Abstract
The first enzyme in the pathway for l-arabinose catabolism in eukaryotic microorganisms is a reductase, reducing l-arabinose to l-arabitol. The enzymes catalyzing this reduction are in general nonspecific and would also reduce d-xylose to xylitol, the first step in eukaryotic d-xylose catabolism. It is not clear whether microorganisms use different enzymes depending on the carbon source. Here we show that Aspergillus niger makes use of two different enzymes. We identified, cloned, and characterized an l-arabinose reductase, larA, that is different from the d-xylose reductase, xyrA. The larA is up-regulated on l-arabinose, while the xyrA is up-regulated on d-xylose. There is however an initial up-regulation of larA also on d-xylose but that fades away after about 4 h. The deletion of the larA gene in A. niger results in a slow growth phenotype on l-arabinose, whereas the growth on d-xylose is unaffected. The l-arabinose reductase can convert l-arabinose and d-xylose to their corresponding sugar alcohols but has a higher affinity for l-arabinose. The Km for l-arabinose is 54 ± 6 mm and for d-xylose 155 ± 15 mm.
Keywords: Aspergillus, Enzyme Catalysis, Fungi, Metabolism, Metabolic Regulation, Reductase, EC.1.1.1.21, Aldose Reductase, Pentose Catabolism
Introduction
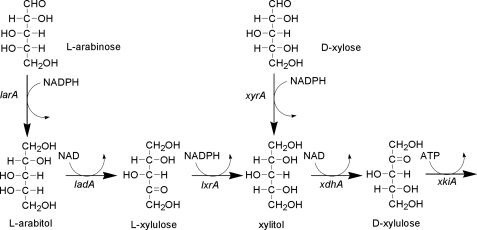
l-Arabinose is an abundant carbon source present in the plant cell wall of a range of plant tissues in form of arabinoxylan. Many saprobic microorganisms, including Aspergillus niger, are able to efficiently hydrolyze the plant material and utilize the released saccharides. In the case of A. niger, l-arabinose and d-xylose are catabolized through the pentose metabolic pathway (1), which is responsible for the conversion of these sugars to d-xylulose-5-phosphate. d-Xylulose-5-phosphate enters the central metabolism via the pentose phosphate pathway. The pentose metabolic pathway can be split into three parts (Fig. 1); l-arabinose-specific part, consisting of l-arabinose reductase (LarA)2, l-arabitol dehydrogenase (LadA), and l-xylulose reductase (LxrA); d-xylose specific part consisting with d-xylose reductase (XyrA); and a common part consisting of xylitol dehydrogenase (XdhA), and d-xylulose kinase (XkiA). l-Arabinose is converted to l-arabitol by the NADPH-specific l-arabinose reductase (2). l-Arabitol is then further metabolized via l-xylulose to xylitol in the reactions carried by the NAD-specific enzyme l-arabitol dehydrogenase (3, 4) and the NADPH-specific l-xylulose reductase, respectively. Xylitol, a shared intermediate in the metabolism of l-arabinose and d-xylose, is converted to d-xylulose by the NAD-specific xylitol dehydrogenase (5) and finally to d-xylulose-5-phosphate by xylulokinase (6).
FIGURE 1.
l-arabinose/d-xylose catabolic pathways in A. niger. l-Arabinose and d-xylose go through several reductions and oxidations. The reductions are NADPH-linked and the oxidations NAD-linked. The metabolites are in Fischer-projection, except for the l-xylulose which is upside down. The last step is a phosphorylation resulting in d-xylulose 5-phosphate, which is a metabolite of the pentose phosphate pathway. The gene names are indicated. All the genes have been identified except for the lxrA and the larA, which is subject of the present communication.
d-Xylose and l-arabinose are usually present together in nature and both these sugars are able to induce the expression of the genes of the pentose metabolic pathway. It has been reported for instance that the presence of d-xylose in the medium leads to induction of the l-arabinose pathway genes such as ladA in A. niger (7). However, there are significant differences in the mechanisms of the induction of the two specific parts of the pentose metabolic pathway on the molecular level. The d-xylose-dependent induction works through the transcription factor xlnR (8), which is regulating the xyrA (9). The l-arabinose-dependent induction is mediated by a yet unknown regulatory protein AraR (3). Two A. niger mutants, araA and araB, had been identified that were suggested to regulate the larA, ladA, and the xkiA (10). In addition, the compound responsible for the transcriptional activation of the genes encoding enzymes of the l-arabinose-specific part of the metabolic pathway seems to be l-arabitol (11). The overexpression of xlnR led to an up-regulation of the genes involved in l-arabinose metabolism, including ladA in Aspergillus oryzae (12). Deletion of the gene in Aspergillus niger, on the other hand, seems to have little effect on expression of the genes of l-arabinose pathway (3).
The l-arabinose metabolism of A. niger had been studied for decades and all the enzymatic activities in the pathway have been identified (1). However some of the corresponding genes are still missing. The missing are the genes coding for the l-arabinose reductase and the l-xylulose reductase. In some filamentous fungi, for instance in Hypocrea jecorina, it seems that only one enzyme XYL1 is responsible for the reduction of l-arabinose and d-xylose, because the deletion of xyl1 in H. jecorina results in significantly impaired growth on both sugars (13, 14). In A. niger, however, two specific pentose reductases are present, XyrA and LarA. The l-arabinose reductase had been purified from A. niger and basic biochemical properties characterized (2). Here we set out to identify the l-arabinose reductase of A. niger, and its corresponding gene, larA.
