Abstract
Previous studies have demonstrated that the proteasome inhibitor, lactacystin, suppresses cytokine production and induction of other inflammatory mediators by LPS-stimulated macrophages. The purpose of the present studies was to determine the effect of lactacystin upon the function of activated human Jurkat T cells and murine splenocytes. Lactacystin treatment suppressed interleukin (IL)-2, interferon (IFN)γ, and IL-13 production similarly in both activated Jurkat cells and primary splenocytes. Interestingly, lactacystin produced differential effects on IL-4 transcription in the two models. While lactacystin inhibited IL-4 mRNA transcription in primary splenocytes, it induced IL-4 mRNA in a concentration-dependent manner in Jurkat cells. The increase in IL-4 mRNA levels by lactacystin did not correlate with increases in TH2-specific transcription factors, avian musculoaponeurotic fibrosarcoma AS42 oncogene homolog (c-maf) or GATA binding protein 3 (GATA-3). In addition, the binding of both GATA-3 and t-bet to their respective response elements was essentially unchanged by lactacystin treatment in both splenocytes and Jurkat T cells, suggesting the induction of IL-4 is due to other mechanisms. Collectively, the current studies suggest proteasomal activity has differential effects on IL-4 transcription in activated Jurkat cells and primary splenocytes.
Keywords: Jurkat, proteasome, splenocyte, T cell, IL-2, IL-4, IFNγ, IL-13
1. Introduction
The proteasome is a cytoplasmic organelle with multiple protease activities, including chymotrypsin-like (X, LMP7), post-glutamase (Y, LMP2), and trypsin-like (Z, MECL-1) activities. Structurally, proteasomes exist as multi-subunit complexes, consisting of a number of distinct, well-characterized, proteins [1]. The so-called 26S proteasome complex (2.5 MDa) is comprised of a 20S proteasome, which exhibits proteolytic activity, and a 19S proteasome, which provides regulatory functions [2]. Distinct subunits of the proteasome have been shown to be induced by IFNγ, including LMP2, LMP7, and MECL-1 of the 20S proteasome [3, 4].
In addition to its role in antigen processing, the proteasome impacts various other immunological processes. Previous studies from this laboratory have demonstrated that the proteasome regulates numerous macrophage activities, including production of TNFα, IL-6, IL-12 and induction of cyclooxygenase-2 and inducible nitric oxide synthase [5–9]. It has also been reported that proteasome inhibition induces apoptosis and inhibits progression from G0 to S phase in human Jurkat T cells [10–12]. Although lactacystin ultimately causes apoptosis in human T cells, the cells are viable for 8 – 16 hr after administration with onset of apoptosis occurring 24 – 64 hr after treatment [11]. Interestingly, lactacystin inhibits T cell proliferation at lower concentrations than that at which it induces apoptosis suggesting that the effects upon proliferation may be due to mechanisms independent of apoptosis. The purpose of the present studies was to determine the effect of lactacystin upon T cell cytokine production.
2. Materials and methods
2.1. Materials
Lactacystin was purchased from A.G. Scientific (San Diego, CA). All other reagents were purchased from Sigma Chemical Co. (St. Louis, MO).
2.2. Animals and cell culture
C57BL/6 mice, 6 weeks of age, were purchased from Jackson Laboratories (Bar Harbor, ME). Studies requiring animals were conducted in accordance with the Guide for the Care and Use of Laboratory Animals as adopted by the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee (IACUC) at University of Missouri Kansas City. Spleens were isolated aseptically and processed into single-cell suspensions (5 × 106 c/ml). Cells were cultured in RPMI 1640 supplemented with 100 units penicillin/ml, 100 units streptomycin/ml, 50 µM 2-mercaptoethanol (2-ME), and 2% bovine calf serum (BCS). Jurkat E6-1 T cells were purchased from the American Type Culture Collection (Manassas, VA). Jurkat cells were cultured in RPMI 1640 supplemented with 100 units streptomycin/ml, 100 units penicillin/ml, 10 mM nonessential amino acids, 100 mM sodium pyruvate, and 10% BCS.
2.3. Real-time PCR
Jurkat cells (5 × 105 c/ml) or freshly isolated splenocytes (5 × 106 c/ml) were treated with lactacystin as described in the figure legends. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA), following the manufacturer’s protocol. The relative expression levels of the target genes were determined by Sybr Green real-time PCR using predesigned Quantitect Primer Assays (Qiagen, Valencia, CA). Relative mRNA expression for the target genes were normalized to either TATA-binding protein (Jurkat cells) or ribosomal protein L13a (splenocytes) and calculated as previously described [13].
