Abstract
Certain bacteria have emerged as biological gene vectors with natural tumor specificity, capable of specifically delivering genes or gene products to the tumor environment when intravenously (i.v.) administered to rodent models. We show for the first time that oral administration of bacteria to mice resulted in their translocation from the gastrointestinal tract (GIT) with subsequent homing to and replication specifically in tumors. The commensal, nonpathogenic Bifidobacterium breve UCC2003 harboring a plasmid expressing lux fed to mice bearing subcutaneous (s.c.) tumors were readily detected specifically in tumors, by live whole-body imaging, at levels similar to i.v. administration. Reporter gene expression was visible for >2 weeks in tumors. Mice remained healthy throughout experiments. Cytokine analyses indicated a significant upregulation of interferon-γ (IFN-γ) in the GIT of bifidobacteria-fed mice, which is associated with increases in epithelial permeability. However, B. breve feeding did not increase systemic levels of other commensal bacteria. The presence of tumor was not necessary for translocation to systemic organs to occur. These findings indicate potential for safe and efficient gene-based treatment and/or detection of tumors via ingestion of nonpathogenic bacteria expressing therapeutic or reporter genes.
Introduction
Challenges for oncology practitioners and researchers include the specific treatment and detection of tumors. Genetically-modified, pathogenic, and nonpathogenic bacteria have emerged as potential biological agents with natural tumor specificity.1,2,3,4,5,6 Several bacterial genera (e.g., Escherichia coli, Bifidobacterium, attenuated Salmonella typhimurium, Clostridium, Vibrio cholera, Listeria monocytogenes) have been demonstrated to localize to and replicate in tumor tissue when intravenously (i.v.) administered in rodent models.1,3,4,5,7 Live imaging of mice has shown i.v. administered attenuated pathogenic bacteria encoding light-emitting proteins replicating locally in tumors.5,8,9 Hypoxic or necrotic regions are characteristic of solid tumors in many murine and human cancers. In the case of anaerobic bacteria, tumor targeting has been proposed to be achieved through specific localization of anaerobes such as members of the Bifidobacterium genus to hypoxic regions of tumors following i.v. application.1,3 This property potentially enables targeting of the primary tumor and systemic metastases. By transfection with plasmids that are suitable for bacterial expression of heterologous genes, such bacteria can home to tumors, replicate within them and locally express therapeutic proteins. Most bifidobacteria are a native, harmless resident of the human gut, and certain bifidobacterial strains have been shown to have health-promoting or probiotic benefits.10 A number of bifidobacterial strains that harbor plasmids expressing therapeutic agents, such as endostatin- or prodrug-activating enzymes, have been shown to induce regression in rodent tumor models when administered i.v.11,12,13
No imaging of bifidobacteria in tumors has been published to date, and localization of any bacterial species to tumors has been described only with i.v. administration in preclinical models. In this study, we investigated oral administration of bifidobacteria for the purpose of targeting systemic tumors. The term bacterial translocation refers to trafficking of bacteria from the gastrointestinal tract (GIT), and investigations into this phenomenon are normally confined to pathogenic bacterial sepsis associated with various conditions.14,15 Indeed, several studies have investigated the link between bifidobacterial colonization of the GIT and their ability to inhibit translocation of pathogens.16 Here, using a murine model and employing a lux luminescence-based tagging system, we demonstrate translocation of a nonpathogenic bacterium, Bifidobacterium breve UCC2003, with subsequent homing to and growth specifically in tumors at levels equal to i.v. administration.
Results
In vivo imaging of bifidobacteria in tumors
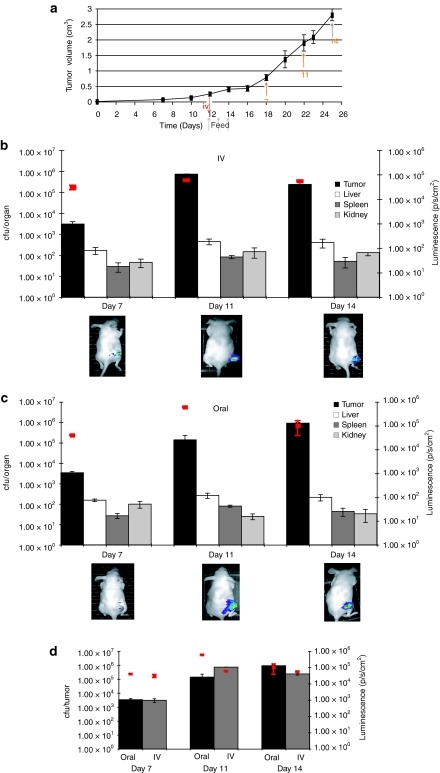
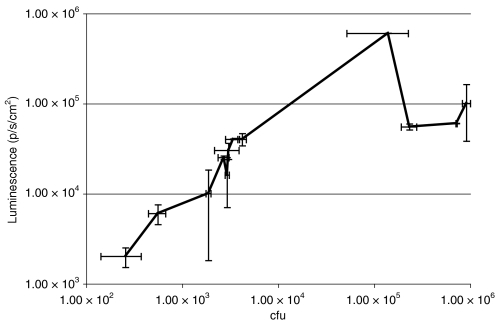
We examined the possibility of imaging bifidobacteria in subcutaneous (s.c.) tumors utilizing B. breve UCC2003 carrying the plasmid pLuxMC3, which expresses the luminescent bacterial luxABCDE operon under the control of the bacterial Phelp promoter.17 We administered this strain by tail vein injection to athymic MF1 nu/nu mice bearing s.c. B16-F10 murine melanoma tumors (Figure 1). Bacterial luminescence was readily detected specifically in s.c. tumors by live whole body imaging (IVIS) (Figure 1b). Ex vivo bacterial quantification by culture of tumors and other organs on selective agar confirmed the presence of UCC2003 in all tissues examined. Numbers of B. breve were low in organs [ ≤100 colony forming units (cfu)/organ] relative to the bacterial numbers present in tumor, and decreased over time. In contrast, high-level replication was observed in tumors, reaching cfu levels of 106 tumor by day 14 (Figure 1b). Tumor volume curves indicated no significant difference in tumor growth rates between bifidobacterial administered mice and controls (data not shown). Live imaging of luminescence from lux-expressing bifidobacteria provided a robust system for indication of bacterial numbers in tumor masses as well as providing evidence of plasmid gene expression (Figure 2).
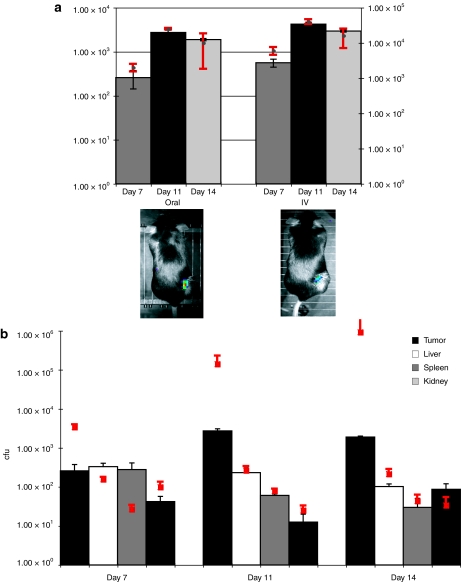
Figure 1.
