Abstract
Cellular prostatic acid phosphatase (cPAcP), an authentic tyrosine phosphatase, is proposed to function as a negative growth regulator of prostate cancer (PCa) cells in part through its dephosphorylation of ErbB-2. Nevertheless, the direct interaction between cPAcP and ErbB-2 has not been shown nor the specific dephosphorylation site of ErbB-2 by cPAcP. In this report, our data show that the phosphorylation level of ErbB-2 primarily at Tyr1221/2 correlates with the growth rate of both LNCaP and MDA PCa2b human PCa cells. Further, cPAcP reciprocally co-immunoprecipitated with ErbB-2 in a non-permissive growth condition. Expression of wild type cPAcP, but not inactive mutant, by cDNA in cPAcP-null LNCaP C-81 cells results in decreased tyrosine phosphorylation of ErbB-2 including Tyr1221/2. Concurrently, Tyr317 phosphorylation of p52Shc, proliferating cell nuclear antigen expression, and cell growth are decreased in these cells. Conversely, decreased cPAcP expression by short hairpin RNA in LNCaP C-33 cells was associated with elevated phosphorylation of ErbB-2 initially at Tyr1221/2. Its downstream p52Shc, ERK1/2, Akt, Src, STAT-3, and STAT-5 were activated, and cell proliferation, proliferating cell nuclear antigen, and cyclin D1 expression were increased. Stable subclones of C-33 cells by small interfering PAcP had elevated Tyr1221/2 phosphorylation of ErbB-2 and exhibited androgen-independent growth and increased tumorigenicity in xenograft female animals. In summary, our data together indicate that in prostate epithelia, cPAcP interacts with and dephosphorylates ErbB-2 primarily at Tyr1221/2 and hence blocks downstream signaling, leading to reduced cell growth. In PCa cells, decreased cPAcP expression is associated with androgen-independent cell proliferation and tumorigenicity as seen in advanced hormone-refractory prostate carcinomas.
Keywords: Phosphotyrosine Receptor, Phosphotyrosine Signaling, Protein-Protein Interactions, Steroid Hormone, Protein-Tyrosine Phosphatase (Tyrosine Phosphatase), Androgen, ErbB-2, Prostatic Acid Phosphatase, Prostate Cancer, Growth Regulation
Introduction
Because testicular androgens are involved in prostate carcinogenesis, systematic androgen ablation therapies are usually performed in advanced stages of prostate cancer (PCa)3 (1, 2). In most cases, however, patients eventually relapse into the highly aggressive form of cancer, which acquires a hormone-refractory (HR) or castration-resistant phenotype, leaving limited modalities in its treatment currently. Understanding the molecular mechanism of HR PCa can lead to improve its therapy.
Clinical studies show that AR plays a critical role in most HR PCa progression. For example, hypersensitivity of AR pathway may circumvent ablation therapy by reducing the threshold of androgens required for cell proliferation (3). Elevated AR protein can convert the action of some AR antagonists to agonists (4). AR mutation may allow AR to be activated by non-androgenic molecules (5). However, elevated expression and mutation of AR together only account for a subpopulation of HR PCa. Thus, other mechanisms such as cross-talk with other signaling pathways could also be involved in the development of HR phenotype (6–8). Recent results indicate that ErbB-2 plays a critical role in AR activation and HR PCa progression. ErbB-2 expression is increased in an androgen-independent xenograft mouse model (9), and elevated ErbB-2 expression allows PCa cells to grow (10) as well as prostate-specific antigen secretion (9–11) in androgen-reduced conditions. Further, ErbB-2 can activate Akt in the absence of androgen, and subsequently, Akt phosphorylates AR at Ser213 and Ser791 sites. Abrogation of Akt signaling abolishes the androgen-independent survival and growth of these cells (12). Nevertheless, in prostate carcinomas, the ErbB-2 gene is not amplified, and ErbB-2 protein is elevated only in a subset of advanced HR PCa (12–16). Other mechanisms including ErbB-2 activation exist for HR PCa progression.
Several lines of evidence suggest that cellular prostatic acid phosphatase (cPAcP), a prostate epithelial differentiation antigen, is involved in regulating the growth of prostate epithelia. The expression and the activity of cPAcP in prostate carcinoma cells is lower than that in normal and benign hypertrophic prostate tissue despite that secreted PAcP may be elevated in circulation (17–20). It is noted that cPAcP and secreted PAcP exhibit different biochemical properties including different isoelectric points and sensitivities to endoglycosidases (20). The function of cPAcP in growth regulation is at least in part contributed by its intrinsic PTP activity (20–23), although it does not contain the signature motif, HCXXGXXR, found in cysteine-dependent PTPs (24, 25). The overall tyrosyl phosphorylation level, the tyrosyl kinase specific activity, and the cell growth rate are inversely correlated with cPAcP activity in PCa cell lines under various growth conditions (26–28). Further studies reveal that the tyrosyl phosphorylation level of ErbB-2/HER-2/neu protein inversely correlates with the activity of cPAcP (29–31). Ectopic expression of WT cPAcP results in decreased ErbB-2 tyrosyl phosphorylation (32). Intratumor injection of PAcP cDNA expression vector suppresses xenograft tumor progression and decreased ErbB-2 tyrosyl phosphorylation (33). Supportively, in PAcP knock-out mice, the prostate develops carcinomas spontaneously and increased tyrosine phosphorylation (34). In addition, ERK1/2 are activated by phosphorylation in prostate carcinomas and PCa cells in which cPAcP is decreased, suggesting that decreased cPAcP resulted in activating ErbB-2 and ERK1/2 signaling (11, 32). Therefore, cPAcP plays a regulatory role of controlling the activation of ErbB-2, and in cPAcP-null PCa cells, activated ErbB-2-induced signaling can be involved in ligand-independent AR activation, leading to HR progression of these cells.
Due to the importance of cPAcP and ErbB-2 involvement in regulating HR PCa progression, it is imperative to clarify the direct interaction of cPAcP and ErbB-2 because until now, only correlative results have been reported. Further, the identification of a ErbB-2 dephosphorylation site by cPAcP can help delineate the biological outcome of PAcP and ErbB-2 interaction and the mechanism involved in the development of HR PCa. Restoring cPAcP expression thus may represent an alternative therapeutic approach in treating PCa patients.
EXPERIMENTAL PROCEDURES
Materials
RPMI 1640 medium, gentamicin, FBS, trypsin/EDTA, and Lipofectamine PLUSTM reagents were purchased from Invitrogen. Charcoal/dextran-treated FBS was from HyClone (Logan, UT) and Atlanta Biologicals (Lawrenceville, GA). Acrylamide, protein molecular weight standard markers, and protein estimation kit were obtained from Bio-Rad. ECL reagent was purchased from Pierce. Anti-phospho-HER-2/ErbB-2 (Tyr877) Ab, anti-phospho-HER-2/ErbB-2 (Tyr1112) Ab, anti-phospho-HER-2/ErbB-2 (Tyr1221/1222) Ab, anti-phospho-STAT3 (Tyr705) Ab, anti-phospho-STAT5 (Tyr694) Ab, anti-phospho-Src (Tyr416) Ab, anti-phospho-Akt (Ser473) Ab, anti-STAT3 Ab, anti-STAT5 Ab, and anti-Src Ab were from Cell Signaling Technology (Beverly, MA). Anti-phospho-Erk1/2 (Thr202/Tyr204) Ab was from United States Biological (Swampscott, MA). Anti-PCNA Ab, anti-ErbB-2/Neu Ab (C-18) (9G6), anti-cyclin D1Ab, anti-Erk1/2 Ab, anti-SHP1 (SH-PTP1) Ab, anti-SHP-2 (SH-PTP2) Ab, anti-Akt Ab, anti-PAcP Ab (045), and horseradish peroxidase-conjugated polyclonal secondary Abs were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-Tyr (4G10) Ab, anti-phospho-HER-2/ErbB-2 (Tyr1248) Ab, anti-phospho-p52Shc (Tyr317) Ab, and anti-Shc Ab were from Millipore (Billerica, MA). Anti-β-Actin (AC-15) Ab and anti-PAcP Ab (P-9808) were from Sigma. Anti-α-tubulin Ab was from NeoMarkers (Fremont, CA). Anti-PAcP antiserum (ATM-3) was obtained as described previously (22).
