Abstract
In 2004, Sudan adopted artesunate + sulfadoxine/pyrimethamine (SP) combination as the first-line drug, in response to the high level of falciparum resistance to antimalarials. In 2007, a molecular study on antimalarial resistance linked genes, pfcrt, pfmdr1, pfdhfr, pfdhps, and pfATPase6, was conducted on 198 isolates from central and eastern Sudan. We observed a high frequency of point mutations at almost all loci analyzed, mainly of pfcrt 76T (72.7%), pfdhfr 51I (75.3%), and pfdhfr 108N (72.7%) alleles. The MARK III in vitro test for chloroquine sensitivity in 45 P. falciparum isolates showed that 37.8% of the isolates were low resistant and 6.7% were fully resistant. This study represents the most recent molecular investigation on antimalarial resistance in this area after the adoption of artemisinin-based combination therapy (ACT), and underlines the importance of the analysis of SP resistance evolution to monitor the efficacy of ACT therapy in endemic areas.
Introduction
Malaria is still one of the most threatening diseases for people living in tropical and subtropical areas and its control is seriously hampered by the quick development of antimalarial drug resistance by Plasmodium falciparum.
During the second half of the 20th century, chloroquine (CQ) was the antimalarial treatment of choice, because it was safe, cheap, and highly effective against susceptible malaria parasites. The CQ resistance arose more than 40 years ago in southeast Asia and South America, and increasing rates of CQ resistance contribute to rising morbidity and mortality from malaria in Africa.
The CQ resistance is determined by the major point mutation at codon 76 of the P. falciparum CQ resistance transporter (pfcrt) gene,1 which is highly correlated with increased clinical CQ tolerance and treatment failure.1–3 In addition, point mutations in P. falciparum multi-drug resistance gene 1 (pfmdr1) (e.g., N86Y, Y183F, S1034C, N1042D, and D1246Y) have been shown to modulate CQ resistance4 and possibly lumefantrine resistance.5
Similarly, resistance of malaria parasites to antifolates is conferred by mutation in dihydrofolate reductase (DHFR) and dihydropteroate syntase (DHPS), two enzymes involved in the parasite's folate synthesis.6–10 In particular, a combination of five point mutations, pfdhfr N51I, C59R, S108N, and pfdhps A437G and K540E, have been shown to be strongly associated with sulfadoxine/pyrimethamine (SP) resistance in Africa.11–13
Adding artemisinin derivatives (i.e., artesunate) to standard antimalarial drugs (i.e., SP) is a strategy to reduce treatment failure and transmission potential, because artemisinin derivatives act rapidly and kill malaria parasites that are resistant to other drugs.14 In fact, many malaria endemic countries have lately adopted artemisinin-based combination therapy (ACT) as the first-line drug for treating uncomplicated malaria, as recommended by the World Health Organization (WHO).15
Although ACT should represent the best option available to prevent the induction of resistance parasites, the risk of development of resistance in P. falciparum to artemisinin compounds is serious, given the recent observation of two cases of clinical resistance in Cambodia16 and cases of diminished P. falciparum susceptibility to arthemeter.17,18
Recently, it was observed that variations in susceptibility profiles of malaria parasites to artemisinin derivatives have been associated with single nucleotide polymorphisms (SNPs) in sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA)-type of P. falciparum ATPAse6 (pfATPase6) gene, the putative target for artemisinins,17,19,20 suggesting a possible role of pfATPase6 gene as a molecular marker of artemisinin resistance.
Sudan is the largest country in Africa, comprising more than 8% of the entire continent. The total population is estimated to be 39.2 million inhabitants, of whom 75% live in rural areas. Malaria is a leading cause of morbidity and mortality, resulting in 3.1 million cases and 2,500 deaths annually.21 The entire country is at risk of malaria, although with different levels of endemicity. In the northern, eastern, and western states malaria endemicity is mainly low to moderate with predominantly seasonal transmission and epidemic outbreaks. In southern Sudan, malaria is moderate to high or highly intense, generally with perennial transmission. Plasmodium falciparum is by far the predominant parasite species.
The CQ has been the most used drug as a first line for years in Sudan. Some studies showed that in five hospitals of central Sudan, 85.6% of patients admitted for severe malaria had received CQ before admission22 and that CQ was also the commonly prescribed drug by health care providers.23 First cases of CQ resistance were reported in 1978,24 and several in vivo and in vitro studies have subsequently documented a high presence of CQ resistance in different Sudanese areas (central Sudan25–27; eastern Sudan28,29).
Sulfadoxine/pyrimethamine resistance was reported in Sudan since the early 1990s30 and was documented by in vivo studies conducted in eastern Sudan just before ACT deployment.31–33 The presence of highly resistant pfdhfr/pfdhps quintuple mutants have been previously reported in studies conducted in different areas of southern34 and eastern Sudan.31
In 2004, because of CQ and SP resistance, the national antimalarial drug policy changed from monotherapy to artemisinin-based combination therapy. The National Malaria Control Program decided to replace CQ with artesunate + sulfadoxine/pyrimethamine as the first-line drug and artemether-lumefantrine has replaced SP as the second-line treatment.35 The aim of this study was to assess the effect of the current antimalarial drug policy, i.e., ACT, on prevalence of mutations in target genes, in two malarious areas of Sudan where to the best of our knowledge no recent investigations on the same subject have been carried out.
Materials and Methods
Study sites.
The fieldwork was carried out in health facilities located in and around Wad Medani–Gezira State (central Sudan) and Elfau locality–Gedarif State (eastern Sudan) (Figure 1) between June and December 2007, corresponding to the period of high malaria transmission. These study sites are located in an irrigated agricultural scheme, where transmission is seasonal as identified by epidemiological strata of malaria in Sudan and are inhabited by different ethnic groups and tribes from all regions of Sudan. Wad Medani is the main town of the irrigated area of the Gezira Agricultural Scheme where malaria is classified as mesoendemic to hyperendemic with an unstable transmission pattern. Elfau is located in the west part of Gedarif State neighboring Gezira State. It is the main town of the Erahad irrigation scheme.
Figure 1.