2.4. DNA/protein binding assay (ELISA-based)
GATA-3 and t-bet binding were quantified using the TransAm GATA and t-bet assay kits respectively from Active Motif (Carlsbad, CA), following the manufacturer’s protocol. The wild-type competitor (provided by the manufacturer) is an unlabeled consensus oligonucleotide which competes with the biotinylated consensus oligonucleotide to confirm specificity. The mutated competitor is included as an additional control, which should not compete with the biotinylated consensus oligonucleotide.
2.4. Statistical Analysis
The mean ± SE was determined for each treatment group in the individual experiments. Homogeneous data were evaluated by one-way parametric analysis of variance and the Dunnett’s test was used to compare treatment groups to the vehicle (VH) control when significant differences were observed. Statistical analyses were performed using SigmaStat 3.0 (Systat Software, Inc., San Jose, CA).
3. Results
3.1. Effect of the proteasome inhibitor, lactacystin, upon cytokine production by Jurkat T cells and primary splenocytes
To determine the role of the proteasome in the induction of cytokine production in activated T cells and splenocytes, the cells were pretreated with lactacystin (1–10 µM) prior to activation. Interestingly, lactacystin treatment produced differential effects upon IL-4 transcription in activated Jurkat T cells and primary splenocytes. While lactacystin inhibited IL-4 production in freshly isolated splenocytes, a concentration-dependent induction of IL-4 by lactacystin occurred in activated Jurkat cells (Fig. 1). The highest concentration of lactacystin (10 µM) induced IL-4 transcription approximately 40-fold over background levels in Jurkat cells. In contrast to IL-4, lactacystin had similar effects upon IL-13 transcription in activated Jurkat T cells and primary splenocytes with 10 µM lactacystin producing approximately 60–70% inhibition. Likewise, lactacystin produced similar effects upon TH1 cytokines in Jurkat cells and splenocytes (Fig. 2). At 10 µM, lactacystin had a more potent effect upon IL-2 transcription (approximately 75–80 % inhibition) as compared to IFNγ transcription (55 – 65% inhibition). While direct comparisons in overall cytokine induction between splenocytes and Jurkat cells were not measured, the data suggest that the magnitude of induction of TH2 cytokines is greater in splenocytes compared to Jurkat cells, while induction of TH1 cytokines is greater in Jurkat cells.
Fig. 1.
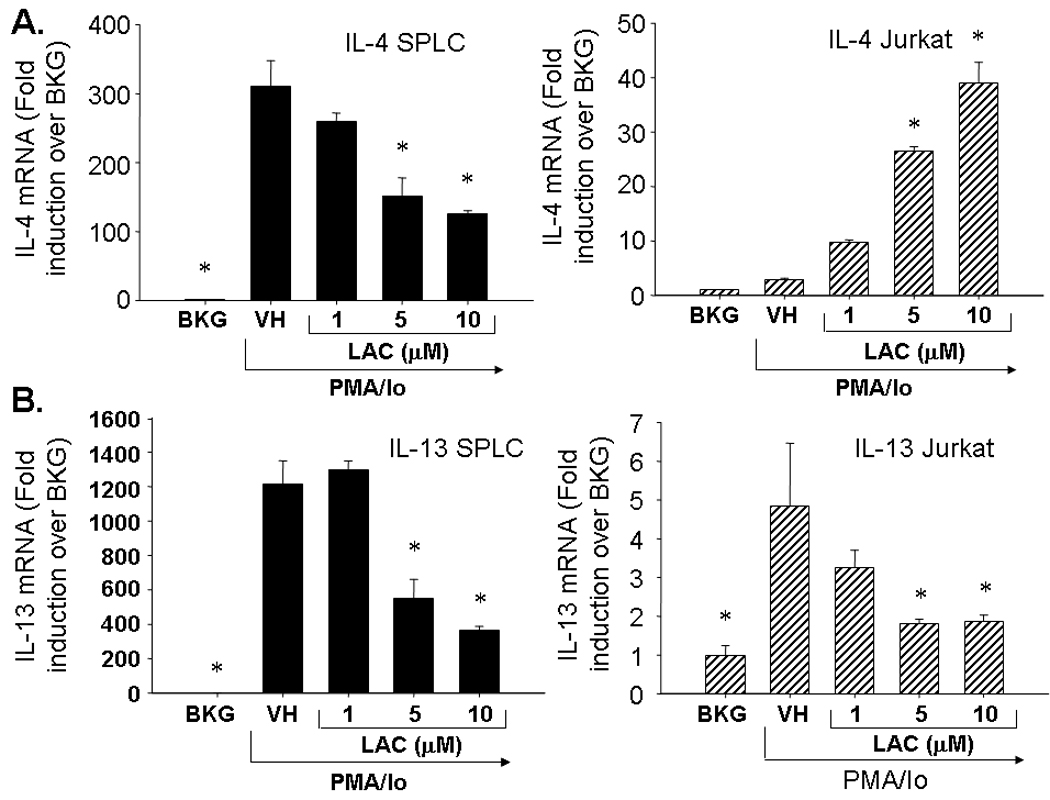
Effect of lactacystin upon PMA/Io-stimulated TH2 cytokine production in activated Jurkat T cells and primary splenocytes. Jurkat T cells (5 × 105 cells/ml) and murine splenocytes (5 × 106 cells/ml) were treated with lactacystin (1–10 µM) or vehicle (VH, 0.1% ethanol) for 30 min followed by activation of the cells with 40 nM/0.5 µM PMA/ionomycin (PMA/Io). Cells were harvested 6h later, the RNA isolated and analyzed for either A.) IL-4 or B.) IL-13. Cellular viability was ≥ 80% for all treatment groups as assessed by trypan blue exclusion. The results are presented as fold induction over BKG and are the mean ± standard error of triplicate cultures. * denotes p<0.05 compared to the VH group as determined by two-tailed Dunnett’s analysis.
