Abstract
Biosafety and efficacy considerations that impede clinical application of gene therapy could be addressed by nonviral ex vivo cell therapy, utilizing transgenic cells that have been comprehensively pre-evaluated for genotoxic potential and transgene expression. We evaluated the genotoxic potential of phiC31 bacteriophage integrase-mediated transgene integration in cord-lining epithelial cells (CLECs) readily cultured from the outer membrane of human umbilical cords, by sequencing and mapping integration sites, spectral karyotyping, high-resolution genome copy number, transcriptome, and transgene copy number analyses and in vivo tumorigenicity. Of 44 independent integration events, <5% were exonic and 85% of modified cells had integrated ≤2 transgene(s). Expression of 95.6% of genes was unaltered in modified cells. Only three small regions showed genome copy number changes that did not correlate with altered gene expression or integration sites. Spectral karyotyping revealed rare nonrecurrent occurrence of three different translocations. Integrase-modified cells were not tumorigenic in immunocompromised mice for at least 4 months. Stable integration of a human factor VIII (FVIII) construct conferred durable FVIII secretion in vitro. Xenoimplantation of FVIII-secreting CLECs in immunocompetent hemophilic mice achieved significant phenotypic correction. Pre-evaluated clonal populations of phiC31 integrase–modified CLECs could be useful as bioimplants for monogenic diseases such as hemophilia.
Introduction
The umbilical cord is a source of different cell types with stem-like characteristics.1,2 The outer cord lining is a monolayer of amniotic epithelium derived from the embryonic epiblast.3 Primary cells with a cobblestone epithelial morphology readily grow out from outer lining membrane explants in vitro and can be cryopreserved indefinitely.4 These cord-lining epithelial cells (CLECs) express Oct-4, Nanog, and type I and II keratins. We have shown that CLECs can be obtained consistently from the amniotic epithelium in clinically relevant quantities, i.e., about 6 × 109 cells per cord even before passaging in culture,5 a somewhat less limiting yield than cord blood stem/progenitor cells.6,7,8 Moreover, cord-derived cells are readily modified ex vivo to express transgenes stably9 and do not form teratomas in vivo.10 These characteristics, together with their noninvasive and ethically uncontroversial derivation, prompted us to investigate the feasibility of developing CLECs as cellular carriers of therapeutic transgenes.
Nonhematopoietic ex vivo cell therapy to correct specific gene deficiencies may overcome major hurdles of conventional gene therapy that, after >40 years, has yet to become standard of clinical care.11 Unlike the known risks of administering randomly integrating gene transfer vectors in vivo, ex vivo modification of somatic cells could be more efficient and allows stably modified cells to be screened in vitro for evidence of therapeutic efficacy. Stable genomic modification of hematopoietic stem cells by randomly integrating vectors has been shown to be genotoxic and associated with adverse clinical events.12,13 The nature of the expressed transgene and the specific disease treated appear to influence the risk of adverse clinical effects, as no untoward outcomes were noted in similar clinical trials for ADA-deficient SCID.14 Techniques that mediate site-directed transgene integration should mitigate genotoxic risk, particularly if the modification is performed ex vivo. Such an approach enables transgenic cells to be comprehensively evaluated for genotoxic potential, assuming cell characteristics are not altered by the in vivo environment. In the event that adverse genome-disruptive and/or likely oncogenic changes are identified, plans for in vivo implantation of transgenic cells can be abandoned without risking iatrogenic complications.
The bacteriophage phiC31 integrase catalyzes unidirectional integration of plasmid-encoded, attB-bearing transgenes into a limited number of pseudo-attP sites in the human genome.15 PhiC31 integrase–mediated transgene insertion has demonstrated efficacy in animal models, e.g., deficiencies of fumarylacetoacetate hydrolase and dystrophin.16,17 Transgenic animals generated using this technique are healthy and not cancer-prone.18 Human embryonic stem cells retained their ability to differentiate normally into all three germ layers after stable transgene integration mediated by phiC31 integrase.19 Thus, this technique of quasi-site-directed transgenesis would be expected to have superior biosafety than randomly integrating viral vectors.15 Although transgene integration sites have been identified in phiC31 integrase–modified cells, integrase-treated cells have not been comprehensively evaluated for genome-disruptive alterations because insertion of exogenous DNA into chromosomes is potentially mutagenic. This concern merits investigation as microdeletions, microinsertions, and chromosomal rearrangements have been reported at phiC31 integrase insertion sites in the mouse and human genome.15,20 Furthermore, primary fibroblasts from two human embryos co-transfected with phiC31 integrase, and puromycin-resistance expression plasmids showed different breakpoints on Q-banded chromosomes (observed in 3 out of 9 clones), whereas control embryonic fibroblasts transfected without integrase remained cytogenetically normal.21 Chromosomal abnormalities, albeit mainly nonrecurrent, were also detected in primary adult human fibroblasts in a similar study.22
The ready availability and potential of CLECs as cellular carriers of therapeutic transgenes together with emerging techniques for stable site-directed transgene integration motivated us to undertake a comprehensive evaluation of the genotoxicity of phiC31 integrase in primary CLECs. We show here that phiC31 integrase induced minimal genomic and transcriptomic alterations in CLECs. Using hemophilia as a model for cell therapy, we show that phiC31 integrase-mediated integration of a human factor VIII (FVIII) transgene in CLEC induced stable FVIII secretion and corrected the phenotype of FVIII-deficient hemophilic mice.
Results
CLEC characteristics
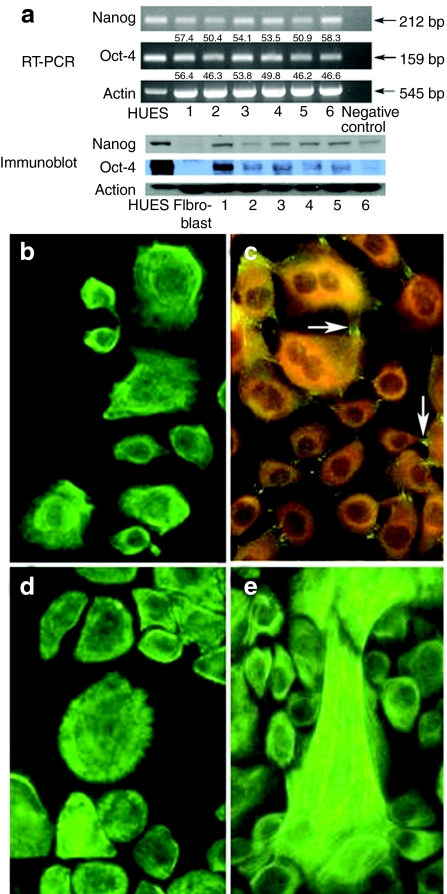
Primary cells cultured from explants of the outer lining membrane of umbilical cords expressed markers of pluripotency, i.e., Oct-4 [159 base pairs (bp)] and Nanog (212 bp) transcripts (at levels ranging from 46 to 56% of a positive control human embryonic stem cell line) and the cognate proteins shown by immunoblots of CLEC protein lysates (Figure 1a). CLECs also expressed epithelial cell markers (universal keratins, keratins 18 and 19) and desmoplakin (Figure 1b–e), and could be propagated for at least 40 passages in vitro without loss of proliferative capacity or multipotency.
