Abstract
Recent studies suggest that active resolution of the inflammatory response in animal models of arthritis may involve leukotriene B4 (LTB4)-dependent stimulation of “intermediate” prostaglandin production, which in turn favors the synthesis of “downstream” anti-inflammatory and pro-resolving lipoxins, resolvins, and protectins. We explored a putative mechanism involving LTB4-dependent control of cyclooxygenase-2 (COX-2) expression, the rate-limiting step in inflammatory prostaglandin biosynthesis. Indeed, LTB4 potently up-regulated/stabilized interleukin-1β-induced COX-2 mRNA and protein expression under conditions of COX-2 inhibitor-dependent blockade of PGE2 release in human synovial fibroblasts (EC50 = 16.5 ± 1.7 nm for mRNA; 19 ± 2.4 nm for protein, n = 4). The latter response was pertussis toxin-sensitive, and semi-quantitative reverse transcription-PCR confirmed the quantitative predominance of the BLT2 receptor. Transfection experiments, using human COX-2 promoter plasmids and chimeric luciferase-COX-2 mRNA 3′-untranslated region (3′-UTR) reporter constructs, revealed that LTB4 exerted its stabilizing effect at the post-transcriptional level through a 116-bp adenylate/uridylate-rich sequence in the proximal region of the COX-2 3′-UTR. Using luciferase-COX-2 mRNA 3′-UTR reporter constructs and Ras/c-Raf expression and mutant constructs, we showed that the Ras/c-Raf/MEK1/2/ERK1/2 signaling pathway mediated LTB4-dependent COX-2 mRNA stabilization. Knockdown experiments with specific short hairpin RNAs confirmed that LTB4 stabilization of COX-2 mRNA was apparently mediated through the RNA-binding protein, p42 AUF1. The nuclear export of p42 AUF1 was driven by c-Raf/MEK1/2/ERK1/2 signaling and sensitive to leptomycin B treatment, suggesting a CRM1-dependent mechanism. We conclude that LTB4 may support the resolution phase of the inflammatory response by stabilizing COX-2, ensuring a reservoir of ambient pro-resolution lipid mediators.
Keywords: Cyclooxygenase (COX) Pathway, Eicosanoid Function, ERK, Gene Regulation, Leukotriene, RNA-binding Protein, shRNA, Signal Transduction
Introduction
Inflammation and the inflammatory response involve the concerted and exquisitely timed interactions of cytokines, chemokines, growth factors, and lipid-derived mediators with inflammatory cells (1–6). Coordinated control of the inflammatory response involves tightly regulated gene expression at transcriptional, post-transcriptional, and translational levels, the latter two manifested in part by sequence-specific RNA-binding proteins that regulate mRNA stability and translation (reviewed in Refs. 7, 8). mRNA turnover mediated by the adenylate/uridylate (AU)2-rich elements (ARE, also known as Shaw-Kamen sequences) (9), which can act in cis or through binding of ARE-binding proteins (BP) in trans, consists of rapid shortening of the poly(A) tail (deadenylation) and 5′-decapping, followed by decay of the mRNA body (8, 10–15). In this way, inflammatory levels of mRNAs and their translation products can rapidly attain newly programmed steady-state levels following the appropriate homeostatic signal, which may form a critical mechanism in the active resolution of acute inflammation (reviewed in Ref. 16).
Leukotriene B4 (LTB4), a 5-lipoxygenase-derived metabolite of arachidonic acid, is a potent lipid mediator released from activated neutrophils, macrophages, mast cells, and arthritis-affected synovial fibroblasts (17, 18). The bioactive lipid activates leukocyte chemotaxis, degranulation, and production of superoxide anions through cognate G protein-coupled receptors, BLT1 and -2 (19–21). Thus LTB4 plays a defined and important role in host defense during the acute inflammatory phase, although recent studies have suggested that LTB4 may also act in the transition to the resolution of inflammation, a return to homeostasis and preservation of tissue integrity (16). It has been hypothesized that the latter transition is made possible by the induction of pro-resolution bioactive lipids like prostaglandins (PGE2, -D2, and -J2) and the directed downstream synthesis of lipoxins, resolvins, and protectins (22). Although the precise molecular and cellular events involved are not fully understood, it has been observed that prostaglandins, for example, stimulate the biosynthesis of pro-resolving mediators that favor local phagocytic activity and the removal of apoptotic cells and tissue debris from the site of inflammation (e.g. the synovium in acute inflammatory arthritis (22, 23)).
Our previous work demonstrated that PGE2 has potent anti-cytokine and anti-catabolic activities in macrophages and synovial fibroblasts, and we recognize that its inflammomodulatory effects depend on the phase context (24–26). Given the putative role of LTB4 in the transition phase and the fact that the latter may be proven to be a “tipping” point where acute inflammation becomes chronic (see Ref. 27 for mast cell, leukotriene, and inflammatory arthritis link), we hypothesized that LTB4 controls the expression and synthesis of COX-2 in target cells at the site of inflammation (e.g. synovial fibroblasts). The latter enzyme forms the rate-limiting step in the synthesis of eicosanoids/PGE2 and would be the likely target for LTB4 action, notwithstanding the prostaglandin synthases (28, 29). We have used COX-2 and cytokine expression as models for studying inflammatory gene expression in arthritis-affected synovial fibroblasts and have established feasibility for the proposed experiments (24, 30).
In this study, we observed that signal activation through the leukotriene B4 BLT receptors by LTB4 and the BLT2-specific ligand (12S)-hydroxyheptadeca-5Z,8E,10E-trienoic acid (12-HHT, a COX-1 product) up-regulated the expression of COX-2 through a Ras/c-Raf/ERK1/2-p42 AUF1 cascade. This resulted in an increased nucleo-cytoplasmic shuttling of p42 AUF1, which, when bound to the proximal AU-rich region of the COX-2 3′-UTR, markedly increased the stability of COX-2 mRNA and COX-2 protein expression. The mechanism was predominantly post-transcriptional as the ligand had little or no direct effects on COX-2 promoter/transcriptional activation.
EXPERIMENTAL PROCEDURES
Chemicals
Sodium fluoride, leupeptin, aprotinin, pepstatin, phenylmethylsulfonyl fluoride, actinomycin D, dithiothreitol, sodium orthovanadate, and bovine serum albumin were products of Sigma. Leukotriene B4 (LTB4) (5S),(12R)-dihydroxy-6,14-cis-8,10-trans-eicosatetraenoic acid, 12-HHT ((12S)-hydroxyheptadeca-5Z,8E,10E-trienoic acid), cysteinyl leukotrienes LTC4, LTD4, LTE4, NS-398 (N-[2-(cyclohexyloxy)-4-nitrophenyl]-methanesulfonamide), U-75302, and LY255283 were products of Cayman Chemical (Ann Arbor, MI). l-NIL, Bay-11-7082, SB202190, U0126, staurosporine, Nonidet P-40, leptomycin B, and wortmannin were purchased from EMD/Calbiochem. SDS, acrylamide, bisacrylamide, ammonium persulfate, and Bio-Rad protein reagent were from Bio-Rad. Tris-base, EDTA, MgCl2, NaCl, CaCl2, chloroform, DMSO, anhydrous ethanol (95%), methanol (99%), formaldehyde, and formamide were obtained from Fisher. Dulbecco's modified Eagle's medium (DMEM, Invitrogen), phosphate-free DMEM, TRIzol reagent, heat-inactivated fetal bovine serum (FBS), an antibiotic mixture (10,000 units of penicillin (base), 10,000 μg of streptomycin (base)), phosphate-buffered saline, TEMED, and Taq polymerase were products of Invitrogen. Puromycin was purchased from Cedarlane Laboratories (Hornby, Ontario, Canada), and human recombinant IL-1β (rhIL-1β) was from R&D Systems (Minneapolis, MN).
Specimen Selection and Cell Culture
Synovial lining cells (human synovial fibroblasts (HSF)) were isolated from synovial membranes (synovia) obtained at necropsy from donors with no history of arthritic disease (mean age 30 ± 27). Additional experiments were conducted (where indicated) with HSF specimens obtained from osteoarthritic and rheumatoid arthritic (RA) patients undergoing arthroplasty who were diagnosed based on the criteria developed by the American College of Rheumatology Diagnostic Subcommittee for osteoarthritic/RA (mean age 67 ± 19) (31, 32). Human synovial fibroblasts were released by sequential enzymatic digestion with 1 mg/ml pronase (Roche Applied Science) for 1 h, followed by 6 h with 2 mg/ml collagenase (type IA, Sigma) at 37 °C in DMEM supplemented with 10% heat-inactivated FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (33). Released HSF were incubated for 1 h at 37 °C in tissue culture flasks (Primaria catalog no. 3824, Falcon, Lincoln Park, NJ), allowing the adherence of nonfibroblastic cells possibly present in the synovial preparation, particularly from osteoarthritic and RA synovia. In addition, flow cytometric analysis (Epic II, Coulter, Miami, FL), using the anti-CD14 (fluorescein isothiocyanate) antibody, was conducted to confirm that no monocytes/macrophages were present in the synovial fibroblast preparation (30). The cells were seeded in tissue culture flasks and cultured until confluence in DMEM supplemented with 10% FBS and antibiotics at 37 °C in a humidified atmosphere of 5% CO2, 95% air. The cells were incubated in fresh medium containing 0.5–1% FBS for 24 h before the experiments, and only second or third passaged HSF was used. HeLa cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and were grown in DMEM supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a humidified atmosphere with 5% CO2, 95% air.