Administration of Bifidobacterium to tumor bearing mice. (a) Protocol for animal trials. Subcutaneous tumors were induced in mice, and bifidobacteria administered upon tumor development. For oral administration, each animal received 109 B. breve UCC2003 on three consecutive days (time points indicated on graph). For i.v. administration, each animal received 104 cells injected directly into the lateral tail vein. Mice were imaged (IVIS) immediately before culling with subsequent recovery of viable bacteria (cfu) from tissues (orange arrows). (b,c) Trafficking of UCC2003 to subcutaneous tumors B. breve UCC2003 following either (b) intravenous or (c) oral delivery, displaying recovery from s.c. tumor tissue and other organs (bars, y-axis) and lux expression in vivo in live MF1 nu/nu mice (red squares, z-axis and images). Increase in bacterial numbers and plasmid gene expression specifically in tumors was observed over time. There was no detectable luminescence in organs of treated animals. (d) Comparative recovery of UCC2003 from tumors following either oral or intravenous delivery UCC2003 cfu counts were similar on day 7 (P = 0.360), with i.v. counts higher on day 11 (P = 0.038) and oral counts higher on day 14 (P = 0.021). cfu, colony forming units; i.v., intravenous; s.c., subcutaneous.
Figure 2.
Correlation between bacterial counts and luminescence. A linear relationship between luminescence and bacterial counts was observed up to 105 cfu, whereas luminescence underestimated higher bacterial concentrations in tumor. Because the activity of bioluminescence imaging systems are dependent on both ATP and oxygen availability,57,58 lower luminescence (than expected based on bacterial cell counts) may be the result of decreased metabolic activity and/or reduced oxygen availability at higher bacterial concentrations and/or larger more hypoxic tumors. ATP, adenosine triphosphate; cfu, colony forming units.
B. breve UCC2003 translocates from the GIT, and colonizes tumors at levels equal to i.v. administration
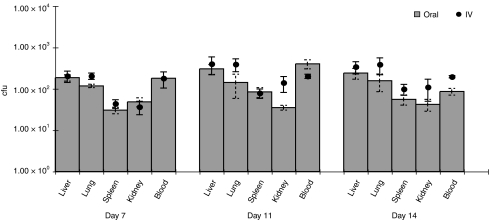
Previous studies have reported bacterial localization to tumors only following i.v. administration.11,12,13 We have previously shown that oral feeding of B. breve UCC2003 results in colonization of the caecum of immunocompetent mice for at least 1 month.18 We repeated the above experiments, but using oral administration rather than i.v. An analogous pattern of tumor and organ colonization was observed (Figure 1c), indicating migration of B. breve from the GIT and systemic spread to internal tissues. Comparisons indicated that oral administration was as effective in achieving targeted tumor-located growth as the well-described i.v. route, using our optimized protocols, and indeed, maximum tumor cfu levels were observed with orally administered mice, on day 14 (Figure 1d). No significant differences were observed between oral and i.v. administration in organs (P > 0.058) (Figure 3). In order to establish that the phenomenon was not tumor-type specific, we repeated the study in MF1 nu/nu mice bearing s.c. MCF7 human breast tumor xenografts or C57 mice bearing s.c. Lewis lung carcinoma tumors and observed similar results (data not shown).
Figure 3.
Systemic localization of UCC2003 administered orally or intravenously. Bacterial counts (cfu) in organs were compared between oral and i.v. administered tumor-free athymic mice. Organ colonization was similar in all cases (P > 0.072). cfu, colony forming units; i.v., intravenous.
Pattern of B. breve UCC2003 growth within tumors
UCC2003 levels within central or peripheral regions of s.c. tumors were assessed utilizing quantitative PCR (specific for lux) on total DNA isolated from tissue taken from either the periphery or centre of 1 cm3 tumors resected from athymic mice 11 days postfeeding. Vector was detectable only in the central region, at 7.28 × 108 (±1.87 × 107) bacterial cells/µg total DNA, suggesting preferential growth within anaerobic regions (data not shown).
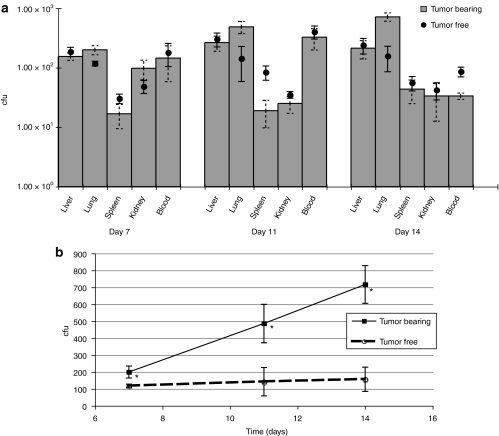
The B16 tumor model spontaneously metastasizes from s.c. tumors to lungs. Significantly increasing bacterial load was observed over time in lungs of tumor-bearing athymic mice fed UCC2003 (P < 0.045), indicating bifidobacterial colonization and replication in small (potentially prehypoxic) metastatic nodules (Figure 4b). No increase in luminescence was observed, suggesting these bacterial levels were below the luminescence detection limit.
Figure 4.
Influence of tumor on systemic bifidobacterial spread cfu levels in blood and organs were compared between tumor-bearing and tumor-free athymic mice orally administered UCC2003. (a) The presence of tumor did not significantly alter levels of cfu recovery from liver, kidney, or blood (P > 0.240). (b) Bacterial colonization of B16 pulmonary metastases. Significantly increasing bacterial load was observed over time in lungs of tumor-bearing animals (*day 7 P = 0.045, day 11 P = 0.030, day 14 P = 0.006) indicating bifidobacterial colonization and replication in metastatic nodules. cfu, colony forming units; i.v., intravenous.
Immune responses to vector
Comparison between bacterial levels in athymic and immunocompetent C57 mice indicated the involvement of T cells in bifidobacterial containment in tumors, whereas translocation to organs occurred at similar levels (P > 0.107) (Figure 5). UCC2003 levels in tumors were significantly increased specifically in tumors in the absence of T cells, with both oral and i.v. administration. To investigate the effect of UCC2003 on T cell levels in tumors, flow cytometry analysis was used to quantify CD3+ T cells in s.c. tumors from C57 mice orally administered UCC2003 or phosphate-buffered saline (PBS) (Figure 6a). A 25.8 (±5.8) % increase in total T cell numbers was observed in UCC2003 colonized tumors 11 days postfeeding, compared with unfed mice (P = 0.06, n = 3). Nonetheless, even in immunocompetent animals, bacteria were readily detected by IVIS imaging specifically in tumors at levels comparable to i.v. administration (Figure 5a). The finding that organ cfu levels were not higher in athymic mice suggests that T cells do not inhibit translocation of bifidobacteria, but do significantly inhibit tumor-associated replication locally.