Figure 1.
Characterization of primary human umbilical cord–lining epithelial cells (CLECs). (a) RT-PCR and western blot analysis of different CLEC samples (1–6), human embryonic stem cell line (HUES, positive control), and human primary dermal fibroblasts (negative control) for expression of Oct-4 and Nanog. Negative control for RT-PCR was a minus template PCR. Shown below RT-PCR gel images are quantitative levels of Oct-4 and Nanog transcripts (normalized to actin) relative to the HUES sample. Indirect immunofluorescence staining for (b) universal keratins; (c) desmoplakin (positive expression is seen as bright green fluorescence (arrows) and negative expression as dull orange); (d) keratin 18 and (e) keratin 19 in cultured CLECs. Original magnification ×400. bp, base pair.
Assessing genotoxicity of stably modified CLEC
Transfection efficiency and integration frequency. The percentage of green fluorescent protein (GFP) positive cells, measured by fluorescence-activated cell sorting (FACS) analysis, was 33 ± 1.5% and 42 ± 2.5% (mean ± SEM, n = 3) for CLECs electroporated with pEGFPattB plasmid with or without phiC31 integrase plasmid, respectively. The efficiency of gene transfer without electroporation was <1% with or without phiC31 integrase plasmid.
From an initial seeding of 5,000 FACS-sorted EGFP-expressing CLECs (in triplicate), scoring of GFP-expressing colonies after G418 selection for 7 days yielded 134 ± 11 colonies (mean ± SD) when co-transfection was performed with integrase, compared with 8 ± 5 cells when transfection was performed without integrase. Data from parallel seeding of 2,000 FACS-sorted EGFP-expressing CLECs (also in triplicate) showed 68 ± 12 stable integrations with integrase, and 8 ± 4 without integrase. These data indicated an average integration frequency of 3.0% with integrase compared to 0.3% without integrase.
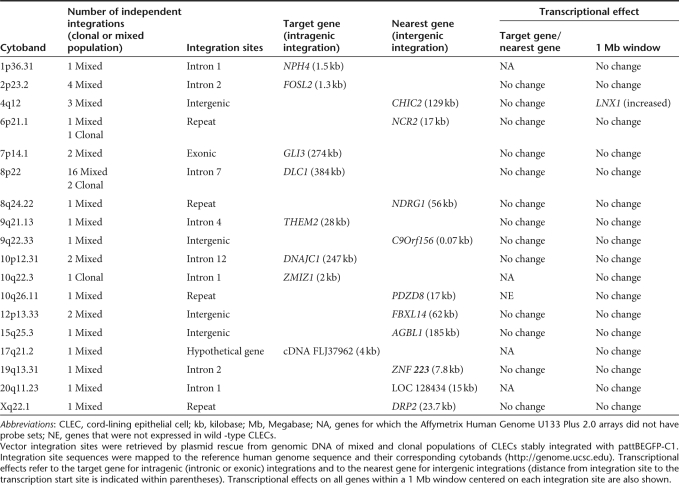
Site-directed integration. We integrated a reporter plasmid, pattBEGFP-C1, into low-passage CLECs by co-expressing phiC31 integrase to determine the profile of integration sites in a primary human diploid cell type. We documented 44 independent integration events that mapped to 18 cytobands by sequencing 90 and 200 plasmid clones from a mixed and clonal population, respectively, of transgenic CLECs (Table 1). An integration was considered to be independent if chromosomal sequences flanking the integration site were different from all other integrations retrieved. Alignment of integration site sequences using the Multiple EM for Motif Elicitation (MEME) program revealed a shared motif among 12 cytobands (E = 5.9 × 10−10) (Supplementary Figure S1) that was 75% identical to a 28-base motif previously identified.15 Integrations into the 8p22 site accounted for >40% of all integrations.
Table 1. PhiC31 integrase–mediated transgene integration sites in CLECs.
We next analyzed genomic site categories of phiC31 integrase-mediated integrations. Of the 44 integration events, 29 were intronic, 7 intergenic, 2 exonic, 5 within repeat elements, and 1 within a hypothetical transcriptional unit. Most integrations were intronic (29 of 31 events) and only 2 were exonic (both were 7p14.1 integrations into exon 15 of GLI3). Moreover, >70% of these integrations were >50 kilobases (kb) away from transcription start sites, unlike retroviral vectors that have a predilection for integrating in close proximity to transcriptional start sites.23 Other reports have suggested that vector integrations close to promoters alter gene expression.24 However, our data showed that the expression of nearly all genes located within a 1 megabase (Mb) window centered on each integration site was comparable to wild-type CLECs. The sole exception was a twofold increase in LNX1 expression, located 476 kb from the 4q12 integration site (Table 1).
Transgene integrations close to or within known oncogenes or tumor suppressor genes would be cause for serious concern. In our study, three independent integration events were within potential oncogenes or tumor suppressor genes (DLC1, FOSL2, and GLI3). Fifteen oncogenes and tumor suppressor genes were located within a 1 Mb window among 44 independent integration sites at a median distance of 224 kb (range 2–463 kb). Despite this, none of these genes showed significantly altered expression by transcriptional profiling.
Transcriptional profile of stably modified CLEC
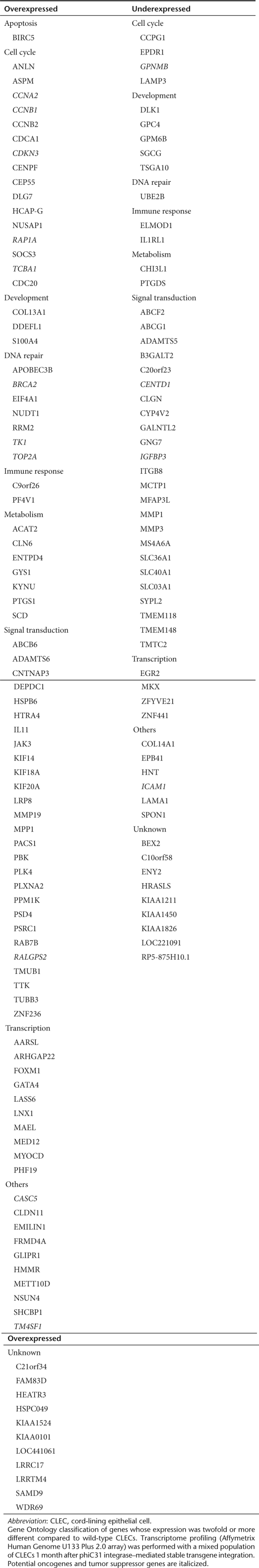
Comparison of the transcriptomes of wild-type and a mixed population of stably integrated CLECs showed no difference in the expression levels of >96.5% transcripts. Of 11,947 CLEC-expressed genes, 94 (0.8%) showed increased expression and 57 (0.5%) showed decreased expression (defined for both as more than twofold difference) in transgenic CLECs compared to wild-type CLECs (Table 2). Functional annotation using DAVID did not reveal significant association of these dysregulated genes to specific pathways except for activation of p53 signaling (CDK2, CCNB1, and IGFBP3) (Fisher's exact P value modified for gene enrichment = 0.02).
