Abstract
New photomap of Anopheles (Nyssorhynchus) darlingi Root, 1926, is described for a population from Guajará-Mirim, State of Rondonia, Brazil. The number of sections in the previous A. darlingi reference map was maintained and new subsections were added to the five chromosome arms. Breakage points of paracentric inversions had been previously incorporated into the photomap of this species. An additional inversion is reported, called 3Lc, totaling 14 inversions in the A. darlingi chromosome arms. The proposed photomap is potentially useful for further evolutionary studies in addition to physical and in silico chromosome mapping using A. darlingi genomic and transcriptome sequences. Furthermore, in our attempt to compare sections of the 2R chromosome arm of A. darlingi with Anopheles funestus, Anopheles stephensi, and Anopheles gambiae, we found great differences in the arrangement of the polytene chromosome bands, which are consistent with the known phylogenetic divergence of these species.
Introduction
Anopheles darlingi Root, 1926, has historically been considered the primary vector of malaria and the most anthropophillic and endophagous Anopheles species in the Americas.1 In the Brazilian Amazon Basin, this mosquito is the most important vector of human malaria.2 Anopheles darlingi has been found in clear water such as lakes, ponds, and streams, and in smaller water courses such as artificial deposits and puddles.2,3 The relevance of A. darlingi as vector of malaria, its isoenzymes, hehavior, genetic structure, biology, metaphase karyotype, mapping, and chromosome polymorphism were demonstrated in several studies.4–14
In South America, 17 Anopheles species are known as important vectors of malaria. Of those, more than two-thirds belong to the subgenus Nyssorhynchus, which contains 29 named species throughout the Neotropics.15 They are of primary importance for transmission of human malaria and include the A. darlingi. Several areas in Amazonia have shown significant differences in A. darlingi incidence.2 At the outer edge of the Ariquemes locality in Rondonia State, in Brazil, the registered index of adult Anopheles species has been 30.6 mosquitoes per person per hour, and A. darlingi is abundant. This mosquito has been observed almost exclusively at the modified sites, especially around and into the houses, a fact that confirms its high degree of association with human dwellings.16
Karyotypes of the studied Anopheles species are relatively conserved. Morphological data obtained from metaphase karyotypes of six chromosomes in brain cells of A. darlingi describe a pair of metacentric (chromosome 2) and a pair of submetacentric (chromosome 3) autosomes, and sex chromosomes (female XX; male XY). The X chromosome is acrocentric in females and the Y chromosome is punctiform in males.11 This pattern is observed in the majority of the species in the Nyssorhynchus subgenus, including Anopheles strodei, Anopheles albimanus, Anopheles aquasalis, Anopheles noroestensis, and Anopheles argyritarsis.17 Morphological variation was detected in the X chromosome, which was metacentric in A. aquasalis and A. noroestensis, A. strodei, A. albimanus, acrocentric in A. argyritarsis,17,18 A. darlingi,11 and A. albitarsis sensu latu,19 whereas it is metacentric in A. aquasalis and A. noroestensis,17 and submetacentric in A. nuneztovari.11 This pattern is observed also in Anopheles cruzii and Anopheles bellator of subgenus Kerteszia,20,21 A. subpictus,22 and A. gambiae species complex, serie Pyretophorus, subgenus Cellia.23 The morphological variation of the sexual chromosome pair suggest selective pressure in these anopheline species, which is apparently occurring only at a genetic level. In the larval salivary gland cells, the well-developed and well-banded polytene chromosome X, 2 and 3 represent this diploid set.
Currently, a new photomap description of A. darlingi polytene chromosomes in substitution of the drawn reference map5 would be highly helpful.The first study on polymorphism of inversions of A. darlingi polytene chromosomes was reported.5 The authors described nine independent inversions and a complex array of references on a drawn map of polytene chromosomes of populations from the regions of Manaus and Itacoatiara, in the State of Amazonas, Northern Brazil. These populations were highly polymorphic compared with a studied A. darlingi population from the Southeast of the country, in the region of Araraquara, in the State of São Paulo. Subsequently, the previously mentioned inversions and described three new inversions, one on chromosome 2 (2Rd), one on chromosome 3 (3Rc), and one on chromosome X (Xb), all belonging to the A. darlingi population from the vicinities of the BR-174 highway, Manaus-Boa Vista, in the State of Amazonas were analyzed.6,7 In three populations of this same region, Tadei and Santos6 conducted a comprehensive study on frequency variation in most of the chromosome inversions. These populations were characterized as highly polymorphic, with up to 60% of individuals presenting three to five inversions, depending on what time of the year the specimens were sampled. High frequencies of inversions in the heterozygous state were also found in this same population.
The major advantage of the A. darlingi photomap is to better define chromosome regions and subsections, allowing structural analysis by means of traditional methods and computer resources. It will be a useful tool in identifying the large-scale physical mapping using probes of expressed sequence tags (EST) from the A. darlingi transcriptome (Rafael MS and others, unpublished data). The polytene chromosome photomap will also offer a better understanding of the anchoring of the complete genome of A. darlingi for the Drosophila 11 Genomes.24
The objective of this work was to construct a polytene chromosome photomap for A. darlingi based on a population with low chromosomal polymorphism from the Northern region of the Amazon, aiming at contributing to the understanding of the A. darlingi genome structure and organization, and of the evolutionary diversification and chromosomal variability of this important vector of Neotropical malaria.
Materials and Methods
Sampling and identification of mosquitoes.
Anopheles darlingi females were sampled in August 1999, in the region of Guajará-Mirim (10°47′0²S, 65°21′0²W), in the State of Rondonia, Brazil. They were captured when resting on stable walls or biting humans in inner houses, between 6:30 pm and 9:00 pm. The Guajará-Mirim population was chosen because of its low chromosomal polymorphism, which was inferred from rearrangement patterns. Wild-captured individuals were transported in moist chambers to the Laboratory of Vectors of Malaria at the National Institute for Amazonian Research (INPA) in Manaus. Morphological identification of specimens was carried out according to the taxonomic keys.25,26 The sampled females were confined individually in plastic cups for egg laying. Offsprings were fed with finely ground liver and fish powder, and reared to the fourth instar larvae.