Cell Culture
Human PCa cell lines LNCaP and MDA PCa2b were purchased from the American Type Culture Collection (Manassas, VA). LNCaP cells were routinely maintained in the regular medium, i.e. phenol red-positive RPMI 1640 medium supplemented with 5% FBS, 2 mm glutamine, and 50 μg/ml gentamicin. The LNCaP cell model was described originally by Lin et al. (30) and further characterized by Igawa et al. (36). The C-33 cells are androgen-sensitive, whereas C-81 cells behave androgen independently. MDA PCa2b cells were maintained in BRFF-HPC1 medium supplemented with 20% FBS, 2 mm glutamine, and 50 μg/ml gentamicin (37).
Stable Transfectants with PAcP Expression Vector
LNCaP C-81 cells were transfected with PAcP expression vector containing WT PAcP cDNA with LipofectamineTM and PlusTM reagents in OptiMEM medium and followed the accompanying protocol. Two stable subclones designated as LNCaP-28 and -40 were described previously (30, 31). LNCaP-CMV is a subline of LNCaP C-81 cells transfected with the pCMV-neo vector alone.
Generation of cPAcP siRNA and Establishment of cPAcP Knockdown Stable Subclones
The pSUPER system-based siRNA approach and the protocol from OligoEngine (Seattle, WA) were utilized. Briefly, four different oligonucleotides were synthesized: siPAcP-78, 5′-GCCTTAGCCTTGGCTTCTT-3′; siPAcP-126, 5′-GTGTACTAGCCAAGGAGTT-3′; siPAcP-183, 5′-GTCCCATTGACACCTTTCC-3′; and siPAcP-236, 5′-GGATTTGGCCAACTCACCC-3′. A pair of 64-nucleotide oligonucleotides containing 19 nucleotides targeting to PAcP mRNA sequence was inserted into the mammalian expression vector pSUPER at the BglII/HindIII restriction sites. Because siPAcP-126 consistently showed the high efficiency of PAcP knockdown, stable subclones were established in LNCaP C-33 cells by transfecting with siPAcP-126 pSUPER plasmid. Positive clones were selected by G418 (600 μg/ml) and screened for cPAcP expression by Western blotting. Three subclones C-3, C-11, and C-17, were selected for further analyses. For the control, pSUPER vector containing scramble oligonucleotide was transfected into C-33 cells, and clone V-3 was established. The transfection was performed as described above.
Immunoblotting and Immunoprecipitation
Subconfluent cells were harvested, and the immunoblotting was performed as described in previous reports (11, 30). For rehybridization, the membranes were stripped as described previously (11, 30, 31), blocked, and reprobed with specific Abs. For immunoprecipitation, cells were harvested and lysed in ice-cold cell lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, plus phosphatase and protease inhibitors). An aliquot of 250 μg of lysate protein was incubated with primary Ab at 4 °C overnight, and then protein A-agarose beads were added (20 μl of 50% bead slurry) for 3 h at 4 °C. The immunocomplexes were spun at 700 × g for 1 min, washed three times with ice-cold lysis buffer, and suspended in SDS-PAGE sample buffer.
In Vitro Growth Kinetics Analysis
Cells were seeded at densities as indicated in each set of experiments and cultured in the corresponding regular culture medium. The maintenance and determination of cell growth were conducted as described (30, 31). The cell number was counted with a Z1 model Coulter counter (Coulter Corp.). To determine androgen-independent growth, cells were maintained in a steroid-reduced medium for 4 days before cell number analysis (30, 33).
Subfractionation of Cellular Proteins
To subfractionate cellular proteins, subconfluent or confluent LNCaP C-33 cells were fed with a steroid-reduced medium for 2 days. Cell membrane, cytoplasmic, and nuclear proteins were fractionated following the protocol of the subcellular protein fractionation kit (Thermal Scientific). LNCaP C-33 cells were also subfractionated by ultracentrifugation. Briefly, confluent cells were lysed in a hypotonic Buffer A (20 mm acetate buffer, pH 5.0, containing 1 mm dithiothreitol and protease inhibitors), homogenized, and followed by centrifugation at 2000 × g. The obtained soluble fraction was spun at 160,000 × g for 1 h, and the supernatant was collected as the cytosol fraction. The pellets were suspended in Buffer B (i.e. Buffer A plus 0.15 m NaCl) and spun at 100,000 × g for 45 min. The supernatant was designated as the membrane-associated fraction. The pellet was then solubilized in Buffer C (i.e. Buffer B plus 0.5% Nonidet P-40) and centrifuged at 40,000 × g. The supernatant was designated as the membrane-bound fraction.
Acid Phosphatase Assay
The AcP activity was performed as described previously with para-nitrophenyl phosphate as the substrate in a citrate buffer, pH 5.5 (22, 30, 38). l(+)-Tartrate is a classical inhibitor of PAcP activity (21, 22, 26, 38), and in LNCaP C-33 cells, l(+)-tartrate-sensitive AcP activity is routinely used to represent PAcP activity (26, 27). Alternatively, PAcP-specific AcP activity was analyzed in the immunocomplex by anti-PAcP Ab (22, 26).
Xenograft Animal Experiments
To compare the tumorigenicity of PAcP knockdown stable sublines, 1 × 106 cells at the exponential growth phase were suspended in 0.1 ml of medium, mixed with 0.1 ml of Matrigel (BD Biosciences), and injected subcutaneously in the hind flank of female nude mice (Charles River Laboratories) (19, 33). Animal maintenance and tumor measurement were done according to National Institutes of Health guidelines and the specific guidelines at our medical center. The protocol was approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee.
Statistical Analyses
Experimental results were expressed as the mean ± S.E. Paired two-tailed Student's t tests were used for comparison between each groups. p < 0.05 was considered statistically significant.