Preparation of Cell Extracts and Western Blotting
Fifty-100 μg of cellular protein extracted in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml each of aprotinin, leupeptin, and pepstatin, 1% Nonidet P-40, 1 mm sodium orthovanadate, and 1 mm NaF) or hot SDS-PAGE loading buffer, from control and treated cells, were subjected to SDS-PAGE through 10% gels (16 × 20 cm, final concentration of acrylamide) under reducing conditions and transferred onto nitrocellulose membranes (GE Healthcare Amersham Biosciences). Following blocking with 5% BLOTTO for 2 h at room temperature and washing, the membranes were incubated overnight at 4 °C with polyclonal anti-human COX-2 (Cayman Chemical, 1:7500 dilution) in TTBS containing 0.25% BLOTTO. The second anti-rabbit antibody-horseradish peroxidase conjugate (Cell Signaling, Danvers, MA; 1:10,000 dilution) was subsequently incubated with membranes for 1 h at room temperature and washed extensively for 30–40 min with TTBS and a final rinse with TTBS at room temperature. Following incubation with an ECL chemiluminescence reagent (Amersham Biosciences), membranes were prepared for autoradiography, exposed to Kodak X-Omat film, and subjected to digital imaging system (Alpha G-Imager 2000; Canberra Packard Canada, Mississauga, Ontario, Canada) for semi-quantitative measurements. In addition to the anti-COX-2 antisera (Cayman Chemical), the following polyclonal antibodies were used: pan-AUF1 antibody (Upstate Biotechnology, Inc., Lake Placid, NY), goat anti-actin, rabbit anti-GAPDH, and donkey anti-goat horseradish peroxidase conjugate (Santa Cruz Biotechnology, Santa Cruz, CA); anti-histone H3, anti-c-Raf, total (independent of phosphorylation state) and anti-phospho-p38 MAPK (Thr-180/Tyr-182), total and anti-phospho-c-Jun N-terminal kinase (JNK)/stress-activated protein kinase (Thr-183/Tyr-185), total and anti-phospho-p44/42 (Thr-202/Tyr-204), and monoclonal anti-c-Jun (Cell Signaling Ltd., Danvers, MN).
Northern Blot Analysis of mRNA
Total cellular RNA was isolated (1 × 106 cells = 10–20 μg of RNA) using the TRIzol (Invitrogen) reagent. Generally, 5 μg of total RNA were resolved on 1.2% agarose-formaldehyde gel and transferred electrophoretically (30 V overnight at 4 °C) to Hybond-NTM nylon membranes (Amersham Biosciences) in 0.5× tris/acetate/EDTA (TAE) buffer, pH 7. After prehybridization for 24 h, hybridizations were carried out at 50–55 °C for 24–36 h, followed by high stringency washing at 68 °C in 0.1× SSC, 0.1% SDS. The following probes, labeled with digoxigenin (DIG)-dUTP by random priming, were used for hybridization: human COX-2 cDNA (1.8 kb, Cayman), initially cloned into the EcoRV site of pcDNA 1 (Invitrogen), was released by PstI and XhoI digestion, resulting in a 1.2-kb cDNA fragment; a 780-bp PstI/XbaI fragment from GAPDH cDNA (1.2 kb; American Type Culture Collection) that was initially cloned into a PstI site of pBR322 vector. This latter probe served as a control of RNA loading as GAPDH is constitutively expressed in cells used in these experiments. All blots were subjected to a digital imaging system (Alpha G-Imager 2000; Canberra Packard Canada) for semi-quantitative measurements, and changes in COX-2 expression were always considered as a ratio, COX-2:GAPDH mRNA.
Plasmids and Transfection Experiments
Transient transfection experiments were conducted in 4-, 6-, or 12-well cluster plates as described previously (30, 33). Transfections were conducted using FuGENETM 6 (Roche Applied Science) or Lipofectamine 2000 (Invitrogen) reagents for 6 h according to the manufacturers' protocols with cells at around 30–40% confluence. Cells were re-exposed to a culture medium with 1% FBS for 2 h prior to the addition of the biological effectors. Transfection efficiencies were controlled in all experiments by co-transfection with 0.2–0.5 μg of pCMV-β-gal, a β-galactosidase reporter vector under the control of the CMV promoter (Stratagene, La Jolla, CA), or a pHSV-TK-driven Renilla luciferase construct (Promega Corp., Madison, WI). A BSu36I COX-2 promoter (−415 to + 34)-LUC and the full-length COX-2 promoter (−2390 to + 34)-LUC plasmid were kindly provided by Dr. Stephen Prescott, University of Utah. Chimeric luciferase reporter plasmids fused with the human COX-2 mRNA 3′-UTR (1451 bp; 22 Shaw-Kamen sequences), AU-rich elements (429 bp, of which the first 116 bp contain a 6-AU cluster), the 3′-UTR minus the AU-rich element cluster, or a construct completely devoid of the COX-2 3′-UTR but containing the SV40 poly(A) signal were employed (34). The plasmids are designated LUC-3′-UTR, LUC-ARE, LUC-ΔARE, and LUC-Δ3′-UTR (pZEO-LUC3), respectively, and were a kind gift of Dr. D. Dixon, University of Utah.
Ha-Ras (p21) and c-Raf-1 (c-Raf) expression construct sets were obtained from Clontech and were composed of pCMV-driven wild-type Ras, constitutively active RasV12 (G12V), dominant negative RasN17 (S17N), pCMV-driven wild-type c-Raf-1, constitutively active RafCAAX that localizes to the cell membrane when overexpressed, and the dominant negative RafS621A, which lacks the critical Ser-621 phosphorylation site. Expression plasmids for p37, p40, p42, and p45 AUF1 were generated by cloning AUF1 cDNAs into KpnI and NotI (p42 and p45) and KpnI and ApaI (p37 and p40) sites of pcDNA3. They were kindly provided by Dr. Gary Brewer (Department of Molecular Genetics, Microbiology, and Immunology, University of Medicine and Dentistry of New Jersey). The HuR and TTP expression plasmids (cloned into pcDNA3) were kindly provided by Dr. Imad Gallouzi (McGill University, Montreal, Canada) and Dr. William Rigby (Dartmouth Medical School, Hanover, NH), respectively.
RNA interference and gene knockdown were achieved through the use of short hairpin RNAs targeted to particular sequences of the AUF1 gene, including all four isoforms, p37, p40, p42, and p45. The short hairpins were cloned into BamHI/HindIII sites and consisted of a 29-nucleotide target sequence, a 7-nucleotide loop, and the 29-nucleotide reverse complementary sequence followed by a termination sequence for RNA polymerase III. The expression was driven by a U6 small nuclear RNA gene promoter as part of the pRS (retroviral silencing) vector. Targeted sequences include the following: those from exon 9, shRNA-1; exon 1, shRNA-2; exon 3, shRNA-3, and exon 3/4, shRNA-4 (OriGene Technologies, Rockville, MD). The empty vector (pRS) served as a negative control. For stable transfectants, HeLa cells were grown to 50% confluence in DMEM supplemented with 10% FBS and transfected with AUF1-shRNAs using FuGENE 6 according to the manufacturer's instructions. After 48 h, cells were cultured in fresh medium containing puromycin (1 μg/ml), and the puromycin-containing medium was changed every 3–4 days. After 2–4 weeks, resistant isolated colonies appeared, subjected to limiting dilution procedures, and cultured further using standard culture conditions.
Luciferase values, expressed as enhanced relative light units, were measured in a Lumat LB 9507 luminometer (EG&G, Stuttgart, Germany) and normalized to the levels of β-galactosidase activity (absorbance at 450 nm after 24 h of incubation) and cellular protein (bicinchoninic acid procedure; Pierce).
RT-PCR
The oligonucleotide primers for the PCRs were prepared with the aid of a DNA synthesizer (Cyclone model, Biosearch Inc., Montreal, Quebec, Canada) and used at a final concentration of 200 nmol/liter. The sequences for the luciferase primers were as follows: 5′-ACGGATTACCAGGGATTTCAGTC-3′ and 5′-AGGCTCCTCAGAAACAGCTCTTC-3′ (antisense) for the luciferase fragment of 367 bp (30, 34). The sequences for the GAPDH (which served as a standard of quantitation) primers were 5′-CAGAACATCATCCCTGCCTCT-3′, which corresponds to positions 604–624 bp of the published sequence, and 5′-GCTTGACAAAGTGGTCGTTGAG-3′, from positions 901 to 922 bp, for an amplified product of 318 bp (30, 34). For BLT1, we used nested PCR as follows: nested-1 (sense) 5′-GCCCAAGGCACCTGGA-3′ and (antisense) 5′-GCGGCGAAGGTCAGGATGAT-3′ (460 bp: 6720–7180 bp); nested-2 (sense) 5′-GTCTGCGGAGTCAGCATGTA-3′ and (antisense) 5′-TTCGTTTTCCAGGGCACTAC-3′ (206 bp), corresponding to positions 6771–6976 bp of the published sequence (GenBankTM accession number AB008193). For the BLT2 receptor, standard RT-PCR was sufficient: (sense) 5′-AGACTCTGACCGCTTTCGTG-3′ and (antisense) 5′-AAGGTTGACTGCGTGGTAGG-3′ (182 bp), corresponding to positions 2059 to 2241 bp of the published sequence (20, 21).
Two μg of total RNA, extracted with the TRIzol reagent, was reverse-transcribed and then subjected to PCR as described previously (30). RT and PCR assays were carried out with the enzymes and reagents of the GeneAmP RNA PCR kit manufactured by PerkinElmer Life Sciences. Both the RT and PCR reactions were done in a Gene ATAQ Controller (GE Healthcare). The amplification process was conducted over 10–30 cycles to define the linear range of product amplification as follows: the first cycle consisted of a denaturation step at 95 °C for 1 min, followed by annealing and elongation at 60 °C for 30 s and 72 °C for 1.5 min, respectively. All subsequent cycles were executed under the same conditions, with the exception of the last cycle, where the elongation step was extended to 7 min. We found a linear range (log luciferase/GAPDH versus log cycle number) between 10 and 17 cycles; as such, we chose 11–13 cycles depending on the type of experiment. In the case of nested PCR for BLT1, the conditions were essentially the same except cycle number was increased to 30–40 cycles.
