Abstract
Lentiviral vectors have remarkable cell entry and gene delivery properties that make them highly attractive for gene therapy. However, all integration-competent gene delivery systems have come under scrutiny for possible adverse insertional events. Circumventing the risk of insertional mutagenesis, integration-deficient human immunodeficiency virus (HIV)-1–derived vectors have been shown to support durable transcription of transgenes in certain nonmitotic cell lineages. In mitotic cell populations, such nonintegrated viral forms are lost during cell division and so have time-limited effects. Hybrid lentiviral vectors that harness the cell entry properties of HIV to facilitate carriage of alternative DNA modification systems into cells may allow durable genetic modification with more favorable integration profiles. Thus, systems, which have previously been plasmid-based such as those based on nuclease-enhanced homologous recombination (HR) and artificial transposons, have been incorporated into the viral genome to allow them to “hitch-hike” into cells that are difficult to transfect. Here, we review recent progress in the development of such hybrid lentiviral systems and consider potential applications of such vectors.
Introduction
The broad tropism of human immunodeficiency virus (HIV-1)–derived lentiviral vectors and their ability to infect hard-to-transfect cells make them highly attractive for clinical gene therapy applications. Stable transduction of target cell populations makes them particularly useful for the genetic modification of dividing cells. Conventional lentiviral vectors undergo reverse transcription shortly after cell entry and form a preintegration complex comprising linear viral DNA, integrase, and matrix and cellular proteins. The complex is localized to genomic DNA through an interaction with lens epithelium–derived growth factor/p75 and the viral enzyme integrase subsequently mediates integration into host DNA.1,2,3 The process of integration site selection is not random, but favors transcriptionally active regions. Site selection is influenced by a variety of factors and as for other retroviral vectors, cellular factors including higher order chromatin structures are likely to govern accessibility to target DNA.4,5 Thus around 70% of HIV-1 integration sites occur in genes compared to a predicted level of around 30% if the process was purely random.6,7 Although there is evidence for the targeting of certain chromosomal hotspots, lentiviral vectors (unlike γ-retroviral vectors) do not appear to exhibit particular preferences for transcriptional start sites, areas close to DNAase1 hypersensitivity sites, or CpG islands. Viral integrase plays a key role in integration site selection, and this was demonstrated in experiments where substitution of HIV integrase with murine leukemia virus integrase resulted in redirection toward a murine leukemia virus–like integration profile.8 Overall, HIV-1 based lentiviral vectors may partially obviate some of the concerns linked to γ-retroviral vectors that more frequently target gene regulatory regions (accounting for ~20% of integrants) and have been linked to insertional mutagenesis. Murine leukemia virus–derived retroviral vectors used in two independent studies of autologous stem cell gene therapy for X-linked severe combined deficiency have caused T-cell leukemiagenesis.9,10,11 In addition, retroviral insertional transactivation caused clonal expansion of myeloid cells and myelodysplasia following retroviral modification of hematopoietic stem cells (HSCs) in patients with chronic granulomatous disease.12 Ahead of clinical studies of lentiviral vectors, studies in mice have been reassuring. In Cdkn2a−/− tumor–prone mice, there was a reduced risk of insertional mutagenesis for self-inactivating (sin) lentiviral vectors following transduction and grafting of murine bone marrow–derived HSCs compared to long terminal repeat (LTR)–intact murine retroviruses.13,14 However, it should also be borne in mind that recent in vitro studies suggested that there was only a threefold reduction in transforming activity of primary murine HSCs when using lentiviral rather than a γ-retroviral backbone with the same SIN configuration.14 Clinical studies using lentiviral vectors in humans have recently been initiated but only limited information is available about the longer term consequences of viral integration. To date, there have been no reports of adverse insertional events in patients with HIV