Map of Sudan. On the right are the sites of this study. The geographical coordinates are indicated in the figure.
Patients and blood samples.
Participants were enrolled in the study after appropriate informed consent was obtained. Ethical approval for this study was obtained from the Ethical Committee of the Blue Nile National Institute for Communicable Disease/University of Gezira and from the State Health Authority in Gezira.
Blood samples were collected from 212 (central Sudan) and 63 (eastern Sudan) microscopically confirmed P. falciparum-infected patients. Of 275 patients screened, 235 patients (42% males, 58% females, age range: 4–45 years) met the criteria for inclusion in the analysis: patients had P. falciparum monoinfection, asexual parasitaemias in excess of 1,000 parasites, but less than 80,000 parasites (mean of 29,520 parasites) per μL blood, and had not received quinine and artemisinin within the last 7 days, 4-aminoquinolines within the last 14 days, pyrimethamine and/or sulfonamide within the last 28 days, or mefloquine within the last 56 days.
For molecular analysis, infected blood was collected by finger prick, blotted in triplicate on a filter paper, and air dried. Filter papers were wrapped separately in a plastic bag and stored at room temperature. Total DNA was extracted from 149 samples using the phenol/chloroform method9 and from 86 by using PureLink Genomic DNA Kit (Invitrogen, Carlsbad, CA), following the manufacturer's instructions.
For the in vitro test, 2 mL of blood were collected from subjects into a sterile EDTA container after cleaning the site of puncture with 70% ethanol.
Real-time PCR assay.
All samples were genotyped at pfcrt 76 codon, pfdhfr 51-59-108 codons, and pfdhps 436-437 codons using a 5′-nuclease real-time polymerase chain reaction (PCR) assay. Primers and probes sequences were previously described36–37 and were synthesized by Roche-Diagnostics (Mannheim, Germany). Wild-type and mutant-type probes were labeled with a FAM and HEX fluorescent reported at their 5′ ends, respectively. Real-time PCR was performed with a LightCycler 480 System (Roche) according to the following steps: pre-incubation 95°C for 10 min; amplification 95°C for 10 sec, 55°C for 20 sec, and 72°C for 5 sec for 45 cycles. The PCR assays were optimized according to the LightCycler 480 Probes Master Kit, and 0.5 μM of each primer and 0.2 μM of each probe, used in combination, were used in this study. All reactions were done in duplicate, in a final volume of 20 μL. The DNAs from W2 and 3D7 laboratory strains and from characterized field isolates were used as reference standards.
PCR-restriction fragment length polymorphism (RFLP) analysis.
Detection of pfmdr1 86 and pfdhps 540 polymorphisms was performed by the PCR-RFLP method. Pfmdr1 amplification was achieved under conditions described in Duraisingh and others.38 The PCR products were digested at 37°C with 1U of AflIII (New England Biolabs, Ipswich, MA) restriction enzyme. Detection of pfdhps540 polymorphism was performed using primers described in Alker and others.36 The PCR products were digested at 37°C with 1U of FokI (Biolabs) restriction enzyme, as described in Duraisingh and others.39
Sequencing.
Assessment of single nucleotide polymorphisms for the pfATPase6 gene was based on PCR and the sequencing method, as described previously in Menegon and others.40
Statistical analysis.
Differences in mutation frequency of the studied genotypes by sampling sites or by sampling period were analyzed by the two-sample test of proportions (STATA Statistical Software: Release 8.1, 2003, Stata Corp., College Station, TX). The same statistical test was adopted to compare the 59R mutation frequency observed in 200331 and in this study. Results are showed as z(test) and p(z) values.
In vitro test.
The micro-technique test (Mark III: supplied by WHO Division of Control of Tropical Disease 2001, England) was used to evaluate in vitro sensitivity to CQ of P. falciparum malaria parasites. Sixty-nine malaria cases were selected for this study, according to the in vitro protocol described in Mark III-WHO.41 The test was performed in tissue culture plates pre-dosed with drugs in increasing concentrations; briefly, 0.9 mL of RPMI 1640 medium were taken into the sterile falcon tube and 100 μL from well-mixed blood mentioned previously were added. All wells of appropriate columns were dosed with 50 μL of blood-medium mixture using the 50 μL fixed volume Eppendorf pipette and a disposable sterile tip, as provided with the test kit. Plates were incubated at 37.5°C in a candle jar for 24 to 30 h. At the end of incubation, blood from each well was harvested and a thick film was prepared; thick films were stained for 30 min in a Giemsa stain at a dilution of 1% (vol/vol) in buffered water of pH 6.8. After drying, films were examined through a light microscope with oil immersion lens, and the number of schizonts with three or more nuclei of a total of 200 asexual parasites (i.e., schizonts and trophozoites) was counted. For an acceptable test, schizont maturation in control must be 10% or more (i.e., 20 schizonts with three or more nuclei per 200 asexual parasites). Average results for EC50, EC90, EC95, and EC99 (i.e., drug concentrations producing 50%, 90%, 95%, or 99% inhibition of schizont maturation, respectively) were calculated using the WHO log probit program (www.who.int/csr/drugresist/malaria/en/probit.xls) in all isolates.
Results
Between June and December 2007, a total of 235 blood samples from P. falciparum-infected individuals were examined for the presence of mutations in five genes related to antimalarial resistance: in particular, we investigated one codon of pfcrt gene (K76T), one codon of pfmdr1 (N86Y), three codons of the pfdhfr gene (N51I, C59R, S108N), three codons of the pfdhps gene (S436A, A437G, K540E), and SNPs in two regions of the pfATPase6 gene, from nucleotide (nt) 652 to 1422 and from nt 1744 to 2402 (nt number is according to the ATG start codon). Among the blood samples examined, 198 samples gave PCR-positive results for each of the genes included in our study, exactly 27 samples from Gedarif State (eastern Sudan), and 171 samples from Gezira State (central Sudan). Thirty-seven patients (16%) with PCR-negative results were excluded from analysis.