Chromosome preparation.
Salivary glands of fourth stage A. darlingi larvae were dissected and used to prepare chromosome slides.12,13,27 Preparations of well-spread polytene chromosomes on 11 slides were photographed using a light microscope (Carl Zeiss Axioplan, Ulm, Germany), for further analysis and construction of a photomap.
Construction of the photomap and description of inversions.
The photomap of the A. darlingi analyzed population was constructed using carefully selected photos of different chromosomes considered representative of the banding pattern. The best images were captured with the aid of a scanner and processed using Adobe Photoshop (Adobe, 2002 7.0, v 701; Adobe Systems Inc., San Jose, CA) and Corel Draw (Corel-DRAW 10 2000, 10.410, Corel Corp., Ottawa, Canada). All chromosomes were divided into sections represented by numbers and into subsections represented by capital letters, starting with A in the distal region. Chromosomes were divided into 45 sections, but the previous name of each paracentric inversion was changed following the system.6 Thus, the diverse arrangements of heterozygous inversions in the chromosome X and autosomes 2 (chromosomal arms 2R and 2L) and 3 (chromosomal arms 3R and 3L) were identified by lower case letters, in alphabetical order according to their previous descriptions. Wherever possible, the correspondence was maintained between our descriptions and the earlier studies.5–7 The former authors studied a highly polymorphic A. darlingi population from the Central region of the Amazon and described nine independent inversions and a complex chromosome arrangement. Tadei and others7 studied populations from the vicinities of the BR-174 highway, between Manaus and Boa Vista, in the State of Amazonas and described two previously unknown independent inversions, increasing to 12 the number of chromosome inversions.
A comparative analysis of chromosome arm tips, except of the X chromosome, was done for the major malaria vectors Anopheles gambiae Patton, Anopheles stephensi Liston, Anopheles funestus Giles, and Anopheles darlingi. The Anopheles gambiae and A. funestus species are the major malaria vectors in Africa, A. stephensi is the major vector in Indo-Pakistan and Middle East, and A. darlingi is the primary vector of Netropical malaria. The species Anopheles gambiae, A. stephensi, and A. funestus belong to the subgenus Cellia and A. darlingi belongs to the subgenus Nyssorhynchus.17,28,29 Homologies of bands in the chromosome arm tips of A. darlingi was compared with the photomaps previously described for A. gambiae,30 A. stephensi,31 and A. funestus.3
Results
A photomap of polytene chromosomes from salivary glands of A. darlingi was developed for a low polymorphic population from the region of Guajará-Mirim, in the Northern Brazilian Amazon (Figure 1). The photomap is oriented so that the centromere of each chromosome is positioned on the right and the chromosome distal region is on the left. The photomap was divided into 45 sections, following the original draw reference map.5 Sections were numbered from the starting point of the distal part of the X chromosome (section 1) ending at the base of the distal part chromosome 3L (section 45). Correspondence of the sequences followed, to the possible extent, the division proposed in the draw reference map.5 Alternativelly, photos were used in this work to visualize the nature of the banding pattern of chromosomes and their sizes. Thus, new subsections were included in several of the sections, so that each subsection includes approximately 3 to 4 bands of similar size.
Figure 1.
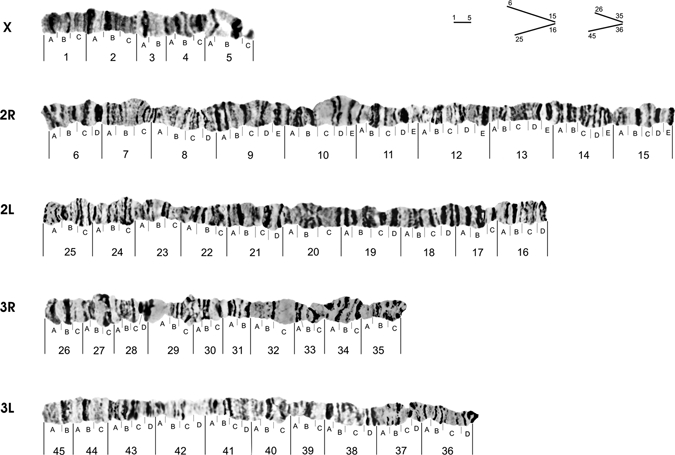
Photomap of polytene chromosomes of Anopheles darlingi from the Brazilian Amazon.
New divisions were added, as sections 2 and 4 of the X chromosome, subdivided into three subsections (Table 1). In the chromosome arm 2R, sections 6, 7, 8, 9, 10, 12, 13, and 15 were subdivided into 4, 3, 4, 5, 5, 5, 5, and 5 subsections, respectively. In the chromosome arm 2L, sections 16, 17, 19, 20, 21, 22, 23, and 24 were subdivided into 4, 3, 4, 4, 3, 4, 3, 3, and 3 subsections, respectively. Chromosome arm 3R shows the sections 26, 27, 28, 29, 30, 33, and 34, which were subdivided into 3, 3, 4, 3, 3, 3, and 3 subsections, respectively. In the chromosome arm 3L, sections 36, 37, 38, 39, 40, 41, 42, 43, and 44 were subdivided into 4, 4, 4, 3, 3, 4, 4, 4, and 3 subsections, respectively. Other sections in which not all chromosome arms were modified, the number of subdivisions was maintained according to the reference map.5 The final result was a photomap with well-distributed sections, subsections, and total size of all chromosomes together.
Table 1.