The PCR products were analyzed and verified by electrophoresis on 1.15% agarose gels in a Tris borate/EDTA buffer system as described previously (30). All gel photos were subjected to a digital imaging system (see above) for semi-quantitative measurements, and where indicated, results were expressed as a ratio of luciferase/GAPDH PCR fragments.
Statistical Analysis
All results were expressed as the mean ± S.D. or mean and the coefficient of variation (CV) of 3–5 separate experiments as indicated. Transfection experiments were performed in triplicate. Statistical treatment of the data was performed parametrically (Student's t test) or by nonparametric (Mann-Whitney) analysis if Gaussian distribution of the data could not be confirmed. Where appropriate, ANOVA analysis was used to compare the statistical difference between multiple mean values. Significance was acknowledged when the probability that the Null Hypothesis was satisfied at <5%.
RESULTS
LTB4 Up-regulated COX-2 mRNA and Protein in HSF
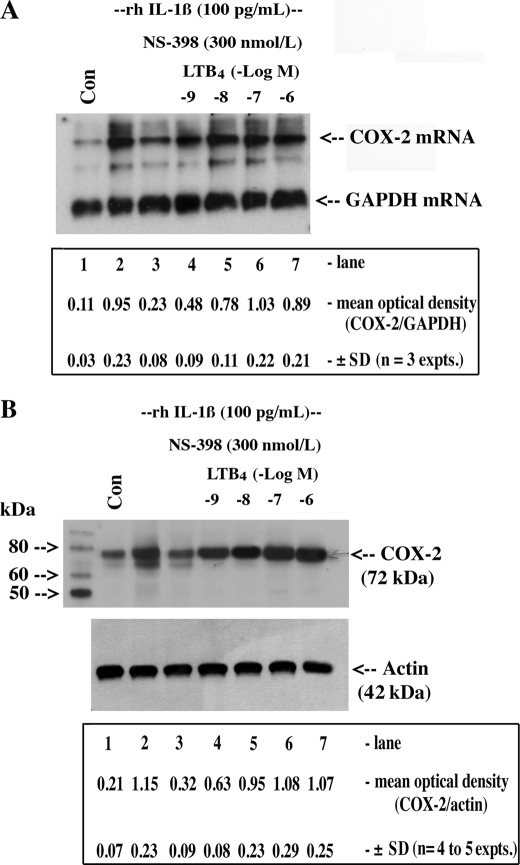
Previous studies in our laboratory showed that COX-2 expression in HSF is controlled in part by a positive feedback loop involving PGE2-mediated stabilization of COX-2 mRNA and that ambient PGE2 may explain persistently high levels of COX-2 expression in RA-affected synovium (30). However, COX-2 mRNA and protein levels remained evident in explanted arthritis-derived synovial membranes exposed to COX-2 inhibitors (COXib) (25), suggesting that other COXib-insensitive inflammatory mediators may be responsible. Notwithstanding cytokine activity, and given the recent appreciation for the impact of neutrophils/mast cells and their cell-derived inflammatory mediators (e.g. LTB4) in the biology of the inflammatory response (27), we tested the hypothesis that activated neutrophil/mast cell products may play a role in the regulation of the prototypic inflammomodulatory gene COX-2. We examined a number of such products (LTC4, LTD4, and LTE4, data not shown) and found that LTB4, added in increasing concentrations, potently up-regulated/stabilized rhIL-1β-induced COX-2 mRNA and protein expression under conditions of COXib-dependent inhibition of PGE2 release in HSF (Fig. 1, A and B, EC50 = 16.5 ± 1.7 nm for mRNA; 19 ± 2.4 nm for protein, mean ± S.D., n = 3–5). Because LTB4 functions in target tissues through cognate G protein-coupled receptors BLT1 and -2, we performed semi-quantitative RT-PCR to determine, in a preliminary fashion, the expression profile in HSF. We observed that the apparent level of BLT2 mRNA was greater than BLT1 given that BLT1 mRNA detection required nested (two rounds of cycling) PCR procedures, whereas BLT2 mRNA PCR fragments could be clearly detected after 30 cycles of standard RT-PCR (data not shown).
FIGURE 1.
LTB4 up-regulated/stabilized COX-2 mRNA and protein in HSF. Cultured confluent HSF (1.2 × 106 cells in 6-well cluster plates) were preincubated overnight in DMEM supplemented with 1% FBS plus antibiotics at 37 °C to ensure synchrony and quiescence. Cells were then treated with vehicle (Con) or with 5.7 pmol/liter (100 pg/ml) of rhIL-1β for 16 h in the presence or absence of the COX-2 inhibitor NS-398 and increasing concentrations of LTB4 as indicated. Monolayers were extracted for RNA (A) and protein (B) as follows: 5 μg of total RNA was analyzed for COX-2 mRNA and GAPDH mRNA by Northern hybridization using specific DIG-labeled cDNA probes, and 50 μg of protein was analyzed for COX-2 and actin (loading control) protein by Western blotting using specific polyclonal antisera as described under “Experimental Procedures.” ANOVA of means for densitometric analysis in A and B is as follows: IL-1β + NS-398 versus IL-1β + NS-398 + LTB4 (−log M −9 to −6), p < 0.007; IL-1β versus IL-1β + NS-398 + LTB4 (−log M −8 to −6), not significant (NS).
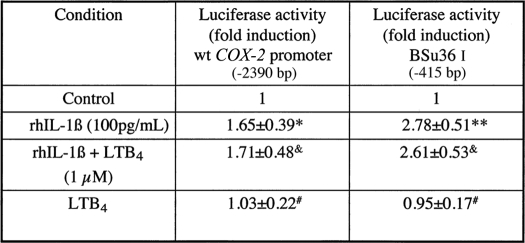
To assess the contribution of transcriptional and post-transcriptional mechanisms to the LTB4-dependent up-regulation of COX-2, we initially conducted experiments with COX-2 promoter constructs in transfected HSF; the wild-type and Bsu36I constructs were used as a representative study (see Table 1). Recombinant human IL-1β induced a 1.65–2.78-fold increase in promoter activity as measured by the luciferase reporter activity, whereas LTB4 co-incubated with rhIL-1β or alone was without effect. Judging by these observations, it seemed unlikely that LTB4 modified COX-2 expression in HSF at the transcriptional level. As such, we examined post-transcriptional mechanisms involving strictly message stabilization and protein translation. As a first approach, we employed classical techniques involving measuring COX-2 mRNA in transcriptionally arrested cells (actinomycin D) in the absence or presence of LTB4 (30, 33). When HSF were activated with rhIL-1β for 2 h (steady state) followed by wash-out and a fresh change of medium, the elevated levels of COX-2 mRNA declined rapidly such that within 1–2 h the levels were similar to control unstimulated cells (Fig. 2A, lanes 1, 2, and 7–10). The inclusion/exclusion of actinomycin D (1 μg/ml) had no effect on the rate of COX-2 mRNA decay in these experiments (data not shown). However, if LTB4 (50 nm, saturation kinetics) was added to fresh medium (in the presence of actinomycin D), COX-2 mRNA levels declined slowly such that COX-2 messages were discernible for up to 8 h (Fig. 2A, lanes 1–6). Parenthetically, these results could be reproduced in both isolated neutrophil and macrophage populations in short term culture conditions (data not shown).
TABLE 1.
Transcriptional activation of the COX-2 promoter, effect of LTB4
HSFs at 30–50% confluence were transiently transfected using FuGENE 6 for 6 h with 1 μg/well human COX-2 promoter (wild type and BSu361)-luciferase constructs plus 200 ng of pCMV-β-gal expression vector (transfection efficiency control marker). Fold-induction values were expressed as mean ± S.D. from three to five determinations in duplicate. Typical values for promoter induction by rhIL-1β ranged from 2.45 × 104 to 5.6 × 104 relative light units.
*p < 0.025.
** p < 0.001 versus controls.
& Not significant (NS) versus rhIL-1β.
# Not significant (NS) versus controls.
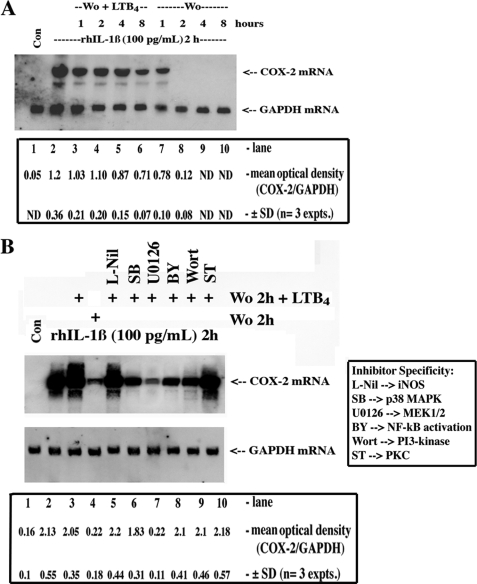
FIGURE 2.