Fig. 2.
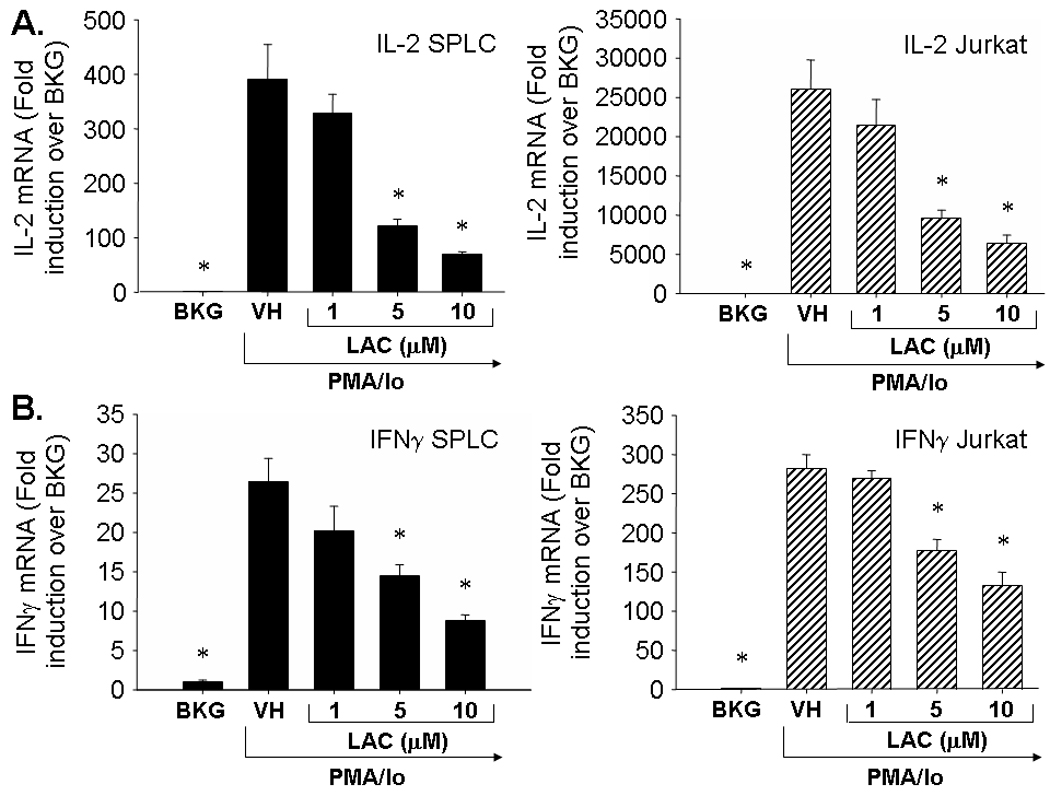
Effect of lactacystin upon PMA/Io-stimulated TH1 cytokine production in activated Jurkat T cells and primary splenocytes. Jurkat T cells (5 × 105 cells/ml) and murine splenocytes (5 × 106 cells/ml) were treated with lactacystin (1–10 µM) or vehicle (VH, 0.1% ethanol) for 30 min followed by activation of the cells with 40 nM/0.5 µM PMA/ionomycin (PMA/Io). Cells were harvested 6h later, the RNA isolated, and analyzed for either A.) IL-2 or B.) IFNγ. Cellular viability was ≥ 80% for all treatment groups as assessed by trypan blue exclusion. The results are presented as fold induction over BKG and are the mean ± standard error of triplicate cultures. * denotes p<0.05 compared to the VH group as determined by two-tailed Dunnett’s analysis.