RESULTS
Tyrosyl Phosphorylation of ErbB-2 in Different Passages of LNCaP and MDA PCa2b Cells
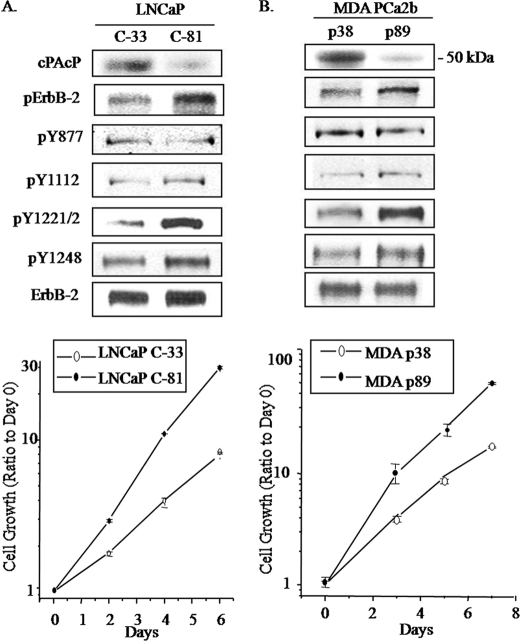
To investigate the correlation of cPAcP expression with ErbB-2 tyrosyl phosphorylation, we examined the phosphorylation status of specific Tyr residues of ErbB-2 with commercially available Abs in both LNCaP and MDA PCa2b cells. As shown in Fig. 1A (upper panel), in regular culture medium, the expression of cPAcP decreased in LNCaP C-81 cells; concurrently, the overall Tyr(P) level of ErbB-2 elevated, correlating with increased cell growth rate by about 2-fold (Fig. 1A, lower panel). Notably, phosphorylation of Tyr1221/2 and Tyr1248, but not Tyr877 or Tyr1112, was elevated in C-81 cells, higher than that in C-33 cells. In parallel, in MDA PCa2b p89 cells, cPAcP decreased, cell growth increased by about 2-fold, and Tyr1221/2 phosphorylation was greatly elevated (Fig. 1B). The phosphorylation of Tyr1112 and Tyr1248 was only slightly elevated with no significant change at Tyr877 (Fig. 1B). The data collectively imply Tyr1221/2 and possibly also Tyr1248 as potential dephosphorylation sites by cPAcP in PCa cells, resulting in growth regulation.
FIGURE 1.
Analyses of tyrosine phosphorylation of ErbB-2 in different LNCaP and MDA PCa2b cells. Upper panels, A, LNCaP C-33 and C-81 cells were seeded in regular culture medium at a density of 5 × 105/T25 flask for 3 days. p indicates phosphorylation. B, MDA PCa2b cells (passages 38 and 89) were seeded with a density of 1 × 106/T25 flask in regular culture medium. Cells were fed with fresh medium and cultured for another 2 days. Immunoblottings were performed with Abs to different tyrosine phosphorylation sites of ErbB-2. After stripping, the membrane was hybridized with Ab to detect ErbB-2 protein. Similar results were obtained from over three sets of independent experiments. Lower panels, growth rates of different LNCaP and MDA PCa2b cells. A, LNCaP cells were seeded at a density of 3 × 104 cells/well in 6-well plates in the regular culture medium. 3 days after plating, one set of attached cells was harvested and counted as day 0. The remaining cells were refreshed with the regular medium, and the total cell numbers were counted on the indicated days. B, for MDA PCa2b cell growth kinetics, cells were seeded in regular medium at a density of 1 × 105 cells/well in 6-well plates and counted on days 3, 5, and 7. Data shown are the ratios of cell numbers normalized to the corresponding number on day 0. Bars = the range of duplicates. Similar results were obtained from at least three sets of independent experiments. Y, tyrosine.
Interactions of cPAcP and ErbB-2 Protein
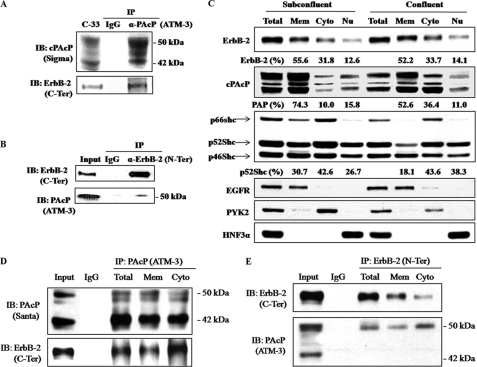
To determine the interaction of cPAcP and ErbB-2, we performed co-IP of cPAcP with ErbB-2 in a non-permissive growth condition. As shown in Fig. 2A, ErbB-2 co-immunoprecipitated with cPAcP by anti-PAcP Abs. The 50-kDa form of PAcP is the mature glycosylated subunit, and the 42-kDa protein is an intermediate form of processing product (20). Both forms of PAcP proteins are recognized by anti-PAcP Ab (20). This co-IP was further validated by reciprocal co-IP (Fig. 2B). Significantly, only the mature 50-kDa cPAcP was seen to co-immunoprecipitate with ErbB-2 by anti-ErbB-2 Ab, even after prolonged exposure (Fig. 2B). The data together indicate that cPAcP and ErbB-2 are in the same functional complex.
FIGURE 2.
Co-immunoprecipitation of cPAcP and ErbB-2. A, confluent C-33 cells were maintained in a steroid-reduced medium for 48 h. Cells were then harvested, lysed, and immunoprecipitated by PAcP Ab (ATM-3) (36). The immunoprecipitate was electrophoresed and blotted (IB) with anti-ErbB-2 Ab (C-terminal (C-Ter) Ab) and anti-PAcP Ab (Sigma catalog number P-9808). B, C-33 cells in a steroid-reduced medium for 48 h were harvested and lysed for immunoprecipitation by ErbB-2 Ab (N-terminal (N-Ter) Ab). The immunoprecipitate was immunoblotted with anti-PAcP Ab (ATM-3) and anti-ErbB-2 Ab (C-terminal Ab). Similar results were obtained from three sets of independent experiments. C, LNCaP C-33 cells were seeded at a density of 0.6 × 106/T75 (Subconfluent) and 4.5 × 106/T75 (Confluent) for 3 days and then maintained in steroid-reduced medium for 2 days. Cells were harvested and separated into membrane (Mem), cytoplasmic (Cyto), and nuclear (Nu) protein subfractions using the subcellular protein fractionation kit. Immunoblottings were performed with respective Abs to detect ErbB-2, cPAcP, Shc proteins, EGFR, PYK2, and HNF3α. The intensity of the hybridization band was semiquantified and calculated for a total of 100% for each group. EGFR, PYK2, and HNF3α served as markers for the respective subcellular fractionation. D and E, confluent LNCaP C-33 cells were harvested after being maintaining in a steroid-reduced medium for 2 days. The total cell lysates, membrane, and cytoplasmic fractions, separated by the subcellular protein fractionation kit, were immunoprecipitated by cPAcP Ab (ATM-3) or ErbB-2 Ab (N-terminal Ab). The immunoprecipitate was immunoblotted with anti-PAcP Ab (Santa Cruz Biotechnology (Santa), sc-80908) and anti-ErbB-2 Ab (C-terminal Ab). Similar results were obtained from three sets of independent experiments.
To clarify the co-subcellular localization of cPAcP and ErbB-2, we subfractionated membrane, cytosol, and nuclear fractions of LNCaP C-33 cells from both subconfluent and confluent cells under a non-permissive growth condition. Immunoblotting revealed that ErbB-2 was in all three fractions, and the distribution was independent of cell density (Fig. 2C). Interestingly, in subconfluent cells, the majority of cPAcP protein was localized in the membrane fraction, whereas in confluent cells, cPAcP was greatly enriched in the cytosolic fraction by about 3-fold (Fig. 2C). Unexpectedly, we observed a subgroup of cPAcP in the nuclear fraction (Fig. 2C). Further, the membrane fraction of p52Shc that mediates receptor tyrosine phosphorylation signaling was reduced by about 40% in confluent cells (Fig. 2C). EGFR, PYK2, and HNF3α served as markers for membrane, cytosol, and nucleus, respectively (Fig. 2C).