Table 2. Transcriptionally altered genes in stably integrated CLECs.
Cross-referencing 151 transcriptionally altered genes in transgenic CLEC identified 15 in a database of 1,650 possible oncogenes and tumor suppressor genes. Of these, three were tumor suppressor genes (BRCA2, RAP1A, and TOP2A) whose increased expression could be expected to promote cell death rather than proliferation. The remaining 12 genes were mainly involved in cell cycle regulation or cell adhesion.
Genomic copy number changes in stably modified CLEC
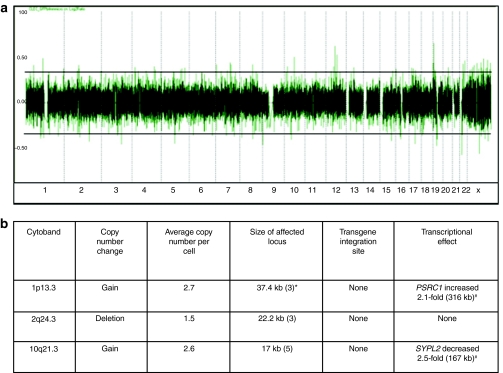
High-resolution genome copy number analyses of genomic DNA of naive and mixed population of stably integrated cells showed that transgene integration into CLECs had quite minimal effects on genome copy number. Modest copy number gain was identified in two loci and copy number loss in one locus—none of which was a transgene integration site (Figure 2a,b). Moreover, genes residing within 1 Mb intervals centered on each copy number change region were unaltered in their expression. We did, however, detect a 2.1-fold and 2.5-fold increase in the expression of PSRC1 and SYPL2, respectively. Given the distance of these genes from the regions of copy number gain, these transcriptional changes were probably independent of copy number gains. The data above are consistent with infrequent copy number changes in stably integrated CLECs, in contrast to multiple changes commonly observed in cancer cells.25
Figure 2.
High-resolution genome copy number analysis. (a) Genome-wide copy number profile of a mixed population of cord-lining epithelial cells (CLECs) stably integrated with pattBEGFP-C1 generated on Affymetrix Human Mapping 500K Array Set. Human chromosomes are shown on the horizontal axis. Log2 ratios are on the vertical axis. Horizontal lines are normal copy number boundaries. Dotted vertical lines demarcate individual chromosomes. (b) Characteristics of copy number change loci. Effects on transcription (twofold or more difference compared to wild-type CLECs) of genes in a 1 Mb window centered on each copy number change locus is shown. *The number of consecutive probe sets for each copy number change locus. #Distance of gene from copy number change locus.
Spectral karyotyping of stably modified CLECs
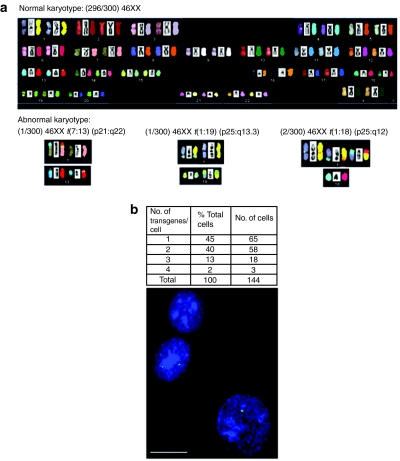
We spectrally karyotyped mixed populations of naive and stably integrated CLECs to determine whether phiC31 integrase induced chromosomal rearrangements. Although no chromosomal translocations or aneuploidy was detected in naive cells (40 metaphases), four of 90 metaphases from a mixed population of stably integrated cells had chromosomal translocations. Two translocations were observed only once [46XX t(7:13) (p21:q22); 46XX t(1:19) (q25, q13.3)]. A third translocation was observed twice [46XX t(1:18) (q25, q12)] (Figure 3a). These led us to further analyze eight clonal populations of stably integrated cells in which we found no structural or numerical chromosomal abnormalities in >210 metaphases. The presence of nonrecurrent translocations is consistent with the known low background of chromosome aberrations in normal human somatic cells.26,27 That the mixed population of stably modified CLECs showed no evidence of clonal expansion of cells harboring translocations also indicated that such affected cells, when present, had no cellular growth or survival advantage.
Figure 3.
Copy number of integration and spectral karyotype of phiC31 integrase–modified cord-lining epithelial cells (CLECs). (a) Normal spectral karyotype in 296 of 300 metaphases of integrase-modified CLECs (upper panel). Rare chromosomal translocations (lower panel), t(7:13) (p21:q22) and t(1:19) (q25, q13.3), were each observed in 1 of 300 metaphases. A third translocation, t(1:18) (q25, q12), was observed in 2 of 300 metaphases. (b) Frequency distribution of copy number of integrated transgene in CLECs determined by fluorescence in situ hybridization of a fluorescein-labeled vector-specific probe. Representative image of integrated transgene (green signals) in DAPI-stained interphase nuclei of CLECs. Original magnification ×1,000. Bar = 5 µm.
Transgene copy number by fluorescence in situ hybridization
Interphase fluorescence in situ hybridization using probes specific to the integrated vector determined the number of integrations per cell. Examination of >200 stably modified CLECs revealed that >85% harbored either one or two integrations per cell (Figure 3b). The technique employed did not differentiate monoallelic from biallelic integrations, where cells had >1 integrations. The low number of integrations per cell is advantageous as it reduces the likelihood of integration into high-risk sites and paves the way for selecting single cell clones with safe integration sites that could make this approach acceptable for clinical application.
Tumorigenic potential of modified CLEC
PhiC31 integrase–mediated transgene integration did not alter the proliferative behavior of CLECs as assessed by in vitro colony-forming assays [26.0 ± 0.5 (wild-type) versus 25.7 ± 0.3 (transgene-integrated); data are mean colony counts and standard error of triplicates; P = 0.643].
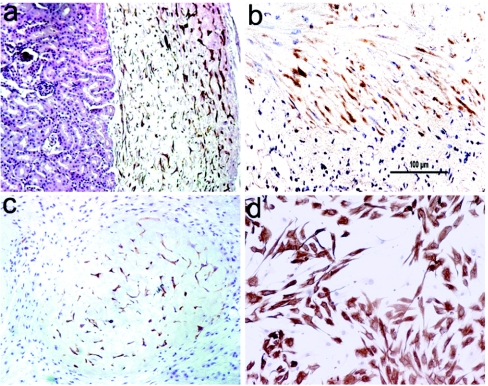
The innate oncogenic activity of integrase-modified CLECs was evaluated by implantation into the nuchal subcutaneous region (n = 6) and renal subcapsular space (n = 4) of NOD-SCID mice (Figure 4a–c). Tumors did not develop in any site for at least 4 months after implantation, although CLECs had engrafted and were recovered by reculturing from the excised implants (Figure 4d).