There were 14 samples (7%) that showed mixed infections of both wild-type and mutant alleles. These mixed wild/mutant infections were considered as mutants. Moreover, a total of 69 samples were analyzed in vitro for their susceptibility to CQ, however only 45 (65.2%) isolates with an adequate schizonts growth were successfully tested.
Polymorphism analysis.
Our results showed that 185 samples (93.4%) out of 198 analyzed carried a mutant allele of at least one gene associated with resistance to antimalarial drugs chloroquine and sulfadoxine-pyrimethamine. The results are summarized in Table 1 and Table 2. The majority of the isolates were found to carry the mutant codons pfdhfrI51 (75.3%) and pfdhfrN108 (72.7%), whereas pfdhfrR59 was found in 15 isolates (7.6%). With respect to pfdhps gene, pfdhpsA436, pfdhpsG437, and pfdhpsE540 were detected in 8.1%, 20.7%, and 16.7% of the isolates, respectively.
Table 1.
Frequency of specific point mutations in isolates analyzed in this study*
| Table 1A | Prevalence of point mutations | Z† | p(z)† | Table 1B | Prevalence of point mutations | z‡ | p(z)‡ | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Gedarif | Gezira | Gezira | Gezira | ||||||
| No. = 198 | No. = 27 | No. = 171 | No. = 71 | No. = 100 | ||||||
| June–December | November–December | June–December | June–July | November–December | ||||||
| No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | ||||||
| Pfcrt76T | 144 (72.7) | 24 (88.8) | 120 (70.1) | 2.03 | 0.043 | Pfcrt76T | 54 (76) | 66 (66) | 1.41 | 0.159 |
| Pfmdr186Y | 110 (55.5) | 23 (85.2) | 87 (50.8) | 3.34 | 0.001 | Pfmdr186Y | 36 (50.7) | 51 (51) | −0.04 | 0.969 |
| Pfdhfr51I | 149 (75.3) | 21 (77.7) | 128 (74.8) | 0.32 | 0.0746 | Pfdhfr51I | 58 (81.6) | 70 (70) | 1.72 | 0.085 |
| Pfdhfr59R | 15 (7.6) | 0 | 15 (8.8) | −1.52 | 0.128 | Pfdhfr59R | 7 (9.9) | 8 (8) | 0.43 | 0.665 |
| Pfdhfr108N | 144 (72.7) | 24 (88.8) | 120 (70.1) | 2.03 | 0.043 | Pfdhfr108N | 52 (73.3) | 68 (68) | 0.75 | 0.455 |
| Pfdhps436A | 16 (8.1) | 5 (18.5) | 11 (6.4) | 2.15 | 0.032 | Pfdhps436A | 4 (5.6) | 7 (7) | −0.37 | 0.713 |
| Pfdhps437G | 41 (20.7) | 8 (29.6) | 33 (19.3) | 1.24 | 0.214 | Pfdhps437G | 16 (22.5) | 17 (17) | 0.9 | 0.369 |
| Pfdhps540E | 33 (16.7) | 6 (22.2) | 27 (15.7) | 0.84 | 0.399 | Pfdhps540E | 15 (21.2) | 12 (12) | 1.62 | 0.104 |
| PfATPase6-402V | 8 (4) | 1 (3.7) | 7 (4) | −0.07 | 0.941 | PfATPase6-402V | 7 (9.8) | 0 | 3.2 | 0.001 |
| PfATPase6-431K | 35 (17.7) | 12 (44.5) | 23 (13.4) | 3.94 | < 0.001 | PfATPase6-431K | 13 (18.3) | 10 (10) | 1.57 | 0.117 |
1A = allele frequencies and z test values among 198 Plasmodium falciparum isolates from Gedarif State (eastern Sudan) and Gezira State (central Sudan); 1B = allele frequencies and z test values among 171 P. falciparum isolates from Gedarif State (eastern Sudan) sampled in two different period of the year; z = two-sample test of proportion (z test); p(z): significance of z test.
Comparison between Gedarif and Gezira.
Comparison between Gezira isolates sampled in June–July and in November–December. Significance value p(z) < 0.05.
Table 2.
Types and frequency of haplotypes identified in Sudanese isolates by analysis of four molecular markers of CQ and SP resistance*
| No. of point mutations | Haplotype* | No. isolates from Gezira | No. isolates from Gedarif | Total % | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | KNNCSSAK | 13 | 0 | 6.6 | ||||
| 1 | TNNCSSAK | 8 | 0 | 4.1 | ||||
| 1 | KYNCSSAK | 2 | 0 | 1 | ||||
| 1 | KNICSSAK | 4 | 0 | 2 | ||||
| 1 | KNNCSAAK | 2 | 0 | 1 | ||||
| 1 | KNNCSSGK | 1 | 0 | 0.5 | ||||
| 2 | TYNCSSAK | 8 | 1 | 4.6 | ||||
| 2 | TNICSSAK | 2 | 0 | 1 | ||||
| 2 | TNNCNSAK | 0 | 1 | 0.5 | ||||
| 2 | TNNCSAAK | 0 | 1 | 0.5 | ||||
| 2 | KYNCNSAK | 1 | 1 | 1 | ||||
| 2 | KYNCSSGK | 0 | 1 | 0.5 | ||||
| 2 | KNICNSAK | 9 | 0 | 4.6 | ||||
| 2 | KNNRNSAK | 4 | 0 | 2 | ||||
| 3 | TNICNSAK | 22 | 2 | 12.2 | ||||
| 3 | TYNCSSGK | 1 | 0 | 0.5 | ||||
| 3 | TYICSSAK | 3 | 0 | 1.5 | ||||
| 3 | TYNCSSAE | 1 | 0 | 0.5 | ||||
| 3 | TNIRSSAK | 1 | 0 | 0.5 | ||||
| 3 | TNNCSAAE | 1 | 0 | 0.5 | ||||
| 3 | TNNCSSGE | 1 | 0 | 0.5 | ||||
| 3 | KYICNSAK | 5 | 0 | 2.5 | ||||
| 3 | KNIRNSAK | 3 | 0 | 1.5 | ||||
| 3 | KNICNAAK | 2 | 0 | 1 | ||||
| 3 | KNICNSGK | 2 | 0 | 1 | ||||
| 3 | KNICSSGE | 1 | 0 | 0.5 | ||||
| 4 | TYICNSAK | 38 | 9 | 23.8 | ||||
| 4 | TNIRNSAK | 1 | 0 | 0.5 | ||||
| 4 | KYICNAAK | 1 | 0 | 0.5 | ||||
| 4 | KYICNSGK | 1 | 0 | 0.5 | ||||
| 5 | TYIRNSAK | 2 | 0 | 1 | ||||
| 5 | TYICNAAK | 4 | 4 | 4 | ||||
| 5 | TYICNSGK | 3 | 1 | 2 | ||||
| 5 | TYICNSAE | 1 | 0 | 0.5 | ||||
| 5 | TYICSSGE | 1 | 0 | 0.5 | ||||
| 5 | TYNCNSGE | 0 | 1 | 0.5 | ||||
| 5 | KYICNSGE | 0 | 1 | 0.5 | ||||
| 5 | TNICNSGE | 8 | 0 | 4 | ||||
| 6 | TYICNSGE | 10 | 4 | 7.1 | ||||
| 6 | TNIRNSGE | 1 | 0 | 0.5 | ||||
| 7 | TYIRNSGE | 3 | 0 | 1.5 | ||||
| Reference | Crt 76 | Mdr1 86 | Dhfr 51 | Dhfr 59 | Dhfr 108 | Dhps 436 | Dhps 437 | Dhps 540 |
| Wild-type | K aaa | N aat | N aat | C tgt | S agc | S tct | A gct | K aaa |
| Mutant | T aca | Y tat | I att | R cgt | N aac | A gct | G ggt | E gaa |
The construction of haplotype involved pfcrt76, pfmdr1-86, pfdhfr51-59-108, and pfdhps 436-437-540. Mutant codon is in boldface.
