L, Zhang H, Lee AV, O'Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–1885. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg RA. The Biology of Cancer. Garland Science, Taylor & Francis Group; New York: 2007. pp. 587–654. [Google Scholar]
- 35.Singh RR, Kumar R. MTA family of transcriptional metaregulators in mammary gland morphogenesis and breast cancer. J Mammary Gland Biol Neoplasia. 2007;12:115–125. doi: 10.1007/s10911-007-9043-7. [DOI] [PubMed] [Google Scholar]
- 36.Myers E, Hill AD, Kelly G, McDermott EW, O'Higgins NJ, Young LS. A positive role for PEA3 in HER2-mediated breast tumour progression. Br J Cancer. 2006;95:1404–1409. doi: 10.1038/sj.bjc.6603427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol. 2001;11:1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.