We examined the interaction of cPAcP with ErbB-2 in the membrane and the cytosolic fractions of confluent LNCaP C-33 cells under non-permissive growth condition. Reciprocal co-IP showed that cPAcP interacts with ErbB-2 preferentially in cytosol than in membrane fraction (Fig. 2, D and E). Additionally, we quantified AcP activity in each fraction of confluent cells after ultracentrifugation. As shown in supplemental Table 1, both AcP and PAcP activities were detected in cytosol and membrane-bound fractions with ∼40% PAcP activity in cytosol, similar to that in Fig. 2C. Thus, the data clearly showed the presence of cytosolic cPAcP in LNCaP C-33 cells, which is enriched in confluent cells under a non-permissive growth condition and interacts with ErbB-2 protein in the same functional complex.
Effect of Ectopic cPAcP Expression on ErbB-2 Tyrosyl Phosphorylation and Downstream Signaling in LNCaP C-81 Cells
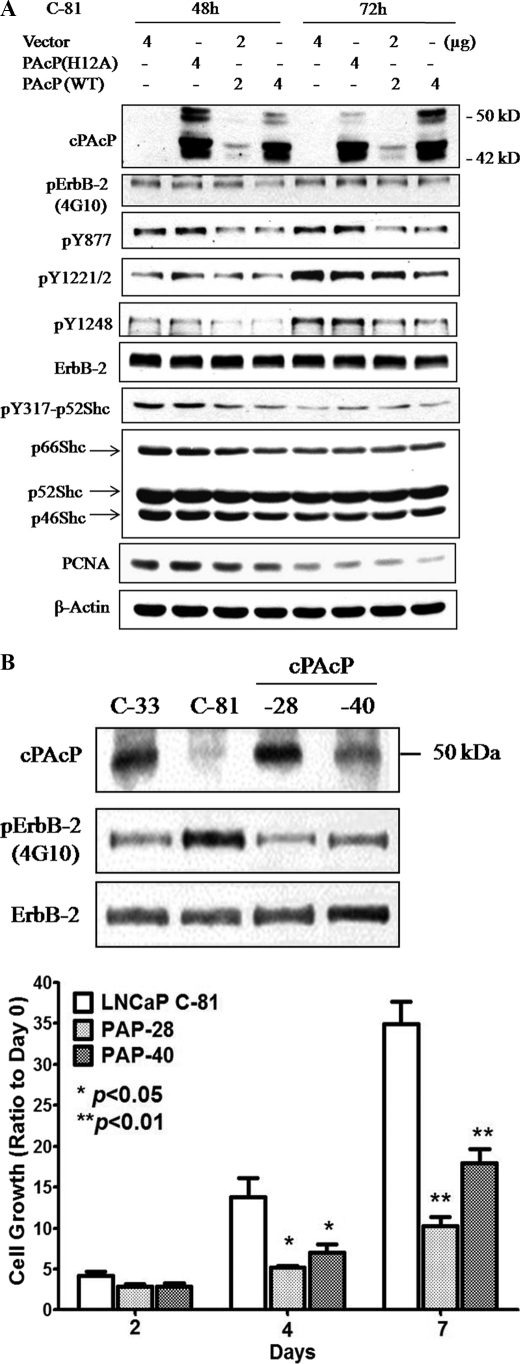
We explored the interaction of cPAcP with ErbB-2 by transiently transfected LNCaP C-81 cells with PAcP cDNA encoding WT PAcP protein. Control cells were transfected with the vector alone or the expression vector encoding mutant PAcP (H12A) that lacks PTP activity (32). Preliminary data (supplemental Fig. 1) showed that when compared with C-33 cells (lane 1), the overall tyrosine phosphorylation of ErbB-2 and Tyr877, Tyr1221/2, and Tyr1248 phosphorylation levels were highly elevated in control C-81 cells transfected with vector alone (lane 2) or in the expression of inactive H12A PAcP (lanes 3 and 4). In cells expressing WT cPAcP, phosphorylation levels of Tyr1221/2 and Tyr1248 decreased in a dose-dependent manner and had the same low level as that in C-33 cells (supplemental Fig. 1, lane 1 versus lane 6).
The effect of cPAcP expression on the tyrosine phosphorylation of ErbB-2 was seen at 48 and 72 h after transfection (Fig. 3A). The overall tyrosine phosphorylation of ErbB-2 and its Tyr877, Tyr1221/2, and Tyr1248 phosphorylation levels were decreased in cells expressing WT cPAcP but not mutant PAcP. The phosphorylation at Tyr317 of p52Shc, an ErbB-2 downstream signaling protein (39), and PCNA protein, a positive cell cycle-associated protein, also decreased in these cells. Kinetic analyses revealed that decreased phosphorylation of Tyr1221/2 and Tyr1248 of ErbB-2 could be seen at 30 h after cDNA transfection (data not shown). To examine the effect of cPAcP expression on cell growth, we established cPAcP-28 and cPAcP-40 stable sublines in C-81 cells by transfecting with WT PAcP cDNA expression vector. As shown in Fig. 3B, the expression of cPAcP resulted in decreased overall tyrosyl phosphorylation of ErbB-2, lower than that in C-81 parental cells. Concurrently, the proliferation of cPAcP-28 and cPAcP-40 cells diminished (Fig. 3B, lower panel). These results together indicate that cPAcP dephosphorylates ErbB-2 and blocks its downstream signaling via p52Shc tyrosine phosphorylation, resulting in decreased cell proliferation.
FIGURE 3.
Effect of cPAcP expression on tyrosine phosphorylation of ErbB-2 in LNCaP C-81 cells. A, LNCaP C-81 cells were transiently transfected with WT PAcP, mutant PAcP H12A cDNAs, or vector alone. Cells were harvested after 48 and 72 h, respectively, and an equal amount of cellular lysates was separated by SDS-PAGE for Western blot analyses. cPAcP, the total and the specific tyrosine phosphorylation (p) level of ErbB-2, and phosphorylation of p52Shc and PCNA were detected by the corresponding Ab. The protein levels of ErbB-2, Shc, and β-actin were examined with the respective Ab after the membranes were stripped. B, cPAcP, ErbB-2, and total tyrosine phosphorylation of ErbB-2 protein in WT PAcP cDNA-transfected LNCaP-28 and LNCaP-40 stable sublines were detected with anti-PAcP, anti-ErbB-2, and 4G10 anti-Tyr-phosphorylation Abs, respectively. For cell growth kinetics, cPAcP-28, cPAcP-40, and LNCaP C-81 cells were seeded at a density of 5 × 104 cells/well in 6-well plates in regular medium for 3 days. One set of attached cells was harvested and counted as day 0. The remaining cells were fed with regular medium. Cells were harvested on days 2, 4, and 7 for counting. Similar results were obtained in two sets of independent experiments in duplicate wells. *, p < 0.05 versus LNCaP C-81 cells; **, p < 0.01 versus LNCaP C-81 cells.