infection who have undergone ex vivo gene modification of autologous T cells.15,16 A smaller number of patients have successfully undergone lentiviral modification of autologous HSCs, for the inherited metabolic disorder Adrenoleukodystrophy without significant vector-related toxicity.17 However, in another study of HSC lentiviral transduction for the blood disorder β-thalassemia, a clonal expansion of erythroid precursors have recently triggered alerts from regulatory bodies.18 The vector used encoded elements derived from the β-globin locus control region and had insulator sequences incorporated within the LTRs. The underlying mechanism of the clonal expansions is being investigated further but is likely to be related to integration within the gene locus for high mobility group A2 proteins. Thus, although the integration profile of lentiviral vectors may be inherently more favorable than γ-retroviral vectors, the potency and specificity of internal regulatory elements are likely to have a greater influence on biosafety. Even with the inclusion of insulator elements, risks associated with disrupted gene expression need to be carefully considered. How can the safety profile of lentiviral vectors be further improved? One option which is highly attractive for nonmitotic target cells is to avoid vector integration into genomic DNA entirely through the use of integration-deficient (nonintegrating) lentiviruses (NILV). Such an approach relies on disabling conventional integration pathways, for example through mutations in integrase. The biology of NILV vectors has been reviewed in detail recently in this journal19 and others20 and therefore only the salient features are summarized here. Mutations of integrase such as D64V are designed to disrupt its role in proviral integration, but not to compromise its other functions, including virus packaging and nuclear translocation of the preintegration complex.21 Thus, following reverse transcription, lentiviral DNA fails to integrate into chromosomal DNA, and is predisposed to forming DNA circles as a consequence of host DNA repair proteins. Lentiviral DNA circularizes by nonhomologous end-joining forms 2-LTR circles or can undergo homologous recombination (HR) of the 5′ and 3′LTRs to form a single LTR form. It has been estimated that approximately one-third of linear lentiviral DNA forms either 1- or 2-LTR circles in target cells and these episomal forms remain transcription competent.22 It is also apparent that a greater proportion of HIV DNA forms episomal circles from integrase-deficient vectors compared to vectors produced with intact integrase. Levels of background integration (through nonintegrase pathways) are extremely low and in nonmitotic cells, gene expression from episomal lentiviral circular DNA has been sustained for many months.
Clearly this approach is less useful for the sustained correction of dividing cell systems such as the hematopoietic cells or skin tissue because episomal, nonreplicating, lentiviral forms rapidly dilute as cells turnover, and thus transgene effects are lost over a period of days or weeks. Gene expression from NILV may also not always be as efficient as from an equivalent integrated proviral form.23,24 However, it has been shown there is a window of opportunity during which nonintegrating lentiviral vectors can be used to express alternative machinery for the modification of genomic DNA. Marrying the cell entry properties of lentivirus with previously plasmid-based gene therapy tools could produce synergistic improvements. Nonintegrating vectors have recently been used to deliver recombinase enzymes, meganucleases, and zinc finger nucleases (ZFNs) (Table 1).25,26,27 Importantly, the nonintegrated lentiviral DNA forms can also act as a suitable donor template for DNA recombination and although efficiency in primary cells is currently limited, the prospect of generating single viral constructs capable of encoding both the integration/repair enzymes and corrected DNA for repair is a realistic ambition. Experience from a variety of different hybrid systems is reviewed and the generic issues relating to the use of NILV in these experiments, and problems with reduced transcriptional activity and background integration events are considered.
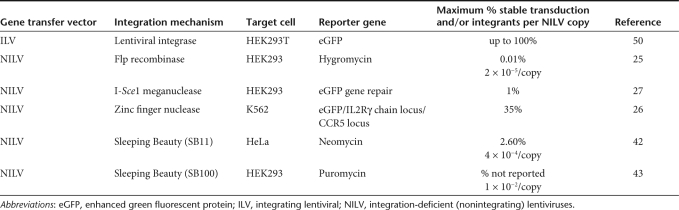
Table 1. Integration-deficient lentiviral vectors (NILV) used to deliver alternative DNA-modifying systems and published efficiencies of gene transfer compared to integrating lentiviral vectors.