3.2. Effect of lactacystin on TH2 transcription factors in Jurkat cells and freshly isolated splenocytes upon activation
In order to determine whether the differential effects of proteasome inhibition on TH2 cytokines are due to effects on inducible TH2-specific transcription factors, the effects of lactacystin on GATA binding protein 3 (GATA-3) and avian musculoaponeurotic fibrosarcoma AS42 oncogene homolog (c-maf) were assessed. Pretreatment of Jurkat cells with lactacystin suppressed GATA-3 transcription. In contrast, there was a trend toward increased c-maf transcription with 5 µM lactacystin, although the difference was not statistically significant (Fig. 3). In primary splenocytes, lactacystin had little effect on GATA-3 transcription and modestly increased c-maf transcription, suggesting the differential effects of lactacystin upon IL-4 transcription are independent of c-maf and GATA-3 induction.
Fig. 3.
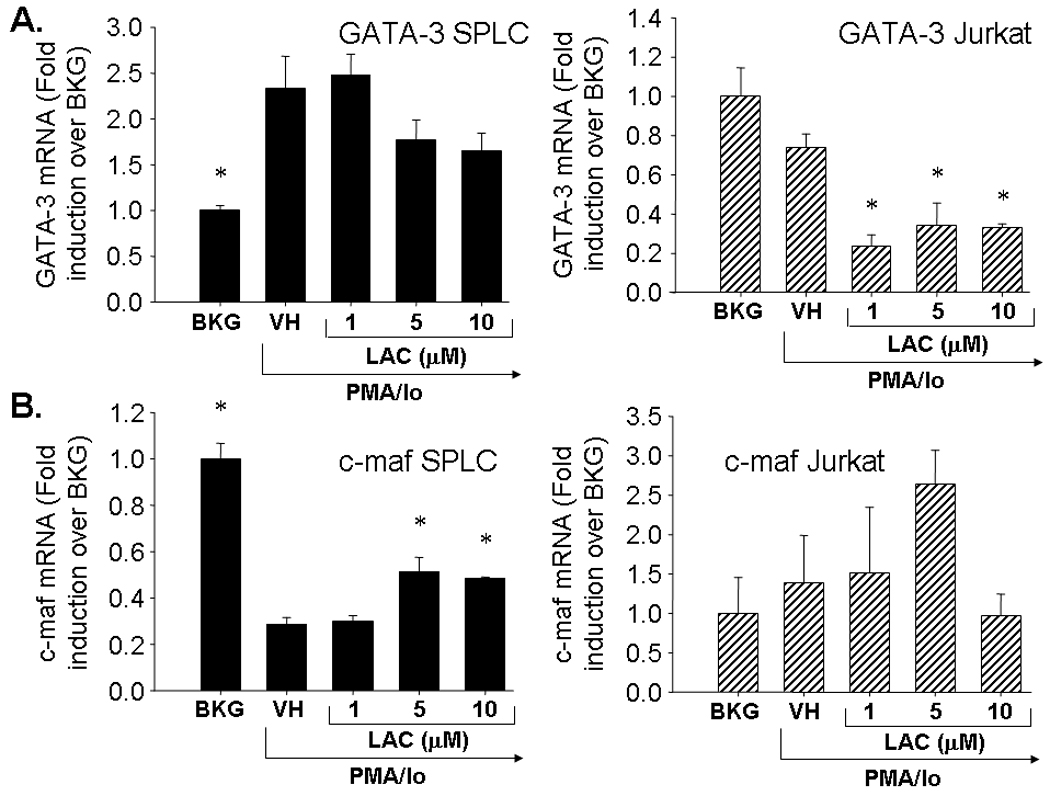
Effect of lactacystin upon PMA/Io-stimulated TH2 cytokine production in activated Jurkat T cells and primary splenocytes. Jurkat T cells (5 × 105 cells/ml) and murine splenocytes (5 × 106 cells/ml) were treated with lactacystin (1–10 µM) or vehicle (VH, 0.1% ethanol) for 30 min followed by activation of the cells with 40 nM/0.5 µM PMA/ionomycin (PMA/Io). Cells were harvested 6h later, the RNA isolated, and analyzed for either A.) GATA-3 or B.) c-maf. Cellular viability was ≥ 80% for all treatment groups as assessed by trypan blue exclusion. The results are presented as fold induction over BKG and are the mean ± standard error of triplicate cultures. * denotes p<0.05 compared to the VH group as determined by two-tailed Dunnett’s analysis.