LTB4-dependent stabilization of COX-2 mRNA, effects of chemical kinase inhibitors. A, quiescent HSF were treated with vehicle (Con) or with 100 pg/ml (5.7 pmol/liter) of rhIL-1β for 2 h (steady state), after which time cells were washed out (Wo) and treated with actinomycin D (1 μg/ml) for 30 min, and then fresh medium was added containing either vehicle (Wo) or LTB4 (50 nmol/liter). After an additional 1, 2, 4, or 8 h of incubation, monolayers were extracted for RNA at each time point, and 5 μg of total RNA was analyzed for COX-2 mRNA and GAPDH mRNA by Northern hybridization using specific DIG-labeled cDNA probes. B, cells were treated with vehicle (Con) or with 100 pg/ml (5.7 pmol/liter) of rhIL-1β for 2 h, washed out (Wo), and treated with actinomycin D (1 μg/ml) for 30 min, and then fresh medium was added containing either vehicle (washed out 2 h) or LTB4 (50 nmol/liter) alone or in the presence of l-Nil (1 μm), SB202190 (SB, 1 μmol/liter), U0126 (2 μmol/liter), Bay-11-7082 (BY, 5 μmol/liter), wortmannin (Wort, 200 nmol/liter), or staurosporine (ST, 10 nmol/liter) for 2 more h. 5 μg of total RNA was analyzed for COX-2 mRNA and GAPDH mRNA by Northern hybridization using specific DIG-labeled cDNA probes. ANOVA of the means for densitometric analysis is as follows: A, IL-1β versus IL-1β + washed out (Wo) + LTB4 (1–4 h), not significant (NS), and ND, not determined; IL-1β versus washed out (1, 2 h), p < 0.001; B, Student's t test; IL-1β versus IL-1β + washed out 2 h, p < 0.003; IL-1β versus IL-1β + washed out 2 h + LTB4, not significant; IL-1β versus IL-1β + washed out 2 h + LTB4 + U0126, p < 0.006; IL-1β versus IL-1β + washed out 2 h + LTB4 + other kinase inhibitors, not significant.
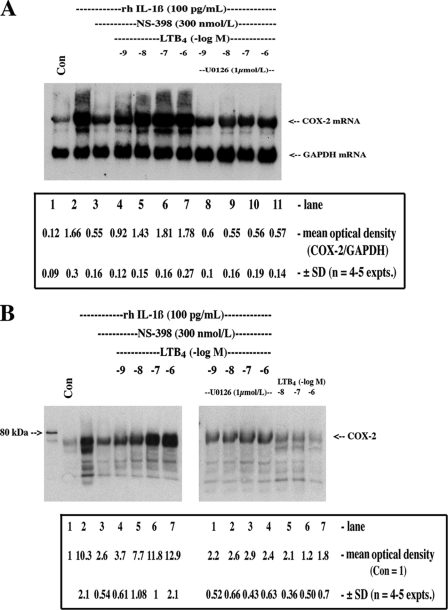
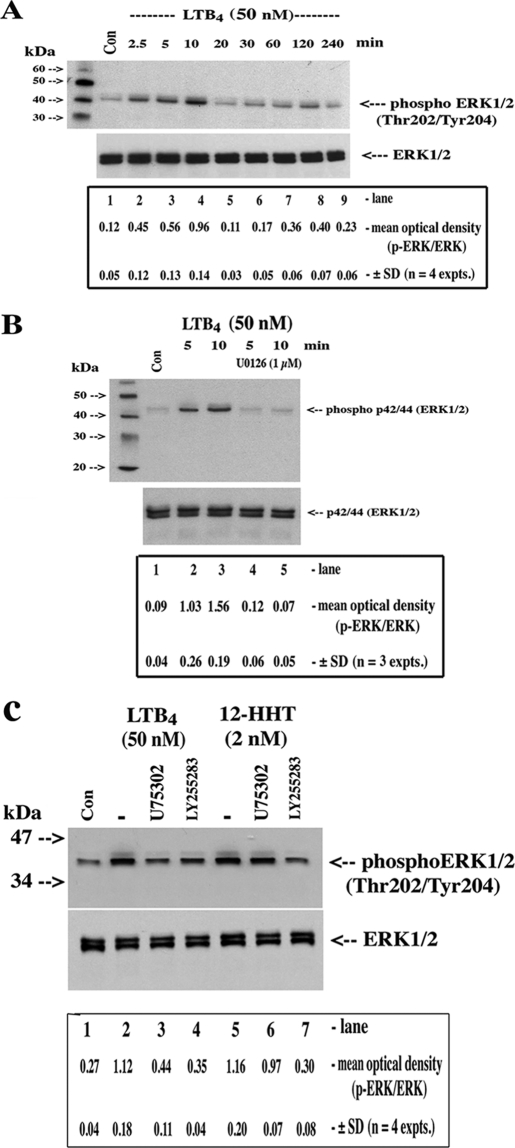
Because the BLT receptors mediate both pertussis toxin-sensitive and -insensitive cellular responses to LTB4 and likely associate with Gαi, Gαq, and Gα11/14 subunits (35), we used, in a preliminary context, cell-permeable inhibitors of key kinase signaling cascades and found that U0126, a MEK1/2 inhibitor, abolished LTB4-dependent up-regulation/stabilization of induced COX-2 mRNA (Fig. 2B); p38 MAPK, NF-κB, protein kinase C, phosphatidylinositol 3-kinase inhibitors were far less active in this regard. Indeed, the inclusion of cell-permeable chemical inhibitors into the basic protocol (i.e. Fig. 1) supported the previous observations regarding the role of ERK1/2 cascade, as U0126 substantially reversed the stabilization effects of LTB4 on induced COX-2 mRNA and protein (Fig. 3, A and B). LTB4 stabilized basal levels of COX-2 (Fig. 3B, last 3 lanes) but, in the absence of induced COX-2 gene transcription, the levels are necessarily low. Finally, we used a Western blotting screening protocol of kinase activation using specific anti-phospho antibodies (data not shown) and verified the predominant activation of the ERK1/2 pathway by LTB4 (Fig. 4, A and B). Interestingly, the LTB4-dependent activation of ERK1/2 phosphorylation was blocked by both a BLT1-specific antagonist (U75302) and the BLT2 antagonist (LY255283) (Fig. 4C). The specificity of the antagonists was verified with 12-HHT, a preferential, COX-1/thromboxane synthase-derived BLT2 receptor agonist; 12-HHT-dependent activation of ERK1/2 phosphorylation was reversed exclusively by the BLT2 antagonist (Fig. 4C).
FIGURE 3.
LTB4-dependent stabilization of COX-2 mRNA was mediated through MAPK signaling. Cells were treated with vehicle (Con) or with 5.7 pmol/liter (100 pg/ml) of rhIL-1β for 16 h in the presence or absence of the COX-2 inhibitor NS-398 and increasing concentrations of LTB4 with or without U0126 (1 μmol/liter). Monolayers were extracted for RNA and protein as follows: A, 5 μg of total RNA was analyzed for COX-2 mRNA and GAPDH mRNA by Northern hybridization using specific DIG-labeled cDNA probes; B, 50 μg of protein was analyzed for COX-2 protein by Western blotting using specific polyclonal antisera as described under “Experimental Procedures.” ANOVA of means for densitometric analysis in A and B is as follows: IL-1β + NS-398 versus IL-1β + NS-398 + LTB4 (− log M −9 to −6), p < 0.001; IL-1β versus IL-1β + NS-398 + LTB4 (− log M −8 to −6), not significant; IL-1β + NS-398 + LTB4 (− M −9 to −6) versus IL-1β + NS-398 + LTB4 (− log M −9 to −6) + U0126, p < 0.002.
FIGURE 4.
LTB4-dependent activation of ERK1/2 phosphorylation. A, cultured confluent serum-starved HSF (1.2 × 106 cells in 6-well cluster plates) were treated with vehicle (Con) or for varying time periods (2.5–240 min) with LTB4 (50 nm). B, cells were treated with vehicle (Con) or with LTB4 (50 nm) in the presence of U0126 as indicated. C, cells were treated with vehicle (Con), LTB4, or with 12-HHT in the absence or presence of the BLT1 antagonist U75302 or the BLT2 antagonist LY255283 for 10 min as indicated. Monolayers were extracted for protein, and 50 μg was analyzed for total and phospho-ERK1/2 by Western blotting using specific rabbit polyclonal antisera as described under “Experimental Procedures.” ANOVA of means for densitometric analysis in A, control versus LTB4 (2.5 to 10 min), p < 0.009; B, LTB4 5–10 min versus LTB4 5–10 min + U0126, p < 0.0003; C, Student's t test; LTB4 versus LTB4 + U75302, p < 0.05; LTB4 versus LTB4 + LY255283, p < 0.02; control versus 12-HHT, p < 0.023; 12-HHT versus 12-HHT + U75302, not significant; 12-HHT versus 12-HHT + LY255283, p < 0.021.
LTB4 Stabilized COX-2 mRNA through the Proximal 116-bp AU-rich Region of the COX-2 3′-UTR
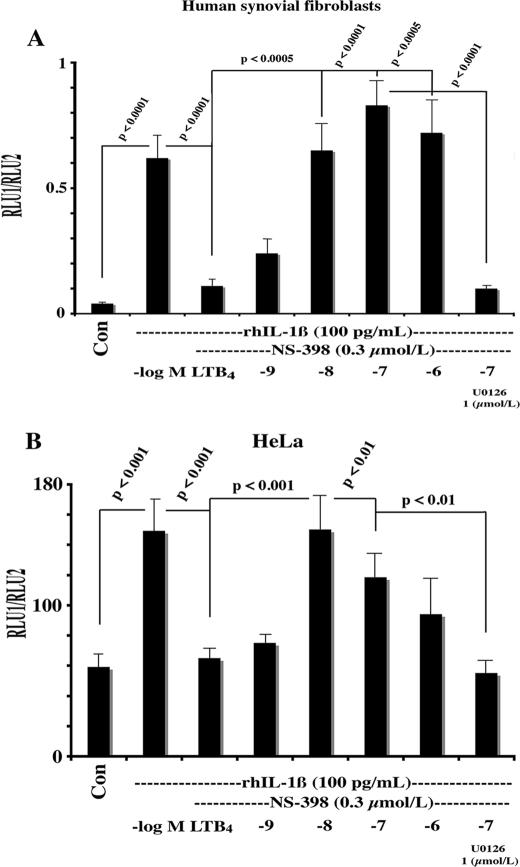
The COX-2 mRNA has multiple copies of the Shaw-Kamen AU-rich sequences that are believed to influence message stability (34, 36). Studies (34, 37) have provided evidence that the six AU-rich elements in the first 116 bp of the 3′-UTR may mediate COX-2 mRNA instability by a number of biological effectors, although PGE2 functions via distal Shaw-Kamen sequences in the 3′-UTR (30). Furthermore, specific cytoplasmic AU-binding proteins (AUBPs) were identified that may initiate message degradation (8, 37, 38). To determine whether LTB4/ERK1/2-dependent COX-2 mRNA stabilization was manifested through the 3′-UTR and AU-rich sequences, we transfected HSF with CMV-driven chimeric expression constructs containing luciferase cDNA (reporter) fused to the COX-2–3′-UTR (Luc+3′-UTR). Using an identical experimental design as in Fig. 1, we observed that rhIL-1β stimulated luciferase mRNA after 16 h of incubation by more than 3-fold using Luc+3′-UTR constructs in both HSF and HeLa cell cultures (Fig. 5, A and B). The addition of NS-398 COXib markedly reduced luciferase activity, whereas LTB4 reversed NS-398-dependent inhibition and stimulated reporter activity to levels similar to rhIL-1β; the LTB4-dependent effect was blocked by the U0126 in both HSF and HeLa cultures (Fig. 5, A and B).