New divisions for polytene chromosome photomap of Anopheles darlingi of Guajara-Mirim locality
| Chromosome X | Chromosome 2 | Chromosome 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sections | Subsections | Right arm (2R) | Left arm (2L) | Right arm (3R) | Left arm (3L) | ||||
| Sections | Subsections | Sections | Subsections | Sections | Subsections | Sections | Subsections | ||
| 2 | A, B, C | 6 | A, B, C, D | 16 | A, B, C, D | 26 | A, B, C | 36 | A, B, C, D |
| 4 | A, B, C | 7 | A, B, C | 17 | A, B, C | 27 | A, B, C | 37 | A, B, C, D |
| 8 | A, B, C, D | 18 | A, B, C, D | 28 | A, B, C, D | 38 | A, B, C, D | ||
| 9 | A, B, C, D, E | 19 | A, B, C, D | 29 | A, B, C | 39 | A, B, C, | ||
| 10 | A, B, C, D, E | 20 | A, B, C | 30 | A, B, C | 40 | A, B, C, | ||
| 12 | A, B, C, D, E | 21 | A, B, C, D | 33 | A, B, C | 41 | A, B, C, D | ||
| 13 | A, B, C, D, E | 22 | A, B, C | 34 | A, B, C | 42 | A, B, C, D | ||
| 15 | A, B, C, D, E | 23 | A, B, C | 43 | A, B, C, D | ||||
| 24 | A, B, C | ||||||||
The relative size of each chromosome arm in the photomap was carefully estimated from individual measurements with a digital caliper. Several nuclei were measured and only photomicrographs displaying all chromosomes together were chosen, to minimize experimental errors. It can be observed in the photomap of A. darlingi (Figure 1), the X chromosome is 10% of the total size of all chromosomes together. Chromosome 2 corresponds to 49% (2R = 28% and 2L = 21%) and chromosome 3 corresponds to 41% (3R = 18% and 3L = 23%). These results agree with Kreutzer and others5 who reported the approximate proportions of 10% for each of the X chromosomes, 52% for chromosome 2 (2L = 30% and 2R = 22%), and 40% for chromosome 3 (3R = 20% and 3L = 20%).
In general, all chromosomes are quite stable regarding polymorphism and presence of puffs and constrictions. However, the X chromosome was an exception in the analyzed population. Its variation in levels of distension and compression, especially in sections 1A and between 5A and 5C, is shown in Figure 2. Figure 3 shows the X chromosome pattern and the breakpoints of the heterozygous inversion Xa.5,7
Figure 2.

Possible configuration of homozygous X chromosomes of Anopheles darlingi.
Figure 3.
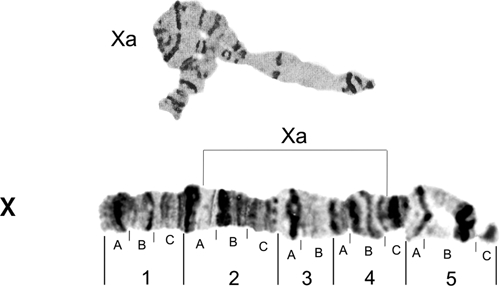
Photomap of Anopheles darlingi X chromosome. Lines over the chromosome arm indicate probable breakpoints of the heterozygous inversion Xa that involves regions 2A and 4C.4
The photomap of chromosome arm 2R is shown in Figure 4, and the chromosome arms 2L is shown in Figure 5. The probable breakpoints of the inversions 2Ra (between sections 7B and 8C), 2Rb (between 9D and 10A), and 2Rc (between 11A and 12E) are indicated in Figure 4. They were first described by Kreutzer and others5 and later by Tadei and others,7 who also described a new reversal in the 2Rd (from 12E to 14B). Figure 5 shows the photomap of the chromosome arm 2L, probable breakpoints of the heterozygous inversion 2La (between sections 20A and 23C),7 and a complex of inversions 2La + b, with breakpoints between sections 23C and 21B.5 The latter authors also described an inversion homozygote 2Lb present only in a population from the region of Araraquara, in the State of São Paulo.
Figure 4.

Photomap and arrangements of Anopheles darlingi 2R chromosome arm. Lines over the chromosome arm indicate probable breakpoints of the heterozygous inversion 2Ra (between sections 7B and 8C), found in our study and first described,4 in addition to inversions 2Rb (between sections 9D and 10A) and 2Rc (between 11A and 12E),4 and inversion 2Rd (between section 12E and 14B) plus 2Rc.7 Grid lines represent chromosome arm continuation.
Figure 5.

Photomap and arrangements of the Anopheles darlingi 2L chromosome arm. Lines over the chromosome arm indicate probable breakpoints of the heterozygous inversion 2La (between sections 23C and 20A),7 and the complex inversion 2La + b (inversion 2Lb with a breakpoint between sections 23C and 21B).4 The inversion 2Lb was also described by the latter authors as a homozygous inversion described only in the population from Araraquara. Grid lines represent chromosome arm continuation.
Figures 6 and 7 show, respectively, the photomap of chromosome arms 3R and 3L. For the inversion in chromosome arm 3R, Figure 6 shows the probable breakpoints of the heterozygous inversions 3Ra (between sections 28D and 32C) and 3Rc (between sections 34A and 35A),7 and the inversions 3Rb (between sections 35A and 35C) and 3Rd (between the sections 31A, 28B).5 The probable breakpoints of the heterozygous inversions 3La (between sections 43C and 42C) and 3Lb (between 40A and 37A)5 and renamed7 are shown in Figure 7. A new inversion 3Lc in A. darlingi, with breakpoints between sections 42B and 37D, is presented here, for the first time, occurring in the Guajará-Mirim population. This inversion is the largest of all the investigations already described for A. darlingi. It was found only in heterozygous state and represents approximately 57% of the length of chromosome arm 3L. Thus, the A. darlingi chromosomes comprise at least 14 inversions, with one inversion on the X chromosome, 4 inversions in the chromosome arm 2R, 2 in 2L, 4 in 3R, and 3 in 3L.
Figure 6.
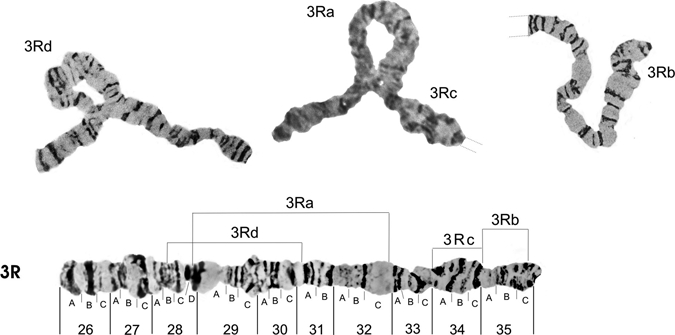
Photomap and arrangements of the Anopheles darlingi 3R chromosome arm. Lines over the chromosome arm indicate probable breakpoints of the heterozygous inversion 3Ra (between sections 28D and 32C) and inversion 3Rc (between sections 34A and 35A),7 as well as inversions 3Rb (between sections 35A and 35C) and 3Rd (between sections 28B and 31A).4 Grid lines represent chromosome arm continuation.