3.3. Effect of lactacystin on GATA-3 and t-bet DNA binding activity in Jurkat cells and freshly isolated splenocytes upon activation
In order to determine whether proteasome inhibition impacts GATA-3 activity, the effects of lactacystin on GATA-3 DNA binding were assessed in activated Jurkat cells and murine splenocytes. As shown in Fig. 4a, the wild-type competitor successfully competed with labeled GATA-3 consensus sequence, whereas the mutant competitor did not, confirming the specificity of the assay. Lactacystin treatment had little effect on GATA-3 binding in activated splenocytes, which was consistent with the GATA-3 transcription which was also unaffected by lactacystin (Figs. 3–4). The effects of lactacystin on GATA-3 binding in Jurkat cells were tested at multiple time points (2.5 and 6h after PMA/Io activation). GATA-3 binding was essentially unchanged at either time point tested, suggesting that the effect of lactacystin on IL-4 is independent of GATA-3. Whereas GATA-3 transcription in Jurkat cells was significantly inhibited by lactacystin 6h after activation, GATA-3 activity was not. This is likely because the effects on transcription will precede effects on translation and ultimately, activity.
Fig. 4.
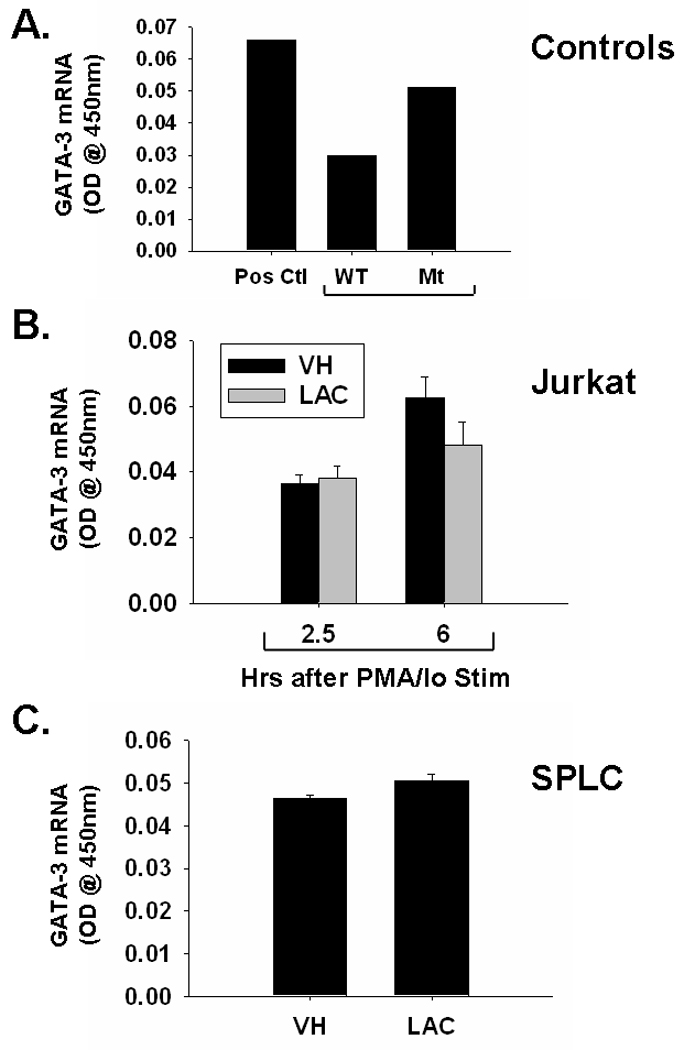
Effect of lactacystin upon GATA-3 DNA binding in activated Jurkat T cells and primary splenocytes. A.) GATA-3 binding by activated splenocytes (Pos. Ctl) is reduced by unlabeled wild-type competitor (WT Comp), but not mutant competitor (Mt Comp). B.) Jurkat T cells (5 × 105 cells/ml) and C.) murine splenocytes (5 × 106 cells/ml) were treated with lactacystin (10 µM) or vehicle (VH, 0.1% ethanol) for 30 min followed by activation of the cells with 40 nM/0.5 µM PMA/ionomycin (PMA/Io). Cells were harvested 2.5h or 6h later (only 6h for SPLC), the nuclear protein isolated, and DNA binding of GATA-3 was quantified by TF binding ELISA as described in the Methods section. The results are presented as optical density at 450nm and are the mean ± standard error of triplicate cultures (with the exception of the control samples in panel A, which were not run in triplicate).
Because t-bet is known to suppress IL-4 transcription, the effect of lactacystin on t-bet activity was also assessed [14]. The inhibition of binding by the wild-type, but not the mutant, competitor confirms the specificity of the assay. Similar to GATA-3, lactacystin had little effect on binding of t-bet to its consensus sequence in either splenocytes or Jurkat cells, suggesting the differential effects of lactacystin on IL-4 mRNA levels are independent of t-bet activity (Fig 5).
Fig. 5.