Prevalence of the CQ resistance-related mutant alleles was relatively high: pfcrtT76 allele was present in 72.7% of the isolates, whereas 86Y mutation of the pfmdr1 gene was detected in 55.5% of the isolates.
Isolates from eastern Sudan (Gedarif) showed a higher prevalence of all but one mutation when compared with isolates from central Sudan (Gezira). This difference has been confirmed as significant by statistical analysis for pfcrt76T, pfmdr186Y, pfdhfr108N, and pfdhps436A mutated codons (Table 1A). None of the isolates from eastern Sudan had pfdhfr59R mutation.
Moreover, when we compared samples from central Sudan collected during June–July (N = 71) and those collected during November–December (N = 100), we observed that the rate of almost all mutations is lower in the isolates collected in November–December. However, statistical analysis showed that there were no significant differences between Plasmodium genotype frequencies from parasite populations collected in two different periods within the same sampling area (Table 1B).
By the analysis of allele combinations of these four resistance markers, we observed 41 different haplotypic conformations in our isolates (Table 2). In particular, we found 5 distinct haplotypes in isolates that carried a single point mutation (8.6% of the isolates analyzed), 8 haplotypes in double mutant isolates (14.7%), 12 haplotypes in triple mutant isolates (22.7%), 4 haplotypes in mutants isolates carrying quadruple mutations (25.3%), 8 haplotypes in quintuple mutant isolates (13%), and 2 and 1 haplotypes in mutant isolates that carried 6 (7.6%) and 7 (1.5%) point mutations, respectively (Table 2, Figure 2). The wild-type sequence K76N86N51C59S108S436A437K540 was present in 6.6% of isolates (Table 2, Figure 2).
Figure 2.
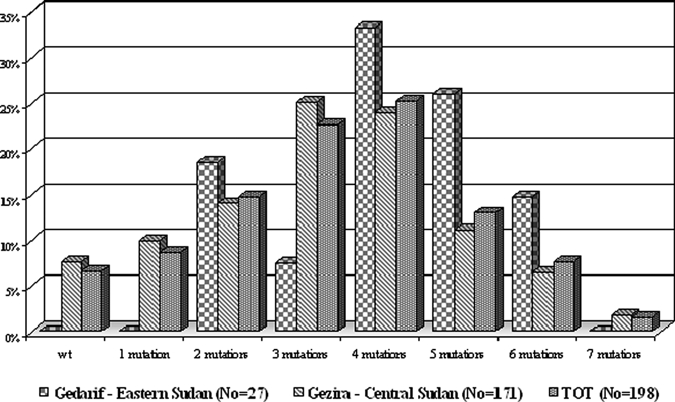
Overall prevalence of Sudanese isolates carrying point mutations observed in a total of eight codons in the pfcrt, pfmdrl, pfdhfr, and pfdhps genes.
With regard to the haplotypes, isolates from eastern Sudan did not exhibit wild-type, single mutant type, and 7-fold mutant genotypes but they displayed a high prevalence of mutant alleles with four mutations (33.3%), whereas the majority of isolates from central Sudan were found to carry the triple mutant haplotype (25.1%) (Figure 2).
P. falciparum ATPase6 SNPs.
In this study, we analyzed the prevalence of single nucleotide polymorphisms in two regions of the pfATPase6 gene. Most of the Sudanese parasite population harbored wild-type allele. Only two previously reported mutations were detected: 431K in 35 isolates (17.7%) and 402V in 8 isolates (4%) (Table 1).
Chloroquine susceptibility.
In vitro activity of CQ was successfully evaluated in 45 isolates. The majority (55.6%) of tested isolates were fully sensitive to CQ, 37.8% had a low resistance level, and 6.7% were marked resistant (Table 3). The test results processed by log concentration–response probit analysis for determination of 50%, 90%, 95%, and 99% effective concentrations (EC50, EC90, EC95, and EC99) were: EC50:312.1808 nmol/L, EC90:930.0186 nmol/L, EC95:1267.3088 nmol/L, and EC99:3633.1583 nmol/L.
Table 3.