Effect of PAcP Knockdown on ErbB-2 Tyrosyl Phosphorylation and LNCaP C-33 Cell Proliferation
To mimic clinical phenomena of decreased cPAcP expression in PCa (18, 19), we knocked down endogenous PAcP in LNCaP C-33 cells using shRNA approach. Four oligonucleotides were designed, and scrambled oligonucleotides were also generated and used as a control. As shown in supplemental Fig. 2, transient transfection of siPAcP shRNA resulted in decreased cPAcP expression in C-33 cells, and siPAcP-126 consistently exhibited more efficiency on knocking down cPAcP than siPAcP-78, siPAcP-183, and siPAcP-236. In these same transfected cells, siPAcP shRNA did not have an off-target effect on SHP-1 or SHP-2 proteins, demonstrating the specificity of these siPAcP shRNA. The decreased cPAcP protein concurred with an elevation of overall ErbB-2 tyrosyl phosphorylation.
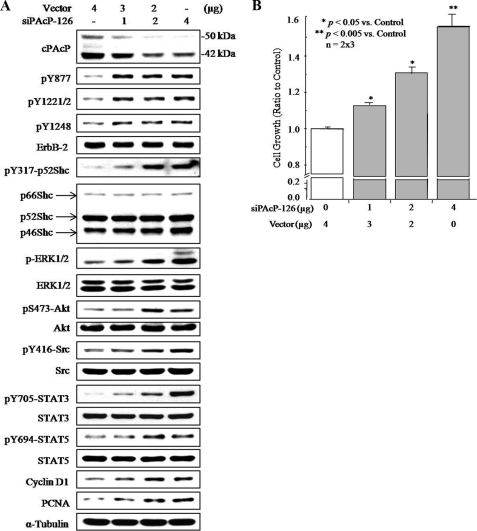
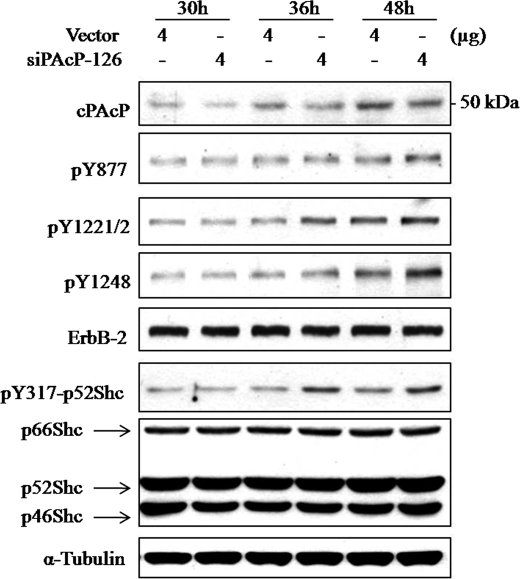
siPAcP-126 shRNA was used for further experiments. As shown in Fig. 4A, siPAcP-126 shRNA knocked down cPAcP in a dose-dependent manner in C-33 cells, which inversely correlated with increased phosphorylation at Tyr877, Tyr1221/2, and Tyr1248 of ErbB-2. Significantly, the downstream p52Shc, ERK1/2, Akt, Src, STAT-3, and STAT-5 were also activated by elevated phosphorylation. The cell proliferation was promoted as indicated by increased level of cyclin D1 and PCNA (Fig. 4A) and cell number counting (Fig. 4B) in regular culture condition. Kinetic analyses revealed that after siPAcP transfection, Tyr1221/2 of ErbB-2 and Tyr317 of p52Shc were activated by phosphorylation seen at 36 h, whereas elevated phosphorylation of Tyr877 and Tyr1248 was observed until 48 h (Fig. 5).
FIGURE 4.
Effect of transient knockdown of cPAcP by siRNA in LNCaP C-33 cells on tyrosine phosphorylation of ErbB-2. A, lysate proteins of transiently transfected cells with different amounts of PAcP siRNA-126 plasmids for 48 h were analyzed for cPAcP, cyclin D1, PCNA, and the phosphorylation (p) levels of ErbB-2, p52Shc, ERK1/2, Akt, Src, STAT3, and STAT5 proteins. After stripping the membrane, the protein levels of ErbB-2, Shc, ERK1/2, Akt, Src, STAT3, STAT5, and α-tubulin were detected, respectively. Similar results were obtained from five sets of independent experiments. B, LNCaP C-33 cells were plated at a cell density of 3 × 105/well in 6-well plates for 48 h and then transfected with different amounts of PAcP siRNA-126 plasmids. Control cells were transfected with the vector containing scramble oligonucleotides. Cell numbers were counted 3 days after transfection. The ratio of cell growth was calculated by normalizing the cell number to that of the control. The result shown is the average from three sets of independent experiments in duplicates (n = 2 × 3). Bars = S.E.
FIGURE 5.
Kinetic effects of knockdown of cPAcP on tyrosine phosphorylation of ErbB-2. LNCaP C-33 cells were transiently transfected with siPAcP-126 plasmid or vector alone and harvested at 30, 36, and 48 h after transfection. An equal amount of cellular lysates was separated by SDS-PAGE for Western blot analyses. cPAcP, the specific tyrosine phosphorylation (p) level of ErbB-2, and phosphorylation of p52Shc were detected by the corresponding Ab. The protein levels of ErbB-2, Shc, and α-tubulin were examined after the membranes were stripped. Similar results were obtained from three sets of independent experiments.
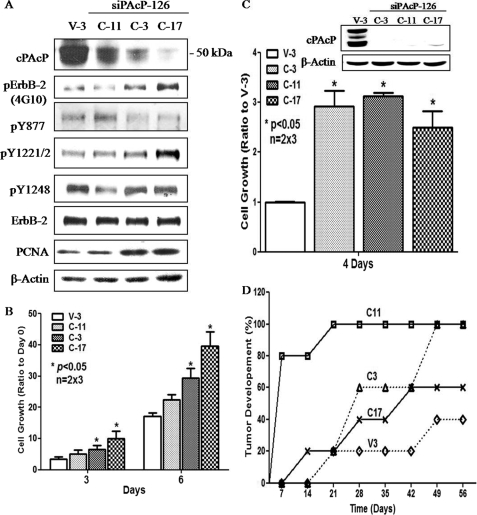
To explore the biological significance of decreased cPAcP expression in PCa cells, we established PAcP knockdown stable subclones, C-3, C-11, and C-17, in siPAcP-126-transfected LNCaP C-33 cells. V-3 cells, a stable subline transfected with pSUPER vector-containing the scrambled oligonucleotides, had cPAcP expression similar to that of parental C-33 cells (data not shown) and thus were used as a control. As shown in Fig. 6A, in regular culture condition, C-17 cells showed the greatest extent of cPAcP knockdown; concurrently, the overall Tyr(P) level of ErbB-2 as well as the phosphorylation of Tyr1221/2 in these cells were the highest among the four subclones examined. Additionally, C-17 cells exhibit the fastest growth rate and express the highest PCNA level among the subclones examined at the exponential growth phase (Fig. 6B).
FIGURE 6.