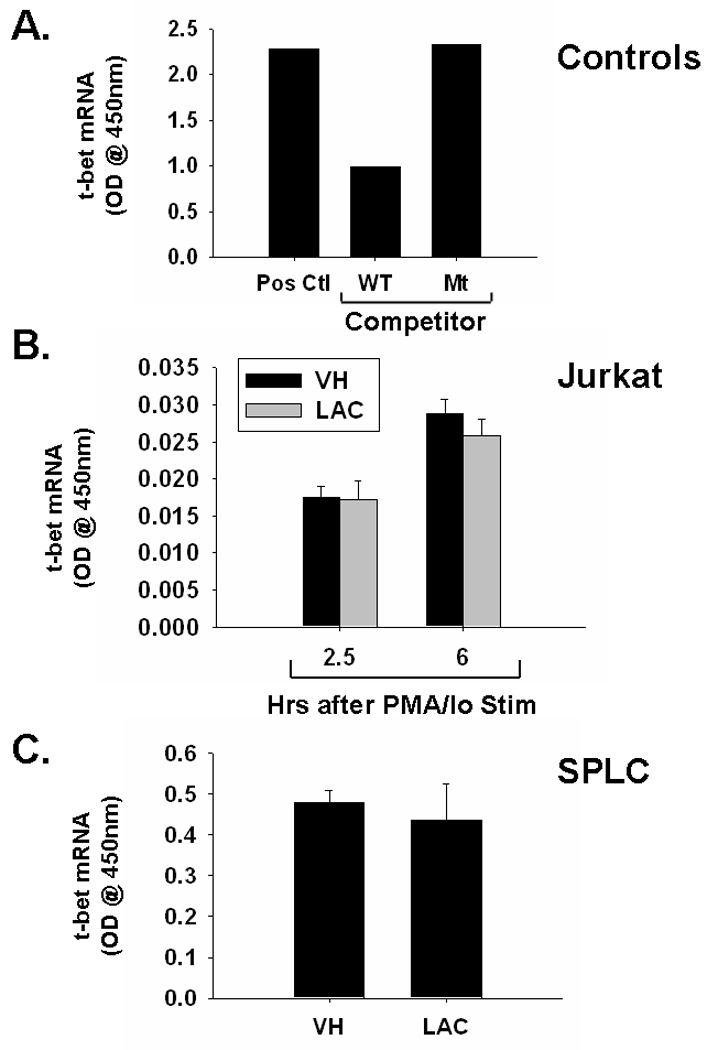
Effect of lactacystin upon t-bet DNA binding in activated Jurkat T cells and primary splenocytes. A.) t-bet binding in t-bet transfected Cos-7 cells (Pos. Ctl) is reduced by unlabeled wild-type competitor (WT Comp), but not mutant competitor (Mt Comp). B.) Jurkat T cells (5 × 105 cells/ml) and C.) murine splenocytes (5 × 106 cells/ml) were treated with lactacystin (10 µM) or vehicle (VH, 0.1% ethanol) for 30 min followed by activation of the cells with 40 nM/0.5 µM PMA/ionomycin (PMA/Io). Cells were harvested 2.5h or 6h later (only 6h for SPLC), the nuclear protein isolated, and DNA binding of GATA-3 was quantified by TF binding ELISA as described in the Methods section. The results are presented as optical density at 450nm and are the mean ± standard error of triplicate cultures (with the exception of the control samples in panel A, which were not run in triplicate).
4. Discussion
The present studies are the first to demonstrate the differential effects of the proteasome inhibitor, lactacystin, upon IL-4 mRNA levels in activated Jurkat cells and primary splenocytes. Whereas lactacystin increased IL-4 mRNA levels in Jurkat cells, it caused a decrease in IL-4 mRNA levels in primary splenocytes, demonstrating that the proteasome contributes to the induction of IL-4 in activated splenocytes, but suppresses IL-4 production in activated Jurkat cells. The aforementioned studies also show that lactacystin has a suppressive effect on IL-2 and IFNγ production in both Jurkat cells and splenocytes, consistent with the reported effects of another proteosome inhibitor, bortezomib, on human CD4+ cells [15]. In addition, lactacystin also suppresses IL-13 transcription in both models.
The current studies suggest that the increase in IL-4 transcription by the proteasome inhibitor, lactacystin, is not due to effects on transcription factors involved with TH1/ TH2 differentiation. This finding was unexpected because such transcription factors are a logical way in which IL-4 could be differentially induced, while production of other cytokines is inhibited. Because many of the other transcription factors involved in IL-4 transcription (e.g. AP-1 and NFAT) also regulate numerous other cytokines, the present data suggest the differential increase in IL-4 mRNA levels may not be due to differences in transcriptional regulation. Alternatively, lactacystin treatment may impact IL-4 mRNA stability.