MATERIALS AND METHODS
Strains and Plasmids
A. niger, strain ATCC 1015 (CBS 113.46), and the strains derived from it were maintained as conidia in 20% (v/v) glycerol, 0.8% (w/v) NaCl, and 0.025% (v/v) Tween 20 at −80 °C. Saccharomyces cerevisiae, strain FY834, was obtained from Jay C. Dunlap (Dartmouth College, Hanover, NH) and stored at −80 °C in 20% (v/v) glycerol with 0.9% (w/v) NaCl.
Plasmid pRSET-A was obtained from Invitrogen. The pCL2-Amds vector was constructed as described previously (15). The gpdA promoter of A. nidulans was replaced by the gpdA promoter of A. niger in the pCL2-Amds plasmid. The gpdA promoter was amplified from the A. niger ATCC1015 genomic DNA using primers An_gpdA-pr_NotI_F, and An_gpdA-pr_R (Table 1) and inserted to the pCL2-Amds plasmid after digestion with NotI and SpeI. The ORF of the larA gene (JGI47818) was inserted between PacI and AscI sites. Plasmid pYX212 was obtained from R&D systems. The ORFs of xyrA (JGI51997), larA (JGI47818), and putative reductase (purA; JGI46249) were synthesized, codon optimized for expression in Saccharomyces cerevisiae, at GeneArt (Regensburg, Germany). The genes were inserted into the plasmid pYX212 between EcoRI and SalI sites allowing the expression under control of the TPI1 promoter. For the expression of the C-terminal His6-tagged larA, the gene was amplified by PCR from the synthetic codon optimized ORF with the primers larA-Sc-ORF_F and larA-HIS-C_R (Table 1). The PCR product was inserted between EcoRI and SalI sites of the pYX212 plasmid and the resulting construct was verified by sequencing.
TABLE 1.
Primers used for PCR
| Primer name | Sequence |
|---|---|
| pyrG-del-F_n | TATACCCGGGTGATTGAGGTGATTGGCGAT |
| pyrG-del-R_n | TATACCCGGGTTATCACGCGACGGACAT |
| xyrA-5-F | TATAGAATTCACGCAAGATCAAAAGATGGC |
| xyrA-5-R | TATACCCGGGAAGGTTTGGAGGGGTAATA |
| xyrA-3-F | TATACCCGGGTAGACCATTCAGCTATGAGTG |
| xyrA-3-R | TATAGAATTCTTTGAAGAATCTGGAGGAGC |
| xyrA-ORF-F | ATGGCCTCTCCCACAGTAAAG |
| xyrA-ORF-R | GAAAATAGGAGCGTAGAGTC |
| larA-5-F | TATAGGATCCACCAGACTAGTATTGATCGGC |
| larA-5-R | TATAGAGCTCATAGGAGGAGAGATGGAAGGA |
| larA-3-F | TATAGAGCTCCACTCTCGCTTGTATGATGATG |
| larA-3-R | TATAGAATTCGGCGATGCAGACAACGTCTT |
| larA-ORF-F | GAAGGTTACTCTCAACTCCGG |
| larA-ORF-R | ACAATCACCTTATGACCAGCA |
| PUR-5-F | TATAGAGCTCTTTAGGGTCGAATCCCTAGTCA |
| PUR-5-R | TATACCCGGGACAAGAGGAAGATAGCAGATGGC |
| PUR-3-F | TATACCCGGGCCGATCGCAACGTACTTTTA |
| PUR-3-R | TATAAAGCTTGCATTTCCGCAAACATTTCC |
| PUR-ORF-F | TTTACCATCGCACTTCACCCT |
| PUR-ORF-R | TAACCTTATTACTGGGGCCTG |
| larA(Sc)_F | CAATGTCTTTGGGTAAAAAGG |
| larA(Sc)_R | GGGTCGACGCGTAACAATAA |
| larA-Sc-ORF_F | TTAAGAATTCACAATGTCTTTGGG |
| larA-HIS-C_R | TAATGTCGACTCAGTGATGGTGATGGTGATGCCCAACAATAACCTTATGACCAGC |
Media and Cultivation
A. niger strains were grown on potato dextrose agar (Beckton Dickinson) to produce conidia, and in YPG medium containing 10 g of yeast extract liter−1 and 2 g of Bacto peptone l−1, and 3% DifcoTM gelatin (Beckton Dickinson) to produce mycelium. Carbon sources were l-arabinose (Merck, 1.01492), l-arabitol (Sigma Aldrich, A3506), d-xylose (Sigma Aldrich, X1500), and d-glucose (VWR, AnalaR Normapure).
To assess growth on l-arabinose, d-xylose, and l-arabitol, the spores of the A. niger strains were grown on agar plates containing 6.7 g of yeast nitrogen base liter−1 (YNB, Becton Dickinson), synthetic complete amino acid mixture (16), 20 g of agar liter−1, and 20 g liter−1 of l-arabinose or d-xylose or l-arabitol, respectively.