Figure 7.
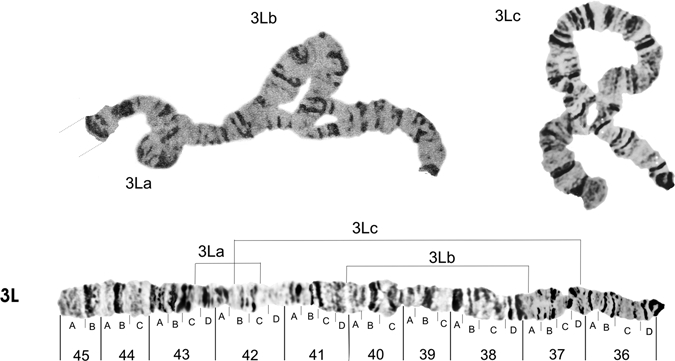
Photomap and arrangements of the Anopheles darlingi 3L chromosome arm. Lines over the chromosome arm indicate probable breakpoints of the heterozygous inversion 3La (between sections 43C and 42C) and inversion 3Lb (between sections 40A and 37A),4 besides the inversion 3Lc (between sections 42B and 37D) here described as a new inversion for the A. darlingi species. Grid lines represent the chromosome arm continuation.
Discussion
Autosome arm tips of A. darlingi were compared with the species A. gambiae, A. stephensi, and A. funestus. Figure 8 shows the probable chromosome homology of the 2R arm tip between A. darlingi and A. gambiae, and of A. stephensi compared with A. gambiae and A. funestus. Figure 9 shows part of the homology in the 2R chromosome arm tip of A. gambiae and 2R of A. funestus.30 The homology in the 2R arm tip of both species was unclear. Therefore, we suggest another possibility of homology between A. funestus and A. gambiae, plus A. stephensi, as presented in Figure 8, where A. stephensi and A. gambiae are very similar in relation to the 2R arm tip. Figures 9–15 present the probable homology between tips of the chromosome arms 2L, 3R, and 3L, except X chromosome of A. darlingi that was quite divergent.
Figure 8.
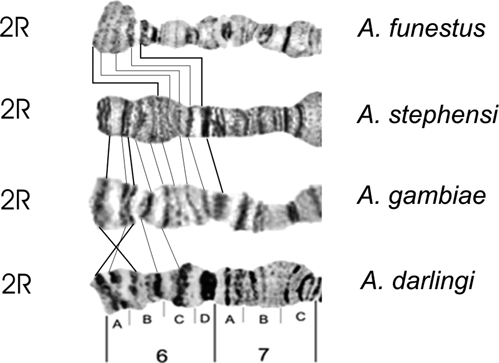
Homology in the 2R chromosome arm tip of four main Anopheles malaria vectors in the world, according to the photomaps of A. darlingi (present work), A. gambiae and A. funestus,30 and A. stephensi.31
Figure 9.
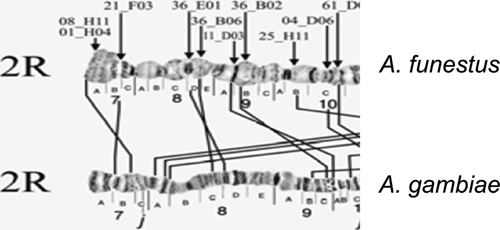
Homology in the 2R chromosome arm tip of A. gambiae and A. funestus.30
Figure 15.
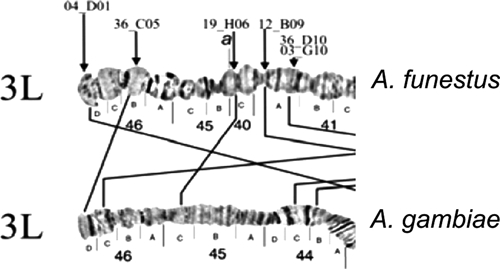
Homology between the chromosome arm tips 3L of A. gambiae and 3L of A. funestus.30
Figure 10.
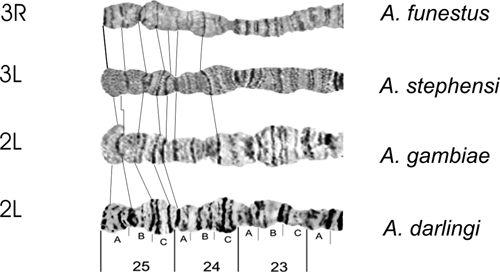
Homology in the 3R chromosome arm tip of four main Anopheles malaria vectors in the world, according to the photomaps of A. darlingi (present work), A. gambiae and A. funestus,30 and A. stephensi.31
Figure 11.
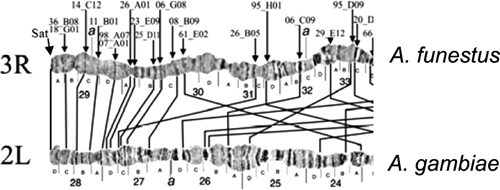
Homology between the chromosome arm tips 3R of A. funestus and 2L of A. gambiae.30
Figure 12.
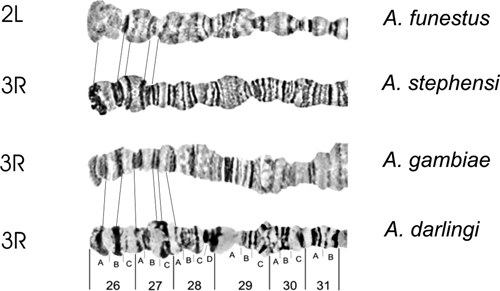
Homology between the 2L and 3R chromosome arm tips in four main Anopheles malaria vectors in the world, according to the photomaps of A. darlingi (present work), A. gambiae and A. funestus,30 and A. stephensi.31
Figure 13.