FIGURE 5.
LTB4-stabilized luciferase activity expressed from a luciferase-COX-2–3′-UTR fusion chimeric construct; effect of the MEK1/2 inhibitor U0126. Both HSF (A) and HeLa cells (B) were plated at 40% confluence in DMEM supplemented with 10% heat-inactivated FBS and antibiotics. One μg of the luciferase-COX-2–3′UTR chimeric fusion construct and 50 ng of a pHSV-TK-driven Renilla luciferase plasmid were co-transfected for 6 h using FuGENE 6 according to the manufacturer's instructions. Cells were incubated overnight in complete medium; following a change to medium containing 1% FBS (2 h), transfected cells were incubated for 16 h with vehicle (Con) or with rhIL-1β (5.7 pmol/liter), NS-398 (300 nmol/liter), LTB4 (1–1000 nmol/liter), and U0126 (1 μmol/liter) as per the figure. Cells were lysed, and the luciferase activity and protein content were determined as described under “Experimental Procedures.” Values were expressed as the mean of the ratio of RLU1 (Firefly)/RLU2(Renilla) normalized to protein content.
Role of Ras/c-Raf/MEK1/2/ERK1/2 Signaling Pathway in LTB4-mediated COX-2 mRNA Stabilization
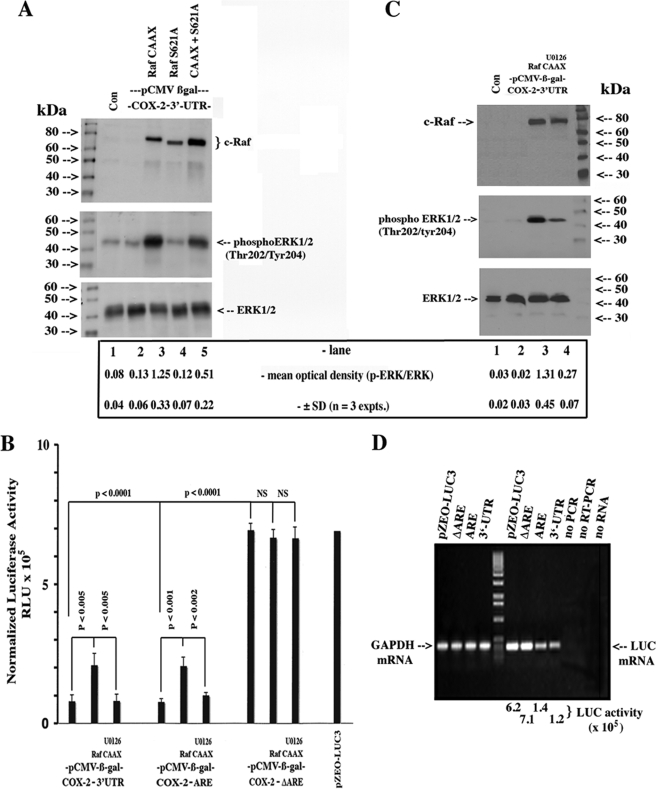
Given the results with the MEK1/2 inhibitor, a role for the MAPK cascade in the LTB4 stabilization of COX-2 mRNA was strongly suggested. In principle, signal transducers and kinases upstream of MEK1/2 and ERK1/2 should recapitulate the LTB4-dependent effects. To this end, we used an overexpression strategy with c-Raf expression constructs together with their respective dominant negative mutants to substantiate our observations with the chemical inhibitor. Indeed, overexpression of the RafCAAX construct strongly stimulated ERK1/2 phosphorylation, whereas co-transfection with the RafS621A dominant negative mutant largely abrogated the effect (Fig. 6A), although it was not as efficient as U0126 (data not shown). We incorporated another dimension to this series of experiments by using deletion mutants of the 3′-UTR (see “Experimental Procedures”) fused to the luciferase reporter to isolate the Shaw-Kamen sequences mediating the putative LTB4-dependent stabilization effects. As shown in Fig. 6B, the overexpression of the constitutively active RafCAAX induced luciferase reporter activity in the full-length construct and the construct harboring the ARE in the first 116 bp; the distal sequences were largely refractive to the c-Raf-induced effect. Fig. 6C confirmed the expression patterns of RafCAAX and activation of ERK1/2 phosphorylation under the experimental conditions of Fig. 6B. The effect of 3′-UTR sequences on basal levels of luciferase activity and mRNA is highlighted in Fig. 6B (1st column versus 10th column) and Fig. 6D (6th lane from left versus 9th lane), respectively.
FIGURE 6.
Role of Ras/c-Raf/MEK1/2/ERK1/2 signaling pathway in the stabilization of luciferase activity expressed from a luciferase-COX-2–3′-UTR fusion chimeric construct. A, HeLa cells were plated at 40% confluence in DMEM supplemented with 10% heat-inactivated FBS and antibiotics. 200 ng of the luciferase-COX-2–3′-UTR chimeric fusion construct, 400 ng each of pCMV, pRafCAAX, or pRafS621A, and 200 ng of a pCMV-β-gal were co-transfected for 6 h using FuGENE 6 according to the manufacturer's instructions. Cells were incubated overnight in complete medium and following a change to medium containing 1% FBS (2 h), and monolayers were extracted for protein, and 50 μg were analyzed for c-Raf, total, and phospho-ERK1/2 by Western blotting using specific rabbit polyclonal antisera as described under “Experimental Procedures.” Untransfected cells were cultured and extracted in the same way (Con). B, cells were transfected with 10 ng of pCMV-luciferase-Δ3′-UTR (pZEO-LUC3), pCMV-luciferase-COX-2–3′-UTR, pCMV-luciferase-COX-2-ARE, or pCMV-luciferase-COX-2-ΔARE with or without 400 ng of pRafCAAX, 200 ng of pCMV-β-gal, and U0126 (1 μmol/liter) as indicated. Values were expressed as the mean ± S.D. of luciferase reporter activity normalized to β-galactosidase activity and protein content. Monolayers from tandem experiments, including untransfected cells (Con), were extracted for protein, and 50 μg were analyzed for c-Raf, total, and phospho-ERK1/2 by Western blotting (C). D, either HeLa cells or HSFs were transfected with 1 μg of pCMV-luciferase-Δ3′-UTR (pZEO-LUC3), pCMV-luciferase-COX-2–3′-UTR, pCMV-luciferase-COX-2-ARE, or pCMV-luciferase-COX-2-ΔARE for 6 h using FuGENE 6. Cells were incubated overnight in complete medium and, following a change to medium containing 1% FBS (2 h), monolayers were extracted for total RNA and subjected to RT-PCR using specific primers for luciferase and GAPDH as per “Experimental Procedures.” Densitometric analysis in A, Student's t test; pCMV versus pRafCAAX, p < 0.022; pRafCAAX versus pRafS621A, p < 0.05; C, β-gal versus p RafCAAX, p < 0.001; pRafCAAX versus pRafCAAX + U0126, p < 0.021.
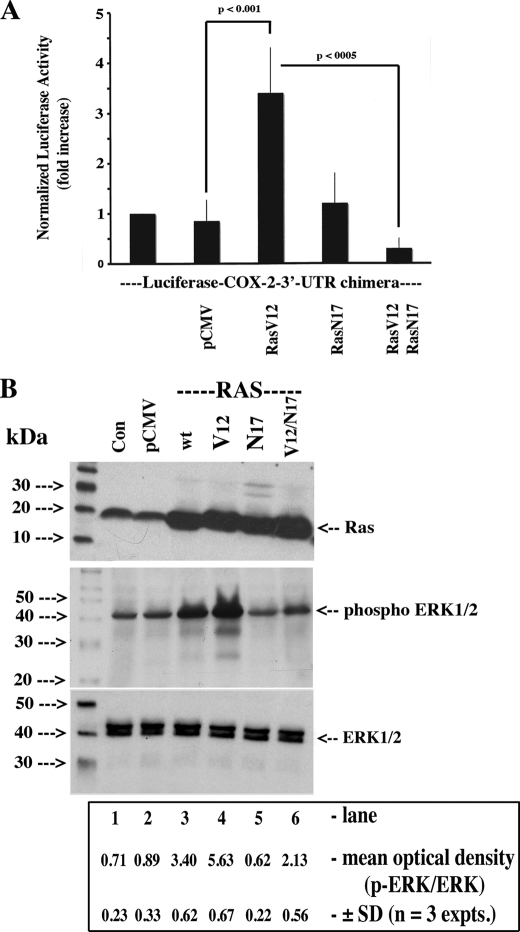
Our RafCAAX construct harbors a K-Ras C-terminal localization signal targeting the kinase to the plasma membrane where a Ras-independent mechanism completes the activation process through phosphorylation (39). A number of receptor-associated kinases can phosphorylate c-Raf resulting in its interaction/activation of pathways not necessarily associated with the G-protein-coupled receptors(BLT)/Ras pathway (39, 40). As such, we investigated whether a constitutively activated form of p21 Ras (and a wild-type construct) could activate endogenous c-Raf (Raf-1) and propagate the same signal required to stabilize COX-2 mRNA. The intent was to validate our observations in Fig. 6 and link our findings to Gi- and/or Gq-coupled receptors. As shown in Fig. 7A, co-transfection of RasV12 with the luciferase-COX-2–3′-UTR fusion construct resulted in a greater than 3-fold (3.3 ± 0.9; n = 5) increase in reporter activity; the addition of RasN17 (3rd column versus 5th column) to the latter transfection mixture abrogated the inductive effect. Data in Fig. 7B confirmed the expression patterns of the plasmids RasV12 (Ha-Ras) and RasN17 and the activation/inhibition of ERK1/2 phosphorylation under the experimental conditions of Fig. 7A.