Lentiviral-Mediated Flp Recombination
In proof of principle studies, cells were infected with two nonintegrating vectors—one for expression of an enhanced Flp recombinase and the other providing a hygromycin-resistance transgene flanked by a flippase recognition target (Flp recombination) site.25 The mechanism of Flp-directed recombination was such that only circularized lentiviral molecules were able to integrate successfully. Sustained gene transfer could be demonstrated following co-transduction of HEK293 cells, which had been engineered to contain three chromosomal flippase recognition target sites. Importantly, only low levels of background integration were observed because the vector was designed so that hygromycin resistance could only be expressed following Flp-mediated insertion at the target site. Sequence analysis confirmed that 1- and 2-LTR circles were the major substrate for Flp-mediated insertion. The authors used quantitative PCR to compare the efficiency of plasmid transfection or viral transduction, and found that the rate of Flp-mediated integration from 2-LTR circle was around 60 times more efficient compared to plasmid. Possible reasons for the greater efficiency of 2-LTR-circle-derived insertion include preferential nuclear localization of nonintegrated viral DNA and important structural differences in the nature of the substrate DNA, including the degree of supercoiling. The Flp-based system is not therapeutically useful as artificial sites for recombination are required, but these experiments supported the principle of nonintegrating lentiviral–mediated delivery of alternative integration machinery.
Lentivirus Delivery of Nucleases and DNA Repair by Homologous Recombination
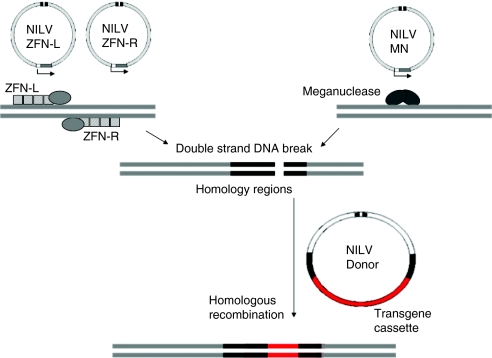
The creation of a targeted double-stranded DNA break stimulates DNA repair machinery and results in increased HR (Figure 1). The meganuclease I-Sce1 binds to an 18-bp recognition site and causes double-strand breaks in genomic DNA, triggering gene repair by HR. Cornu et al. described a lentiviral vector able to undergo gene targeting by HR after I-Sce1 activity.27 An I-Sce1 expression cassette and a homologous repair template were cloned into independent NILVs. Gene conversion at a chromosomal enhanced green fluorescent protein target site was observed in ~1% of HEK293 cells transduced with both NILVs compared to 0.03% of cells transduced with the template NILV only. Interestingly, gene conversion occurred in 12% of HEK293 cells when I-Sce1 was expressed from an integrating lentiviral vector. The authors suggest that low expression from NILVs was a limiting factor in the gene targeting frequency. As discussed below, reduced levels of transgene expression from NILV have been reported elsewhere and strategies to increase expression are being investigated.28 Recently, engineered heterodimeric variants of homing endonucleases have been shown to be capable of targeting genes relevant to human disease such as the human xeroderma pigmentosa group C gene.29 Xeroderma pigmentosum arises from a defect of nucleotide excision repair and engineered variants of the homing endonuclease I-Cre1 mediated high levels of specific gene targeting without genotoxicity. This example raises the prospect of NILV delivery of further custom-engineered endonucleases capable of highly specific DNA cleavage.
Figure 1.
Schematic representation of nonintegrated lentiviral (NILV) circles in which 2-LTRs have joined by nonhomologous end-joining in the absence of integrase activity. These episomal forms support transcription and one attractive application has been the delivery of nucleases capable of site-specific DNA modification. In the case of zinc finger nucleases (ZFNs), the zinc finger motifs bind to opposite strands of target DNA sites and this allows dimerization of Fok1 endonuclease resulting in double-stranded cleavage of genomic DNA. Alternatively, meganucleases (MN) mediate precise DNA cleavage following highly specific DNA recognition and binding. Provision of a third NILV, carrying sequences homologous to the cleavage site, provides a template for homologous recombination and repair of cleaved genomic DNA, and this can allow the insertion of a minimal promoter/transgene cassette if flanked by appropriate homology sequences.