Stability of mRNA has been shown to be impacted by particular structural features, such as the presence of AU-rich elements in the untranslated region (UTR). Such sequences allow binding of RNA-stabilizing proteins. One such protein is HuR, which has been shown to specifically interact with and stabilize IL-4 mRNA [16, 17]. Furthermore, it has been reported that the binding motif for HuR is found in human IL-4 mRNA. It is not known whether murine IL-4 has this motif. Moreover, HuR is induced in activated human T cells. In addition, it has been shown that IL-4 itself triggers binding of HuR to IL-4 mRNA, which stabilizes IL-4 mRNA.
In addition to HuR, other proteins have been implicated in the stabilization of IL-4 transcripts, including p38 MAP kinase [18]. Treatment of T cells with ionomycin causes induction of p38 phosphorylation, which is calcium-dependent. Inhibition of p38 with SB530280 in ionomycin-treated T cells causes a decrease in IL-4 mRNA stability. Interestingly, proteasome inactivation also induces p38 phosphorylation, which correlates with increased IL-8 mRNA levels in retinal pigment epithelial cells [19].
While great progress has been made in elucidating the sequence of cell signaling events and chromatin remodeling that determine T cell fate and lineage decisions, there is still much that is unknown. While it is clear that the TH2-specific transcription factors, GATA-3 and c-maf, contribute to IL-4 transcription in TH2-differentiated cells and that IL-4 itself is pivotal to differentiation of TH2 cells [20], the cellular and molecular mechanisms of early IL-4 production remain unclear. As such, it is likely that there are other, as yet undescribed mechanisms regulating IL-4, which may include the proteasome. In addition, the marked differences in IL-4 regulation between primary splenocytes and Jurkat T cells, may provide a useful tool in pinpointing some of these mechanisms. Furthermore, with the expansion in the clinical use of proteasome inhibitors in the treatments of different types of lymphoma, it will be necessary to know the effects of proteasomal inhibition upon cytokine production by both malignant as well as non-malignant immune cells.
Whereas the proteasome inhibitor, bortezomib, is currently approved for the treatment of refractory multiple myeloma and mantle cell lymphoma in the U.S., its use in other cancers and a variety of other diseases is currently under investigation. Of relevance to the present study, bortezomib treatment is currently being investigated as a novel therapy for peripheral T cell non-Hodgkins lymphomas [21]. Consistent with the current studies, a case study of a patient with myelodysplasia/myeloproliferative syndrome treated with bortezomib recently reported elevated IL-4 levels in bone marrow, whereas TNFα and IL-6 levels were decreased [22]. Collectively, the current studies and recent clinical observations suggest further studies are needed to fully characterize the effects of bortezomib on IL-4 production in humans.
The present studies are the first to describe the differential effects of proteasome inhibition on cytokine production in Jurkat T cells and primary splenocytes. Specifically, the current studies show that while lactacystin treatment has similar effects upon IL-2, IL-13, and IFNγ in splenocytes and Jurkat cells, IL-4 is differentially affected by lactacystin in the two models. In addition, the aforementioned studies suggest that the role of the proteasome in IL-4 transcription may be dependent upon species and/or differentiation status of the cell population. It is also possible that some other cell type in the splenocyte population (not present in the homogeneous Jurkat T cell population) is exerting a negative effect on IL-4 transcription after lactacystin treatment. While it has long been recognized that numerous factors contribute to the complex regulation of IL-4 in T cells under various activation/differentiation states, the current studies suggest that in addition to these known IL-4 regulators, the proteasome also plays an important role in IL-4 transcription.