Figure 4.
Lack of tumorigenicity of phiC31 integrase–modified cord-lining epithelial cells (CLECs) in SCID mice. Immunostaining with antihuman vimentin antibody shows renal subcapsular engraftment of CLECs (a) 1 month and (b) 3 months after implantation in SCID mice. (c) Engraftment of CLECs, 1 month after injection into nuchal subcutaneous region. Original magnification ×200. (d) Immunostaining of cultured CLECs from explants recovered from subcutaneous regions of mice 1 month after implantation. Original magnification ×100. Bar = 100 µm.
FVIII secretion and phenotypic correction of hemophilic mice
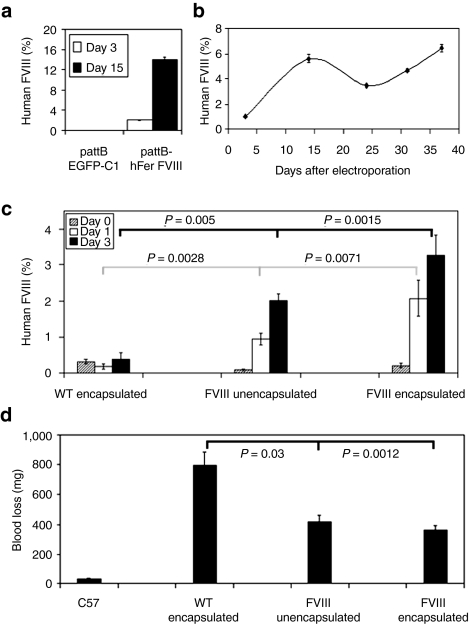
Stable secretion of FVIII in vitro. We coelectroporated CLECs with pattBhFer-FVIII and pCMV-Int to derive FVIII-expressing cells. FVIII was readily detectable in conditioned media on day 3 (19.9 ± 2.0 mU/ml per 2 × 105 cells per 24 hours) and day 15 (140 ± 5.3 mU/ml per 2 × 105 cells per 24 hours; mean ± SEM, n = 3) after electroporation (Figure 5a). CLECs stably integrated with pattBhFer-FVIII secreted FVIII unabated for at least 5 weeks in vitro (Figure 5b), whereas FVIII secretion from CLECs transfected with pattBhFer-FVIII without pCMV-Int never lasted >1 week.
Figure 5.
Stable secretion of factor VIII by transfected CLECs in vitro and demonstration of FVIII bioactivity in vivo. (a) FVIII secretion in vitro by transgenic CLECs shows FVIII activity in the conditioned media of CLECs 3 (open bar) and 15 days (black bar) after transfection with pattBhFer-FVIII. Control CLECs were transfected with pattBEGFP-C1. Data are mean ± SEM, n = 3. (b) Durable in vitro secretion of FVIII by phiC31 integrase–modified CLECs. Data are mean ± SEM, n = 2. (c) Plasma FVIII levels of hemophilic mice implanted subcutaneously with 8 × 106 stably integrated FVIII-secreting CLECs that were either unencapsulated or encapsulated with Matrigel. Control animals were implanted with Matrigel-encapsulated wild-type CLECs. Plasma FVIII antigen levels were measured using an ELISA technique specific for human FVIII. FVIII levels of treated hemophilic mice were significantly higher (P < 0.05) on days 1 (open bar) and 3 (black bar) compared to day 0 values (shaded bar). Data are mean ± SEM; n = 5 per group. (d) Assessment of phenotype correction. The amount of blood loss from a tail clip was determined for FVIII-replete C57BL/6 mice, hemophilic mice implanted subcutaneously with Matrigel-encapsulated wild-type CLECs, hemophilic mice implanted with unencapsulated or Matrigel-encapsulated FVIII-secreting CLECs. Blood loss was significantly reduced in treated mice compared to control hemophilic animals. Data are mean ± SEM; n = 5 per group.
In vivo FVIII secretion following implantation. In vivo secretion of transgenic FVIII was shown when subcutaneous implantation of 8 × 106 Matrigel-encapsulated, stably integrated CLECs significantly raised plasma FVIII antigen levels of hemophilic mice from 3.9 ± 1.8 mU/ml to 32.7 ± 5.6 mU/ml (mean ± SEM, n = 5) 3 days after implantation (P = 0.002 compared to control FVIII-deficient mice) (Figure 5c). These levels significantly improved the bleeding phenotype. Control hemophilic mice implanted with naive CLECs had a mean blood loss of 797 ± 89 mg in the tail-clip assay, whereas hemophilic mice implanted with unencapsulated or encapsulated CLECs stably integrated with FVIII cDNA lost 418 ± 43 mg and 363 ± 28 mg of blood (P = 0.03 and 0.001, respectively) compared to control FVIII-replete mice (Figure 5d).
Discussion
Different cell types are being investigated in current attempts to develop ex vivo cell therapy. We describe here the isolation, culture, characterization, and utilization of a novel cell type that can be consistently and readily obtained in quantity from the amniotic membrane of umbilical cords. These cells were epithelioid and expressed some of the key markers of pluripotency. Other similarities to embryonic stem cells were their capacity for self-renewal and long-term propagation in culture. The ability to derive an estimated 6 × 109 cells from a single healthy cord5 even before expansion in culture indicates the feasibility of clinically relevant quantities of CLECs for cell therapy. These characteristics, coupled with their lack of tumorigenicity and ethically uncontroversial derivation, make CLECs a suitable cell type for therapeutic applications. CLECs augment the range of cord-derived cells beyond cord blood cells, and expand cell therapy to nonhematopoietic and nononcologic applications.
The risk of insertional mutagenesis leading to adverse events28 following the use of integrating vectors is a serious and practical concern that limits enthusiasm for clinical gene therapy. We undertook this study to evaluate the possibility of using gene-modified CLECs as potential bioimplants for safe and long-term secretion of FVIII in hemophilic patients for whom factor replacement therapy is unaffordable. The main objective of this study was to characterize and evaluate whether phiC31 integrase could adversely alter the genomic architecture of transgenic cells, and thus incur unacceptable genotoxic and oncogenic risks. PhiC31 integrase has been shown to mediate site-directed integrations into an estimated 370 pseudo-attP sites in the human genome, although there is evidence for only a small subset of sites.15 In this study, we identified 44 independent integration events and confirm the previously reported conserved sequence motif15 at a majority of integration sites. Forty percent of recovered integrations occurred at 8p22, confirming the observations made when a therapeutic transgene, COL7A1, was integrated into human primary epidermal progenitor cells.29 Our data suggest that the 8p22 integration site is likely to be safe as no chromosomal translocations were detected in >132 metaphases from five clonal populations bearing 8p22 integrations, and no changes were detected in the expression of genes that mapped within 1 Mb intervals centered around 8p22 integrations. This tendency to integrate at 8p22 could be accentuated by developing integrase variants of higher target site specificity30,31 and, if combined with clonal selection to further enhance the biosafety of these modified cells, could advance toward clinical applications.