Tyrosyl phosphorylation of ErbB-2 protein and the growth properties of PAcP knockdown stable subclones. A, siPAcP stable sublines were seeded in regular culture medium with 5 × 105/T25 flask for 3 days. Total lysates from PAcP siRNA-transfected stable subclone cells (C-11, C-3, C-17) and the control subclone cells (V-3) were analyzed by Western blotting. The data shown are representative of three sets of independent experiments. B, for growth analyses, cells were seeded in duplicates at a density of 5 × 104/well in 6-well plates. After 3 days, the numbers of each stable subclone cell were counted at the time points as indicated in the figure. The relative cell growth was represented by the ratio of total cell number normalized to the respective number on day 0. Similar results were obtained from three sets of independent experiments (n = 2 × 3). Bar = S.E. C, Cells were seeded in duplicates in the condition as described above. After 4 days of culturing in steroid-reduced medium, the cell number was counted. The relative cell growth was represented by the ratio of total cell number normalized to that of V-3 control cells. Similar results were obtained from three sets of independent experiments (n = 2 × 3). Bar = S.E. Inset, cPAcP protein was analyzed by Western blotting from one set of experiments. D, xenograft animal model for siPAcP subclone cells. 1 × 106 cells of each subline in 0.1 ml of medium with 0.1 ml of Matrigel were injected into five female nude mice/group. The tumor growth was monitored weekly. *, p < 0.05 versus V-3 control cells.
We examined the HR phenotype of these PAcP knocked down stable subclone cells. In a steroid-reduced condition, C-17 cells unexpectedly expressed a higher level of cPAcP than C-11 and C-3 subclones (Fig. 6C, inset). Importantly, these PAcP knockdown cells, but not V-3 control cells, proliferated rapidly in a steroid-reduced condition, inversely correlated with cPAcP expression (Fig. 6C). Further, cPAcP knockdown subclone cells exhibited higher tumorigenicity than control cells expressing cPAcP in tumor development in female mice (Fig. 6D). These results together support the notion that decreased cPAcP expression in PCa cells leads to increased cell proliferation in androgen-ablated environment and enhanced HR tumor progression, corroborating clinical phenomena.
DISCUSSION
The results of the present study clearly show that cPAcP interacts with ErbB-2 in the same functional complex and down-regulates its tyrosyl phosphorylation in prostate epithelial cells, leading to blocking its downstream signaling and diminished proliferation. Conversely, loss of cPAcP expression as seen in clinical PCa correlates with increased tumorigenicity and the HR phenotype. Importantly, the reciprocal co-IP of cPAcP with ErbB-2 is conducted with different Abs to the same molecule. Further, decreased cPAcP expression correlates with increased ErbB-2 tyrosyl phosphorylation initially at Tyr1221/2, an Shc protein-interacting site. The data together provide a mechanistic explanation on an important clinical phenomenon of HR phenotype.
We previously showed an inverse correlation between cPAcP expression and ErbB-2 tyrosine phosphorylation. However, we were unable to obtain the co-IP of the two proteins in the exponential growth condition with numerous attempts (31). In this report, we are the first to demonstrate clearly that cPAcP interacts with ErbB-2 in PCa cells under a non-permissive growth condition by reciprocal co-IP analyses. Thus, in non-permissive growth condition, cPAcP as a negative growth regulator can efficiently target to its in vivo substrate ErbB-2, and the cPAcP/ErbB-2 interaction occurs effectively.
The differential subcellular localization of cPAcP is biologically important. Immunofluorescent staining has shown that the intensity of PAcP staining in the cytosol area correlates with cell density (23). In confluent cells, a fraction of PAcP was stained around the inner face of the plasma membrane. In this report, our data further show different subcellular localizations of PAcP biochemically (Fig. 2C, supplemental Table 1). The molecular mechanism as to how this translocation is regulated is currently unknown; nevertheless, it would aid in regulating the spatial and temporal interactions between cPAcP and ErbB-2. In the absence of growth stimulus, cPAcP interacts with and dephosphorylates ErbB-2, resulting in growth suppression. It remains unknown regarding how ErbB-2 recruits cPAcP. Further experiments are needed to clarify the domain interaction between cPAcP and ErbB-2 protein and the molecular mechanism of differential subcellular localizations of cPAcP relating to its biological function. The mechanism by which ErbB-2 recruits cPAcP also requires further investigation, and the result may have a potential for clinical applications. Our results further corroborate the notion that histidine-dependent tyrosine phosphatases play a critical role in regulating cell growth and differentiation (25).
Different autophosphorylation sites of ErbB-2 are shown to transmit diverse biological responses. For example, mutation of rat ErbB-2 protein (Tyr882), the homologous site to human ErbB-2 (Tyr877), reduces its intrinsic kinase activity and transforming potential (40). The residue Tyr1248 is also necessary for the transforming activity of ErbB-2 protein (41–43), whereas Tyr1221/2 phosphorylation is possibly a surrogate marker for high grade tumor with shorter disease-free survival in hormone receptor-positive breast cancer (44). Accordingly, cPAcP might dephosphorylate human ErbB-2 at different sites. Phosphopeptide binding analyses show that cPAcP has the most favorable binding energy toward the synthesized peptide DNLpYYWD, corresponding to Tyr1221 phosphorylation, with the possibility of acting on Tyr1248 as the additional site (45). The cPAcP dephosphorylation model supports the notion that dimeric cPAcP dephosphorylates two autophosphorylated residues on an activated receptor simultaneously because the presence of a second phosphorylated tyrosyl residue at the C terminus of ErbB-2 may considerably enhance the binding affinity (45). Alternatively, due to the close proximity of Tyr1221/2 and Tyr1248, elevated phosphorylation on Tyr1248 in PAcP knockdown cells may be secondary to the removal of cPAcP from Tyr1221/2. Although further studies are required to clarify the molecular mechanism, the kinetic studies on ErbB-2 activation in PAcP knockdown cells reveal that phosphorylation of Tyr1221/2 is elevated prior to Tyr1248 activation (Fig. 5).
The interaction between cPAcP and ErbB-2 is also regulated by other factors. The growth condition affects the expression level as well as the subcellular localization of cPAcP (Fig. 2C), and the concentration of cPAcP affects its oligomerization and specific activity (46, 47). Further, ErbB-2 has no known ligand and is activated via either homodimerization or heterodimerization with other members of the family (48–50), which results in autophosphorylation at different sites that act as docking sites for multiple signaling proteins. For example, ErbB-2 and ErbB-3 heterodimerization can activate phosphatidylinositol 3-kinase via its binding to the six docking sites on ErbB-3 (49), and Src will be recruited to Tyr(P)877 of ErbB-2 for its Tyr416 phosphorylation (51). Hence cPAcP may differentially dephosphorylate ErbB-2. The dimerization status of ErbB-2 during cPAcP dephosphorylation is also not known. Because the conformation of ErbB-2 adopts an extended configuration that readily forms homodimers or heterodimers with other ligand-activated ErbB members, cPAcP may dephosphorylate the phosphorylated ErbB-2 dimers and abolish the recruitment of downstream effectors. Interestingly, in a steroid-reduced condition, ErbB-2 inhibitor, but not EGFR inhibitor, effectively reduces cell growth (10, 52). Further experiments by knocking down each member of the ErbB family by the siRNA approach or application of monoclonal antibodies should aid in clarifying their roles in cPAcP-regulated PCa cells.