Another notable hybrid system to date has used NILVs to express ZFNs. These zinc-finger assemblies comprise Cys2His2 zinc fingers, which target specific DNA base triplets. Fusion to the nuclease domain of endonuclease Fok1 has generated artificial ZFNs capable of inducing site-directed double-stranded DNA breaks. Subsequent DNA repair by nonhomologous end-joining or HR induces permanent genomic modification. Initial studies used nucleofection to deliver plasmids expressing ZFNs to target the IL2RG locus in cell lines.30 To improve efficiency, more recent work has focused on viral delivery. Lombardo et al. generated transduced cells with three NILVs: two for expression of ZFN dimerizing pairs and one for delivery of the repair template encoding a transgene flanked by sequences homologous to the target site.26 Gene conversion was observed at the IL2RG locus in 16% of K562 cells infected with all three vectors. Besides gene conversion by substitution, gene addition into the IL2RG locus in K562 cells was achieved with an efficiency of 3.4% by flanking an enhanced green fluorescent protein expression cassette with arms homologous to the IL2RG, indicating site-specific integration of the marker gene. Similarly, ZFNs have been designed to target the CCR5 locus. CCR5 is a coreceptor for HIV-1 entry and a homozygous Δ32 deletion in CCR5 is linked to viral resistance in man. Perez et al. sought to permanently disrupt CCR5 expression and recreate the Δ32-CCR5 null phenotype in human CD4+ T cells using adenoviral vectors to deliver ZFNs targeting the transmembrane domain of CCR5.31 In this situation, the ZFN-mediated double-strand break is repaired by nonhomologous end-joining. The primary aim of these experiments was to inhibit HIV entry by disruption of CCR5 expression. However, in other experiments using NILV encoding a reporter gene/promoter cassette flanked with sequences homologous to those upstream and downstream of the ZFN target site, site-specific gene delivery by HR was demonstrated. Gene addition at the CCR5 locus occurred in 35% of K562 cells with a 2% background integration rate and 0.06% of CD34+ hematopoietic progenitor cells with a 0.005% background integration rate.26 The CCR5 locus is being investigated as a “safe-harbor” site for integration, as disrupted CCR5 expression, present in around 1% of the population, does not appear to result in any significant reduction in immunity. Ideally, safe-harbor sites should be able to support stable gene expression and resist silencing. Levels of transgene expression at the CCR5 locus have been found to be reduced compared to those at another putative safe site, the adeno-associated virus S1 (AAVS1) locus. Such data suggest that levels of transcriptional activity are influenced by as yet undefined site-specific features. It is also not yet clear how promoter/transgene cassette integration into a “safe-site” impacts on the neighboring genes. Although retrovirus-derived promoters have been reported to upregulate neighboring genes by up to 100-fold, similar effects were not detected for human PGK or EF1α promoters.32 Nonetheless, formal studies will need to be undertaken to demonstrate reduced (or absent) potential for insertional mutagenesis in comparison to more conventional γ-retroviral or lentiviral vectors.