FIGURE 7.
Ha-Ras (p21) stabilization of luciferase activity expressed from a luciferase-COX-2–3′-UTR fusion chimeric construct. A, HeLa cells were plated at 40% confluence in DMEM supplemented with 10% heat-inactivated FBS and antibiotics. 500 ng of the luciferase-COX-2–3′-UTR chimeric fusion construct, 500 ng each of pCMV, pRasV12, or pRasN17, and 50 ng of a pCMV-β-gal were co-transfected for 6 h using FuGENE 6 according to the manufacturer's instructions. Cells were incubated overnight in complete medium, and following a change to medium containing 1% FBS (2 h), the cells were lysed, and the luciferase activity, β-galactosidase activity, and protein content were determined as described under “Experimental Procedures.” Values were expressed as the mean ± S.D. of fold induction of luciferase activity normalized to β-galactosidase activity and protein content versus the control (1% FBS for 2 h). B, tandem monolayers, including untransfected cells (Con), were extracted for protein, and 50 μg were analyzed for Ha-Ras and phospho-ERK1/2 by Western blotting using specific rabbit polyclonal antisera as described under “Experimental Procedures.” Densitometric analysis in B, Student's t test; pCMV versus wtRas, p < 0.01; pCMV versus pV12Ras, p < 0.004; pV12Ras versus pV12Ras/RasN17, p < 0.04.
Role of AUF1 (Heterogeneous Nuclear Ribonucleoprotein D) in the LTB4/Ras/c-Raf/MEK1/2/ERK1/2 Signaling Pathway-mediated Stabilization of COX-2 mRNA
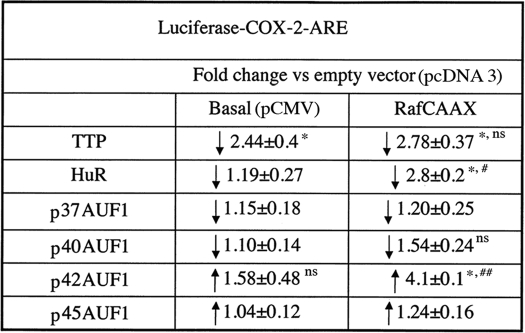
Well characterized AUBPs either stabilize target mRNA through as yet ill-defined mechanisms or enhance the degradation of the target through recruitment of the exosome, a scaffold containing a number of RNA-degrading enzymes (9). Both systems respond rapidly to changes in extracellular signaling patterns (e.g. as in pathological states). The simplest interpretation of our results suggests that LTB4 either suppresses the expression and/or activity of AUBPs that destabilize mRNA or the reverse. In this regard, we used our luciferase reporter screening assay (see above) to evaluate the behavior of TTP, HuR, and AUF1 under basal and MAPK signaling conditions. These AUBPs were chosen based on previous reports regarding COX-2 mRNA stability (13, 14, 34, 36). As shown in Table 2, ectopic expression of TTP precipitated a marked and specific (i.e. minus effects of pcDNA3.1 cloning vector) reduction in reporter activity under basal conditions; TTP was apparently refractive to MAPK signaling because the magnitude of reporter suppression was statistically similar in both basal (pCMV) and RafCAAX-transfected cells. Furthermore, when HuR (ELAVL1), a member of the embryonic lethal abnormal vision (ELAV)-like family of RNA-binding proteins (8, 38), was overexpressed in the presence or absence of RafCAAX, a discernible diminution of reporter activity was observed under basal conditions, but a marked reduction was obtained in RafCAAX-transfected cells. In contrast, under similar experimental conditions, the p42 isoform of AUF1 increased basal but especially RafCAAX-induced reporter activity, whereas the p37, p40, and p45 isoforms were essentially without effect.
TABLE 2.
Effect of RNA-binding protein overexpression on luciferase activity
HeLa cells at 30–50% confluence were transiently transfected using FuGENE 6 for 6 h with 10 ng per well of luciferase-COX-2 ARE fusion chimeric construct together with 500 ng of pcDNA3 or pcDNA3 harboring the TTP, HuR-FLAG, or p37, p40, p42, or p45 AUF1 cDNAs. In addition, 400 ng of pCMV or pRafCAAX constructs and 200 ng of pCMV-β-gal (transfection efficiency control marker) were also included in the mixture. Luciferase reporter activities were measured 24 (data not shown) and 72 h post-transfection. Fold-change values were expressed as mean ± S.D. from three to five determinations. Typical values for luciferase-COX-2-ARE reporter ranged from 7.3 × 103 to 1.29 × 105 relative light units corrected for β-galactosidase expression/activity. ns means not significant.
*Minimum p < 0.01 versus pcDNA3; basal (pCMV) versus pRafCAAX.
#p < 0.007.
##p < 0.005.
shRNA Knockdown of p42 AUF1 and LTB4/c-Raf-dependent COX-2 Stabilization
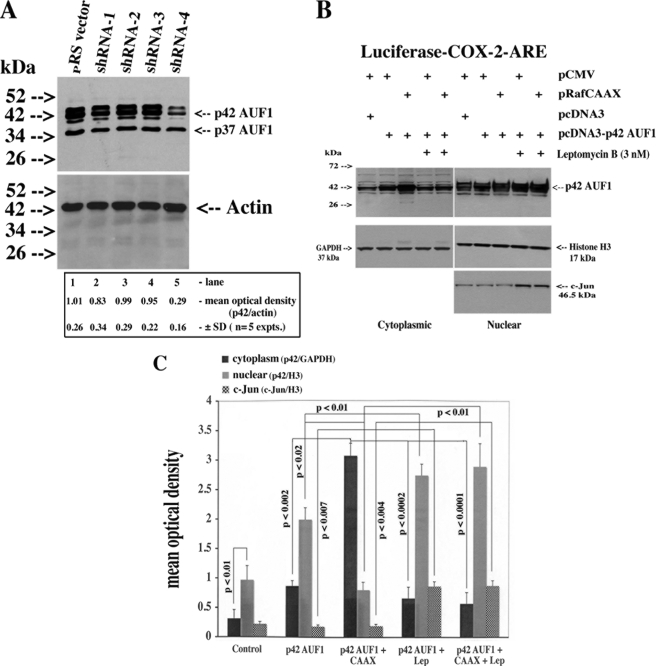
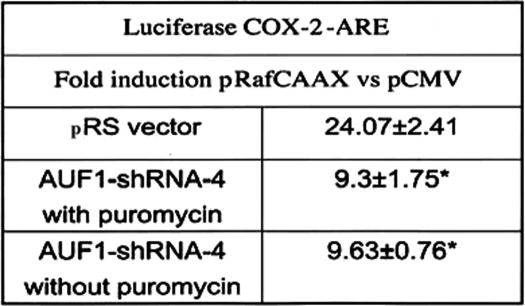
The previous experiments suggested that the p42 AUF1 isoform mediated the LTB4/c-Raf-controlled stabilization of COX-2 mRNA. To confirm this possibility, we chose a gene knockdown strategy involving the use of four different HuSH 29-mer shRNAs targeting exons 1, 3, 4, and 9 of AUF1 stably transfected in HeLa cells. As shown in Fig. 8A, only shRNA-4, spanning the junction of exon 3 and 4, reduced protein expression of AUF1 to a significant extent. Using a pan-AUF1 antibody, it was revealed that the expression levels of isoforms p40, p42, and p45 were reduced a minimum of 3-fold, whereas p37 remained stable (p42, 3.4 ± 0.7-fold versus pRS empty vector, mean ± S.D., n = 5 experiments). Parenthetically, exposure times for film development were increased because of relatively low levels of endogenous AUF1. Stable p42 AUF1 knockdown inhibited RafCAAX activation of luciferase reporter activity by 2.8 ± 0.31-fold (Table 3).
FIGURE 8.
shRNA knockdown of p42 AUF1 and BLT/c-Raf/ERK1/2 signaling induced a leptomycin B-sensitive nuclear export of p42 AUF1. A, HeLa cells were stably transfected through puromycin selection with the pRS vector or four constructs containing shRNA gene-specific target sequences of AUF1 as described under “Experimental Procedures.” Monolayers were extracted for protein, and 50 μg were analyzed for AUF1 and actin by Western blotting using pan-AUF1 and actin antibodies. B, HeLa cells (3 × 105 in 6-well plates) were transiently co-transfected with 10 ng of luciferase-COX-2-ARE, 500 ng of pcDNA 3, or pcDNA3-p42 AUF1, and 400 ng of pCMV or pRafCAAX as per the illustration. After 16 h, 3 nm leptomycin B or vehicle was added to the wells as indicated for an additional 16 h, after which time monolayers were extracted for nuclear and cytoplasmic protein and 30–50 μg were analyzed for AUF1, GAPDH (cytoplasm), histone H3 (nuclear), and c-Jun (nuclear) by Western blotting. Densitometric analysis in A, Student's t test; pRS versus pRS AUF1-shRNA-4, p < 0.001; pRS versus pRSAUF1-shRNA-1–3, NS, not significant. C, densitometric and statistical analysis of B.