One of the possible adverse effects of integrating vectors is dysregulation of the function of genes at or close to integration sites.23,24,28 We determined the influence of transgene integration on the transcriptome of phiC31 integrase–modified CLECs. The technique employed had a detection sensitivity of 1 transcript per 200,000 (ref. 32) and thus could accurately reflect the transcriptional changes in even a small proportion of transgenic CLECs. Our analysis of transcriptome data with reference to retrieved integration events revealed no significant perturbations in the expression of genes close to integration sites or within 1 Mb intervals centered around these sites, with the single exception of LNX1, thus confirming the transcriptionally benign effects of these integrations. It is worth noting that even potential oncogenes and tumor suppressor genes in these regions showed no perturbation of expression.
Global transcriptome analysis did reveal that 1.3% of total expressed genes had significant changes in transgenic CLECs. A major biosafety concern of genome modification is the risk of mutating or dysregulating oncogenes or tumor suppressor genes.28,33 In our study, 15 of 151 transcriptionally altered genes were either potential oncogenes or tumor suppressor genes. Pathway mapping of these 151 transcriptionally altered genes identified only three genes that mapped significantly to a single pathway (p53 signaling; P = 0.02). The likelihood that transgenic CLECs were oncogenically transformed was judged to be low based on unaltered colony-forming activity in vitro and absence of tumor development in immunocompromised mice in vivo.
Previous reports have shown the association of phage integrase-mediated integrations with deletions of up to a few thousand base pairs and insertions of up to a few hundred base pairs at integration sites.34 In our study, however, DNA sequence analysis revealed microdeletions of vector DNA (≤37 bp) at recovered integration junctions. Using high-resolution genome copy number data of a mass culture of transgenic CLECs showed significant copy number changes (two deletions and one gain) in only three genomic regions that were not integration sites. The size of these genomic regions ranged from 17 to 37 kb. Only two genes (PSRC1, SYPL2) within 1 Mb windows centered around these genomic regions showed significant transcriptional changes (Figure 2a,b). Thus, no major deletions or insertions could be ascribed to phiC31 integrase-mediated transgene integrations, and the few copy number changes detected had minimal transcriptional effects of uncertain functional relevance.
Another major concern from studies of phiC31 integrase in human cells to date is the occurrence of chromosomal translocations in up to 15% of stably modified cell lines,34 primary human embryonic and adult fibroblasts.21,22 However, our analysis of >300 spectral karyotypes of phage integrase–modified cells revealed only two cells with nonrecurring chromosomal translocations and two cells harboring the same translocation. These rearranged chromosomes were only detected in mass cultures of CLECs and none was found in eight clonal populations. The frequency of observed translocations in this study (4 of 300 metaphases) was lower compared to other reports (15–30%) and may be related, in part, to the different cell types used by other investigators for phiC31 integrase modification.21,22,34 Surveys of two large series of prenatal genetic screening have shown de novo chromosomal translocations in amniocytes, chorionic villus, and fetal blood samples. In one study, normal infants were born of pregnancies that were not terminated on the basis of abnormal karyotypes,26 whereas in the other study, the risk of congenital malformations was sufficiently close to the background rate that it did not support a relationship of chromosomal rearrangements to somatic abnormalities.27 Although both studies are limited with respect to accurate risk assessment, it remains true that at least some de novo chromosomal translocations are functionally silent and inconsequential.35 Our data are consistent with the proposition that translocations in phiC31 integrase–modified cells are uncommon stochastic events that do not confer either a survival or proliferative advantage to the affected cell(s). Had this not been the case, the translocations we identified in modified CLECs would have been the dominant karyotype in a mass culture, rather than the highly sporadic events actually observed.
Although translocations are a cause for concern, it is axiomatic that malignant transformation results from multiple, rather than single, genetic and genomic alterations.36 Moreover, specific rather than random translocations are associated with hematologic and solid tissue malignancies in order.37 Robust defenses against neoplastic transformation in the form of cell cycle arrest, apoptosis, and senescence are activated by the genotoxic stress of unrestrained cell proliferation induced by mutations and/or genomic aberrations.38 Operation of these innate tumor suppressive mechanisms probably explain the fact that mass cultures of phiC31-modified CLECs did not develop clonal dominance of cells harboring chromosomal translocations.
The benign safety profile of phiC31 integrase–modified CLECs suggested by in vitro analyses was confirmed by lack of in vivo tumorigenicity when these cells were implanted into immunocompromised mice. Careful autopsies performed 4 months later showed a complete absence of tumors, whereas transformed human cells typically form tumors within 3–6 weeks.39 Immunohistochemical staining for human vimentin of implanted cells in situ and after reculturing from explants in vitro showed that these cells were viable in vivo for at least 4 months but did not form tumors.
Implantation of FVIII-secreting CLECs raised plasma FVIII levels and partially corrected the bleeding phenotype in hemophilic mice, suggesting the potential for developing nongenotoxic cellular therapy that could be especially effective for autologous or allogeneic applications. Modest increase in plasma FVIII levels could be attributed to the known short circulating half-life of human FVIII in mice (74 minutes compared to 12 hours in humans)40 and suboptimal engraftment and vascularization of implanted human cells in a xenogeneic host.
Although our data show that phage integrase–modified CLECs expanded as polyclonal mass cultures appear to have sustained few or no potentially oncogenic genomic alterations, this approach could be rendered even safer by implanting clonal populations prescreened ex vivo for biosafety using a range of assays such as those we employed. The use of integrases with greater specificity,30,31 high-throughput screening methods for selecting safe clones,41 and the high proliferation capacity of these cells could make this approach acceptable for clinical trials.
Materials and Methods
CLEC isolation and characterization. Fresh umbilical cords from uncomplicated pregnancies were transported in L-15 medium supplemented with 50 IU/ml penicillin, 50 µg/ml streptomycin, 250 µg/ml Fungizone, and 50 µg/ml gentamicin (Invitrogen, Carlsbad, CA) and processed in sterile conditions. Blood was removed by flushing the cannulated cord with phosphate-buffered saline (PBS) supplemented with 5 IU/ml heparin (Sigma-Aldrich, St Louis, MO). The cord was next cut into 2 cm segments, washed and disinfected with 70% ethanol and washed again with antibiotic-containing PBS. The amniotic membrane was dissected free from other cord contents, cut into 0.5 cm2 squares and placed in a cell culture dish filled with 5 ml of Medium 171 (Cascade Biologics, Portland, OR). Explants were cultured at 37 °C/5% CO2, with medium change every 3 days. Outgrowing cells were trypsinized (0.0125% trypsin/0.05% EDTA) and seeded at a density of 1 × 106 cells/dish in Medium 171 supplemented with 50 µg/ml insulin-like growth factor-1, 50 µg/ml platelet-derived growth factor-BB, 5 µg/ml transforming growth factor-β1, and 5 µg/ml insulin (R&D Systems, Minneapolis, MN). Cells were subcultured at 70% confluency and expanded or cryopreserved.