Figure 5.
Bifidobacterial translocation in immunocompetent mice. (a) B. breve colonization of tumors after oral or i.v. administration to immunocompetent animals. B. breve UCC2003 recovery (y-axis) and luminescence (z-axis) from B16-F10 tumors following either oral or intravenous delivery to immunocompetent C57 mice. Bacterial levels in tumors were comparable whether UCC2003 was orally or i.v administered. In all cases, detectable luminescence and significantly higher bacterial recovery (P < 0.003) were observed in tumors relative to other tissues. Sample IVIS images from day 11 are shown. (b) Comparison between B. breve levels in athymic and immune-competent mice. Following oral administration of UCC2003, there were no significant differences (P > 0.107) observed between cfu levels in organs from C57 mice (bars) and athymic mice (red dots). Tumor cfu levels were significantly higher in athymic mice, with differences of up to 3-log fold by day 14. cfu, colony forming units; i.v., intravenous.
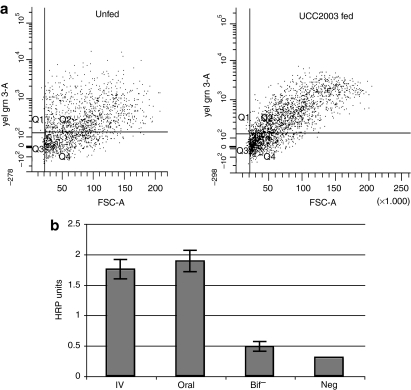
Figure 6.
Cellular and humoral immune responses to UCC2003. (a) T cell levels in bifidobacteria-colonized tumors. Flow cytometry analysis was used to quantify CD3+ T cells in s.c. tumors from C57 mice orally administered UCC2003 or PBS. A 25.8 (±5.8) % increase in total T cell numbers was observed in UCC2003 colonized tumors 11 days postfeeding, compared with unfed mice (P = 0.06, n = 3). (b) Humoral responses. IgG antibody specific for UCC2003 was measured by ELISA from serum taken from immunocompetent C57 mice 14 days postadministration (n = 3). Results indicated that antibodies were raised against UCC2003. There was no significant difference between anti-UCC2003 IgG antibody levels in serum from IV and fed mice (P = 0.359). ELISA, enzyme-linked immunosorbent assay; HRP, horseradish peroxidase; IgG, immunoglobulin G; PBS, phosphate-buffered saline; s.c., subcutaneous.
UCC2003 was found to persist indefinitely in athymic mice (up to 40 cfu/organ and 5 cfu/ml blood at week 5 postfeeding) but was eventually cleared in immunocompetent animals by week 5 after oral administration (data not shown), indicating immune recognition by T cells but the ability to evade other immune effector systems. Immunoglobulin G antibody specific for UCC2003 was detected by enzyme-linked immunosorbent assay in serum taken from immune-competent C57 mice 14 days postadministration (Figure 6b). There was no significant difference between anti-UCC2003 immunoglobulin G antibody levels in serum from IV and fed mice (P = 0.359). Circulating levels of cytokines examined were not significantly increased in serum from fed mice, except for the proinflammatory cytokine interleukin-1β (IL-1β) (946.16 ± 47.31% with respect to unfed mice levels, P < 0.001), suggesting the presence of some, but not a dramatic, degree of immune inflammatory response to systemic UCC2003 cells (data not shown).
Examination of mechanism of gastrointestinal egress
Increased levels of cytokines such as interferon-γ (IFN-γ) in the GIT are associated with increases in epithelial permeability and gut barrier loss in pathogenic bacterial sepsis.19,20 Nonpathogenic bacteria have previously been shown to upregulate cytokines such as IFN-γ and IL-12 in the GIT.21,22 In our experiments, cytokine analyses of IFN-γ, IL-1b, IL-12p70, IL-6, IL-10, tumor necrosis factor–α, and KC levels were performed on GIT samples from UCC2003-fed and PBS-fed B16 tumor-bearing athymic mice. Only IFN-γ levels differed between the groups, with UCC2003 colonized GIT samples found to have significantly higher levels of this cytokine (295 ± 44%, P = 0.003), with respect to unfed mice, suggesting a possible IFN-γ induced increase in epithelial permeability.
In order to examine for the possibility that UCC2003 feeding might result in generic increased translocation of GIT-residing bacteria, we compared systemic bacterial levels between UCC2003-fed and PBS-fed mice. Low levels (5–50 cfu) of indigenous commensal bacteria [subsequently identified as Lactobacilli, Enterobacteriaceae, Staphlococci, Micrococci and Bacteroides by API analysis (BioMérieux, Marcy I'Etoile, France)] were recovered from all organs examined, but not blood, in both immunocompetent and athymic mice (Figure 7). No differences in non-UCC2003 bacterial levels were observed between UCC2003-fed and control (PBS-fed) mice (P > 0.114), except for decreases in Gram− bacteria in lung, liver, and kidney of C57 mice fed UCC2003 (P < 0.044), indicating inhibition of certain species. These data suggest that UCC2003 specifically mediates its own translocation, with no collateral translocation of other commensal bacteria.
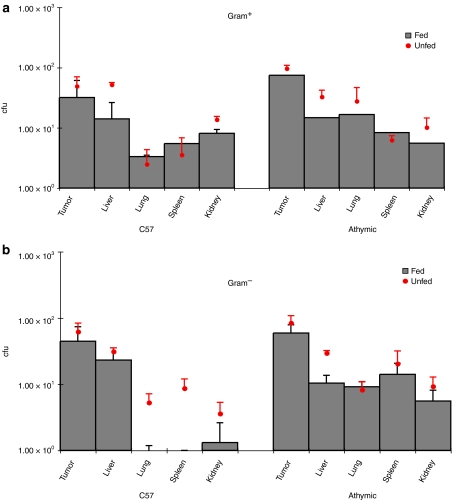
Figure 7.
Recovery of various bacterial genera from murine tissues. Low levels (5–50 cfu) of indigenous commensal bacteria (Lactobacilli, Enterobacteriaceae, Staphlococci, Micrococci, and Bacteroides) were recovered from all organs examined, but not blood, in both immunocompetent and athymic mice. No differences in non-UCC2003 bacterial levels were observed between UCC2003-fed and control (PBS-fed) mice (P > 0.114), except for decreases in Gram− bacteria in lung, liver, and kidney of C57 mice fed UCC2003 (P < 0.044), indicating inhibition of certain species by UCC2003. cfu, colony forming units.
To examine the route of bacterial trafficking from the GIT, various fractions of whole blood were assessed for presence of B. breve. Up to 500 cfu/ml UCC2003 were recovered from serum and not the cellular component of blood, when examined up to day 14 postfeeding, indicating hematogenous trafficking of free bacteria from the GIT, rather than immune cell-mediated phagocytosis and transport (data not shown). Orally administered B. breve numbers in blood remained static over this 14-day period, and were similar to i.v. levels at day 7 (P = 0.296), whereas i.v. UCC2003 numbers decreased over time.