TABLE 3.
AUF1 knockdown suppresses Raf-induced luciferase activity
HeLa cells (3 × 105 cells in 6-well plates), stably transfected with AUF1-shRNA-4, were transiently transfected with 10 ng/well luciferase-COX-2-ARE fusion chimeric construct, 400 ng of pCMV or pRafCAAX, and 200 ng of pCMV-β-gal. Luciferase reporter activities were measured 24 h post-transfection. Fold-induction values were expressed as mean ± S.D. from four determinations. Typical values for luciferase reporter activity ranged from 1.2 × 104 (pCMV) to 4.58 × 105 relative light units corrected for β-galactosidase activity.
* p < 0.0008 versus pRS vector.
BLT/c-Raf/ERK1/2 Signaling Induced a Leptomycin B-sensitive Nuclear Export of p42 AUF1
All AUF1 family members were detected in the nuclear compartment of HeLa cells and HSF under quiescent conditions, and small but discernible levels of p40 and p42 were observed in cytoplasmic extracts (data not shown). In principle, AUF1 proteins act on mRNA stability (and protein translation) in the cytoplasmic compartment and must undergo nucleo-cytoplasmic export through binding to an export receptor like chromosomal region maintenance 1 (CRM1); AUF1 proteins harbor nuclear export sequences in the C-terminal domain (exon 6/7) that promote binding to CRM1 (41–43). In transiently transfected cell populations overexpressing p42 AUF1, the protein was predominantly nuclear. However, in co-transfections with RafCAAX, a 3.95 ± 0.0.45-fold increase (mean ± S.D., n = 5) in the cytoplasmic localization of p42 was observed in comparison with co-transfections with a pCMV vector (Fig. 8, B, compare 2nd versus 3rd lanes from left, and C). This effect was mitigated by co-incubation of leptomycin B, a bacterial toxin that alkylates the Cys-528 residue of CRM1 and inhibits cognate protein binding (Fig. 8, B, compare 3rd versus 5th lanes, and Fig. 8C) (43). Nuclear fractions were further enriched with p42 AUF1 in the presence of leptomycin B as was the nuclear transcription factor c-Jun, further validating our assay procedure.
DISCUSSION
We previously proposed that PGE2 was more akin to a modulator of inflammation as opposed to one of the many inflammatory mediators, based on our studies demonstrating the anti-cytokine and anti-catabolic effects of the most abundant of prostanoids (24–26). These observations were further supported by recent studies on PGE2-dependent production of “downstream” eicosanoids such as lipoxins, resolvins, and protectins, which are believed to be involved in the transition and resolution phase of the acute inflammatory response in animal models (16). This study provided new data demonstrating that “early” mediators like LTB4, synthesized and released by transmigrating neutrophils and resident mast cells, control PGE2 synthesis through direct effects on COX-2 mRNA stability in the synovium. In addition, we provided a detailed mechanism involving BLT-dependent restricted activation of mitogenic pathways with the subsequent nucleo-cytoplasmic shuttling of p42 AUF1 and targeted message stabilization via 3′-UTR ARE sequences.
The BLT1 receptor expression is largely restricted to leukocytes (neutrophils, macrophages, and eosinophils) and functionally associates with pertussis-sensitive and -insensitive G proteins that mediate calcium mobilization, accounting for the high affinity effects of LTB4 (KD ∼1 nm) on target cell chemotaxis (21). In contrast, BLT2 exhibits a wider tissue distribution (e.g. spleen, liver, and ovary) and associates with pertussis-sensitive G proteins (e.g. Gαi) linked to strong MAPK/ERK activation (KD ∼50 nm) but weaker calcium mobilization effects (19, 20, 44, 45). We believe this study demonstrating BLT receptor expression and function in HSF suggests that the latter cell type is integrated into the inflammatory response partly through responses to LTB4. Therefore, in acute inflammatory synovitis, the synovial fibroblast, the preponderant cell type, is the principle target of ambient mediators, and our work supports an important link of the LTB4/receptor system to arthritis. Indeed, mast cell-derived LTB4 may be the trigger for migration of CD8+ effector T-cells into the inflamed lesion/membrane (23, 27). Our RT-PCR data suggested that BLT2 mRNA was the predominant receptor species over BLT1, but functionally speaking, both activated mitogenic signaling. Others reported similar expression profiles in HSF but did not delineate functional activity (46). Further studies using receptor knock-out mice and/or in vitro knockdown experiments would be required at this stage to resolve these uncertainties. BTL1 is an inducible gene with restricted tissue expression, likely the result of promoter activity governed epigenetically through CpG island methylation (21). Because the BLT2 open reading frame encompasses all of the BLT1 promoter and exon 1 on chromosome 14, it is difficult to assess how constitutive promoter methylation might have affected the BLT2 expression profile, particularly in pathological states where the methylation status of any given promoter may be altered.
Heterogeneous nuclear ribonucleoprotein D/AUF1 is transcribed from a single gene but, as a result of alternative splicing, is translated into four distinct isoforms, p37, p40, p42, and p45 AUF1 (pan-AUF1). AUF1 is a bona fide RNA-binding protein with high affinity for ARE sequences found in a variety of target mRNA-3′-UTR, and RNA electrophoretic mobility shift/supershift analysis strongly suggests that AUF1 binds to class II AREs like those found in COX-2 mRNA. There is, however, no consensus as to whether it functions to stabilize or destabilize mRNA (8, 13, 14, 42, 47–49). In our hands, LTB4/BLT markedly stabilized COX-2 mRNA as judged by Northern analysis and COX-2-ARE-luciferase reporter activity through an ERK1/2-dependent pathway with apparent recruitment of p42 AUF1 stabilizing capacity via a CRM1-dependent nuclear export. Furthermore, we incorporated RNA-binding proteins TTP and HuR into our assays (Table 2), and the results confirmed our previous observations that TTP strongly destabilizes COX-2 mRNA through a p38 MAPK-dependent process (50) and was not subject to ERK1/2 modulation as has been suggested by previous work (13, 14). In our cell culture models (HSF and HeLa), HuR exhibited modest effects on COX-2 mRNA stability and on COX-2-ARE-luciferase reporter activity, but we were surprised to see how mitogenic activation by c-Raf/ERK1/2 strongly increased HuR-destabilizing effects as this was not evident from the literature (13, 14). The congruity of our results using different experimental approaches and cell types strengthens the validity of our observations. Nevertheless, we used an overexpression protocol, and although pan-AUF1 (predominantly nuclear) and TTP (cytoplasmic) were modestly expressed in our cell types, HuR is abundantly expressed and partitions between the nuclear and cytoplasmic compartments (50, 51). The results may be subject to interpretation as it is unclear whether ectopically expressed HuR impacts on endogenous levels and whether c-Raf/ERK1/2 activation is targeting endogenous or ectopic HuR.
In a number of studies, p37 and p40 AUF1 isoforms have been ascribed predominant roles in the modulation of target mRNA stability via AU-rich sequences, largely because they contain putative nuclear localization sequences in the C-terminal domains, favoring nuclear “maturation” prior to cytoplasmic localization and polyribosome association (48). Furthermore, because of structural determinants, p42 and p45 AUF1 are predominantly nuclear, although a consensus is emerging that all isoforms can shuttle (49). In quiescent HSF and HeLa cells, the four isoforms were nuclear with roughly equal albeit low level expression, although some cytoplasmic p42 AUF1 staining was observed. A similar partitioning profile was observed when p42 AUF1 was overexpressed, although a marked cytoplasmic accumulation of p42 AUF1 was observed under strong c-Raf/ERK1/2 signaling suggesting that mitogenic signals support active nuclear export. c-Raf/ERK1/2-dependent phosphorylation of p42 AUF1 could increase its association with exportins thereby stimulating the overall shuttling rate. Interestingly, analysis of AUF1 from control and phorbol ester-treated THP-1 cells indicated that post-translational modifications of the major cytoplasmic isoform, p40 AUF1, are altered concomitant with changes in RNA binding activity and stabilization of ARE-containing mRNAs (47). In particular, p40 AUF1 recovered from polysomes is phosphorylated on Ser-83 and Ser-87 in untreated cells but dephosphorylated following 12-O-tetradecanoylphorbol-13-acetate treatment. In addition to post-translational modifications, another possibility may be that, in our cell cultures, CRM1 levels were up-regulated by mitogenic signaling with the accompanying increase in transcription of target genes (e.g. CRM1), an intriguing avenue for future research.
We believe this study to be the first report providing a plausible molecular mechanism whereby LTB4, an early inflammatory mediator, supports the resolution phase of the inflammatory response by stabilizing COX-2 mRNA (protein), providing for an abundance of ambient pro-resolution lipid mediators (see review in Ref. 16). Furthermore, AUF1 function clearly varies with the target mRNAs, and one must consider that AREs may not be the only cognate binding sites and/or that the AUF1 exerts in effects by modulating mRNA transport processes (see above). Finally, AUF1 is evidently functionally integrated into the immune and inflammatory responses, and a better understanding of its role in this regard may lead to targeted therapeutic intervention.