For biomass measurement, the A. niger strains were pregrown overnight in YPG medium and the corresponding amount of 0.1 g l−1 (dry mass) of mycelium was inoculated to 6.7 g of yeast nitrogen base liter−1, synthetic complete amino acid mixture (16), and 20 g liter−1 of l-arabinose or d-xylose or l-arabitol, respectively. After 24 h, the mycelium was collected, washed, and vacuum dried for subsequent biomass measurement. The experiment was done in triplicates.
For the qPCR analysis, strains were cultivated in YPG medium overnight. The mycelia were transferred to fresh medium containing 10 g of yeast extract llter−1, 2 g of Bacto peptone liter−1, and 20 g of liter−1 of d-glucose, d-xylose, or l-arabinose. The mycelia were incubated in this medium for up to 10 h at 28 °C and the samples were taken at different time intervals for RNA isolation and transcription analysis.
S. cerevisiae strains were grown overnight in 6.7 g of yeast nitrogen base liter−1, synthetic complete amino acid mixture without uracil and 4% d-glucose prior to protein extraction, purification, and enzymatic assays.
For pentose reductase activity measurements in A. niger, the wild-type ATCC1015, ΔxyrA, ΔlarA, and ΔpurA strains were cultivated in YPG medium overnight. The mycelia were transferred to fresh medium containing 10 g of yeast extract liter−1, 2 g of Bacto peptone liter−1, and 20 g liter−1 of d-glucose or d-xylose or l-arabinose, respectively, and cultivated for 4 h. The mycelia were collected by filtration and washed with PBS buffer (pH 6.9) prior to cell wall disruption and subsequent enzymatic tests.
Transcription Analysis
To quantify the transcription of the selected genes in A. niger in different conditions, mycelia were grown as described above. After the transfer to media with d-glucose, d-xylose, or l-arabinose, the mycelium was removed by filtration at 0, 1, 2, 4, 6, 8, and 10 h and washed with water. The total RNA was purified with RNeasy®Plant Mini kit (Qiagen). 6 μg of purified total RNA was used in the reverse transcription reaction (SuperScript, Invitrogen).
cDNA produced in the reverse transcription reaction was diluted 1:20 with water and 5 μl of diluted solution was used in qPCR reactions (LightCycler 480 SYBR Green I Master; Roche, Switzerland). The primers are listed in Table 2. The reactions were carried out in a LightCycler 480 Instrument II, and the analysis was performed with the accompanying software (Advance Relative Quantification tool). The signal was normalized to that of actin.
TABLE 2.
Primers used for qPCR
| Primer name | Sequence |
|---|---|
| LAR_qPCR_F | GACTACCTCGATCTCTACCTC |
| LAR_qPCR_R | GTCGTTGATCACGACATCAC |
| PUR_qPCR_F | GCCTACCTGGATCTCTATCTC |
| PUR_qPCR_R | CGGATGTTGAAGTTGGACAC |
| xyrA_qPCR_F | CCAAGAGTAACAACCCTCAG |
| xyrA_qPCR_R | GTAGAGTCCGTATCCCAAAGG |
| LAD_qPCR_F | ATTTGGAGTGTCAAGTTCGG |
| LAD_qPCR_R | TGTCACAAGCTTCTTCAAGTC |
| act_qPCR_F | CAACATTGTCATGTCTGGTGG |
| act_qPCR_R | GGAGGAGCAATGATCTTGAC |
Deletion of the Pentose Reductase Genes in A. niger ATCC 1015 ΔpyrG
ATCC 1015 ΔpyrG strain, which was generated as described previously (15), was transformed by the deletion cassettes for the respective reductase genes.
The cassette for deletion of the xyrA gene (JGI51997) contained 1502 bp from the A. niger xyrA promoter region, 1500 bp from the A. niger xyrA terminator region, and a 1928-bp fragment containing the pyrG gene flanked by its native promoter and terminator. These fragments were obtained by PCR of A. niger ATCC1015 genomic DNA using primers xyrA-5-F, xyrA-5-R, xyrA-3-F, xyrA-3-R, pyrG-del-F_n, and pyrG-del-R_n (Table 1), and the proofreading DNA polymerase Phusion (Finnzymes). The xyrA terminator fragment (xyrA-3) digested with EcoRI (NEB) was inserted into the plasmid pRSET-A (Invitrogen), which was digested with EcoRI and PvuII (both NEB). This intermediary construct was digested with XmaI (NEB) and Ecl136II (Fermentas) and ligated to the XmaI-digested promoter fragment (xyrA-5). The resulting vector was digested with SmaI (NEB) and treated with phosphatase. The pyrG DNA fragment, after digestion with SmaI, was inserted between the two xyrA flanking regions. The resulting plasmid was verified by restriction analysis and sequencing. The deletion cassette, 4930 bp, containing the xyrA flanking regions and the pyrG gene, was released by EcoRI (NEB) digestion and transformed into A. niger ATCC 1015 ΔpyrG.