Patterns of pfcrt and pfmdr1 point mutations from the 45 isolates tested for CQ in vitro susceptibility*
| pfcrt codons | pfmdr1 codons | pfcrt/pfmdr1 codons | ||||||
|---|---|---|---|---|---|---|---|---|
| In vitro test result | Wt K76 | Mut T76 | Wt N86 | Mut Y86 | Wt/wt | Wt/mut | Mut/wt | Mut/mut |
| K76/N86 | K76/Y86 | T76/N86 | T76/Y86 | |||||
| Sensitive (N = 25) | 12 (48%) | 13 (52%) | 14 (56%) | 11 (44%) | 11 (44%) | 1 (4%) | 3 (12%) | 10 (40%) |
| Low resistant (N = 17) | 3 (17%) | 14 (82.4%) | 3 (17.6%) | 14 (82.4%) | 1 (5.9%) | 2 (11.8%) | 2 (11.8%) | 12 (70.6%) |
| Marked resistant (N = 3) | 2 (66.7%) | 1 (33.3%) | 3 (100%) | 0 | 2 (66.7%) | 0 | 1 (33.3%) | 0 |
N = no. isolates; wt = wild-type codon; mut = mutant codon.
Polymorphism analysis of this group of isolates showed high prevalence of single and double mutations pfcrtT76/pfmdr1Y86, related to CQ resistance (Table 3), however we found that isolates showed low correlation between molecular profile of both genes and in vitro susceptibility. In fact, two (66.7%) of three marked CQ-resistant isolates carried wild-type pfcrtK76/pfmdr1N86 allele, whereas double mutant pfcrtT76/pfmdr1Y86 alleles were found in 40% of sensitive isolates. Concerning low-resistant isolates, they showed a high incidence of single (11.8%) or double (70.6%) point mutations at pfcrtT76/pfmdr1Y86 codons; consequently, the correlation between in vitro outcome and mutation rate was relatively higher.
Discussion
We believe that the current study is one of the few recent reports of drug resistance molecular markers in these malarious areas of Sudan. In this view, it is interesting to compare our results with those obtained in studies previously carried out in this malaria endemic region.
In Sudan, malaria has a scattered distribution and varies a lot according to the different regions of the country: there are regions with high, stable malaria transmission (southern Sudan), others with a high seasonal transmission pattern (central/eastern Sudan), and finally, in the northern part of the country, spotted areas characterized by riverine and urban transmission. Furthermore, the level of falciparum resistance to the most used antimalarials differs from region to region,31,34,42 making a countrywide drug policy difficult.
In 2004, the National Control Malaria Program decided to adopt artesunate + sulfadoxine/pyrimethamine as the first-line drug and artemether-lumefantrine as the second line. However, the implementation of this decision and consequent real withdrawal of CQ from the local market were still in agenda in the year 2005. As reported by Malik and others,42 manufacturing and importation of CQ tablets and vials was stopped completely in January 2006 only. As a consequence, we might expect that CQ has been available in the local market for a long period after that date. This is an important consideration in evaluating the result of the molecular marker investigation.
In fact, frequency of pfcrt 76T mutation observed in our study is still relatively high (72.7%, N = 144/198) and it is consistent with a current or recent presence of drug pressure, even if it is lower than previously reported from other Sudanese studies.31,43,44 A-Elbasit and others,31 for example, reported a pfcrt 76T frequency close to fixation of mutated allele (pfcrt 76T = 93%). In this scenario, the results of the in vitro tests are quite surprising and the observed high percentage of isolates showing full sensitivity to CQ is difficult to explain. Recent data from literature have already discussed the potential inaccuracies of in vitro drug susceptibility tests45,46 and also some conflicting results between the frequency of pfcrt mutations and in vitro/in vivo tests.47–49 However, we can speculate that in vitro tests data obtained in our study could have resulted from the limited number of tests performed since, according to the WHO protocol, a correct MARK III outcome can be obtained by performing at least 100 tests. If we take into account the recognized strong link between presence of pfcrt mutations and CQ resistance and we look at the frequency of pfcrt 76T mutation from this study, we can assume that CQ is definitely an unsuitable drug for this area. These data suggested that prevalence of molecular markers for CQ should be monitored (while this drug is being replaced by combination therapies) to verify if the level of CQ resistance is decreasing in this area. In fact, as previously reported,50,51 recession of chloroquine-resistant parasites preceded the complete withdrawal of CQ in areas with high malaria transmission.
As far as ACT implementation is concerned, unfortunately falciparum tolerance/resistance to artemisinin derivatives has been recently reported in Cambodia.16 This recent report increases the interest in monitoring pfATPase6 point mutations that have been suggested to be involved in modulating artemisinins sensitivity, even if their role is still under debate.
In our investigation on pfATPase6 gene polymorphism, we found two mutated codons (431K and 402V) only and at low frequencies (17.7% and 4%, respectively). In particular, E431K substitution was previously detected at higher frequences both in Western (Cameroon, 37%52) and Eastern (Zanzibar 31%, Tanzania 39%53) Africa. Moreover, we previously identified 431K substitution in 17% of 73 isolates from nine African countries, as described in Menegon and others.40 Taking into consideration that the first-line treatment currently adopted in Sudan is AS + SP, it is important to look carefully at the dhfr/dhps mutation prevalence observed in our study. An important issue is the very low frequency of mutant IRN-GE (2%, Table 2), haplotype that is commonly indicated as a good predictor of treatment failure, therefore, the SP combination is expected to be still effective in this region. However, it is necessary to note the frequency of pfdhfr51/59/108 point mutations singularly (Table 1), in fact, while prevalence of the 51 and 108 mutations, observed in our study, are substantially comparable to the one reported in previous studies carried out in the same Sudanese regions (characterized by a seasonal malaria transmission and dated 2002/200331,33,54), frequency of 59R mutations significantly increased from 0.6%31 to 7.6% (z = 3.255, p(z) = 0.001). This increase of pfdhfr59R frequency in these last 3–4 years could be considered a signal of spread of SP resistance in this area. In view of this, we believe that in vivo tests assessing falciparum sensitivity to pyrimetamine and sulfadoxine drug combination are needed, because the lack of efficacy of SP will adversely affect the outcome of ACT treatment.