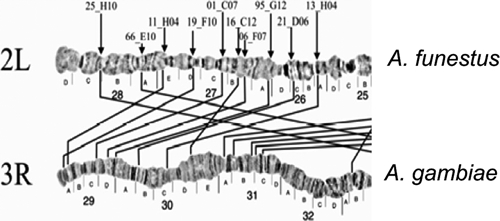
Homology between the chromosome arm tips 3R in A. gambiae and 2L in A. funestus.30
Figure 14.
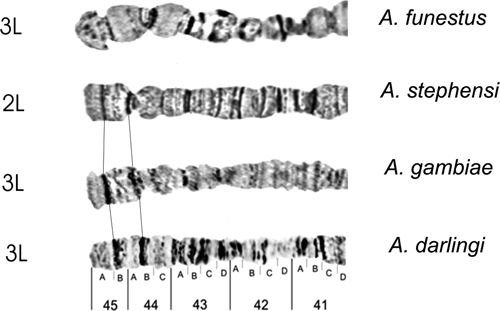
Homology in the 3L the chromosome arm tip of four main Anopheles malaria vectors in the world, according to the photomaps of A. darlingi (present work), A. gambiae and A. funestus,30 and A. stephensi.31
A standard polytene chromosome photomap is one of the essential requirements for cytogenetic investigation of Dipteran insects, including the anopheline mosquitoes. The data described herein show that A. darlingi is no exception. The construction of a photomap of A. darlingi polytene chromosomes was motivated by the need to map, in silico and in situ, the ESTs generated from the genome sequencing of this species (Rafael and others, unpublished data). This was triggered by the first mapping of ribosomal genes12 and genes of single copy (Hsp70) in polytene chromosomes of A. darlingi populations of Manaus and Macapá regions, in the Brazilian Amazon.13
In previous studies, photomaps were described for other species as Anopheles funestus,29 Anopheles stephensi,31–33 Anopheles gambiae,34,35 Anopheles subpictus,36 Anopheles albimanus,37 Anopheles triannulatus,38 Anopheles cruzii, and Anopheles belattor.20,21 The photomaps have been useful for identifying species with high chromosome polymorphism and monomorphic species35 constructing maps of low resolution. Furthermore, chromosome arm translocations have been described for the important vectors of malaria A. gambiae, A. funestus, and A. stephensi, of the subgenus Cellia.28,29 For example, chromosomes X and 2R are homologous in the three species. The 2L arm of A. gambiae corresponds to the 3R of A. funestus and the 3L of A. stephensi. The 2L arm of A. stephensi corresponds to the 3L arms of A. funestus and A. gambiae. The 2L arm of A. funestus corresponds to the 3R arms of A. gambiae and A. stephensi.29,31 In contrast, among the important Neotropical vectors of malaria, A. darlingi does not show homologies in the X chromosomes, 3R or 3L similarly to A. aquasalis, A. albimanus, or A. nuneztovari.5 However, there is homology in the 2R arms of A. darlingi, and in A. argyritarsis, A. aquasalis, A. albimanus, and A. nuneztovari.18 The morphologically alike species are expected to and do share more banding pattern similarity than species with low morphological similarity.39
In this study, we showed probable chromosome homology between the 2R arm tips of A. darlingi and A. gambiae, and the probable homology between 2R arms of A. funestus and A. stephensi. In these four species, the 2R chromosome arm is the longest and contrasts with the X chromosome, the smallest. Despite chromosome length, the heat shock protein gene Hsp70 was mapped on the 2R chromosome arm of A. gambiae40 and A. darlingi.13 The Hsp70 gene mapped on 12A and 14A bands of the 2R chromosome arm of A. darlingi populations in Manaus (AM) and Macapá (AP) in the Amazonian region. The Hsp70-12A and Hsp70-14A genes were useful in analysis of their fixed inversion and chromosome evolution. Anopheles darlingi is very divergent from A. gambiae, possibly because of one little inversion only in the 2R chromosome tip of A. darlingi. Furthermore, it was infeasible to compare all 2R chromosome arm sections of A. darlingi with A. funestus, A. stephensi, and A. gambiae, because of the great differences in the order of the polytene chromosome bands, which seems consistent with the phylogenetic divergence of these species. Despite the probable tip homology among the chromosome arms 2L, 3R, and 3L, except the X chromosome of A. darlingi, they were very divergent comparing A. funestus, A. stephensi, and A. gambiae. In addition, phylogenetic relationships between A. gambiae, A. funestus, and A. stephensi have been estimated by polymorphism on ribosomal DNA (rDNA) (18S and 28S) sequence data.41 Accordingly, A. funestus and A. gambiae are more distant species from each other than from A. stephensi. In this latter mosquito, genetic studies indicated chromosomal positions for the breakpoints of the 19 paracentric inversions.28,42,43 In addition, polymorphic inversions are not distributed evenly across the A. stephensi genome. More than two-thirds of them are distributed among two arms, such as five inversions are on 2R and nine on 3L. Nonrandom distribution of polymorphic inversions has been described for other species. For instance, of eight polymorphic inversions described in A. gambiae s.s., seven occur on chromosome 2R35 and 11 of 17 polymorphic inversions involve the 2R of A. funestus.44
Similar to the previous species, the A. darlingi complement of polytene chromosomes consists of five chromosome arms (X, 2L, 2R, 3L, and 3R). It includes one inversion on the X chromosome, four inversions in the chromosome arm 2R, two in 2L, four in 3R, and three in 3L, totaling 14 inversions. Moreover, chromosome markers and morphological traits of A. darlingi populations revealed significant genetic differentiation among populations from Northern and Southern Brazil,5,6,9 and along with its geographic distribution from Southern Mexico to Northern Argentina, respectively,26,45 but A. darlingi consists of a single taxon.46 Microssatellite data suggest little genetic structure for A. darlingi across the Eastern,10 Central, and Western Amazonian regions.47 The latter authors described seven microssatelite loci of A. darlingi populations from nine localities in Central and Western Amazonian Brazil. Genetic differentiation was low and not significant among populations separated by distances less than 152 km, with exceptions for Castanho-Puraquequara (CAS-PUR), Ramal do Brasileirinho (CAS-RBR) localities, both in the state of Amazonas, and the Nm values suggested extensive gene flow among A. darlingi populations.