It is also important to identify the downstream effectors of ErbB-2 that mediate growth regulation of PCa cells. The activation of Tyr317 of p52Shc and ERK1/2 is inversely correlated with the expression of cPAcP in PCa cells (11), supporting the involvement of Tyr1221/2 and/or Tyr1248 in ErbB-2 dephosphorylation by cPAcP. Importantly, ERK1/2 is activated in clinical HR prostate carcinomas (53). Phosphorylation of p52Shc at Tyr317 is involved in mediating androgen-stimulated PCa proliferation (39) and also estrogen-stimulated breast cancer cells (54). In parallel, p52Shc may play a critical role in the early stages of breast cancer progression (55). Additionally, Src can be activated by docking to Tyr877 of ErbB-2 and subsequently phosphorylates the downstream effectors STAT3 (Tyr705) (49, 56) and EGFR (Tyr845) (57, 58). Although Tyr845 of EGFR is not an autophosphorylation site, its phosphorylation by Src is required for further recruitment of Src to this site for advanced phosphorylation of STAT3 and STAT5 (59). STAT5 can also be activated by directly docking to phosphorylated Tyr998 of EGFR (49). STAT3 is indicated to induce metastatic behavior of human PCa in vitro and in vivo (60), whereas the STAT5 signaling pathway is involved in the transition from androgen-sensitive to hormone-refractory diseases (61). Ligand-activated AR interacts with active STAT5 and enhances its nuclear translocation, and STAT5 can in turn increase the nuclear translocation of AR in these PCa cells (62). Further, active STAT5 is positively correlated with breast tumor progression (63). Cyclin D1 has been identified as one of STAT5 target genes in PCa cells (64). STAT3 and the phosphatidylinositol 3-kinase signaling pathway are also involved in cyclin D1 expression (35, 65). A thorough study of the ErbB-2 phosphorylation pattern and its downstream signaling cascade would help distinguish diverse ErbB-2-modulated bioactivities and clarify the significance of its dephosphorylation by cPAcP in each specific function. The results may lead to the development of specific targeting therapy for both prostate and breast cancers.
Supplementary Material
Acknowledgment
We thank Renee Garcia for technical support in ultracentrifugation.
This work was supported, in whole or in part, by National Institutes of Health Grants 2R01CA88184 (to M.-F. L.) and P20RR018759 (to S.-J. C.). This study was also supported by United States Department of Defense Grants PC074289 and PC050769 and the Nebraska Research Initiative.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- PCa
- prostate cancer
- AcP
- acid phosphatase
- PAcP
- prostatic acid phosphatase
- cPAcP
- cellular PAcP
- PCNA
- proliferating cell nuclear antigen
- AR
- androgen receptor
- HR
- hormone refractory
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA
- EGFR
- epidermal growth factor receptor
- PTP
- protein-tyrosine phosphatase
- co-IP
- co-immunoprecipitation
- WT
- wild type
- PYK2
- proline-rich tyrosine kinase 2
- Ab
- antibody
- FBS
- fetal bovine serum.
REFERENCES
- 1.Huggins C., Hodges C. V. (1972) CA Cancer J. Clin. 22, 232–240 [DOI] [PubMed] [Google Scholar]
- 2.Seidenfeld J., Samson D. J., Hasselblad V., Aronson N., Albertsen P. C., Bennett C. L., Wilt T. J. (2000) Ann. Intern. Med. 132, 566–577 [DOI] [PubMed] [Google Scholar]
- 3.Isaacs J. T., Isaacs W. B. (2004) Nat. Med. 10, 26–27 [DOI] [PubMed] [Google Scholar]
- 4.Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 5.Rahman M., Miyamoto H., Chang C. (2004) Clin. Cancer Res. 10, 2208–2219 [DOI] [PubMed] [Google Scholar]
- 6.Culig Z., Hobisch A., Cronauer M. V., Radmayr C., Trapman J., Hittmair A., Bartsch G., Klocker H. (1995) Eur. Urol. 27, (Suppl. 2), 45–47 [DOI] [PubMed] [Google Scholar]
- 7.Seaton A., Scullin P., Maxwell P. J., Wilson C., Pettigrew J., Gallagher R., O'Sullivan J. M., Johnston P. G., Waugh D. J. (2008) Carcinogenesis 29, 1148–1156 [DOI] [PubMed] [Google Scholar]
- 8.Malinowska K., Neuwirt H., Cavarretta I. T., Bektic J., Steiner H., Dietrich H., Moser P. L., Fuchs D., Hobisch A., Culig Z. (2009) Endocr. Relat. Cancer 16, 155–169 [DOI] [PubMed] [Google Scholar]
- 9.Craft N., Shostak Y., Carey M., Sawyers C. L. (1999) Nat. Med. 5, 280–285 [DOI] [PubMed] [Google Scholar]
- 10.Meng T. C., Lee M. S., Lin M. F. (2000) Oncogene 19, 2664–2677 [DOI] [PubMed] [Google Scholar]
- 11.Lee M. S., Igawa T., Yuan T. C., Zhang X. Q., Lin F. F., Lin M. F. (2003) Oncogene 22, 781–796 [DOI] [PubMed] [Google Scholar]
- 12.Wen Y., Hu M. C., Makino K., Spohn B., Bartholomeusz G., Yan D. H., Hung M. C. (2000) Cancer Res. 60, 6841–6845 [PubMed] [Google Scholar]
- 13.Signoretti S., Montironi R., Manola J., Altimari A., Tam C., Bubley G., Balk S., Thomas G., Kaplan I., Hlatky L., Hahnfeldt P., Kantoff P., Loda M. (2000) J. Natl. Cancer Inst. 92, 1918–1925 [DOI] [PubMed] [Google Scholar]
- 14.Osman I., Scher H. I., Drobnjak M., Verbel D., Morris M., Agus D., Ross J. S., Cordon-Cardo C. (2001) Clin. Cancer Res. 7, 2643–2647 [PubMed] [Google Scholar]
- 15.Calvo B. F., Levine A. M., Marcos M., Collins Q. F., Iacocca M. V., Caskey L. S., Gregory C. W., Lin Y., Whang Y. E., Earp H. S., Mohler J. L. (2003) Clin. Cancer Res. 9, 1087–1097 [PubMed] [Google Scholar]
- 16.Shi Y., Chatterjee S. J., Brands F. H., Shi S. R., Pootrakul L., Taylor C. R., Datar R., Cote R. J. (2006) BJU Int. 97, 170–178 [DOI] [PubMed] [Google Scholar]
- 17.Foti A. G., Herschman H., Cooper J. F. (1977) Clin. Chem. 23, 95–99 [PubMed] [Google Scholar]
- 18.Vihko P., Virkkunen P., Henttu P., Roiko K., Solin T., Huhtala M. L. (1988) FEBS Lett. 236, 275–281 [DOI] [PubMed] [Google Scholar]
- 19.Lin M. F., Lee M. S., Zhou X. W., Andressen J. C., Meng T. C., Johansson S. L., West W. W., Taylor R. J., Anderson J. R., Lin F. F. (2001) J. Urol. 166, 1943–1950 [PubMed] [Google Scholar]
- 20.Veeramani S., Yuan T. C., Chen S. J., Lin F. F., Petersen J. E., Shaheduzzaman S., Srivastava S., MacDonald R. G., Lin M. F. (2005) Endocr. Relat. Cancer 12, 805–822 [DOI] [PubMed] [Google Scholar]