We have also taken into consideration the frequency of each mutated codon by sampling areas (Gedarif versus Gezira) and by sampling periods (June/July versus November/December, Gezira). The results of statistical analysis showed that the differences in mutations frequencies between the two sampling periods are not significant, thus indicating that probably the sampling period encompassed the same malaria transmission season. Conversely, when comparing the two sampling areas, there are significant differences in frequency for four mutated codons (pfcrt76T, pfmdr186Y, pfdhfr108N, and pfdhps436A). However, we have to be cautious in drawing conclusions if we consider that there are some differences between the two batches of samples (samples size and sampling period) and that information about the malaria treatment implementation in the two study areas is lacking. Therefore, we can only speculate that probably falciparum populations in Gedarif are exposed to a higher drug pressure than parasite populations from Gezira.
Finally, interesting issues arise from analysis of the different haplotypes generated by the association of all identified mutations in four analyzed genes. In fact, the most prevalent haplotype is quadruple mutant Tcrt76Ymdr186Idhfr51Ndhfr108 (23.8%), but also haplotypes showing five or six mutations are well represented in the falciparum isolates circulating in this study area, in particular sextuple mutant Tcrt76Ymdr186Idhfr51Ndhfr108Gdhps437Edhps540 that accounts for 7.1%. Moreover, we found also three isolates showing seven mutations Tcrt76Ymdr186Idhfr5 Rdhfr59Ndhfr108Gdhps437Edhps540. The presence of multiple haplotypes in African falciparum isolates has been already described and discussed in previous studies.43,55,56 The reason for these observed patterns of mutations in specific genes scattered on four different chromosomes and linked to the resistance to a different class of antimalarials is still to be elucidated. We can speculate that this association could occur in endemic areas where the probably high fitness cost of multiple mutations is balanced by simultaneous pressure of different drugs. The current situation in Sudan could represent an example: as stated previously, because in this country CQ is probably still available and used (or has been used until sometime ago) especially in rural setting and, as a part of ACT, SP combination is used as well. This could explain the detection in the isolates analyzed in our study of the TYIN, TYINGE, and TYIRNGE haplotypes. The circulation of these isolates harboring multiple CQ and SP resistance mutations in an endemic area is not to be underestimated, because, as previously reported,57 selection of a high degree of resistance toward a given antimalarial, in this case the pyrimethamine, could be accompanied by increase of a total number of mutations in P. falciparum. Hence, the Sudanese isolates of P. falciparum carrying multiple mutations are expected to be somewhat resistant and in case of accumulation of further mutations could become really difficult to be treated.
In conclusion, this study supports the usefulness of molecular marker screening to be carried out in the frame of country surveillance activities. Monitoring evolution of P. falciparum drug resistance by analyzing the prevalence of drug resistance mutations in an endemic area as Sudan, where both logistical and political problems make the implementation of the Efficacy Therapeutic Tests difficult, could provide key information for the deployment of an effective countrywide drug policy.
Acknowledgments
We thank Simone Cacciò and Giuseppe La Rosa for their helpful assistance in performing Real-Time experiments. We are grateful to Ivano Iavarone for his help in the statistical analysis.
Disclaimer: None of the authors has a conflict of interest to declare.
Footnotes
Financial support: Part of this work has been carried out with the financial support from a Technical Service Agreement (HQ/07/100294) between the World Health Organization, Global Malaria Programme, Geneva, Switzerland, and Istituto Superiore di Sanità, Roma, Italy.
Authors' addresses: Michela Menegon, Carlo Severini, and Giancarlo Majori, Department of Infectious, Parasitic and Immunomediated Diseases and WHO Collaborating Centre for Research and Training in Tropical Diseases Control, Istituto Superiore di Sanità, Rome, Italy, E-mails: michela.menegon@iss.it, carlo.severini@iss.it, and giancarlo.majori@iss.it. Albadawi A. Talha and Ahmed A. Mohamedani, Faculty of Medical Laboratory Sciences, University of Gezira, WadMedani, Sudan. E-mails: badawiat@yahoo.com and a_mohamedani@hotmail.com. Sayed M. Elbushra and Bakri Y. M. Nour, Blue Nile Research National Institute for Communicable Diseases, University of Gezira, Wad Medani, Sudan, E-mails: sayedelbushra@yahoo.com and bakrinour@hotmail.com. Elfatih M. Malik, Directorate of Communicable Diseases Control–Federal Ministry of Health, Khartoum, Sudan, E-mail: fatihmmalik@hotmail.com. Tarig A. Mohamed, National Malaria Control Program–Federal Ministry of Health, Khartoum, Sudan, E-mail: tarigmohmed_ali@hotmail.com. Walther H. Wernsdorfer, National Malaria Control Program–Federal Ministry of Health, Khartoum, Sudan, Institute of Specific Prophylaxis and Tropical Medicine, Center for Physiology and Pathophysiology, Medical University, Vienna, Austria, E-mail: walter.wernsdorfer@meduniwien.ac.at.