The importance of chromosomal inversions in ecological adaptations of anopheline mosquitoes has been well established,48 suggesting that the differences in chromosomal rearrangments are indicators of adaptation to different ecological habitats. The data on chromosomal polymorphism of A. darlingi that are related to behavioral aspects may be associated to genetic structure of this species, possibily affecting their capacity to adapt to different environments. Ecological observations of several anopheline vectors of malaria in the Brazilian Amazon describes low density and infectivity indexes, which is caused by control methods used against adult mosquitoes. However, only a few A. darlingi individuals infected with Plasmodium vivax and Plasmodium falciparum, according to enzyme-linked immunosorbent assay (ELISA) tests49 are needed to maintain the malaria circulating in a population.2 Studied parameters consisted of their breeding sites, samples distribution, incidence, feeding preferences, hour of maximum activity of adult samples seasonality, resting places, and the presence of Plasmodium species. They support the hypothesis that seasonality, deforestation, and associated ecological alterations may contribute to the behavior,2,50 chromosome differentiation, and are conducive to A. darlingi presence, thereby they increase malaria risk.
Another important aspect of A. darlingi chromosomal polymorphism refers to the identification of populations based on the degree of heterozygosity. Comparisons of chromosomal variability among A. darlingi populations from Northern and Southern Brazil have revealed a higher frequency of heterozygous inversions, and significant genetic differentiation in the Northern populations.5–7,14 Salivary polytene chromosome analysis of A. darlingi have shown high frequencies of inversions in the heterozygous state and in two random regions, one near the centromere of the X chromosome and two on chromosome 2 in A. darlingi population along the Highway BR-174, Manaus-Boa Vista.7 In subsequent studies,7,14 populations of the central axis of the Amazon showed a number of heterozygous inversions per individual that was almost the double in the marginal populations. These inversions represent 50% reduction in the values of mean inversion per individual in the heterozygous state. In addition, this work describes a new inversion, named 3Lc, which is located in chromosome arms of the A. darlingi population from Guajará-Mirim in the State of Rondonia. These high polymorphism inversions for A. darlingi populations could be a consequence of a high degree of variability that can be seen in this species, giving it sufficient adaptive plasticity to exploit the different habitat. Furthermore, its extensive geographic distribution, and in accordance with similar morphological characteristics support independent speciation processes for this mosquito.
These aspects confirm the important heterozygosity of A. darlingi populations found in Amazon region. This region is considered the hub of inhabitation in the Brazilian Amazon, as are the vicinities of the Highway BR-174, km 137, in the State of Amazonas, Highway BR-319 from Manaus to Porto Velho and Ariquemes, in the State of Rondonia. In these areas, heterozygosity of A. darlingi populations averaged between 3.20 ± 0.22 and 4.45 ± 0.34 were found.51 In the Marabá and Tucurui marginal populations, in the State of Pará, and in Cruzeiro do Sul, in the State of Acre, the averages ranged from 2.07 ± 0.19 to 2.19 ± 0.15. The heterozygosity averages were low in specimens from Cachoeira Porteira and Trombetas River, in the State of Pará, and in Maruanum, in the State of Amapá. The data, however, are based on the reference maps. Errors of interpretation may occur caused by the non-existence of a photomap default, or detailed pictures of heterozygous or homozygous inversions. Such limitations justified the investment on the construction of the A. darlingi photomap, which represents a unique opportunity to surpass limitations for more precise studies in this species. The photomap presented here is a major tool to standardize the genetic studies on chromosomal and ecology polymorphisms, to map genes and to open new research fields for comparative genomics and evolutionary studies in chromosomes of A. darlingi.
Footnotes
Financial support: We thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES (PROCAD – Process no. 023/2006), Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq, Ministério da Ciência e Tecnologia – MCT, Instituto Nacional de Pesquisas da Amazônia – INPA (Process no. 014/2008), the Fundação de Amparo à Pesquisa do Estado do Amazonas – FAPEAM (PIPT and Infraestructure projects), PIATAM/Petrobrás, Ana Tereza Vasconcelos from the Laboratório Nacional de Computação Científica – LNCC, Coordinator of the Complete Genome Project A. darlingi and the Fundação de Amparo à Pesquisa do Estado de Pernambuco – FACEPE (APQ-0757-2.02/06 process), for financial support.
Authors' addresses: Míriam S. Rafael and Wanderli P. Tadei, Coordenação de Pesquis as em Ciências da Saúde, Laboratório de Vetores de Malária e Dengue, Instituto Nacional de Pesquisas da Amazônia, Manaus, AM, Brazil, E-mails: msrafael@inpa.gov.br and tadei@inpa.gov.br. Cláudia Rohde, Centro Acadêmico de Vitória, Núcleo de Biologia, Universidade Federal de Pernambuco, Vitoria de Santo Antão, Pernambuco, Brazil, E-mail: claudiarohde@yahoo.com. Letícia C. Bridi, Programa de Pós-Graduação em Genética, Conservação e Biologia Evolutiva, Instituto Nacional de Pesquisas da Amazônia, Manaus, AM, Brazil, E-mail: lcbridi@gmail.com. Vera Lúcia da Silva Valente Gaiesky, Departamento de Genética, Universidade Federal do Rio Grande do Sul, Porto Alegre, Rio Grande do Sul, Brazil, E-mail: 00004887@ufrgs.br.