The cassette for deletion of the larA gene (JGI47818) contained 1525 bp from the A. niger larA promoter region, 1575 bp from the A. niger larA terminator region, and a 1928-bp fragment containing the pyrG gene flanked with its native promoter and terminator. These fragments were obtained by PCR of A. niger ATCC 1015 genomic DNA using primers larA-5-F, larA-5-R, larA-3-F, larA-3-R, pyrG-del-F_n, and pyrG-del-R_n (Table 1), and the proofreading DNA polymerase Phusion (Finnzymes). The larA terminator fragment (larA-3) digested with EcoRI (NEB) was inserted into the plasmid pRSET-A (Invitrogen), which was digested with EcoRI and PvuII (both NEB). This intermediary construct was digested with NdeI (NEB), treated with the Klenow polymerase, purified and then digested with SacI (NEB). The resulting linear fragment was ligated to the SacI digested promoter fragment (larA-5). The resulting vector was digested with Ecl136II (Fermentas) and treated with phosphatase. The pyrG DNA fragment, after digestion with SmaI, was inserted between the two larA flanking regions. The resulting plasmid was verified by restriction analysis and sequencing. The deletion cassette, 4861 bp, containing the larA flanking regions and the pyrG gene, was released by BssSI and BlnI (both NEB) digestion and transformed into A. niger ATCC1015 ΔpyrG.
The cassette for deletion of the gene encoding a putative reductase (purA; JGI46249) contained 1554 bp from the A. niger purA promoter region, 1542 bp from the A. niger purA terminator region, and a 1928-bp fragment containing the pyrG gene flanked with its native promoter and terminator. These fragments were obtained by PCR of A. niger ATCC1015 genomic DNA using primers pur-5-F, pur-5-R, pur-3-F, pur-3-R, pyrG-del-F_n, and pyrG-del-R_n (Table 1), and the proofreading DNA polymerase Phusion (Finnzymes). The purA terminator fragment (purA-3) digested with HindIII (NEB) was inserted into the plasmid pRSET-A (Invitrogen), which was digested with HindIII and PvuII (both NEB). This intermediary construct was digested with BglII (NEB), treated with the Klenow polymerase, purified, and then digested with XmaI (NEB). The resulting linear fragment was ligated to the XmaI-digested promoter fragment (purA-5). The resulting vector was digested with SmaI (NEB) and treated with phosphatase. The pyrG DNA fragment, after digestion with SmaI (NEB), was inserted between the two purA flanking regions. The resulting plasmid was verified by restriction analysis and sequencing. The deletion cassette, 4888 bp, containing the purA flanking regions and the pyrG gene, was released by BssSI and BlnI digestion and transformed into A. niger ATCC1015 ΔpyrG.
All the above described transformants were selected by the ability to grow in the absence of uracil. Strains with successful deletions were verified by PCR. The primers for testing the presence/absence of the respective ORFs are listed in Table 1.
Expression of the larA Gene in A.niger ATCC1015 ΔlarA
The larA ORF, codon optimized for expression in S. cerevisiae at GeneArt (Regensburg, Germany), was inserted to pCL2-Amds via PacI and AscI sites. The resulting construct was linearized with NotI and transformed into A. niger ATCC1015 ΔlarA. Transformants were selected by ability to grow in the presence of acetamide, and the correct strains were verified by PCR with primers larA(Sc)_F and larA(Sc)_R (Table 1). Expression of the gene was also tested by growth in the presence of l-arabinose.
Expression of the Pentose Reductases in S. cerevisiae
S. cerevisiae strain CEN.PK2–1D was transformed with plasmids pYX212 containing xyrA (JGI51997), larA (JGI47818), or putative reductase (purA; JGI46249). The transformants were selected by growth in the absence of uracil, and the expression of the reductases was tested in the cell extracts by enzymatic activity measurements.
Protein Extraction and the Enzyme Activity Measurements
For testing of d-xylose and l-arabinose reductase activities in A. niger after induction with the respective sugar, the mycelium was mixed with 800 μl of PBS buffer with protease inhibitors mixture (Complete, Roche) and 800 μl of acid-washed glass beads (Sigma). The cells were disrupted in two 40-s sessions in Fast Prep (Bio 101). The cell extracts were clarified by centrifugation, and the supernatants were used in the assay. The protein concentration was analyzed by the Protein Assay kit (BioRad).
For specific activity measurements, xyrA, larA, and purA were expressed in S. cerevisiae. The cells from overnight cultures (0,5L SD-URA) were harvested and washed with PBS (pH 6.9). The cell pellets were resuspended in 15 ml of ice-cold PBS supplemented with protease inhibitors (Complete, Roche). The cells were disrupted by Bead-Beater (Biospec), and the extracts were clarified by centrifugation (15 min, 20,000 rcf, 4 °C), the assays were performed on diluted samples. Protein concentration was analyzed by the Protein Assay kit (BioRad).
In the case of larA, the C-terminal 6× histidine version of the gene was expressed in S. cerevisiae strain CEN.PK2–1D. The histidine-tagged LarA protein was purified from the cell extract by Ni-NTA resin (Qiagen). The protein purity was confirmed by SDS-PAGE and silver staining. The specific activity was analyzed with l-arabinose, d-xylose, and d-galactose.
d-Xylose and l-arabinose reductase activities were measured in reaction mixtures containing 0.2 mm NADPH, various concentrations of either d-xylose or l-arabinose, and the cell extract (or purified histidine-tagged LarA) in PBS buffer (pH 6.9). The concentration of d-xylose and l-arabinose was 115 mm in the case of the tests performed with the A. niger protein extracts. The removal of NADPH from the reaction was monitored by measuring absorption at 340 nm in microtiter plates (NUNC) in the spectrophotometer Varioskan (Thermo Electron Corporation). The Km and Vmax was estimated from the Michaelis-Menten equation fitted to the measured data.