References
- 1.Wellems T, Plowe C. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 2.White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 4.Hayton K, Su XZ. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr Drug Targets Infect Disord. 2004;4:1–10. doi: 10.2174/1568005043480925. [DOI] [PubMed] [Google Scholar]
- 5.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Björkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 6.Basco LK, Tahar R, Ringwald P. Molecular basis of in vivo resistance to sulfadoxine-pyrimethamine in African adult patients infected with Plasmodium falciparum malaria parasites. Antimicrob Agents Chemother. 1998;42:1811–1844. doi: 10.1128/aac.42.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Pecoulas PE, Basco LK, Le Bras J, Mazabraud A. Association between antifol resistance in vitro and DHFR gene point mutation in Plasmodium falciparum isolates. Trans R Soc Trop Med Hyg. 1996;90:181–182. doi: 10.1016/s0035-9203(96)90130-3. [DOI] [PubMed] [Google Scholar]
- 9.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 10.Sibley CH, Hyde JE, Sims PF, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 11.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 12.Mberu EK, Mosobo MK, Nzila AM, Kokwaro GO, Sibley CH, Watkins WM. The changing in vitro susceptibility pattern to pyrimethamine/sulfadoxine in Plasmodium falciparum field isolates from Kilifi, Kenya. Am J Trop Med Hyg. 2000;62:396–401. doi: 10.4269/ajtmh.2000.62.396. [DOI] [PubMed] [Google Scholar]
- 13.Nzila AM, Mberu EK, Sulo J, Dayo H, Winstanley PA, Sibley CH, Watkins WM. Towards an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob Agents Chemother. 2000;44:991–996. doi: 10.1128/aac.44.4.991-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balint GA. Artemisinin and its derivatives: an important new class of antimalarial agents. Pharmacol Ther. 2001;90:261–265. doi: 10.1016/s0163-7258(01)00140-1. [DOI] [PubMed] [Google Scholar]
- 15.World Health Organization . Antimalarial Drug Combination Therapy. Geneva: World Health Organization, WHO/CDS/RBM/2001.35; 2001. [Google Scholar]
- 16.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 17.Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, Ekala MT, Bouchier C, Esterre P, Fandeur T, Mercereau-Puijalon O. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- 18.Legrand E, Volney B, Meynard JB, Esterre P, Mercereau-Puijalon O. Resistance to dihydroartemisinin. Emerg Infect Dis. 2007;13:808–809. doi: 10.3201/eid1305.061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlemann AC, Cameron A, Eckstein-Ludwig U, Fischbarg J, Iserovich P, Zuniga FA, East M, Lee A, Brady L, Haynes RK, Krishna S. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat Struct Mol Biol. 2005;12:628–629. doi: 10.1038/nsmb947. [DOI] [PubMed] [Google Scholar]
- 20.Eckstein-Ludwig U, Webb RJ, van Goethem IDA, East JM, Lee AG, Kimura M, O'Neill PM, Bray PG, Ward SA, Krishna S. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 2003;424:957–961. doi: 10.1038/nature01813. [DOI] [PubMed] [Google Scholar]
- 21.Sudan, Country Profile. 2005. http://rbm.who.int/wmr2005/profiles/sudan.pdf Available at. Accessed April 28.
- 22.Malik EM, Nour SM, Hamid IK, Elmardi KA, Mohamed TA, Ahmed ES. From home to hospital: beliefs and practices related to severe malaria in Sudan. J Family Community Medicine. 2005;12:85–90. [PMC free article] [PubMed] [Google Scholar]
- 23.Yousif MA, Adeel AA. Antimalarials prescribing pattern in Gazera State: precepts and practices. East Mediterr Health J. 2000;6:939–947. [PubMed] [Google Scholar]
- 24.Omer AH. Response of Plasmodium falciparum in Sudan to oral chloroquine. Am J Trop Med Hyg. 1978;27:853–857. doi: 10.4269/ajtmh.1978.27.853. [DOI] [PubMed] [Google Scholar]
- 25.Kouznetsov RL, Rooney W, Wernsdorfer WH, El Gaddal AA, Payne D, Abdalla RE. Use of the in vitro microtechnique for the assessment of drug sensitivity of Plasmodium falciparum in Sennar, Sudan. Bull World Health Organ. 1980;58:785–789. [PMC free article] [PubMed] [Google Scholar]
- 26.Ibrahim AM, Ali FR, Ali ME. Assessment of chloroquine resistance of Plasmodium falciparum in children of Wad Medani (central Sudan) J Trop Pediatr. 1992;38:162–166. doi: 10.1093/tropej/38.4.162. [DOI] [PubMed] [Google Scholar]
- 27.Nour BY, Faragalla IA, Saeed OK, Mohamadani AA. In vitro study assessing the response of Plasmodium falciparum malaria to chloroquine, sulfadoxine/pyrimethamine, quinine and mefloquine in Wad Medani District, Sudan. Saudi Med J. 2006;27:808–812. [PubMed] [Google Scholar]
- 28.Bayoumi RA, Babiker HA, Ibrahim SM, Ghalib HW, Saeed BO, Khider S, Elwasila M, Karim EA. Chloroquine resistant Plasmodium falciparum in Eastern Sudan. Acta Trop. 1989;461:157–165. doi: 10.1016/0001-706x(89)90032-6. [DOI] [PubMed] [Google Scholar]
- 29.Babiker HA, Satti G, Ferguson H, Bayoumi R, Walliker D. Drug resistant Plasmodium falciparum in an area of seasonal transmission. Acta Trop. 2005;94:260–268. doi: 10.1016/j.actatropica.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim ME, Awad-el-Kariem FM, el Hassan IM, el Mubarak ER. A case of Plasmodium falciparum malaria sensitive to chloroquine but resistant to pyrimethamine/sulfadoxine in Sennar, Sudan. Trans R Soc Trop Med Hyg. 1991;85:446. doi: 10.1016/0035-9203(91)90210-p. [DOI] [PubMed] [Google Scholar]
- 31.A-Elbasit IE, Khalil IF, Elbashir MI, Masuadi EM, Bygbjerg IC, Alifrangis M, Giha HA. High frequency of Plasmodium falciparum CICNI/SGEAA and CVIET haplotypes without association with resistance to sulfadoxine/pyrimethamine and chloroquine combination in the Daraweesh area, in Sudan. Eur J Clin Microbiol Infect Dis. 2008;27:725–732. doi: 10.1007/s10096-008-0499-1. [DOI] [PubMed] [Google Scholar]
- 32.A-Elbasit IE, Elbashir MI, Khalil IF, Alifrangis M, Giha HA. The efficacy of sulfadoxine-pyrimethamine alone and in combination with chloroquine for malaria treatment in rural eastern Sudan: the interrelation between resistance, age and gametocytogenesis. Trop Med Int Health. 2006;11:604–612. doi: 10.1111/j.1365-3156.2006.01616.x. [DOI] [PubMed] [Google Scholar]