We also assessed whether the presence of a tumor was required for bacterial translocation to occur. When cfu levels in blood and organs were compared between tumor-bearing and tumor-free mice, no significant differences were observed for liver, kidney, and blood (P > 0.240) (Figure 4a), indicating that translocation is not mediated or promoted by some tumor-related factor.
Discussion
We have demonstrated for the first time that a food grade bacterial vector can be ingested, resulting in high-level gene expression over time in systemic tumors. Furthermore, we have developed an imaging system for bifidobacteria which permits their detection in tumors, and consequently the detection of tumors, in real time by luminescence imaging. The nature of the bacterial lux system is such that no exogenous substrate is required for detection. Luminescence is largely uninvestigated at clinical level, as mammalian luminescence reporter genes such as firefly luciferase require additional chemical substrates for function, which are not licensed for clinical use. Thus, the use of bacteria expressing lux in tumors presents a potentially powerful diagnostic clinical tool. Furthermore, this bacterial vector could be engineered to express alternative genes for use with existing clinical diagnostic equipment, such as herpes simplex virus thymidine kinase in combination with positron emission tomography.23
The maximum injectable dose of B. breve UCC2003 cells was used for i.v. injections in our studies. Probably due to exopolysaccharide production by this strain, the viscosity of preparations containing quantities higher than 104 resulted in embolism in mice following tail vein injection. We cannot rule out that if higher quantities could be administered, a proportional increase in tumor bacterial levels may occur. However, given the similarity in B. breve numbers and growth kinetics in tumors between i.v. and oral groups in our studies, it is also plausible that immune containment or nutrient availability are the limiting factors in terms of maximum bacterial levels achieved. In the case of oral administration, doses above 109 per mouse did not increase GIT colonization levels (data not shown). UCC2003 preferentially colonizes the caecum, with peak levels of 106 cfu/g tissue observed 19 days postfeeding, and 105 cfu/g tissue 1 month postfeed.18 It is unlikely that increasing tumor bacterial load over time is due to newly translocated bacteria becoming trapped in tumor microvasculature, as bacterial levels also increased in tumors in i.v. administered mice receiving a single dose of UCC2003. The precise mechanism of tumor-specific bacterial growth has yet to be demonstrated. Several groups have demonstrated homing to and replication in tumors, with many types of bacteria, including bifidobacteria, following i.v. injection.1,2,3,5 Early observations with strictly anaerobic bacteria (clostridia and bifidobacteria) lead to the hypothesis that, unlike normal tissues, the hypoxic environment in tumors provides anaerobic growth conditions.24,25 However, evidence of similar tumor-specific growth of nonanaerobic bacteria, coupled with findings of growth in small, prehypoxic tumors, suggest that lack of oxygen in the necrotic centre of tumors may not be the determining factor for the tumor-specific nature of bacterial growth in these settings.5 In our studies, B. breve UCC2003 was observed to increase over time in lungs containing small (<5 mm3) B16 pulmonary metastatic nodules, also suggesting that hypoxia may not be a requirement for anaerobic bacterial targeting. Although a gross increase in UCC2003 numbers in lungs was shown, we did not demonstrate specific localization of the bacteria within metastatic tumor nodules, and we cannot rule out the possibility of a general enhanced UCC2003 accumulation within lung tissue. Yu et al. proposed that the entry, survival, and replication of bacteria in tumors is dependent on tumor vascularization and the tumor immune microenvironment which provides a sanctuary for a small number of bacteria that will escape the immune system.5 This model involves bacteria entering the tumor's leaky vasculature and escaping the host immunosurveillance due to the immune-privileged nature of solid tumors. Tumor cell line xenografts are known to have different vasculature to spontaneously arising tumors, and investigations in an appropriate spontaneous tumor murine model may yield more clinically relevant data. The nutrient rich environment may also play an important role as evidenced by findings that tumors can support the replication of auxotrophic strains of S. typhimurium.6,26,27 The nature of growth within tumors also appears to be bacterial strain specific. It has been reported for many species, including strains of S. typhimurium, that the bacterial vector growth was confined to the central necrotic regions of tumors whereas in contrast, the S. typhimurium A1 strain has been shown to grew throughout the tumor, including viable malignant tissue in a wide range of tumor models.6,26,27,28,29,30 In our studies, UCC2003 was detected solely in the central necrotic region.
It is unknown why UCC2003 was tolerated by the immune system in our studies. We found that this bacterium persisted indefinitely in athymic mice but was eventually cleared in immunocompetent animals indicating immune recognition by T cells but the ability to evade other immune effector systems. It is noteworthy that bacterial replication was restricted in immune-competent mice specifically in tumors. It is likely that these anti-UCC2003 immune responses are active systemically, but a reduction in organ numbers as evidence by our assays may be masked by the constant influx of newly translocated bacteria, whereas tumor-located bacteria do not exponentially multiply at the same rate in tumors in immune-competent as in athymic mice, resulting in readily measurable differences. The absence of significant upregulation of cytokines characteristic of antibacterial responses in blood further demonstrated immune tolerance to this strain. Other studies in cynomolgus monkeys, dogs, and guinea-pigs have indicated no adverse effects following i.v. administration of Bifidobacterium longum.31 Recent reports indicated differential stimulation of the immune system by bifidobacteria depending on species, and indeed strain, with B. breve shown to have little effect on the immune system, whereas specific B. longum strains influenced the orientation of Th1/Th2 responses differently.32
Fu et al. previously described the use of B. longum to deliver a therapeutic peptide to the GIT33 In that report, B. longum expressing endostatin was administered orally to athymic mice, and the authors reported that subsequent gut absorption of the therapeutic peptide resulted in slowing in the growth of s.c. liver tumors. Translocation of bacteria from the gut to tumors was not investigated or reported in that study. We demonstrated that B. breve UCC2003 survives the upper GIT, colonizes the caecum and translocates to extraintestinal sites. We did not elucidate the precise mechanism of bifidobacterial translocation in these studies. Most bacteria which breach the epithelial barrier are killed by gut-associated lymphoid tissue.14 In healthy animal models in which the intestinal barrier is not physically damaged, indigenous bacteria have been shown to translocate by an intracellular route through the epithelial cells lining the intestines and then travel via the lymph to the mesenteric lymph nodes.34,35 Our data suggest that immune cell uptake or transport was not involved in UCC2003 trafficking as all B. breve bacteria in blood were found in serum only. In animal models exhibiting damage to the mucosal epithelium, indigenous bacteria translocate intercellularly between the epithelial cells to directly access the blood.15 The ability of microorganisms to translocate, survive, and proliferate in extraintestinal tissues involves complex interactions between the host defense mechanisms and the bacterium's ability to invade host tissues. Although the importance of host immune function34,36,37 and the bacterium's intestinal population size34,38,39,40,41 have been implicated as significant contributory factors, the precise mechanisms involved remains unknown.42,43.