References
- 1.Fleming G. Biología y Ecología de los Vectores de la Malaria. Washington, DC: OPS; 1986. [Google Scholar]
- 2.Tadei WP, Dutary-Thatcher B, Santos JM, Scarpassa VM, Rodrigues IB, Rafael MS. Ecologic observations on Anopheles vectors of malaria in the Brazilian Amazon. Am J Trop Med Hyg. 1998;59:325–335. doi: 10.4269/ajtmh.1998.59.325. [DOI] [PubMed] [Google Scholar]
- 3.Deane LM. Malaria studies and control in Brazil. Am J Trop Med Hyg. 1988;38:223–230. doi: 10.4269/ajtmh.1988.38.223. [DOI] [PubMed] [Google Scholar]
- 4.Santos JM, Contel EP, Kerr WE. Biology of Amazonian mosquitoes. III. Esterase isozymes in Anopheles darlingi. Acta Amazon. 1985;15:167–177. [Google Scholar]
- 5.Kreutzer RD, Kitzmiller JB, Ferreira E. Inversion polymorphism in the salivary gland chromosomes of Anopheles darlingi Root. Mosq News. 1972;32:555–565. [Google Scholar]
- 6.Tadei WP, Santos JM. Biologia de anofelinos Amazônicos. VII. Estudo da variação de freqüências das inversões cromossômicas de Anopheles darlingi root (Diptera, Culicidae) Acta Amazon. 1982;12:759–785. [Google Scholar]
- 7.Tadei WP, Santos JM, Rabbani MG. Biologia de anofelinos Amazônicos. V. Polimorfismo cromossômico de Anopheles darlingi root (Diptera, Culicidae) Acta Amazon. 1982;12:353–369. [Google Scholar]
- 8.Rosa-Freitas MG, Broomfield G, Priestmann A, Milligan P, Momen H, Molyneux DH. Studies on cuticular components, isoenzymes and behaviour of 3 populations of Anopheles darlingi from Brazil. J Am Mosq Control Assoc. 1992;8:357–366. [PubMed] [Google Scholar]
- 9.Santos JM, Lobo JA, Tadei WP, Contel EP. Intrapopulational genetic differentiation in Anopheles (N.) darlingi root, 1926 (Diptera: Culicidae) in the Amazon region. Genet Mol Biol. 1999;22:325–331. [Google Scholar]
- 10.Conn JE, Vineis JH, Bollback JP, Onyabe DY, Wilkerson RC, Póvoa MM. Population structure of the malaria vector Anopheles darlingi in a malaria-endemic region of eastern Amazonian Brazil. Am J Trop Med Hyg. 2006;74:798–806. [PubMed] [Google Scholar]
- 11.Rafael MS, Tadei WP. Metaphase karyotypes of Anopheles (Nyssorhynchus) darlingi root and A. (N.) nuneztovari Gabaldón (Diptera, Culicidae) Genet Mol Biol. 1998;21:351–354. [Google Scholar]
- 12.Rafael MS, Tadei WP, Recco-Pimentel SM. Location of ribosomal genes in the chromosomes of Anopheles darlingi and Anopheles nuneztovari (Diptera, Culicidae) from the Brazilian Amazon. Mem Inst Oswaldo Cruz. 2003;98:629–635. doi: 10.1590/s0074-02762003000500008. [DOI] [PubMed] [Google Scholar]
- 13.Rafael MS, Tadei WP, Hunter FF. The physical gene Hsp70 map on polytene chromosomes of Anopheles darlingi from the Brazilian Amazon. Genetica. 2004;121:89–94. doi: 10.1023/b:gene.0000019959.45267.d7. [DOI] [PubMed] [Google Scholar]
- 14.Tadei WP. Biology of Amazonian mosquitoes IX. On chromosome polymorphism of Anopheles (Nyssorhynchus) darlingi and a new arrangement in the X-chromosome. Cienc Cult. 1985;37:1329–1331. [Google Scholar]
- 15.Rúbio-Palis Y, Zimmermann RH. Ecoregional classification of malaria vectors in the neotropics. J Med Vet Entomol. 1997;6:325–334. doi: 10.1093/jmedent/34.5.499. [DOI] [PubMed] [Google Scholar]
- 16.Lourenço-de-Oliveira R, Luz SL. Simian malaria at two sites in the Brazilian Amazon. II. Vertical distribution and frequency of anopheline species inside and outside the forest. Mem Inst Oswaldo Cruz. 1996;91:687–694. doi: 10.1590/s0074-02761996000600005. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber G, Guedes AS. Estudo comparativo do cromosoma X em algumas espécies de Anopheles do sub-gen. Nyssorhynchus (Dipt. Culic.) Cienc Cult. 1959;11:128–129. [Google Scholar]
- 18.Kreutzer RD, Kitzmiller JB, Rabbani MG. The salivary gland chromosomes of Anopheles argyritarsis compared with those of certain other species in the subgenus Nyssorhynchus. Mosq News. 1975;35:354–365. [Google Scholar]
- 19.Rafael MS, Santos Junior IP, Tadei WP, Carvalho KA, Sallum MA, Forattini OP. Cytogenetic study of Anopheles albitarsis (Diptera: Culicidae) by C-banding and in situ hybridization. Hereditas. 2006;143:1–6. doi: 10.1111/j.2006.0018-0661.01926.x. [DOI] [PubMed] [Google Scholar]
- 20.Ramírez CC, Dessen EM. Cytogenetic analysis of a natural population of Anopheles cruzii. Rev Bras Genet. 1994;17:41–46. [Google Scholar]
- 21.Ramírez CC, Dessen EM. The polytene chromosomes of the mosquito Anopheles bellator compared with those of Anopheles cruzii. Rev Bras Genet. 1996;19:555–558. [Google Scholar]
- 22.Seetharam PL, Chowdaiah BN. Chromosome studies on oriental anophelines. III. The salivary gland chromosomes of Anopheles subpictus. Proc Indian Natl Sci Acad. 1974;40B:399–403. [Google Scholar]
- 23.Coluzzi M. Osservazioni comparitive sul cromosoma X nelle specie A e B del complesso Anopheles gambiae. Rend Clas Sci Fis Mat Nat Acc Naz Lacei. 1966;40:671–678. [Google Scholar]
- 24.Schaeffer SW, Bhutkar A, McAllister BF, Matsuda M, Matzkin ML, Rohde C, Valente VL, Aguadé M, Anderson WW, Edwards K, Garcia AC, Goodman J, Hartigan J, Kataoka E, Lapoint RT, Lozovsky ER, Machado CA, Noor MA, Papaceit M, Reed LK, Richards S, Rieger TT, Russo SM, Sato H, Segarra C, Smith DR, Smith TF, Strelets V, Tobari YN, Tomimura Y, Wasserman M, Watts T, Wilson R, Yoshida K, Markow TA, Gelbart WM, Kaufman TC. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forattini OP. Entomologia Médica. Volume 1. São Paulo, Brazil: Faculdade de Higiene e Saúde Pública da USP. Editora da USP; 1962. [Google Scholar]