- 33.Alifrangis M, Enosse S, Khalil IF, Tarimo DS, Lemnge MM, Thompson R, Bygbjerg IC, Rønn AM. Prediction of Plasmodium falciparum resistance to sulfadoxine/pyrimethamine in vivo by mutations in the dihydrofolate reductase and dihydropteroate synthetase genes: a comparative study between sites of differing endemicity. Am J Trop Med Hyg. 2003;69:601–606. [PubMed] [Google Scholar]
- 34.Anderson TJ, Nair S, Jacobzone C, Zavai A, Balkan S. Molecular assessment of drug resistance in Plasmodium falciparum from Bahr El Gazal province, Sudan. Trop Med Int Health. 2003;8:1068–1073. doi: 10.1046/j.1360-2276.2003.01144.x. [DOI] [PubMed] [Google Scholar]
- 35.National Malaria Control Program . The National Protocol for Treatment of Malaria. Khartoum, Sudan: Federal Ministry of Health; 2004. [Google Scholar]
- 36.Alker AP, Mwapasa V, Meshnick SR. Rapid real-time PCR genotyping of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2004;48:2924–2929. doi: 10.1128/AAC.48.8.2924-2929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson PE, Kazadi W, Kamwendo DD, Mwapasa V, Purfield A, Meshnick SR. Prevalence of pfcrt mutations in Congolese and Malawian Plasmodium falciparum isolates as determined by a new Taqman assay. Acta Trop. 2005;93:97–106. doi: 10.1016/j.actatropica.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 39.Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998;89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- 40.Menegon M, Sannella AR, Majori G, Severini C. Detection of novel point mutations in the Plasmodium falciparum ATPase6 candidate gene for resistance to artemisinins. Parasitol Int. 2008;57:233–235. doi: 10.1016/j.parint.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization . In vitro Micro-Test (MARK III) for the Assessment of the Response of Plasmodium falciparum to Chloroquine, Mefloquine, Quinine, Amodiaquine, Sulfadoxine/Pyrimethamine and Artemisinin. WHO/MAL/97.20; 2001. [Google Scholar]
- 42.Malik EM, Mohamed TA, Elmardi KA, Mowien RM, Elhassan AH, Elamin SB, Mannan AA, Ahmed ES. From chloroquine to artemisinin-based combination therapy: the Sudanese experience. Malar J. 2006;5:65. doi: 10.1186/1475-2875-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osman ME, Mockenhaupt FP, Bienzle U, Elbashir MI, Giha HA. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum. Infect Genet Evol. 2007;7:52–59. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- 45.Borrmann S, Binder RK, Adegnika AA, Missinou MA, Issifou S, Ramharter M, Wernsdorfer WH, Kremsner PG. Reassessment of the resistance of Plasmodium falciparum to chloroquine in Gabon: implications for the validity of tests in vitro vs. in vivo. Trans R Soc Trop Med Hyg. 2002;96:660–663. doi: 10.1016/s0035-9203(02)90345-7. [DOI] [PubMed] [Google Scholar]
- 46.Noedl H, Wongsrichanalai C, Wernsdorfer WH. Malaria drug-sensitivity testing: new assays, new perspectives. Trends Parasitol. 2003;19:175–181. doi: 10.1016/s1471-4922(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 47.Daily JP, Roberts C, Thomas SM, Ndir O, Dieng T, Mboup S, Wirth DF. Prevalence of Plasmodium falciparum pfcrt polymorphisms and in vitro chloroquine sensitivity in Senegal. Parasitology. 2003;126:401–405. doi: 10.1017/S0031182003002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas SM, Ndir O, Dieng T, Mboup S, Wypij D, Maguire JH, Wirth DF. In vitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from Senegal. Am J Trop Med Hyg. 2002;66:474–480. doi: 10.4269/ajtmh.2002.66.474. [DOI] [PubMed] [Google Scholar]
- 49.Chen N, Russell B, Fowler E, Peters J, Cheng Q. Levels of chloroquine resistance in Plasmodium falciparum are determined by loci other than pfcrt and pfmdr1. J Infect Dis. 2002;185:405–407. doi: 10.1086/338470. [DOI] [PubMed] [Google Scholar]
- 50.Laufer MK, Plowe CV. Withdrawing antimalarial drugs: impact on parasite resistance and implications for malaria treatment policies. Drug Resist Updat. 2004;7:279–288. doi: 10.1016/j.drup.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 52.Tahar R, Ringwald P, Basco LK. Molecular epidemiology of malaria in Cameroon. XXVIII. In vitro activity of dihydroartemisinin against clinical isolates of Plasmodium falciparum and sequence analysis of the P. falciparum ATPase 6 gene. Am J Trop Med Hyg. 2009;81:13–18. [PubMed] [Google Scholar]
- 53.Dahlström S, Veiga MI, Ferreira P, Mårtensson A, Kaneko A, Andersson B, Björkman A, Gil JP. Diversity of the sarco/endoplasmic reticulum Ca(2+)-ATPase orthologue of Plasmodium falciparum (PfATP6) Infect Genet Evol. 2008;8:340–345. doi: 10.1016/j.meegid.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 54.A-Elbasit IE, Alifrangis M, Khalil IF, Bygbjerg IC, Masuadi EM, Elbashir MI, Giha HA. The implication of dihydrofolate reductase and dihydropteroate synthetase gene mutations in modification of Plasmodium falciparum characteristics. Malar J. 2007;6:108. doi: 10.1186/1475-2875-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mockenhaupt FP, Bousema JT, Eggelte TA, Ehrhardt S, Otchwemah RN, Sauerwein RW, Bienzle U. Concurrence of Plasmodium falciparum dhfr and crt mutations in northern Ghana. Malar J. 2005;4:42. doi: 10.1186/1475-2875-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mockenhaupt FP, Eggelte TA, Till H, Bienzle U. Plasmodium falciparum pfcrt and pfmdr1 polymorphisms are associated with the pfdhfr N108 pyrimethamine-resistance mutation in isolates from Ghana. Trop Med Int Health. 2001;6:749–755. doi: 10.1046/j.1365-3156.2001.00792.x. [DOI] [PubMed] [Google Scholar]
- 57.Toteja R, Nair L, Bhasin V. Genome comparison of progressively drug resistant Plasmodium falciparum lines derived from drug sensitive clone. Mem Inst Oswaldo Cruz. 2001;96:427–433. doi: 10.1590/s0074-02762001000300025. [DOI] [PubMed] [Google Scholar]