Although significant evidence for the translocation of pathogenic bacteria exists,44,45 relatively little information is available on translocation by indigenous species. In animal models, the recovery of Bacteriodes, lactobacilli and enterococci has been reported in healthy pathogen-free mice,15,38,39,46 and Yamazaki et al. also reported B. longum colonization of organs postfeeding.47 Sampling from humans has indicated that bacterial translocation may be a phenomenon that occurs in healthy individuals and may be a normal physiological event without deleterious consequences.48 Clinical studies with probiotic bacteria conducted with healthy subjects have not reported severe disease caused by the bacteria even when shown to translocate from the GIT.42 Lactobacillus, Leuconostoc, Pediococcus, Enterococcus, and Bifidobacterium have been isolated from infected lesions in patients.42,49,50,51 Penn et al.34 reported an increased translocation from the GIT of S-180 tumor-bearing mice, leading to the hypothesis that immune deficiencies associated with progressive tumor growth may be sufficient to permit viable bacteria to translocate from the GIT. We did not observe statistically significant differences in Gram− or Gram+ organ bacterial levels between B16 tumor-bearing and -free athymic or C57 mice, and the presence of tumor was not necessary for UCC2003 translocation to systemic organs to occur.
The population level obtained in the GIT by a particular bacterial species may be a critical factor determining whether or not this bacterial species will translocate to other organs.38 This has been observed for E. coli in the presence of an intact anaerobic flora, where caecal concentrations of >107 E. coli per g are required before this organism reliably translocates to other sites in healthy mice.39,40,41 Our cytokine analyses indicated that UCC2003 colonization altered the local GIT cytokine milieu, perhaps leading to an IFN-γ-induced increase in epithelial permeability. Examination of other commensal bacteria indicate that rather than B. breve inducing a general leakiness of the GIT barrier permitting generic bacterial translocation, UCC2003 specifically mediates its own translocation, with no collateral translocation of other commensal bacteria.
It is unknown whether the observed translocation property is unique to the strain examined. It is plausible that other nonpathogenic, and indeed pathogenic, species could display similar capabilities. We are currently screening a range of both bifidobacterial and non-bifidobacterial species. The B. breve UCC2003 translocated bacteria proved nonpathogenic even in immunocompromised animals and if desired the vector could be easily cleared systemically by antibiotic administration. We have shown that ingestion of these nonpathogenic bacteria carrying a gene of interest, in this case a luminescent reporter, results in high-level expression specifically in tumors. Because up to 106 bacterial cells (with multiple plasmid gene copies/cell) were recovered from tumors, the potential for efficient delivery of genes to the tumor environment compares favorably with existing vectors. Potential for strain optimization exists through reisolating vector with increased efficiencies from experimental tumors, as has been accomplished with other tumor-targeting bacteria.27,26,30 Overall, this strategy represents a novel, safe, and noninvasive vector system, with the potential to deliver therapeutic or diagnostic agents systemically. The route of administration of a therapeutic is important in clinical and commercial settings. Direct intratumoral administration of bacterial vectors restricts usage to accessible tumors, and whereas i.v. administration facilitates targeting of systemic tumor sites, the oral route is amenable to administering much higher doses of bacteria safely (as demonstrated in this study) and is likely to be more appealing when applied to the clinical setting due to ease of drug application. Furthermore, pharmaceutical industry dogma displays a preference for the oral route of administration. Hence, this study opens a new area for oral administration of nonpathogenic bacteria for delivery of therapeutic transgenes.
Materials and Methods
Cell culture. B. breve UCC2003 (UCC Culture Collection) was routinely grown at 37 °C in reinforced clostridial medium (Oxoid, Basingstoke, UK). For bioluminescence assays MRS medium (Oxoid), supplemented with 0.05% (w/v) cysteine-HCl was used. Anaerobic conditions were maintained using an anaerobic chamber [Mac500; Don Whitley Scientific, West Yorkshire, UK (atmosphere 10% H2, 10% CO2, 80% N2)]. B. breve UCC2003/pLuxMC3,18 expressing the luxABCDE operon from the Phelp promoter52 was cultured in the presence of 4 µg/ml chloramphenicol (Cm). To facilitate specific recovery of bifidobacteria from tissue samples, 50 mg mupirocin/l (Oxoid) was included, as previously described.53 B16-F10 (American Type Culture Collection, Manassas, VA) was maintained in Dulbecco's modified Eagle's medium (GIBCO Invitrogen, Paisley, Scotland) supplemented with 10% fetal bovine serum. Cell densities were determined by visual count using a haemocytometer and viable cell counts were conducted using trypan blue dye exclusion (Gibco, Carlsbad, CA).
Animals and tumor induction. All in vivo experiments were approved by the ethics committee of University College Cork (Cork, Ireland). For routine tumor induction, 1 × 106 B16-F10 tumor cells, suspended in 200 µl of serum-free Dulbecco's modified Eagle's medium (Sigma, St Louis, MO), were injected s.c. into the flank of 6–8-week-old female C57 or MF1-nu/nu mice (Harlan Laboratories, Harlan, UK). Tumors were monitored mostly by alternate day measurements in two dimensions using a verniers calliper. Tumor volume was calculated according to the formula V¼ab2P/6, where a is the longest diameter of the tumor and b is the longest diameter perpendicular to diameter a.
Bacterial administration. The inocula were prepared by growing B. breve UCC2003 containing pLuxMC3 anaerobically overnight at 37 °C in 100 ml of MRS broth containing 4 µg/ml Cm. Cultures were harvested by centrifugation (6,000g for 5 minutes), washed with PBS supplemented with 0.05% cysteine-HCl (Sigma), and resuspended in a one-tenth volume of PBS. When tumors reached ~100 mm3 in volume, mice were randomly divided into experimental groups (n = 6) and administered bifidobacteria or an equal volume of PBS as control. For oral administration, 109 cells were administered in 20 µl per mouse on three consecutive days using a micropipette tip placed immediately behind the incisors. For i.v. administration, each animal received 104 cells in 100 µl injected directly into the lateral tail vein, which we found to be the maximum injectable dose with this strain. The viable count of each inoculum was determined by retrospective plating on reinforced clostridial agar containing 4 µg/ml Cm.
Whole body imaging. On day 7, 11, and 14-after first inoculation, animals were anesthetized by intraperitoneal administration of 200 mg xylazine and 2 mg ketamine, and whole-body image analysis was performed in the Xenogen IVIS 100 system (Calipers, Hopkinton, MA) for 5 minutes at high sensitivity.