Reverse transcription PCR. RNA extracted from CLECs and a human embryonic stem cell line (HUES) (RNeasy mini kit; Qiagen, Hilden, Germany) was treated with DNAse I (Fermentas, Hanover, MD), reverse transcribed (Superscript II; Invitrogen) and amplified using GoTaq qPCR master mix (Promega, Madison, WI) and the following PCR primers: Nanog (forward primer 5′ TTCCTTCCTCCATGGATCTG 3′ reverse primer 5′ TCTGCTGGAGGCTGAGGTAT 3′), Oct-4 (forward primer 5′ GGTTCTATTTGGGAAGGTATTCAG 3′ reverse primer 5′ GGTTTCTGCTTTGCATATCTC 3′) and γ-actin (forward primer 5′ ACCACTGGCATTGTCATGGACTCT 3′ reverse primer 5′ ATCTTGATCTTCATGGTGCTGGGC 3′). Amplified products were electrophoresed on 2% agarose gels, imaged and quantified by densitometry measurements using Gel Doc 2000 system (Bio-Rad Laboratories, Hercules, CA).
Western blot analysis. CLECs, HUES, and human primary dermal fibroblasts were lysed with M-PER mammalian protein extraction reagent (Pierce, Waltham, MA); 20–50 µg protein from each cell lysate was separated by 14% SDS-PAGE under reducing conditions, electroblotted onto nitrocellulose membrane (Bio-Rad Laboratories) and probed with specific antibodies against human Oct-4 and Nanog (sc-5279; Santa Cruz Biotechnology, Santa Cruz, CA and ab21624; Abcam, Cambridge, UK, respectively). Antibody binding was visualized by horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Promega and Santa Cruz Biotechnology, respectively) and a chemiluminescence-based photoblot system (Amersham Biosciences, Piscataway, NJ).
Indirect immunofluorescence staining. CLECs in culture dishes were fixed with 4% (vol/vol) paraformaldehyde for 20 minutes, washed with PBS before and after permeabilization with 0.1% Triton X-100, and incubated with monoclonal antibodies against keratins 18 and 19 (Dako, Glostrup, Denmark), pancytokeratin (AE1/AE3; Abcam), and desmoplakin.42 Visualization using fluorescein isothiocyanate-labeled secondary antibodies (Dako) was performed as previously described.43
AttB plasmid constructs. B domain-deleted human FVIII cDNA was assembled by PCR amplification of the desired fragments from the full-length sequence in pSP64-F8 (American Type Culture Collection, Manassas, VA). A 3 kb fragment encoding the A1 and A2 domains and a segment of the B domain was PCR amplified (forward primer 5′ TGTAGCGCTAGCATGCAAATAG 3′ reverse primer 5′ GAATAAGGCGATATCTTTAGTCAA 3′) and ligated to a 2.1 kb fragment encoding part of the B domain, A3, C1 and C2 domains (forward primer 5′ GCAAAGCCCGGGAGGACTGAA 3′ reverse primer 5′ CAGTGGCTCGAGGTCAGTAGAGGT 3′) cloned in pcDNA3.1 (Invitrogen) bearing the CMV promoter. The preceding primer sequences incorporated recognition sites for NheI, EcoRV, SmaI, and XhoI used in cloning. The B domain from amino acids 1,007 to 1,648 (NM 000132) was deleted in this construct.44 F309S substitution in the A1 domain was performed by site-directed mutagenesis using mutagenic primers (forward 5′ AGTTTCTACTGTCTTGTCATATCTCT 3′ reverse 5′ AGAGATATGACAAGACAGTAGAAACT 3′) and PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) and DpnI.45 We assembled pattBCMV-FVIII by ligating the attB sequence as a 300-bp fragment46 upstream of the CMV promoter in pcDNA3.1. Lastly, the CMV promoter was excised with MunI and NheI, and replaced by the human ferritin light chain promoter amplified from pVitro2 (InvivoGen, San Diego, CA) (forward primer 5′ GCAGGCCAATTGTAACTTACGG 3′ reverse primer 5′ TCAGATCGCTAGCACGCCGGTGG 3′) to yield the final construct, pattBhFer-FVIII.
pattBEGFP-C1 was derived by inserting the same attB fragment into the MluI site of pEGFP-C1 (Clontech Laboratories, Mountain View, CA).
Cell culture and electroporation. CLECs were co-transfected by electroporating 2 × 106 cells with 10 µg pattBhFer-FVIII or pattBEGFP-C1 and 1.5 µg of pCMV-Int46 in 400 µl RPMI 1640/10% FCS medium in a 0.4 cm cuvette. Electrotransfer was performed with a single pulse (55 ms, 165V) delivered by BTX ECM 830 electroporator (Genetronics, San Diego, CA). The percentage of GFP+ cells was determined by FACS analysis (BD FACSCalibur; Becton Dickinson, Mountain View, CA). Mixed population of stably integrated CLECs were selected by culture in medium containing 0.6 mg/ml G418 (Invitrogen) for 7 days. Clonal populations of EGFP-expressing CLECs were derived by expanding flow-sorted single cells in culture. Colony-forming assays were performed by low-density seeding of naive or a mixed population of stably integrated CLECs (200 cells/well; 6-well plate) followed by culture for 14 days, with medium change every third day. Cultures were stained with crystal violet (1% wt/vol, 30 minutes; BDH Chemicals, Poole, UK) and washed thrice with PBS.
Integration site retrieval and analysis. The percentage of stable integrants was determined by seeding FACS-sorted EGFP-expressing CLECs (2,000 and 5,000 cells) into 10 cm Petri dishes, followed by scoring the number of GFP+ cells/clones remaining after 7 days of selection with G418. Integration sites were recovered from pooled and mixed populations of stably transfected cells by the plasmid rescue method. Genomic DNA was isolated from transgenic CLECs using the Blood and Cell Culture Miniprep Kit (Qiagen). One microgram of genomic DNA was digested with a combination of either SpeI, XbaI, and NheI, or BamHI and BglII restriction enzymes. Digested DNA was extracted with phenol-chloroform, precipitated in ethanol and ligated under dilute combinations using T4 DNA ligase. Ligated products were electroporated into DH10B E. coli (1.85 kV, 25 µF, and 200 Ω Gene Pulser; Bio-Rad Laboratories) and plated on kanamycin LB-agar plates. All DNA modifying enzymes were from New England Biolabs (Ipswich, MA). Rescued plasmids were sequenced with the CHOSeqR primer (5′TCCCGTGCTCACCGTGACCAC3′).15 DNA sequences were mapped to the reference human genome using the BLAT program (http://genome.ucsc.edu). Crossover junctions were identified by aligning retrieved sequences to the attB sequence. To determine whether the recovered integration sites shared a common motif, 100 bp of genomic DNA sequence flanking the crossover point of integration events in each cytoband was retrieved from the reference genome sequence and analyzed using the motif search program MEME (http://meme.nbcr.net).