Finally the risk of nuclease-mediated toxicity will have to be considered for specific constructs, as it is likely to be dependent on a variety of factors including the specificity of DNA cleavage and the frequency of target-binding sites.33 For example, engineering Fok1 endonuclease to require obligate heterodimerization for effective DNA cleavage can reduce “off-target” effects mediated by conventional Fok1 homodimers.34,35
Hybrid Lentivirus–Transposon Vectors
Sleeping Beauty (SB) lentiviral hybrid vectors have been derived from a plasmid-based Tc1/mariner-type DNA transposon/transposase system,36 which has been widely explored as an integrating vector for transgenesis in eukaryotic cells. The complete wild-type transposon consists of a transposase coding sequence flanked by two nonidentical 230-bp inverted repeats.37 Transposase catalyzes the excision of the transposon from flanking DNA and its reintegration elsewhere in a “cut-and-paste” transposition. Insertion occurs at TA-dinucleotide sites and DNA repair of both the donor and target DNA strands, results in a 3-bp transposon footprint on the donor DNA and duplication of the TA-dinucleotide within the target chromosomal DNA.38,39 This characteristic signature is useful for indentifying transposition events, and integration site analysis in cell lines has shown that integration of SB is almost random with respect to genomic features such as transcription units and CpG islands.40,41,42 This may be advantageous in terms of reduced risk of genotoxicity, but as a plasmid-based system, SB applications may be limited. There have been attempts to create hybrid SB-viral vectors. For example, adenoviral vectors have been used to deliver transposons and transposase cassettes. One insight from such experiments was the realization that efficient transposition from the viral DNA requires circularization of the viral backbone, and in this regard NILVs have the advantage of naturally forming episomal circles. These experiences have led to the development of lentivirus:SB hybrid vectors in various configurations. Incorporation of a transposon cassette, in its correct orientation, between the lentiviral LTRs and provision of transposase in trans, from a second nonintegrating construct, resulted in effective cut-and-paste transposition.42 This configuration has the advantage of inserting a minimal promoter/transgene cassette, with all other virus-derived elements remaining episomal and thus being lost upon cell division. As alternative option, Staunstrup et al. engineered vectors to permit SB-mediated transposition of an entire proviral cassette, including the LTR elements. This was achieved through the inversion and close opposition of the left and right SB inverted repeats between the LTRs in combination with an enhanced SB mutant (SB100).43 Integration site analysis in both cases confirmed that the integration profile was diverted away from that of conventional lentivirus, and toward the profile of SB. Intuitively, this would appear to reduce the risk of adverse insertional events, but the issue of genotoxicity has yet to be formally addressed in suitable tumor models. Furthermore, the issue of transgene silencing within transposon cassettes, and how that relates to integration sites, will also need to be studied. It has recently become clear that SB insertions are subject to a degree of gene silencing and variegation of gene expression, although further studies comparing different promoter/enhancer sequences is warranted. Garrison et al. reported that as few as 1 in 15 chromosomal insertions in cell lines were able to express a fluorescent reporter gene, and treatment of cells with methyltransferase or histone deacetylase inhibitors raised this proportion to almost 100% (refs. 44,45). For future applications, it may be possible to engineer the transposase to target integration to specific sites,46 or to utilize other transposase systems such as piggybac. The latter was derived from the cabbage looper moth Trichoplusia ni and has a carrying capacity of up to 14 kb without loss of transposition efficiency.47 Thus hybrid lentiviral:piggybac vectors may offer additional advantages over SB vectors in terms of larger transgene size, increased transposition efficiency, and resistance to overproduction inhibition effects.
Generic Issues Relating to NILV Hybrid Vectors
Although initial experiments in nondividing cells indicated that NILV can mediate gene transfer at comparable efficiencies to integrating vectors,21 more recently it has become apparent that expression levels may well be reduced compared to those from stable integrants. The effects on vector titer following the incorporation of additional elements in into the lentiviral genome may be unpredictable when including expression cassettes flanked by target site homology sequences or transposable SB cassettes. The consequences of including 3′ PolyA sequences in the sense orientation within transgene cassettes has to be balanced against possible promoter interference from cassettes cloned in the reverse orientation.42
Background integration of NILV is infrequent, in the order of 10−4 to 10−2 depending on cell type and in the range of background integration of plasmid. However, the stable integration of ZFNs, transposases, or other DNA-modifying enzymes even in a small number of cells would be undesirable. In the case of SB, the transposon would be susceptible to “hopping” by repeated cut-and-paste retransposition.48 Thus, measures to eliminate background integration and/or the inclusion of additional safety genes may be required. Thus far, the D64V integrase mutation has proven most efficient in preventing integration, and the inclusion of additional mutations (for example within the attachment site) has not further reduced the frequency of background integration events.22,49 Pharmacological approaches to disrupt integrase and/or lens epithelium–derived growth factor may offer additional security, but perhaps the most robust approach would be to link expression of nuclease or transposase genes to a suicide gene mechanism. This would allow selective prodrug-mediated elimination of cells retaining lentiviral copies as a result of background integration of full-length provirus.