Bacterial recovery from mice. At defined time points (Figure 1) following whole-body imaging, a subset of animals from each group were euthanized by cervical dislocation. Cardiac puncture was immediately performed to obtain blood, and subsequently individual tumors as well as lungs, liver, spleen, and kidneys were aseptically removed and examined for bioluminescence. Ex vivo IVIS imaging detected no luminescence in organs. Following imaging, each tissue was homogenized by fine mincing with a scalpel followed by pushing through a 20-µm pore nylon filter (Falcon; Becton Dickinson, Oxford, England) in sterile PBS supplemented with 0.05% cysteine-HCl. Serial dilutions were plated in duplicate on reinforced clostridial agar containing 4 µg/ml Cm and mupirocin. Resulting colonies were used to calculate the number of UCC2003 cells per tissue sample. To confirm that cfu recovered were B. breve containing pLuxMC3, random isolates were spot inoculated onto reinforced clostridial agar only or reinforced clostridial agar containing Cm (4 µg/ml). Specific identification of UCC2003 pLuxMC3 was confirmed by PCR using primer pairs targeting both the intergenic spacer region54 and the unique apuB gene on the chromosome of UCC2003 (5′-GGTGTGAAAGTCCATCGCT-3′ and 5′-GTCTGCCAAGGCATCCACCA-3′).55
To facilitate the recovery of other bacteria potentially translocating from the GIT, total tissue homogenates were serially diluted and plated on MRS agar (Oxoid) to select for aerobic Gram+ bacteria and on McConkey agar (Oxoid) to select for Gram− bacteria. Plates were incubated for 24–48 hours and cfu enumerated. Morphology of differing colony types was assessed and the colonies restreaked. Following catalase and Gram reaction tests, five colony types were subjected to API analysis (BioMérieux) to determine species.
DNA extraction and quantitative PCR. Tumors (average volume 1 cm3) were excised from MF1 nu/nu mice 11 days after oral administration of UCC2003/pLuxMC3. Each tumor was aseptically dissected to separate the periphery from the central region. Necrosis was evident in the centre of tumors of this size. Total DNA was extracted from each segment using the Sigma GenElute DNA extraction kit (Sigma) with the following modifications; DNA samples were flash frozen in liquid nitrogen and homogenized; digestion of the tissue was in lysis buffer containing 40 mg/ml proteinase K and incubation was increased to overnight at 55 °C. As a UCC2003-free control, DNA was also extracted from the tumor of a naive mouse. The concentration of DNA in all samples was determined using the Nanodrop system (ND-1000 Spectrophotometer; Labtech, East Sussex, UK), and equal concentrations of DNA added to each PCR. The concentration of the mammalian cell housekeeping gene glyceraldehyde-3-phosphate dehydrogenase was also determined for all samples to ensure parity between PCR template DNA samples. Quantitative PCR was performed using the LightCycler system (Roche Diagnostics, West Sussex, UK). A 20 µl reaction contained 0.5 µmol/l of each sense and antisense primer, 4 mmol/l MgCl2, 2 µl LightCycler FastStart DNA Master SYBR Green, and 100 ng of template DNA. PCR conditions were 95 °C for 10 minutes; 95 °C for 10 seconds, 56 °C for 5 seconds for luxA primers (59 °C for glyceraldehyde-3-phosphate dehydrogenase primers), 72 °C for 5 seconds. Melting curve: 95 °C for 0 seconds, 66 °C for 15 seconds, 99 °C for 0 seconds; cooling step: 40 °C for 30 seconds. The concentration of pLuxMC3 in each sample was determined using a standard curve generated with pLuxMC3 plasmid DNA, and corresponding plasmid copy number calculated using the formula (mass DNA/µg = n bp plasmid/3 × 109 bp). pLuxMC3 has an average copy number/cell of 3 ± 0.18 in UCC2003, and resulting plasmid copy numbers were divided by this figure to calculate bacterial cell numbers/µg DNA.
Flow cytometry analysis. Mouse tumors were excised and finely minced using a scalpel. Tissue was subsequently chemically dissociated in Dulbecco's modified Eagle's medium containing Collagenase I (300 U/ml; Sigma) and DNase (0.01%; Sigma-Aldrich) for 40 minutes at 37 °C and then applied to a cell strainer (70-µm mesh size; Becton Dickinson, Oxford, UK). Red blood cells were lysed using ammonium chloride erythrocyte lysis buffer (distilled H2O, 0.15 mol/l NH4Cl, 10 mmol/l KHC03, 0.1 mmol/l Na2EDTA) and cell counts determined using a NucleoCounter (Chemometec, Allerod, Denmark). Cells at a concentration of 1 × 106 cells per 100 µl were washed in PBS and fixed using 70% ethanol. Fluorescence-activated cell sorting analysis was performed using anti-mouse CD3e-PE-Cy5 (145-2C11), anti-mouse Fc block (CD16/32), and relevant isotype control as per manufacturer instructions (eBioscience, Hatfield, UK). Fluorescence-activated cell sorting staining buffer (PBS, 2% fetal bovine serum, 1% bovine serum albumin, and 0.05% sodium azide) was used as wash and staining buffer. Analysis was performed on a FACSDiva (Becton Dickinson) and analyzed using Becton Dickinson FACSDiva software 6.0 (Becton Dickinson).
Antibody detection. Antibody was measured by an indirect-enzyme-linked immunosorbent assay, as described in Yasui et al.56 with the following modifications. B. breve UCC2003 cells (1 × 108 bacteria/well) were coated on the wells of a 96-well enzyme-linked immunosorbent assay plate using carbonate buffer pH 9.6 and incubated at room temperature overnight. Blood samples were collected at necropsy by cardiac puncture from female C57 BL/6 mice administered UCC2003 either orally (n = 3) or i.v. (n = 3), or untreated mice (n = 3), as previously described. To separate plasma, the samples were centrifuged for 10 minutes at 4,000g within 30 minutes of collection. Plates were blocked for 1 hour at 37 °C by the addition of 10% rabbit serum diluted in PBS containing 0.05% NaN3. After washing three times with PBS containing 0.05% NaN3, 50 µl of mouse serum samples (1:1, 1:10, 1:100 dilutions) in blocking buffer (0.05% Tween 20 and 0.25% BSA in PBS containing 0.05% NaN3) were added to the plates and incubated overnight at room temperature. All samples were assayed in triplicate. The plates were then blocked again by 1 hour incubation at 37 °C in 10% sheep serum, and further incubated with goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Sigma) for 5 hours at room temperature. After incubation with the substrate p-nitrophenylphosphate, qualitative hydrolysis of nitrophenylphosphate was detected using a microtiter plate reader (Vmax; Molecular Devices, Sunnyvale, CA) with a 405-nm filter. Antibody levels are expressed as the means and SE of the values for three mice per route.
Cytokine analysis. MF1 nu/nu mice bearing B16 tumors were sacrificed by cervical dislocation at 11 days postfeeding with B. breve UCC2003 pLuxMC3 or PBS. The blood samples were obtained using cardiac puncture, serum separated by centrifugation and used immediately for the cytokine assay. The entire GIT was also extracted, frozen, and then homogenized on ice in tissue lysis buffer containing a denaturing detergent (<0.1% sodium dodecyl sulfate) and reducing agent (<1 mmol/l dithiothreitol) as well as 1% BSA. The MSD Murine Cytokine 7-plex ultrasensitive panel (Meso Scale Discovery, Gaithersburg, MD) was run as per kit recommendations. This plate analyses murine IFN-γ, IL-1β, IL-6, IL-10, IL-12p70, tumor necrosis factor–α and the mouse keratinocyte-derived chemokine, a functional homolog of human IL-8.