Transcriptome and genome copy number profiling. Total RNA from naive and transgenic CLECs (RNeasy Mini Kit; Qiagen) served as starting material for transcript profiling on Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Carla, CA) following the recommended protocols. Transcription expression data were analyzed using GeneChip Operating Software (Affymetrix). Transcripts whose expression levels in transgenic CLECs were significantly altered as determined by the operating software using Wilcoxon signed-rank test and differed more than twofold compared to naive CLECs were selected for analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery) 2.1 Functional Annotation Tool (http://david.abcc.ncifcrf.gov). These genes were referenced to a compilation of known proto-oncogenes and tumor suppressor genes47 (http://microb230.med.upenn.edu/protocols/cancergenes.html).
High-resolution copy number profiling was performed on genomic DNA of naive and transgenic CLECs using the Human Mapping 500K Array Set (Affymetrix) and the data analyzed using GeneChip Chromosome Copy Number Analysis Tool. Regions of copy number gain or loss were defined as having ≥3 consecutive SNPs concordant for significant copy number abnormalities. Log2 signal intensity ratios >0.3 and <−0.3 were criteria for significant copy number gain and loss, respectively.
Spectral karyotyping and fluorescence in situ hybridization. Metaphase spreads of naive and transgenic CLECs were prepared by standard cytogenetic techniques. Spectral karyotyping according to the recommended protocol was performed using chromosome painting probes (Applied Spectral Imaging, Edingen-Neckarhausen, Germany) visualized with an Olympus BX61 epifluorescence microscope (Olympus, Tokyo, Japan) and SkyView ver. 2.1.1 (Applied Spectral Imaging). Fluorescence in situ hybridization was performed on interphase nuclei of naive and transgenic CLECs with an 800 bp probe derived from pattBEGFP-C1. The probe was labeled with fluorescein-12-dUTP (PerkinElmer, Waltham, MA) and the BioPrime DNA Labeling System (Invitrogen). Hybridization and DAPI counterstaining were performed according to standard protocols.48
FVIII and blood loss assays. FVIII activity in culture supernatants of transfected CLECs was assayed using the Coatest SP FVIII kit (Chromogenix, Mölndal, Sweden) and the recommended protocol. Human-specific FVIII levels were quantitated in citrated mouse plasma using an ELISA kit (Affinity Biologicals, Ancaster, Ontario, Canada). Assessment of phenotypic correction was performed by determining the volume of blood loss in exon 16-disrupted FVIII knockout mice (kind gift of H.H. Kazazian, University of Pennsylvania, Philadelphia, PA).49,50 The tail was warmed to 37 °C for 2 minutes, then severed 2 cm from the tip with a sharp blade and immediately placed in a microfuge tube containing 0.5 ml PBS at 37 °C for 15 minutes when bleeding was arrested by cauterization. The difference in the weight of the tube before and after blood collection quantified blood loss.
Animal work. Experimental protocols were approved by the Institutional Animal Care and Use Committee of the Singapore General Hospital. Mice were housed at 27 °C in 12-h light:dark cycles. SCID mice were purchased from Animal Resources Centre, Murdoch, Australia. All procedures were performed under anesthesia induced by intraperitoneal injection of 0.1 ml of a mixture consisting of equal parts of Hypnorm [fluanisone (10 mg/ml) and fentanyl citrate (0.315 mg/ml); Janssen Pharmaceutica, Berchem, Belgium] and Dormicum (midazolam, 5 mg/ml; Roche, Basel, Switzerland), diluted in two parts water. The tumorigenic potential of transgenic CLECs was assessed in SCID mice by subcutaneous nuchal or renal subcapsular implantation of 5 × 106 cells suspended in 50 µl PBS. Mice were visually inspected weekly for the appearance of subcutaneous tumors for 4 months, and killed after 1 and 3 months to check for renal subcapsular tumors. Phenotypic correction was evaluated by implanting 8 × 106 FVIII-secreting CLECs in 300 µl PBS mixed with 300 µl Matrigel (BD Biosciences, San Jose, CA) into the subcutaneous nuchal region of hemophilic male mice. Blood samples for FVIII assays were collected before, 1, 3, 6, and 15 days after cellular implantation. Blood was obtained by puncturing the retro-orbital venous plexus with heparinized capillary tubes and collected in 0.1 volume of 0.1 mol/l sodium citrate. Plasma was obtained by centrifugation at 20,000g and 4 °C for 10 minutes.
Histology. Five-micron paraffin sections of tissues removed from CLECs implantation sites were immunostained with antihuman vimentin antibodies (clone V9; Zymed, San Francisco, CA) followed by visualization using the REAL EnVision kit (Dako) and hematoxylin counterstain.
Statistical analysis. Analysis of variance and the Tukey–Kramer test were used for three or more groups and Student's unpaired t-test for two groups. Fisher's exact test with two-sided P value was used to determine statistical significance between two proportions. P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Site specificity of phiC31 integrase–mediated transgene integration.
Acknowledgments
This work was supported by a research grant from SingHealth Foundation (SHF/FG347P/2006) to O.L.K. We thank Michele P. Calos (Stanford University, Stanford, CA) for gifts of pTA-attb and pCMV-Int, and for advice on plasmid rescue of integration sites; Mark Richards (Nanyang Polytechnic, Singapore) for the gift of HUES cells; Leonard Ang (Singapore Eye Research Institute, Singapore) and Jill Allen (Department of Dermatology, University of Oxford, Oxford, UK) for gifts of antikeratin and antidesmoplakin antibodies, respectively; Mickey Koh, Tsyr Jong Lim, and Madelaine Niam (Health Sciences Authority, Singapore) for cell sorting; and Siew Hong Leong (National Cancer Center, Singapore) for assistance with spectral karyotyping. T.T.P., whose cells were studied in the present work, has a financial interest in CellResearch Corporation.
Supplementary Material
Site specificity of phiC31 integrase–mediated transgene integration.
REFERENCES
- Weiss ML., and, Troyer DL. Stem cells in the umbilical cord. Stem Cell Rev. 2006;2:155–162. doi: 10.1007/s12015-006-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DT., and, Rogers I. Umbilical cord blood: a unique source of pluripotent stem cells for regenerative medicine. Curr Stem Cell Res Ther. 2007;2:301–309. doi: 10.2174/157488807782793790. [DOI] [PubMed] [Google Scholar]
- Miki T., and, Strom SC. Amnion-derived pluripotent/multipotent stem cells. Stem Cell Rev. 2006;2:133–142. doi: 10.1007/s12015-006-0020-0. [DOI] [PubMed] [Google Scholar]
- Ruetze M, Gallinat S, Lim IJ, Chow E, Phan TT, Staeb F, et al. Common features of umbilical cord epithelial cells and epidermal keratinocytes. J Dermatol Sci. 2008;50:227–231. doi: 10.1016/j.jdermsci.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Phan TT., and, Lim IJ.Isolation of stem/progenitor cells from amniotic membrane of umbilical cordUK Patent No. GB 2432166, granted 2 January 2008.