- 21.Li H. C., Chernoff J., Chen L. B., Kirschonbaum A. (1984) Eur. J. Biochem. 138, 45–51 [DOI] [PubMed] [Google Scholar]
- 22.Lin M. F., Clinton G. M. (1986) Biochem. J. 235, 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M. F., Garcia-Arenas R., Xia X. Z., Biela B., Lin F. F. (1994) Differentiation 57, 143–149 [DOI] [PubMed] [Google Scholar]
- 24.Tonks N. K., Neel B. G. (2001) Curr. Opin. Cell Biol. 13, 182–195 [DOI] [PubMed] [Google Scholar]
- 25.Veeramani S., Lee M. S., Lin M. F. (2009) Trends Biochem. Sci. 34, 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin M. F., Lee C. L., Clinton G. M. (1986) Mol. Cell Biol. 6, 4753–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin M. F., DaVolio J., Garcia-Arenas R. (1992) Cancer Res. 52, 4600–4607 [PubMed] [Google Scholar]
- 28.Lin M. F., Garcia-Arenas R., Kawachi M., Lin F. F. (1993) Cell Mol. Biol. Res. 39, 739–750 [PubMed] [Google Scholar]
- 29.Lin M. F., Meng T. C. (1996) Biochem. Biophys. Res. Commun. 226, 206–213 [DOI] [PubMed] [Google Scholar]
- 30.Lin M. F., Meng T. C., Rao P. S., Chang C., Schonthal A. H., Lin F. F. (1998) J. Biol. Chem. 273, 5939–5947 [DOI] [PubMed] [Google Scholar]
- 31.Meng T. C., Lin M. F. (1998) J. Biol. Chem. 273, 22096–22104 [DOI] [PubMed] [Google Scholar]
- 32.Zhang X. Q., Lee M. S., Zelivianski S., Lin M. F. (2001) J. Biol. Chem. 276, 2544–2550 [DOI] [PubMed] [Google Scholar]
- 33.Igawa T., Lin F. F., Rao P., Lin M. F. (2003) Prostate 55, 247–258 [DOI] [PubMed] [Google Scholar]
- 34.Zylka M. J., Sowa N. A., Taylor-Blake B., Twomey M. A., Herrala A., Voikar V., Vihko P. (2008) Neuron 60, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toualbi-Abed K., Daniel F., Güller M. C., Legrand A., Mauriz J. L., Mauviel A., Bernuau D. (2008) Carcinogenesis 29, 536–543 [DOI] [PubMed] [Google Scholar]
- 36.Igawa T., Lin F. F., Lee M. S., Karan D., Batra S. K., Lin M. F. (2002) Prostate 50, 222–235 [DOI] [PubMed] [Google Scholar]
- 37.Chen S. J., Karan D., Johansson S. L., Lin F. F., Zeckser J., Singh A. P., Batra S. K., Lin M. F. (2007) Prostate 67, 557–571 [DOI] [PubMed] [Google Scholar]
- 38.Lin M. F., Lee C. L., Li S. S., Chu T. M. (1983) Biochemistry 22, 1055–1062 [DOI] [PubMed] [Google Scholar]
- 39.Lee M. S., Igawa T., Lin M. F. (2004) Oncogene 23, 3048–3058 [DOI] [PubMed] [Google Scholar]
- 40.Zhang H. T., O'Rourke D. M., Zhao H., Murali R., Mikami Y., Davis J. G., Greene M. I., Qian X. (1998) Oncogene 16, 2835–2842 [DOI] [PubMed] [Google Scholar]
- 41.Mikami Y., Davis J. G., Dobashi K., Dougall W. C., Myers J. N., Brown V. I., Greene M. I. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7335–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akiyama T., Matsuda S., Namba Y., Saito T., Toyoshima K., Yamamoto T. (1991) Mol. Cell Biol. 11, 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Levy R., Paterson H. F., Marshall C. J., Yarden Y. (1994) EMBO J. 13, 3302–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frogne T., Laenkholm A. V., Lyng M. B., Henriksen K. L., Lykkesfeldt A. E. (2009) Breast Cancer Research 11, R11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S., Pirilä P., Kaija H., Porvari K., Vihko P., Juffer A. H. (2005) Proteins 58, 295–308 [DOI] [PubMed] [Google Scholar]
- 46.Luchter-Wasylewska E. (2001) Biochim. Biophys. Acta 1548, 257–264 [DOI] [PubMed] [Google Scholar]
- 47.Luchter-Wasylewska E., Wasylewski M., Röhm K. H. (2003) J. Protein Chem. 22, 243–247 [DOI] [PubMed] [Google Scholar]
- 48.Sweeney C., Carraway K. L., 3rd (2000) Oncogene 19, 5568–5573 [DOI] [PubMed] [Google Scholar]
- 49.Schulze W. X., Deng L., Mann M. (2005) Mol. Syst. Biol. 1, 2005 0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 51.Marcotte R., Zhou L., Kim H., Roskelly C. D., Muller W. J. (2009) Mol. Cell Biol. 29, 5858–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L., Davis J. S., Zelivianski S., Lin F. F., Schutte R., Davis T. L., Hauke R., Batra S. K., Lin M. F. (2009) Cancer Lett. 285, 58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gioeli D., Mandell J. W., Petroni G. R., Frierson H. F., Jr., Weber M. J. (1999) Cancer Research 59, 279–284 [PubMed] [Google Scholar]
- 54.Stevenson L. E., Frackelton A. R., Jr. (1998) Breast Cancer Res. Treat. 49, 119–128 [DOI] [PubMed] [Google Scholar]
- 55.Ursini-Siegel J., Hardy W. R., Zuo D., Lam S. H., Sanguin-Gendreau V., Cardiff R. D., Pawson T., Muller W. J. (2008) EMBO J. 27, 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren Z., Schaefer T. S. (2002) J. Biol. Chem. 277, 38486–38493 [DOI] [PubMed] [Google Scholar]
- 57.Tice D. A., Biscardi J. S., Nickles A. L., Parsons S. J. (1999) Proc. Natl. Acad. Sci. 96, 1415–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biscardi J. S., Maa M. C., Tice D. A., Cox M. E., Leu T. H., Parsons S. J. (1999) J. Biol. Chem. 274, 8335–8343 [DOI] [PubMed] [Google Scholar]
- 59.Olayioye M. A., Beuvink I., Horsch K., Daly J. M., Hynes N. E. (1999) J. Biol. Chem. 274, 17209–17218 [DOI] [PubMed] [Google Scholar]
- 60.Abdulghani J., Gu L., Dagvadorj A., Lutz J., Leiby B., Bonuccelli G., Lisanti M. P., Zellweger T., Alanen K., Mirtti T., Visakorpi T., Bubendorf L., Nevalainen M. T. (2008) Am. J. Pathol. 172, 1717–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Zhang Y., Glass A., Zellweger T., Gehan E., Bubendorf L., Gelmann E. P., Nevalainen M. T. (2005) Clinical Cancer Research 11, 5863–5868 [DOI] [PubMed] [Google Scholar]
- 62.Tan S. H., Dagvadorj A., Shen F., Gu L., Liao Z., Abdulghani J., Zhang Y., Gelmann E. P., Zellweger T., Culig Z., Visakorpi T., Bubendorf L., Kirken R. A., Karras J., Nevalainen M. T. (2008) Cancer Res. 68, 236–248 [DOI] [PubMed] [Google Scholar]
- 63.Cotarla I., Ren S., Zhang Y., Gehan E., Singh B., Furth P. A. (2004) Int. J. Cancer 108, 665–671 [DOI] [PubMed] [Google Scholar]
- 64.Dagvadorj A., Kirken R. A., Leiby B., Karras J., Nevalainen M. T. (2008) Clin. Cancer Res. 14, 1317–1324 [DOI] [PubMed] [Google Scholar]
- 65.Bowman T., Garcia R., Turkson J., Jove R. (2000) Oncogene 19, 2474–2488 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.