Statistical analysis. Two-tailed Student's t-tests were employed to investigate statistical differences. Microsoft Excel 12 (Microsoft, Redmond, WA) was used to manage and analyze data. Statistical significance was defined at the standard 5% level.
Acknowledgments
We thank Dr Gahan for manuscript appraisal and useful discussions. This work was supported through a grant from the Health Research Board of Ireland RP/2007/75. Potential competing financial interests are declared as a patent application relating to this work has been filed (EP Patent Application No. 09165716.3).
REFERENCES
- Fujimori M, Amano J., and, Taniguchi S. The genus Bifidobacterium for cancer gene therapy. Curr Opin Drug Discov Devel. 2002;5:200–203. [PubMed] [Google Scholar]
- Li Z, Fallon J, Mandeli J, Wetmur J., and, Woo SL. A genetically enhanced anaerobic bacterium for oncopathic therapy of pancreatic cancer. J Natl Cancer Inst. 2008;100:1389–1400. doi: 10.1093/jnci/djn308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vassaux G, Nitcheu J, Jezzard S., and, Lemoine NR. Bacterial gene therapy strategies. J Pathol. 2006;208:290–298. doi: 10.1002/path.1865. [DOI] [PubMed] [Google Scholar]
- Wei MQ, Mengesha A, Good D., and, Anné J. Bacterial targeted tumour therapy-dawn of a new era. Cancer Lett. 2008;259:16–27. doi: 10.1016/j.canlet.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Yu YA, Shabahang S, Timiryasova TM, Zhang Q, Beltz R, Gentschev I, et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol. 2004;22:313–320. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci USA. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey D, O'Sullivan GC., and, Tangney M. Tumour targeting with systemically administered bacteria. Curr Gene Ther. 2010;10:3–14. doi: 10.2174/156652310790945575. [DOI] [PubMed] [Google Scholar]
- Tangney M., and, Gahan CGM. Listeria monocytogenes as a vector for anti-cancer therapies. Curr Gene Ther. 2010;10:46–55. doi: 10.2174/156652310790945539. [DOI] [PubMed] [Google Scholar]
- van Pijkeren JP, Morrissey D, Monk IR, Cronin M, Rajendran S, O'Sullivan GC, et al. 2010A novel Listeria monocytogenes-based DNA delivery system for cancer gene therapy Hum Gene Therepub ahead of print). [DOI] [PubMed]
- Leahy SC, Higgins DG, Fitzgerald GF., and, van Sinderen D. Getting better with bifidobacteria. J Appl Microbiol. 2005;98:1303–1315. doi: 10.1111/j.1365-2672.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- Li X, Fu GF, Fan YR, Liu WH, Liu XJ, Wang JJ, et al. Bifidobacterium adolescentis as a delivery system of endostatin for cancer gene therapy: selective inhibitor of angiogenesis and hypoxic tumor growth. Cancer Gene Ther. 2003;10:105–111. doi: 10.1038/sj.cgt.7700530. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Sasaki T, Fujimori M, Yazawa K, Kano Y, Amano J, et al. Cloned cytosine deaminase gene expression of Bifidobacterium longum and application to enzyme/pro-drug therapy of hypoxic solid tumors. Biosci Biotechnol Biochem. 2002;66:2362–2366. doi: 10.1271/bbb.66.2362. [DOI] [PubMed] [Google Scholar]
- Yi C, Huang Y, Guo ZY., and, Wang SR. Antitumor effect of cytosine deaminase/5-fluorocytosine suicide gene therapy system mediated by Bifidobacterium infantis on melanoma. Acta Pharmacol Sin. 2005;26:629–634. doi: 10.1111/j.1745-7254.2005.00094.x. [DOI] [PubMed] [Google Scholar]
- Balzan S, de Almeida Quadros C, de Cleva R, Zilberstein B., and, Cecconello I. Bacterial translocation: overview of mechanisms and clinical impact. J Gastroenterol Hepatol. 2007;22:464–471. doi: 10.1111/j.1440-1746.2007.04933.x. [DOI] [PubMed] [Google Scholar]
- Berg RD. Bacterial translocation from the gastrointestinal tract. Adv Exp Med Biol. 1999;473:11–30. doi: 10.1007/978-1-4615-4143-1_2. [DOI] [PubMed] [Google Scholar]
- Ishibashi N., and, Yamazaki S. Probiotics and safety. Am J Clin Nutr. 2001;73 2 Suppl:465S–470S. doi: 10.1093/ajcn/73.2.465s. [DOI] [PubMed] [Google Scholar]
- Riedel CU, Monk IR, Casey PG, Morrissey D, O'Sullivan GC, Tangney M, et al. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl Environ Microbiol. 2007;73:3091–3094. doi: 10.1128/AEM.02940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin M, Sleator RD, Hill C, Fitzgerald GF., and, van Sinderen D. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol. 2008;8:161. doi: 10.1186/1471-2180-8-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyer MD, Buurman WA, Hadfoune M, Wolfs T, van't Veer C, Jacobs JA, et al. Exposure to bacterial DNA before hemorrhagic shock strongly aggravates systemic inflammation and gut barrier loss via an IFN-γ-dependent route. Ann Surg. 2007;245:795–802. doi: 10.1097/01.sla.0000251513.59983.3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DM, Watson JL, Wang A, Caldwell J, Prescott D, Ceponis PM, et al. Phosphatidylinositol 3'-kinase is a critical mediator of interferon-γ-induced increases in enteric epithelial permeability. J Pharmacol Exp Ther. 2007;320:1013–1022. doi: 10.1124/jpet.106.113639. [DOI] [PubMed] [Google Scholar]
- Haller D, Blum S, Bode C, Hammes WP., and, Schiffrin EJ. Activation of human peripheral blood mononuclear cells by nonpathogenic bacteria in vitro: evidence of NK cells as primary targets. Infect Immun. 2000;68:752–759. doi: 10.1128/iai.68.2.752-759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller D, Bode C, Hammes WP, Pfeifer AM, Schiffrin EJ., and, Blum S. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M, Sato M, Johnson M, Gambhir SS., and, Wu L. Applications of molecular imaging in cancer gene therapy. Curr Gene Ther. 2005;5:607–618. doi: 10.2174/156652305774964695. [DOI] [PubMed] [Google Scholar]
- Lemmon MJ, van Zijl P, Fox ME, Mauchline ML, Giaccia AJ, Minton NP, et al. Anaerobic bacteria as a gene delivery system that is controlled by the tumor microenvironment. Gene Ther. 1997;4:791–796. doi: 10.1038/sj.gt.3300468. [DOI] [PubMed] [Google Scholar]
- Yazawa K, Fujimori M, Amano J, Kano Y., and, Taniguchi S. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 2000;7:269–274. doi: 10.1038/sj.cgt.7700122. [DOI] [PubMed] [Google Scholar]
- Nagakura C, Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, et al. Efficacy of a genetically-modified Salmonella typhimurium in an orthotopic human pancreatic cancer in nude mice. Anticancer Res. 2009;29:1873–1878. [PubMed] [Google Scholar]