RESULTS
Identification of the l-Arabinose Reductase Gene, larA
Previous results suggested that in A. niger, the genes involved in d-xylose or l-arabinose catabolism are up-regulated on d-xylose (7, 17). For instance the l-arabitol dehydrogenase (ladA) is up-regulated in the presence of d-xylose, even though it does not contribute to its catabolism (7, 17). Taking this into account we supposed that also other genes of the l-arabinose pathway might be up-regulated on d-xylose. We tested the transcription after a shift to d-glucose, d-xylose, or l-arabinose for the genes that were previously identified to be up-regulated on d-xylose. The following genes were tested: JGI47818, JGI38546, xyrA (d-xylose reductase), ladA (l-arabitol dehydrogenase) and the JGI46429. The JGI46429 has been previously reported to code for an l-arabinose reductase (7), however no experimental evidence was given. In the following we will call the JGI47818, larA, for l-arabinose reductase and the JGI46429, purA, for putative reductase.
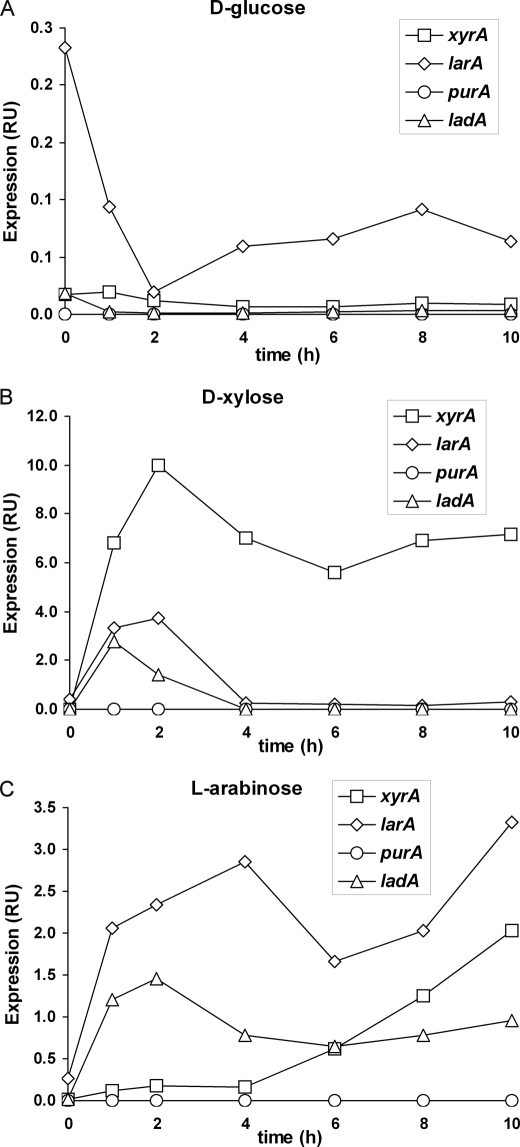
The xyrA is up-regulated on d-xylose as is the larA and the ladA (Fig. 2B). The larA and the ladA are only transiently up-regulated for the first hours whereas the xyrA remained up-regulated during the observed period of 10 h. On l-arabinose the ladA and the larA are up-regulated during the whole period (Fig. 2C), suggesting that ladA and larA are regulated in the same way. The purA is not up-regulated on either carbon source suggesting that it is probably not the l-arabinose reductase as suggested previously. Also the JGI38546 is not up-regulated on l-arabinose (data not shown) indicating that also this gene has no role in l-arabinose catabolism. The xyrA is also up-regulated on l-arabinose, however only after a time lag of about 4 h.
FIGURE 2.
Expression profiles. Transcription analysis of the of selected genes in A. niger. The transcription of xyrA, larA, purA, and ladA was analyzed after the transfer of the mycelia to medium containing d-glucose (A), d-xylose (B), or l-arabinose (C). The mRNA was quantified by using qPCR and normalized to actin mRNA.
Phenotype Analysis of Deletion Strains of A. niger
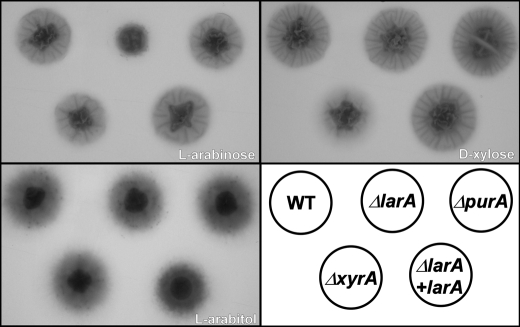
To demonstrate the importance of the genes in question we constructed knock-out strains for the larA, xyrA, and purA genes. The ΔlarA strain showed significantly reduced growth on agar plates in the presence of l-arabinose as a sole carbon source, while its phenotype does not differ from the wild-type strain in the other tested conditions (Fig. 3).
FIGURE 3.
Deletion of larA, purA, and xyrA. Growth on l-arabinose, d-xylose, and l-arabitol. Expression of larA suppress the phenotype of larA deletion.