Acknowledgments
This work was supported in part by National Institute of General Medicine grant, GM050870. The authors thank Julia Reis, Jing Shen, Shary Shelton, Dr. John Robertson, and Xiaoyu Tan for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirsch C, Ploegh HL. Intracellular targeting of the proteasome. Trends Cell Biol. 2000;10:268–272. doi: 10.1016/s0962-8924(00)01768-2. [DOI] [PubMed] [Google Scholar]
- 2.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, Waters JB, Fruh K, Peterson PA. Proteasomes are regulated by interferon gamma: implications for antigen processing. Proc Natl Acad Sci U S A. 1992;89:4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi N, Perera PY, Shen J, Zhang G, Lenschat A, Splitter G, Morrison DC, Vogel SN. The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J Immunol. 2003;171:1515–1525. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 6.Qureshi N, Vogel SN, Van Way C, 3rd, Papasian CJ, Qureshi AA, Morrison DC. The proteasome: a central regulator of inflammation and macrophage function. Immunol Res. 2005;31:243–260. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 7.Shen J, Gao JJ, Zhang G, Tan X, Morrison DC, Papasian C, Vogel SN, Qureshi N. Proteasome-mediated regulation of CpG DNA- and peptidoglycan-induced cytokines, inflammatory genes, and mitogen-activated protein kinase activation. Shock. 2006;25:594–599. doi: 10.1097/01.shk.0000209555.46704.2d. [DOI] [PubMed] [Google Scholar]
- 8.Shen J, Reis J, Morrison DC, Papasian C, Raghavakaimal S, Kolbert C, Qureshi AA, Vogel SN, Qureshi N. Key inflammatory signaling pathways are regulated by the proteasome. Shock. 2006;25:472–484. doi: 10.1097/01.shk.0000209554.46704.64. [DOI] [PubMed] [Google Scholar]
- 9.Reis J, Tan X, Yang R, Rockwell CE, Papasian CJ, Vogel SN, Morrison DC, Qureshi AA, Qureshi N. A combination of proteasome inhibitors and antibiotics prevents lethality in a septic shock model. Innate Immun. 2008;14:319–329. doi: 10.1177/1753425908096855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshansky V, Wang X, Bertrand R, Luo H, Duguid W, Chinnadurai G, Kanaan N, Vu MD, Wu J. Proteasomes modulate balance among proapoptotic and antiapoptotic Bcl-2 family members and compromise functioning of the electron transport chain in leukemic cells. J Immunol. 2001;166:3130–3142. doi: 10.4049/jimmunol.166.5.3130. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Luo H, Chen H, Duguid W, Wu J. Role of proteasomes in T cell activation and proliferation. J Immunol. 1998;160:788–801. [PubMed] [Google Scholar]
- 12.Lu M, Dou QP, Kitson RP, Smith DM, Goldfarb RH. Differential effects of proteasome inhibitors on cell cycle and apoptotic pathways in human YT and Jurkat cells. J Cell Biochem. 2006;97:122–134. doi: 10.1002/jcb.20543. [DOI] [PubMed] [Google Scholar]
- 13.Farraj AK, Harkema JR, Kaminski NE. Allergic rhinitis induced by intranasal sensitization and challenge with trimellitic anhydride but not with dinitrochlorobenzene or oxazolone in A/J mice. Toxicol Sci. 2004;79:315–325. doi: 10.1093/toxsci/kfh112. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura N, Kitamura F, Kaminuma O, Miyatake S, Tatsumi H, Nemoto S, Mori A. IL-4 gene transcription in human T cells is suppressed by T-bet. Int Arch Allergy Immunol. 2007;143 Suppl 1:68–70. doi: 10.1159/000101408. [DOI] [PubMed] [Google Scholar]
- 15.Berges C, Haberstock H, Fuchs D, Miltz M, Sadeghi M, Opelz G, Daniel V, Naujokat C. Proteasome inhibition suppresses essential immune functions of human CD4+ T cells. Immunology. 2008;124:234–246. doi: 10.1111/j.1365-2567.2007.02761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler NS, Monick MM, Yarovinsky TO, Powers LS, Hunninghake GW. Altered IL-4 mRNA stability correlates with Th1 and Th2 bias and susceptibility to hypersensitivity pneumonitis in two inbred strains of mice. J Immunol. 2002;169:3700–3709. doi: 10.4049/jimmunol.169.7.3700. [DOI] [PubMed] [Google Scholar]
- 17.Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol. 2006;177:4426–4435. doi: 10.4049/jimmunol.177.7.4426. [DOI] [PubMed] [Google Scholar]
- 18.Guo L, Urban JF, Zhu J, Paul WE. Elevating calcium in Th2 cells activates multiple pathways to induce IL-4 transcription and mRNA stabilization. J Immunol. 2008;181:3984–3993. doi: 10.4049/jimmunol.181.6.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes AF, Bian Q, Jiang JK, Thomas CJ, Taylor A, Pereira P, Shang F. Proteasome inactivation promotes p38 mitogen-activated protein kinase-dependent phosphatidylinositol 3-kinase activation and increases interleukin-8 production in retinal pigment epithelial cells. Mol Biol Cell. 2009;20:3690–3699. doi: 10.1091/mbc.E08-10-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal S, Viola JP, Rao A. Chromatin-based regulatory mechanisms governing cytokine gene transcription. J Allergy Clin Immunol. 1999;103:990–999. doi: 10.1016/s0091-6749(99)70168-5. [DOI] [PubMed] [Google Scholar]
- 21.Cheson BD. Novel therapies for peripheral T-cell non-Hodgkin's lymphomas. Curr Opin Hematol. 2009;16:299–305. doi: 10.1097/MOH.0b013e32832ad69a. [DOI] [PubMed] [Google Scholar]
- 22.Terpos E, Verrou E, Banti A, Kaloutsi V, Lazaridou A, Zervas K. Bortezomib is an effective agent for MDS/MPD syndrome with 5q- anomaly and thrombocytosis. Leuk Res. 2007;31:559–562. doi: 10.1016/j.leukres.2006.05.018. [DOI] [PubMed] [Google Scholar]