- Zhao M, Yang M, Ma H, Li X, Tan X, Li S, et al. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res. 2006;66:7647–7652. doi: 10.1158/0008-5472.CAN-06-0716. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, et al. Cancer metastasis directly eradicated by targeted therapy with a modified Salmonella typhimurium. J Cell Biochem. 2009;106:992–998. doi: 10.1002/jcb.22078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, et al. Systemic targeting of primary bone tumor and lung metastasis of high-grade osteosarcoma in nude mice with a tumor-selective strain of Salmonella typhimurium. Cell Cycle. 2009;8:870–875. doi: 10.4161/cc.8.6.7891. [DOI] [PubMed] [Google Scholar]
- Zhao M, Geller J, Ma H, Yang M, Penman S., and, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci USA. 2007;104:10170–10174. doi: 10.1073/pnas.0703867104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori M. Genetically engineered Bifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast Cancer. 2006;13:27–31. doi: 10.2325/jbcs.13.27. [DOI] [PubMed] [Google Scholar]
- Ménard O, Butel MJ, Gaboriau-Routhiau V., and, Waligora-Dupriet AJ. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl Environ Microbiol. 2008;74:660–666. doi: 10.1128/AEM.01261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu GF, Li X, Hou YY, Fan YR, Liu WH., and, Xu GX. Bifidobacterium longum as an oral delivery system of endostatin for gene therapy on solid liver cancer. Cancer Gene Ther. 2005;12:133–140. doi: 10.1038/sj.cgt.7700758. [DOI] [PubMed] [Google Scholar]
- Penn RL, Maca RD., and, Berg RD. Increased translocation of bacteria from the gastrointestinal tracts of tumor-bearing mice. Infect Immun. 1985;47:793–798. doi: 10.1128/iai.47.3.793-798.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn RL, Nguyen VQ, Specian RD, Stevens P., and, Berg RD. Interleukin-2 enhances the translocation of Escherichia coli from the intestines to other organs. J Infect Dis. 1991;164:1168–1172. doi: 10.1093/infdis/164.6.1168. [DOI] [PubMed] [Google Scholar]
- Berg RD. Translocation of enteric bacteria in health and disease. Curr Stud Hematol Blood Transfus. 1992. pp. 44–65. [DOI] [PubMed]
- Owens WE., and, Berg RD. Bacterial translocation from the gastrointestinal tract of athymic (nu/nu) mice. Infect Immun. 1980;27:461–467. doi: 10.1128/iai.27.2.461-467.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RD., and, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun. 1979;23:403–411. doi: 10.1128/iai.23.2.403-411.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RD., and, Owens WE. Inhibition of translocation of viable Escherichia coli from the gastrointestinal tract of mice by bacterial antagonism. Infect Immun. 1979;25:820–827. doi: 10.1128/iai.25.3.820-827.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen EK., and, Berg RD. Relationship between cecal population levels of indigenous bacteria and translocation to the mesenteric lymph nodes. Infect Immun. 1983;39:1252–1259. doi: 10.1128/iai.39.3.1252-1259.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen EK, Berg RD., and, Deitch EA. Comparison of translocation rates of various indigenous bacteria from the gastrointestinal tract to the mesenteric lymph node. J Infect Dis. 1988;157:1032–1038. doi: 10.1093/infdis/157.5.1032. [DOI] [PubMed] [Google Scholar]
- Liong MT. Safety of probiotics: translocation and infection. Nutr Rev. 2008;66:192–202. doi: 10.1111/j.1753-4887.2008.00024.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez AV, Baigorí MD, Alvarez S, Castro GR., and, Oliver G. Phosphatidylinositol-specific phospholipase C activity in Lactobacillus rhamnosus with capacity to translocate. FEMS Microbiol Lett. 2001;204:33–38. doi: 10.1111/j.1574-6968.2001.tb10858.x. [DOI] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM., and, Lekkerkerk-van der Wees Colonization resistance of the digestive tract and the spread of bacteria to the lymphatic organs in mice. J Hyg (Lond) 1972;70:335–342. doi: 10.1017/s0022172400022385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CL. Relationship between intestinal microecology and the translocation of intestinal bacteria. Antonie Van Leeuwenhoek. 1990;58:87–93. doi: 10.1007/BF00422722. [DOI] [PubMed] [Google Scholar]
- Berg RD. Bacterial translocation from the intestines. Jikken Dobutsu. 1985;34:1–16. doi: 10.1538/expanim1978.34.1_1. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Machii K, Tsuyuki S, Momose H, Kawashima T., and, Ueda K. Immunological responses to monoassociated Bifidobacterium longum and their relation to prevention of bacterial invasion. Immunology. 1985;56:43–50. [PMC free article] [PubMed] [Google Scholar]
- Sedman PC, Macfie J, Sagar P, Mitchell CJ, May J, Mancey-Jones B, et al. The prevalence of gut translocation in humans. Gastroenterology. 1994;107:643–649. doi: 10.1016/0016-5085(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Pavan S, Desreumaux P., and, Mercenier A. Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin Diagn Lab Immunol. 2003;10:696–701. doi: 10.1128/CDLI.10.4.696-701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancrède CH., and, Andremont AO. Bacterial translocation and Gram-negative bacteremia in patients with hematological malignancies. J Infect Dis. 1985;152:99–103. doi: 10.1093/infdis/152.1.99. [DOI] [PubMed] [Google Scholar]
- Zhou JS, Shu Q, Rutherfurd KJ, Prasad J, Gopal PK., and, Gill HS. Acute oral toxicity and bacterial translocation studies on potentially probiotic strains of lactic acid bacteria. Food Chem Toxicol. 2000;38:153–161. doi: 10.1016/s0278-6915(99)00154-4. [DOI] [PubMed] [Google Scholar]
- Riedel CU, Monk IR, Casey PG, Morrissey D, O'Sullivan GC, Tangney M, et al. Improved luciferase tagging system for Listeria monocytogenes allows real-time monitoring in vivo and in vitro. Appl Environ Microbiol. 2007;73:3091–3094. doi: 10.1128/AEM.02940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Stanton C, Fitzgerald GF., and, Ross RP. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J Bacteriol. 2003;185:2571–2581. doi: 10.1128/JB.185.8.2571-2581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L., and, van Sinderen D. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui H, Mike A., and, Ohwaki M. Immunogenicity of Bifidobacterium breve and change in antibody production in Peyer's patches after oral administration. J Dairy Sci. 1989;72:30–35. doi: 10.3168/jds.S0022-0302(89)79076-7. [DOI] [PubMed] [Google Scholar]
- Moriyama EH, Niedre MJ, Jarvi MT, Mocanu JD, Moriyama Y, Subarsky P, et al. The influence of hypoxia on bioluminescence in luciferase-transfected gliosarcoma tumor cells in vitro. Photochem Photobiol Sci. 2008;7:675–680. doi: 10.1039/b719231b. [DOI] [PubMed] [Google Scholar]
- Wiles S, Pickard KM, Peng K, MacDonald TT., and, Frankel G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect Immun. 2006;74:5391–5396. doi: 10.1128/IAI.00848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]