Also in liquid culture the biomass is reduced. After 24 h the ΔlarA strain produced about 40% less biomass from l-arabinose when compared with the wild type strain. Expression of the larA gene from a plasmid in the ΔlarA background completely rescued the phenotype of the deletion strain. Growth of the ΔxyrA strain, on the other hand, is affected only when d-xylose is present as a sole carbon source. The phenotype of the strain is, however, less pronounced as compared with the ΔlarA strain grown on l-arabinose (Fig. 3). In liquid culture the ΔxyrA strain produced about 30% less biomass from d-xylose than the wild-type strain in 24 h.
The ΔpurA strain does not confer any phenotype dissimilar to the wild-type strain in the tested conditions indicating the product of the gene is not (significantly) involved in the l-arabinose or d-xylose pathways. All tested strains grow comparably well on l-arabitol, which confirms that none of the genes has a role further down the catabolic pathway.
Pentose Reductase Activities in A. niger
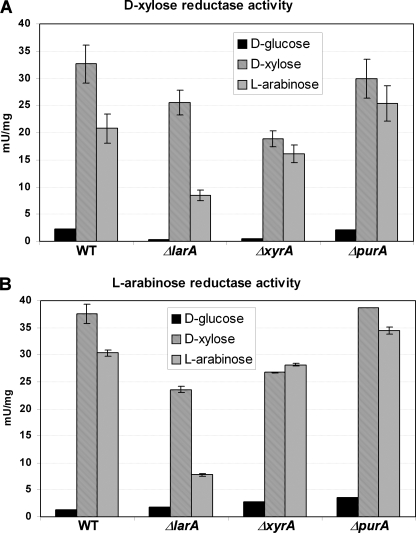
We have analyzed the d-xylose and l-arabinose reductase activities in the protein extracts from the A. niger deletion strains. Three inducing conditions were used; incubation with d-glucose, d-xylose, or l-arabinose in order to up- or down-regulate expression of the genes encoding enzymes involved in metabolism of the sugars. The ΔlarA strain demonstrated significantly lower reductase activity with both, d-xylose and l-arabinose, when l-arabinose is used as the inducing compound. The activity is reduced to 25–40% of the corresponding activity of the wild-type strain (Fig. 4). On the other hand, when grown on d-xylose the strain confers noticeably higher activity with both tested sugars, 60–80% of the wild-type activity. The ΔxyrA strain is especially affected in presence of d-xylose as inducer, when its reductase activity is decreased to 60–70% of the wild-type activity. In the case of l-arabinose-inducing conditions, the reductase activity of the ΔxyrA strain is 80–90% of the wild type. The ΔpurA strain does not differ significantly from the wild type in any tested conditions. In the presence of d-glucose, the d-xylose and l-arabinose reductase activities are diminished in the all tested strains (Fig. 4).
FIGURE 4.
Reductase activity in crude cell extracts of A. niger. d-Xylose reductase activity (A) and l-arabinose reductase activity (B) in crude cell extracts of A. niger grown on the different carbon sources d-glucose, d-xylose, and l-arabinose. Different A. niger strains with deletions in the l-arabinose reductase gene, ΔlarA, the d-xylose reductase gene, ΔxyrA, or in the gene for the putative reductase, ΔpurA, or without a deletion, WT, were tested. The activities are given per mg of total protein in the crude cell extracts.
In Vitro Characterization of the Enzymes
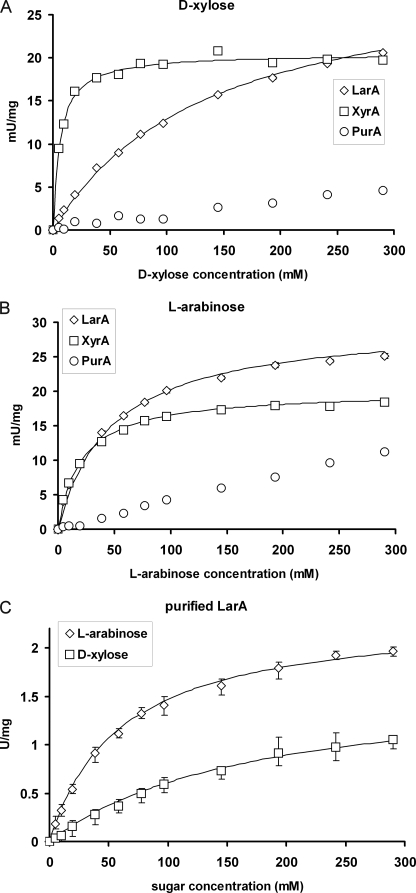
To measure Km values of the tested enzymes, we expressed the genes larA, xyrA, and purA in the yeast S. cerevisiae. The genes were expressed from a strong constitutive promoter as full-length open reading frames without any additional sequences (tags), which could have affected the enzyme activity or specificity. In S. cerevisiae the background reductase activities for d-xylose and l-arabinose are negligible (data not shown), and therefore the expression system allows reliable characterization of the studied proteins in a crude extracts without need of further purification. The l-arabinose reductase (LarA) displays high activities with l-arabinose as well as with d-xylose (Fig. 5, A and B). The Km values of LarA are 54 mm and 155 mm for l-arabinose and d-xylose, respectively. The d-xylose reductase (XyrA) shows comparably high activities with both sugars as well with Km values of 21 mm and 6 mm for l-arabinose and d-xylose, respectively. The putative reductase (PurA), previously suggested being the l-arabinose reductase (7) confers significantly lower activities and affinities to both; d-xylose and l-arabinose. The Km values for the enzyme could not be estimated in the concentration range of the obtained data (Fig. 5).