- 26.Faran ME, Linthicum KJ. A handbook of the Amazonian species of Anopheles (Nyssorhynchus) (Diptera: Culicidae) Mosq Syst. 1981;13:1–91. [Google Scholar]
- 27.French WL, Baker RH, Kitzmiller JB. Preparation of mosquito chromosomes. Mosq News. 1962;22:377–383. [Google Scholar]
- 28.Coluzzi M, Cancrini G, Di Deco M. The polytene chromosomes of Anopheles superpictus and relationships with Anopheles stephensi. Parassitologia. 1970;12:101–112. [Google Scholar]
- 29.Green CA, Hunt RH. Interpretation of the variation in ovarian polytene chromosomes of Anopheles funestus giles, A. parensis gillies, and A. aruni? Genetica. 1980;51:187–195. [Google Scholar]
- 30.Sharakhov IV, Serazin AC, Grushko OG, Dana A, Lobo N, Hillenmeyer ME, Westerman R, Romero-Severson J, Costantini C, Sagnon NF, Collins FH, Besansky NJ. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- 31.Sharakhova MV, Xia A, Mcalister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Sharma GP, Parshad R, Narang SL, Kitzmiller JB. The salivary chromosomes of Anopheles stephensi. J Med Entomol. 1969;6:68–71. doi: 10.1093/jmedent/6.1.68. [DOI] [PubMed] [Google Scholar]
- 33.Mahmood F, Sakai RK. An ovarian chromosome map of Anopheles stephensi. Cytobios. 1985;43:79–86. [PubMed] [Google Scholar]
- 34.Coluzzi M, Sabatini A. Cytogenetics observations on species A and B of the Anopheles gambiae complex. Parassitologia. 1967;9:73–88. [Google Scholar]
- 35.Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 36.Chaudhry S, Neetu GS, Chhilar JS. Salivary polytene chromosome mapping of Anopheles (Cellia) subpictus grassi (Culicidae: Diptera) Genome. 2005;48:241–246. doi: 10.1139/g04-120. [DOI] [PubMed] [Google Scholar]
- 37.Keppler WJ, Kitzmiller JB, Rabanni MG. The salivary gland chromosomal of Anopheles albimanus. J Am Mosq Control Assoc. 1973;33:42–49. [Google Scholar]
- 38.Conn J. A cytogenetic analysis of the polytene chromosomes of Anopheles triannulatus (Diptera: Culicidae) from western Venezuela. Genome. 1991;34:267–272. doi: 10.1139/g91-042. [DOI] [PubMed] [Google Scholar]
- 39.Kitzmiller JB. Chromosomal differences among species of Anopheles mosquitoes. Mosq Syst. 1977;9:112–122. [Google Scholar]
- 40.Cornel AJ, Collins FH. Maintenance of chromosome arm integrity between two Anopheles mosquito subgenera. J Hered. 2000;91:364–370. doi: 10.1093/jhered/91.5.364. [DOI] [PubMed] [Google Scholar]
- 41.Marshall JC, Powell JR, Caccone A. Short report: phylogenetic relationships of the anthropophilic Plasmodium falciparum malaria vectors in Africa. Am J Trop Med Hyg. 2005;73:749–752. [PubMed] [Google Scholar]
- 42.Coluzzi M, DiDeco M, Cancrini G. Chromosomal inversions in Anopheles stephensi. Parassitologia. 1973;15:129–136. [PubMed] [Google Scholar]
- 43.Mahmood F, Sakai RK. Inversion polymorphisms in natural populations of Anopheles stephensi. Can J Genet Cytol. 1984;26:538–546. doi: 10.1139/g84-086. [DOI] [PubMed] [Google Scholar]
- 44.Sharakhov I, Braginets O, Grushko O, Cohuet A, Guelbeogo WM, Boccolini D, Weill M, Costantini C, Sagnon N, Fontenille D. A microsatellite physical map of the African human malaria vector Anopheles funestus. J Hered. 2004;95:29–34. doi: 10.1093/jhered/esh011. [DOI] [PubMed] [Google Scholar]
- 45.Linthicum KJ. A revision of the Argyritarsis section of the subgenus Nyssorhynchus of Anopheles (Diptera: Culicidae) Mosq Syst. 1988;20:101–271. [Google Scholar]
- 46.Sallum MA, Schultz TR, Wilkerson RC. Phylogeny of Anopheles (Diptera: Culicidae) based on morphological characters. Ann Entomol Soc Am. 2000;93:745–775. [Google Scholar]
- 47.Scarpassa VM, Conn JE. Population genetic structure of the major malaria vector Anopheles darlingi (Diptera: Culicidae) from the Brazilian Amazon, using microsatellite markers. Mem Inst Oswaldo Cruz. 2007;102:319–327. doi: 10.1590/s0074-02762007005000045. [DOI] [PubMed] [Google Scholar]
- 48.Aultman KS, Beaty BJ, Walker ED. Genetically manipulated vectors of human disease: a practical overview. Trends Parasitol. 2001;17:507–509. doi: 10.1016/s1471-4922(01)02094-3. [DOI] [PubMed] [Google Scholar]
- 49.Santos RC, Padilha A, Pereira-Costa DP, Maia-Costa E, Dantas-Filho HC, Povoa MM. Vetores de malária em duas reservas indígenas da Amazônia Brasileira. Saúde Pública. 2009;43:859–868. doi: 10.1590/s0034-89102009000500016. [DOI] [PubMed] [Google Scholar]
- 50.Vittor AY, Pan W, Gilman RH, Tielsch J, Glass G, Shields T, Sánchez-Lozano W, Pinedo VV, Salas-Cobos E, Flores S, Patz JA. Linking deforestation to malaria in the Amazon: characterization of the breeding habitat of the principal malaria vector, Anopheles darlingi. Am J Trop Med Hyg. 2009;81:5–12. [PMC free article] [PubMed] [Google Scholar]
- 51.Tadei WP, Santos JM, Cunha AS. Sobre o polimorfismo cromossômico de Anopheles darlingi root (Diptera, Culicidae) Cienc Cult. 1984;36:845. [Google Scholar]