Conclusions and Applications
If generic issues relating to vector titer and levels of transcriptional activity derived from NILV can be addressed, clinical applications of vectors carrying DNA modification systems will follow. The group of conditions most likely to be the first to benefit from such hybrid lentiviral vector technologies are those very same inherited immune disorders in which gene therapy successes were demonstrated using integrating retroviral vectors. In conditions such as X-linked severe combined immunodeficiency, a relatively small number of functional hematopoietic stem or progenitor cells can support immune reconstitution and disease correction because of the strong in vivo survival advantage held by cells expressing functional common γ-chain receptors. In such conditions, HSCs can be manipulated and modified ex vivo, allowing exposure to high multiplicity of infection with viral vectors, increasing the likelihood of targeted gene repair. Even if repair is achieved in only a small fraction of HSC or lymphoid precursor populations, in vivo expansion following reinfusion of corrected cells should provide sustained reconstitution with corrected T cells free of virus-derived components.
Acknowledgments
Research at the Institute of Child Health and Great Ormond Street Hospital for Children NHS Trust benefits from R&D funding received from the NHS Executive. C.A.V. was the recipient of a Child Health Research Appeal Trust award. A.J.T. is a Senior Wellcome Trust Clinical fellow. W.Q. is a Leukaemia Research Fund senior clinical lecturer.
REFERENCES
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdewohld H, Weiher H, Reik W, Jaenisch R., and, Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987;61:336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya S, Steffen DL., and, Robinson HL. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986;60:683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. Targeting retroviral integration. Mol Ther. 2002;6:570–571. [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and γ-retroviral vectors. Mol Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine BL, Humeau LM, Boyer J, MacGregor RR, Rebello T, Lu X, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc Natl Acad Sci USA. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Levine BL, Binder GK, Berry CC, Malani N, McGarrity G, et al. Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells. Mol Ther. 2009;17:844–850. doi: 10.1038/mt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Williams DA. Gene therapy continues to mature and to face challenges. Mol Ther. 2009;17:1305–1306. doi: 10.1038/mt.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanisch K., and, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasik MB., and, McCray PB., Jr Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, et al. Transient gene expression by nonintegrating lentiviral vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Kantor B, Ma H, Webster-Cyriaque J, Monahan PE., and, Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc Natl Acad Sci USA. 2009;106:18786–18791. doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Lu R., and, Engelman A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J Virol. 2001;75:7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Staunstrup NH, Jakobsen M, Yáñez-Muñoz RJ., and, Mikkelsen JG. Genomic insertion of lentiviral DNA circles directed by the yeast Flp recombinase. BMC Biotechnol. 2008;8:60. doi: 10.1186/1472-6750-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Cornu TI., and, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- Bayer M, Kantor B, Cockrell A, Ma H, Zeithaml B, Li X, et al. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Provasi E, Maruggi G, Binini C.et al. (2009Characterization of potential genomic ‘safe harbor' for efficient targeted gene addition with zinc finger nucleases Mol Ther 17S168 [Google Scholar]
- Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ., and, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH., and, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH., and, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- Yant SR., and, Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol Cell Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák Z, Stüwe EE, Fiedler D, Katzer A, Jeggo PA., and, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink CA, Gaspar HB, Gabriel R, Schmidt M, McIvor RS, Thrasher AJ, et al. Sleeping beauty transposition from nonintegrating lentivirus. Mol Ther. 2009;17:1197–1204. doi: 10.1038/mt.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther. 2009;17:1205–1214. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison BS, Yant SR, Mikkelsen JG., and, Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard T, Moldt B, Sharma N, Wolf G, Schmitz A, Pedersen FS, et al. Shielding of sleeping beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Mol Ther. 2009;17:121–130. doi: 10.1038/mt.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S., and, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y., and, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Liang Q, Kong J, Stalker J., and, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]