- Matikainen T., and, Laine J. Placenta–an alternative source of stem cells. Toxicol Appl Pharmacol. 2005;207 2 suppl.:544–549. doi: 10.1016/j.taap.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Kurtzberg J. Update on umbilical cord blood transplantation. Curr Opin Pediatr. 2009;21:22–29. doi: 10.1097/mop.0b013e32832130bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco M, Zucconi E, Vieira NM, Fogaça LL, Cerqueira A, Carvalho MD, et al. Multipotent stem cells from umbilical cord: cord is richer than blood! Stem Cells. 2008;26:146–150. doi: 10.1634/stemcells.2007-0381. [DOI] [PubMed] [Google Scholar]
- Kermani AJ, Fathi F., and, Mowla SJ. Characterization and genetic manipulation of human umbilical cord vein mesenchymal stem cells: potential application in cell-based gene therapy. Rejuvenation Res. 2008;11:379–386. doi: 10.1089/rej.2008.0674. [DOI] [PubMed] [Google Scholar]
- Can A., and, Karahuseyinoglu S. Concise review: human umbilical cord stroma with regard to the source of fetus-derived stem cells. Stem Cells. 2007;25:2886–2895. doi: 10.1634/stemcells.2007-0417. [DOI] [PubMed] [Google Scholar]
- Pfeifer A., and, Verma IM. Gene therapy: promises and problems. Annu Rev Genomics Hum Genet. 2001;2:177–211. doi: 10.1146/annurev.genom.2.1.177. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Kohn DB. Gene therapy for childhood immunological diseases. Bone Marrow Transplant. 2008;41:199–205. doi: 10.1038/sj.bmt.1705895. [DOI] [PubMed] [Google Scholar]
- Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, et al. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP., and, Grompe M. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bertoni C, Jarrahian S, Wheeler TM, Li Y, Olivares EC, Calos MP, et al. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc Natl Acad Sci USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos MP. The phiC31 integrase system for gene therapy. Curr Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Liu Y, Shin S, Lakshmipathy U, Scheyhing K, Xue H, et al. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells. 2008;26:119–126. doi: 10.1634/stemcells.2007-0283. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Xu H, Huang Z, Engler JA., and, Kay MA. A direct comparison of two nonviral gene therapy vectors for somatic integration: in vivo evaluation of the bacteriophage integrase phiC31 and the Sleeping Beauty transposase. Mol Ther. 2005;11:695–706. doi: 10.1016/j.ymthe.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Liu J, Jeppesen I, Nielsen K., and, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- Liu J, Skjørringe T, Gjetting T., and, Jensen TG. PhiC31 integrase induces a DNA damage response and chromosomal rearrangements in human adult fibroblasts. BMC Biotechnol. 2009;9:31. doi: 10.1186/1472-6750-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Recchia A, Bonini C, Magnani Z, Urbinati F, Sartori D, Muraro S, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19:36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Giardino D, Corti C, Ballarati L, Colombo D, Sala E, Villa N, et al. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat Diagn. 2009;29:257–265. doi: 10.1002/pd.2215. [DOI] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, et al. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Sclimenti CR, Thyagarajan B., and, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, et al. Mutational derivatives of PhiC31 integrase with increased efficiency and specificity. Mol Ther. 2009;17:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix, Inc. Design and performance of the genechip human genome U133 plus 2.0 and human genome U133A 2.0 arrays. Technical note . < http://www.affymetrix.com/ support/technical/technote/hgu133_p2_technotes.pdf >. Accessed 15 September2009
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A, Engler JA, Xu H, Cherry AM., and, Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- Varella-Garcia M, Chen L, Powell RL, Hirsch FR, Kennedy TC, Keith R, et al. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med. 2007;176:505–512. doi: 10.1164/rccm.200609-1329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC., and, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B., and, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Cepero E., and, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Mahale AM, Khan ZA, Igarashi M, Nanjangud GJ, Qiao RF, Yao S, et al. Clonal selection in malignant transformation of human fibroblasts transduced with defined cellular oncogenes. Cancer Res. 2008;68:1417–1426. doi: 10.1158/0008-5472.CAN-07-3021. [DOI] [PubMed] [Google Scholar]
- Dwarki VJ, Belloni P, Nijjar T, Smith J, Couto L, Rabier M, et al. Gene therapy for hemophilia A: production of therapeutic levels of human factor VIII in vivo in mice. Proc Natl Acad Sci USA. 1995;92:1023–1027. doi: 10.1073/pnas.92.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher F, Dellambra E, Rico L, Bondanza S, Murillas R, Cattoglio C, et al. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol Ther. 2007;15:1670–1676. doi: 10.1038/sj.mt.6300238. [DOI] [PubMed] [Google Scholar]
- Parrish EP, Steart PV, Garrod DR., and, Weller RO. Antidesmosomal monoclonal antibody in the diagnosis of intracranial tumours. J Pathol. 1987;153:265–273. doi: 10.1002/path.1711530311. [DOI] [PubMed] [Google Scholar]
- Allen J, Phan TT, Hughes MA, Cherry GW., and, Wojnarowska F. The cellular origins of the linear IgA disease target antigens: an indirect immunofluorescence study using cultured human keratinocytes and fibroblasts. Br J Dermatol. 2003;148:945–953. doi: 10.1046/j.1365-2133.2003.05313.x. [DOI] [PubMed] [Google Scholar]
- Miao HZ, Sirachainan N, Palmer L, Kucab P, Cunningham MA, Kaufman RJ, et al. Bioengineering of coagulation factor VIII for improved secretion. Blood. 2004;103:3412–3419. doi: 10.1182/blood-2003-10-3591. [DOI] [PubMed] [Google Scholar]
- Swaroop M, Moussalli M, Pipe SW., and, Kaufman RJ. Mutagenesis of a potential immunoglobulin-binding protein-binding site enhances secretion of coagulation factor VIII. J Biol Chem. 1997;272:24121–24124. doi: 10.1074/jbc.272.39.24121. [DOI] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B., and, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll JH, Lichter P, Bakdounes K., and, Eltoum IE. In situ hybridization and detection using nonisotopic probes. Curr Protoc Mol Biol. 2007;Chapter 14:Unit 14.7. doi: 10.1002/0471142727.mb1407s79. [DOI] [PubMed] [Google Scholar]
- Bi L, Lawler AM, Antonarakis SE, High KA, Gearhart JD., and, Kazazian HH., Jr Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10:119–121. doi: 10.1038/ng0595-119. [DOI] [PubMed] [Google Scholar]
- Rawle FE., and, Lillicrap D. Preclinical animal models for hemophilia gene therapy: predictive value and limitations. Semin Thromb Hemost. 2004;30:205–213. doi: 10.1055/s-2004-825634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Site specificity of phiC31 integrase–mediated transgene integration.