FIGURE 5.
Reductase activities in extracts of S. cerevisiae expressing the larA, xyrA, or purA. The Michaelis-Menten constants were estimated from fitting the Michealis-Menten kinetics to the data points. For the reaction with d-xylose the following constants were obtained: LarA (Km = 155 ± 15 mm; Vmax = 31 units/mg), XyrA (Km = 6 mm; Vmax = 20.5 units/mg). For the reaction with l-rabinose: LarA (Km = 54 ± 6 mm; Vmax = 30 units/mg), XyrA (Km = 21 mm; Vmax = 20 units/mg). The activities are given per mg of total protein in the crude cell extracts (A and B) or as specific activity of the purified histidine-tagged LarA protein (C). The reactions were carried out in PBS buffer with the NADPH concentration of 200 μm.
The LarA was also purified after tagging it C-terminally with a histidine tag. The tagged protein was expressed in S. cerevisiae, and the activity in the crude extract compared with the non-tagged version. The tagged and the non-tagged protein showed the same activity indicating that the tagging of the protein is not compromising the activity. The tagged purified protein showed a single band in SDS-PAGE and was used for a kinetic characterization. The purified enzyme had activity with d-xylose and l-arabinose (Fig. 5C), but not with d-galactose (not shown). The Km and Vmax values for l-arabinose and d-xylose were 54 mm and 2.35 units/mg and 155 mm and 1.65 units/mg, respectively.
DISCUSSION
In the transcription profile of the genome of A. niger and related species during growth on d-xylose 27 genes were up-regulated, among them the ladA (7, 17). The ladA codes for the l-arabinose 4-dehydrogenase, a gene in the pathway for l-arabinose catabolism, that has no function in d-xylose catabolism. This suggested that the regulation of the genes for l-arabinose and d-xylose catabolism might be interrelated. This seems to be different in H. jecorina where the transcription of lad1 coding for the l-arabinitol dehydrogenase is not induced on d-xylose (18). The transcription in H. jecorina was however analyzed 6 h after the transfer to d-xylose medium so that an initial up-regulation of the lad1 cannot be ruled out. This up-regulation of the ladA on d-xylose in A. niger encouraged us to look for other genes of the l-arabinose pathway that might have been up-regulated on d-xylose. And indeed the JGI47818 turned out to be the l-arabinose reductase, larA. The larA gene has 6 introns and the open reading frame codes for a protein with 327 amino acids and a calculated molecular mass of 36.012 Da. The protein belongs to the aldo-keto reductase superfamily, which is a family of monomeric NADPH-dependent oxidoreductases such as aldose reductases (19). Although there are a few enzymes that would have l-arabinose reductase activity, there are a couple of arguments to conclude that the larA is coding for the real l-arabinose reductase.
The larA is transcribed in the presence of l-arabinose but only transiently in the presence of d-xylose. The transcription time course on l-arabinose is reassembling the time course of the ladA that is specific for l-arabinose. The LarA has a higher affinity and specific activity toward l-arabinose than toward d-xylose. The larA deletion results in reduced growth on l-arabinose. The larA deletion results in a reduced l-arabinose reductase activity in the crude cell extract.
When analyzing the time course of the transcription of selected genes on different carbon sources we noticed that the transcription of the l-arabinose-pathway genes (larA and ladA) is only transient in the presence of d-xylose (Fig. 2B). This is different with the xyrA gene. The xyrA is strongly up-regulated on d-xylose and its expression remains high throughout the cultivation. We used d-xylose that was more than 99% pure; however we cannot exclude that it contained small traces of l-arabinose that would be responsible for the transient transcription of the genes of the l-arabinose pathway.
On l-arabinose the larA and ladA are up-regulated throughout the cultivation (Fig. 2C). Also on l-arabinose the transcription of xyrA is low during the first 4 h after which transcription is initiated. This suggests different transcriptional regulations on l-arabinose and d-xylose that are linked to each other. d-Xylose induces the transcription of all genes for pentose catabolism but in the absence of l-arabinose only the d-xylose genes remain up-regulated. l-arabinose induces initially only the l-arabinose genes but later also the d-xylose genes.
Deletion of the larA gene in A. niger leads to a considerable growth defect when l-arabinose is the sole carbon source (Fig. 3), which indicates the importance of the gene for the catabolism of l-arabinose. The ability of the ΔlarA strain to grow on l-arabinose can be explained by the presence of the xyrA gene, which is expressed under these conditions (Fig. 2C). Apparently, the d-xylose reductase (XyrA) activity is not sufficient in the ΔlarA strain to support the full growth on l-arabinose. The growth of the ΔlarA strain in the presence of d-xylose is unaffected (Fig. 3), which is in accordance with the expression analysis, where the xyrA gene is predominantly expressed. It seems that the l-arabinose reductase plays a minor role in the conversion of d-xylose to xylitol apparently due to its low expression level on d-xylose (Fig. 2B). In nature, however, where d-xylose and l-arabinose are usually both present in the typical growth substrates of A. niger, it is likely that l-arabinose reductase contributes to the reduction of d-xylose as well. Deletion of the xyrA gene leads to a growth phenotype only in presence of d-xylose as a sole carbon source. The growth defect is however milder than in the case of the ΔlarA strain on l-arabinose (Fig. 3), indicating insufficient capacity of LarA or other yet unidentified reductase to substitute for the missing XyrA activity under those conditions.