In summary, we propose the following mechanism for the signaling cascade leading to the increase in COX-2 mRNA stability: LTB4, from activated synovial neutrophil/macrophage/mast cell populations, binds BLT1/2:Gαq/Gα11 on synovial fibroblasts resulting in increased [Ca2+]i and downstream activation of the following intermediates: protein kinase C/CaMKii → Ras GRP/GRF → Ras → c-Raf → MEK1/2 → ERK1/2. Nuclear ERK1/2, acting through intermediates, stimulates p42 AUF1 nucleo-cytoplasmic translocation, which binds to COX-2 mRNA in the 3′-UTR region favoring stabilization. Increased levels of COX-2 mRNA/protein promote PGE2 release with feedback suppression of macrophage-derived cytokines and metalloprotease production, principally from synovial fibroblasts and macrophages. In addition, we propose a feedback switch to LXA4 synthesis through PGE2-dependent up-regulation of 15-lipoxygenase with changes in lipid mediator synthesis to anti-inflammatory resolvin/protectin synthesis, blocking further neutrophil recruitment and resolving the inflammatory response.
This work was supported in part by Canadian Institutes for Health Research Grant M11557 (to J. A. D. B.).
- AU
- adenylate/uridylate
- MAPK
- mitogen-activated protein kinase
- ERK1/2 (p42/44)
- extracellular signal-regulated kinase
- PGE2
- prostaglandin E2
- LTB4
- leukotriene B4
- BLT
- leukotriene B4 receptor
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- IL
- interleukin
- rhIL-1β
- recombinant human interleukin-1β
- 3′-UTR
- 3′-untranslated region
- ARE
- AU-rich element
- AUF1
- ARE/poly-U binding/degradation factor 1 (heterogeneous nuclear ribonucleoprotein D)
- TTP
- tristetraproline
- HSF
- human synovial fibroblast
- TEMED
- tetramethylethylenediamine
- shRNA
- short hairpin RNA
- AUBP
- AU-binding protein
- RT
- reverse transcription
- RA
- rheumatoid arthritic
- ANOVA
- analysis of variance
- DIG
- digoxigenin.
REFERENCES
- 1.Burg N. D., Pillinger M. H. (2001) Clin. Immunol. 99, 7–17 [DOI] [PubMed] [Google Scholar]
- 2.O'Garra A., Vieira P. (2004) Nat. Med. 10, 801–805 [DOI] [PubMed] [Google Scholar]
- 3.Yoshimura A., Naka T., Kubo M. (2007) Nat. Rev. Immunol. 7, 454–465 [DOI] [PubMed] [Google Scholar]
- 4.Luster A. D. (1998) N. Engl. J. Med. 338, 436–445 [DOI] [PubMed] [Google Scholar]
- 5.Viola A., Luster A. D. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 171–197 [DOI] [PubMed] [Google Scholar]
- 6.Metz M., Grimbaldeston M. A., Nakae S., Piliponsky A. M., Tsai M., Galli S. J. (2007) Immunol. Rev. 217, 304–328 [DOI] [PubMed] [Google Scholar]
- 7.Dreyfuss G., Kim V. N., Kataoka N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 8.Dean J. L., Sully G., Clark A. R., Saklatvala J. (2004) Cell. Signal. 16, 1113–1121 [DOI] [PubMed] [Google Scholar]
- 9.Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. (2001) Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 10.Wilusz C. J., Wormington M., Peltz S. W. (2001) Nat. Rev. Mol. Cell Biol. 2, 237–246 [DOI] [PubMed] [Google Scholar]
- 11.Lunde B. M., Moore C., Varani G. (2007) Nat. Rev. Mol. Cell Biol. 8, 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. (2005) Nat. Struct. Mol. Biol. 12, 1054–1063 [DOI] [PubMed] [Google Scholar]
- 13.Saklatvala J. (2004) Curr. Opin. Pharmacol. 4, 372–377 [DOI] [PubMed] [Google Scholar]
- 14.Clark A., Dean J., Tudor C., Saklatvala J. (2009) Front. Biosci. 14, 847–871 [DOI] [PubMed] [Google Scholar]
- 15.Sandler H., Stoecklin G. (2008) Biochem. Soc. Trans. 36, 491–496 [DOI] [PubMed] [Google Scholar]
- 16.Serhan C. N., Chiang N., Van Dyke T. E. (2008) Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serhan C. N., Haeggström J. Z., Leslie C. C. (1996) FASEB J. 10, 1147–1158 [DOI] [PubMed] [Google Scholar]
- 18.Zhai B., He Q. W., Di Battista J. A. (2008) Osteoarthritis Cartilage 16, Suppl. 4, S189 [Google Scholar]
- 19.Yokomizo T., Izumi T., Chang K., Takuwa Y., Shimizu T. (1997) Nature 387, 620–624 [DOI] [PubMed] [Google Scholar]
- 20.Yokomizo T., Kato K., Terawaki K., Izumi T., Shimizu T. (2000) J. Exp. Med. 192, 421–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato K., Yokomizo T., Izumi T., Shimizu T. (2000) J. Exp. Med. 192, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy B. D., Clish C. B., Schmidt B., Gronert K., Serhan C. N. (2001) Nat. Immunol. 2, 612–619 [DOI] [PubMed] [Google Scholar]
- 23.Campbell E. L., Louis N. A., Tomassetti S. E., Canny G. O., Arita M., Serhan C. N., Colgan S. P. (2007) FASEB J. 21, 3162–3170 [DOI] [PubMed] [Google Scholar]
- 24.Faour W. H., Alaaeddine N., Mancini A., He Q. W., Jovanovic D., Di Battista J. A. (2005) J. Biol. Chem. 280, 9536–9546 [DOI] [PubMed] [Google Scholar]
- 25.He W., Pelletier J. P., Martel-Pelletier J., Laufer S., Di Battista J. A. (2002) J. Rheumatol. 29, 546–553 [PubMed] [Google Scholar]
- 26.Faour W. H., He Q., Mancini A., Jovanovic D., Antoniou J., Di Battista J. A. (2006) J. Biol. Chem. 281, 19849–19860 [DOI] [PubMed] [Google Scholar]
- 27.Lee D. M., Friend D. S., Gurish M. F., Benoist C., Mathis D., Brenner M. B. (2002) Science 297, 1689–1692 [DOI] [PubMed] [Google Scholar]
- 28.Rocca B., FitzGerald G. A. (2002) Int. Immunopharmacol. 2, 603–630 [DOI] [PubMed] [Google Scholar]
- 29.Smyth E. M., Grosser T., Wang M., Yu Y., FitzGerald G. A. (2009) J. Lipid Res. 50, S423–S428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faour W. H., He Y., He Q. W., de Ladurantaye M., Quintero M., Mancini A., Di Battista J. A. (2001) J. Biol. Chem. 276, 31720–31731 [DOI] [PubMed] [Google Scholar]
- 31.Hochberg M. C., Chang R. W., Dwosh I., Lindsey S., Pincus T., Wolfe F. (1992) Arthritis Rheum. 35, 498–502 [DOI] [PubMed] [Google Scholar]
- 32.Altman R., Asch E., Bloch D., Bole G., Borenstein D., Brandt K., Christy W., Cooke T. D., Greenwald R., Hochberg M., et al. (1986) Arthritis Rheum. 29, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 33.Faour W. H., Mancini A., He Q. W., Di Battista J. A. (2003) J. Biol. Chem. 278, 26897–26907 [DOI] [PubMed] [Google Scholar]
- 34.Dixon D. A., Kaplan C. D., McIntyre T. M., Zimmerman G. A., Prescott S. M. (2000) J. Biol. Chem. 275, 11750–11757 [DOI] [PubMed] [Google Scholar]
- 35.Okuno T., Iizuka Y., Okazaki H., Yokomizo T., Taguchi R., Shimizu T. (2008) J. Exp. Med. 205, 759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Appleby S. B., Ristimäki A., Neilson K., Narko K., Hla T. (1994) Biochem. J. 302, 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawaoka H., Dixon D. A., Oates J. A., Boutaud O. (2003) J. Biol. Chem. 278, 13928–13935 [DOI] [PubMed] [Google Scholar]
- 38.Sengupta S., Jang B. C., Wu M. T., Paik J. H., Furneaux H., Hla T. (2003) J. Biol. Chem. 278, 25227–25233 [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann S., Moelling K. (1999) Science 286, 1741–1744 [DOI] [PubMed] [Google Scholar]
- 40.Yuryev A., Wennogle L. P. (2003) Genomics 81, 112–125 [DOI] [PubMed] [Google Scholar]
- 41.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 42.Mazan-Mamczarz K., Kuwano Y., Zhan M., White E. J., Martindale J. L., Lal A., Gorospe M. (2009) Nucleic Acids Res. 37, 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meissner T., Krause E., Vinkemeier U. (2004) FEBS Lett. 576, 27–30 [DOI] [PubMed] [Google Scholar]
- 44.Kamohara M., Takasaki J., Matsumoto M., Saito T., Ohishi T., Ishii H., Furuichi K. (2000) J. Biol. Chem. 275, 27000–27004 [DOI] [PubMed] [Google Scholar]
- 45.Okuno T., Yokomizo T., Hori T., Miyano M., Shimizu T. (2005) J. Biol. Chem. 280, 32049–32052 [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto A., Endo H., Hayashi I., Murakami Y., Kitasato H., Kono S., Matsui T., Tanaka S., Nishimura A., Urabe K., Itoman M., Kondo H. (2003) J. Rheumatol. 30, 1712–1718 [PubMed] [Google Scholar]
- 47.Wilson G. M., Lu J., Sutphen K., Sun Y., Huynh Y., Brewer G. (2003) J. Biol. Chem. 278, 33029–33038 [DOI] [PubMed] [Google Scholar]
- 48.Chen C. Y., Xu N., Zhu W., Shyu A. B. (2004) RNA 10, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar B., Lu J. Y., Schneider R. J. (2003) J. Biol. Chem. 278, 20700–20707 [DOI] [PubMed] [Google Scholar]
- 50.Mancini A., Di Battista J. A. (2008) Osteoarthritis Cartilage 16, Suppl. 4, S16 [Google Scholar]
- 51.Mancini A., Di Battista J. A. (2007) Inflamm. Res. 56, Suppl. 3, S374 [Google Scholar]