The l-arabinose reductase activity in the crude extract of A. niger is similar, whether the mycelia were grown on d-xylose or on l-arabinose. For the d-xylose reductase activity this is different. Here the difference between d-xylose and l-arabinose grown mycelia is more pronounced (Fig. 4). This is in good agreement with the previous findings of Witteveen et al. (1). When deleting the larA, the l-arabinose and d-xylose reductase activities are lowered for l-arabinose grown mycelia as compared with d-xylose grown. This shows that the LarA is normally responsible for a large fraction of the pentose reductase activity during growth on l-arabinose. The deletion of the putative reductase purA did not affect the reductase activities. The deletion of the xyrA lowered the reductase activity altogether. This is in good agreement with the transcription profile in Fig. 2 and the growth experiment in Fig. 3.
The remaining relatively high reductase activity of the ΔxyrA strain grown in presence of d-xylose indicates that LarA (or another reductase) can compensate for the loss of the XyrA activity. We tested whether the transcription of the larA is up-regulated in the the ΔxyrA strain but could not observe a difference between the larA expression in the wild type and the ΔxyrA strain on d-xylose (data not shown). The XyrA is the main reductase during growth on d-xylose and has the highest affinity for d-xylose while the LarA is the main reductase during growth on l-arabinose and has the highest specific activity for l-arabinose.
This work was supported by the Academy of Finland through the Finnish Centre of Excellence in White Biotechnology, Green Chemistry, and an Academy Research Fellowship (to P. R.).
- LarA
- l-arabinose reductase
- PBS
- phosphate-buffered saline
- ORF
- open reading frame
- LadA
- l-arabitol dehydrogenase
- LxrA
- l-xylulose reductase
- XyrA
- d-xylose reductase.
REFERENCES
- 1.Witteveen C. F., Busink R., van de Vondervoort P. J., Dijkema C., Swart K., Visser J. (1989) J. Gen. Microbiol. 135, 2163–2171 [DOI] [PubMed] [Google Scholar]
- 2.de Groot M. J., Prathumpai W., Visser J., Ruijter G. J. (2005) Biotechnol. Prog. 21, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 3.de Groot M. J. L., van den Dool C., Wösten H. A. B., Levisson M., vanKuyk P. A., Ruijter G. J. G., de Vries R. P. (2007) Food Technol. Biotechnol. 45, 134–138 [Google Scholar]
- 4.Richard P., Londesborough J., Putkonen M., Kalkkinen N., Penttilä M. (2001) J. Biol. Chem. 276, 40631–40637 [DOI] [PubMed] [Google Scholar]
- 5.Witteveen C. F., Weber F., Busink R., Visser J. (1994) Microbiology 140, 1679–1685 [Google Scholar]
- 6.vanKuyk P. A., de Groot M. J., Ruijter G. J., de Vries R. P., Visser J. (2001) Eur. J. Biochem. 268, 5414–5423 [DOI] [PubMed] [Google Scholar]
- 7.Andersen M. R., Vongsangnak W., Panagiotou G., Salazar M. P., Lehmann L., Nielsen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4387–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Peij N. N., Visser J., de Graaff L. H. (1998) Mol. Microbiol. 27, 131–142 [DOI] [PubMed] [Google Scholar]
- 9.Hasper A. A., Visser J., de Graaff L. H. (2000) Mol. Microbiol. 36, 193–200 [DOI] [PubMed] [Google Scholar]
- 10.de Groot M. J., van de Vondervoort P. J., de Vries R. P., vanKuyk P. A., Ruijter G. J., Visser J. (2003) Microbiology 149, 1183–1191 [DOI] [PubMed] [Google Scholar]
- 11.de Vries R. P., Flipphi M. J., Witteveen C. F., Visser J. (1994) FEMS Microbiol. Lett. 123, 83–90 [DOI] [PubMed] [Google Scholar]
- 12.Noguchi Y., Sano M., Kanamaru K., Ko T., Takeuchi M., Kato M., Kobayashi T. (2009) Appl. Microbiol. Biotechnol. 85, 141–154 [DOI] [PubMed] [Google Scholar]
- 13.Seiboth B., Gamauf C., Pail M., Hartl L., Kubicek C. P. (2007) Mol. Microbiol. 66, 890–900 [DOI] [PubMed] [Google Scholar]
- 14.Akel E., Metz B., Seiboth B., Kubicek C. P. (2009) Eukaryot. Cell 8, 1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojzita D., Wiebe M., Hilditch S., Boer H., Penttilä M., Richard P. (2010) Appl. Environ. Microbiol. 76, 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman F., Fink G., Hicks J. B. (1983) Methods in Yeast Genetics. A Laboratory Manual, Cold Springs Harbor Laboratory, Cold Springs Harbour, NY [Google Scholar]
- 17.Jørgensen T. R., Goosen T., Hondel C. A., Ram A. F., Iversen J. J. (2009) BMC Genomics 10, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiboth B., Hartl L., Pail M., Kubicek C. P. (2003) Eukaryot. Cell 2, 867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohren K. M., Bullock B., Wermuth B., Gabbay K. H. (1989) J. Biol. Chem. 264, 9547–9551 [PubMed] [Google Scholar]