Abstract
The molecular mechanisms underlying microtubule participation in autophagy are not known. In this study, we show that starvation-induced autophagosome formation requires the most dynamic microtubule subset. Upon nutrient deprivation, labile microtubules specifically recruit markers of autophagosome formation like class III-phosphatidylinositol kinase, WIPI-1, the Atg12-Atg5 conjugate, and LC3-I, whereas mature autophagosomes may bind to stable microtubules. We further found that upon nutrient deprivation, tubulin acetylation increases both in labile and stable microtubules and is required to allow autophagy stimulation. Tubulin hyperacetylation on lysine 40 enhances kinesin-1 and JIP-1 recruitment on microtubules and allows JNK phosphorylation and activation. JNK, in turn, triggers the release of Beclin 1 from Bcl-2-Beclin 1 complexes and its recruitment on microtubules where it may initiate autophagosome formation. Finally, although kinesin-1 functions to carry autophagosomes in basal conditions, it is not involved in motoring autophagosomes after nutrient deprivation. Our results show that the dynamics of microtubules and tubulin post-translational modifications play a major role in the regulation of starvation-induced autophagy.
Keywords: Autophagy, JNK, Kinesin, Microtubules, Tubulin, Acetylation
Introduction
Macroautophagy (simply referred to here as autophagy) occurs at low basal levels to perform homeostatic functions such as protein and organelle turnover. It is also an adaptive catabolic response to different metabolic stresses, including nutrient deprivation, growth factor depletion, or hypoxia (1, 2). Newly assembled multilayer membranes expand and sequester parts of the cytoplasm to form autophagosomes that subsequently fuse with lysosomes to degrade their content (1, 2). When cells lack nutrients, inhibition of the mammalian target of rapamycin activates the ULK complex (ULK1 and ULK2 are the mammalian orthologs of the yeast Atg1) to initiate the cascade of events leading to the formation of autophagosomes (reviewed in Ref. 3). Building of isolation membranes or phagophores involves a complex comprising Beclin 1 (Beclin 1 is the mammalian homolog of the yeast Atg6) and the class III PI3K4 (PI3K(III), also called hVps34) (4). Newly synthesized phosphatidylinositol 3-phosphate then recruits effectors such as WIPI-1, the homolog of yeast Atg18 (5, 6), to allow the recruitment of other autophagosomal building bricks (4). Atg9 contributes to membrane shuttling that elongates the pre-autophagosomal membrane (reviewed in Ref. 7). Such elongation also involves ubiquitin-like machineries, which in turn allow Atg12-to-Atg5 conjugation, and the modification of LC3 (LC3 is the mammalian ortholog of the yeast Atg8) (8) prior to their recruitment to autophagosomal membranes. LC3 is a light chain of MAP1 first identified in neurons (9, 10). After autophagy induction, the C-terminal region of native LC3 (pro-LC3) is cleaved by Atg4, yielding LC3-I. Atg7 and Atg3 then conjugate LC3-I to phosphatidylethanolamine on its C-terminal glycine, yielding LC3-II that attaches to the autophagosomal membrane (11).
Microtubules (MTs) participate in autophagosome formation and underlie their dynein-dependent, centripetal movement prior to fusion with lysosomes (12–21). However, the mechanisms that underlie their participation in autophagosome formation and dynamics are not clear. MT differentiation into a labile highly dynamic subset and a stable long lived subset (22) might have caused an underestimation of the MT role in autophagosome dynamics. Indeed, tubulin from stable MTs may undergo post-translational modifications like detyrosination and/or acetylation (23, 24) that may regulate the recruitment of molecular motors (25–27) and MT-bound proteins (28). In this study, we examined how MT dynamics and tubulin acetylation are involved in regulating the formation of pre-autophagosomal structures and in influencing MT-based autophagosome movements.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmids
Rabbit polyclonal anti-LC3 was used as described previously (29) for Western blotting. Anti-Glu-tubulin was a kind gift from Dr. L. Lafanechère (INSERM U366, Commissariat à l'Energie Atomique, Grenoble, France). Mouse monoclonal antibody against α-tubulin (clone DM1A), acetyl-tubulin (clone 6–11B1), c-Myc (clone 9E10), and anti-IgG conjugated to fluorescein isothiocyanate, TRITC, or to horseradish peroxidase were from Sigma. Rabbit polyclonal antibodies against Bcl-2, JNK, and phospho-JNK were from Cell Signaling (Beverly, MA). Goat polyclonal antibodies against Atg5, Beclin 1, KHC, and donkey anti-goat conjugated to horseradish peroxidase were from Santa Cruz Biotechnology. Rabbit polyclonal antibody anti-PI3K(III) was from Abgen (San Diego). Anti-p62 was from BD Biosciences. GFP-LC3 and tandem mRFP-GFP-LC3 expression vectors were kind gifts from T. Yoshimori (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan) (30). mCherry-tubulin control vector was kindly provided by Dr. R. Y. Tsien (Department of Pharmacology, Howard Hughes Medical Center, University of San Diego) (31). The mCherry-α tubulin K40A mutated vector was a kind gift from Dr. F. Saudou (Institut Curie Orsay France) (26). Myc-tagged WIPI-1 was used as described previously (5). The kinesin light chain 2 GFP-tagged tetratricopeptide repeat (KLC-TPR) cargo-binding domain was kindly provided by Dr. M. Way (Cell Motility Laboratory, Cancer Research, London, UK) (32). The p50-dynamitin construct was a gift from Dr. S. Etienne-Manneville (Institut Pasteur, Paris, France) (33). The MKK7-JNK1 construct was the kind gift from R. Davis (Howard Hughes Medical Institute and Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, MA) (34).
Cell Culture
HeLa cells stably expressing GFP-LC3 or GFP-CD63 (kindly provided by Dr. Tolkovsky, Department of Biochemistry, University of Cambridge, Cambridge UK) (35) and transiently transfected cells were cultured in Dulbecco's minimum essential medium containing an antibiotic/antifungal mixture, supplemented with 10% fetal calf serum (Dutscher, Rungis, France), with 2 mm sodium pyruvate and 0.5 mg/ml geneticin when appropriate. FuGENE 6 (Roche Diagnostics) was used for transfections according to the manufacturer's instructions. U2OS cell clones stably expressing GFP-WIPI-1 (5) were isolated by standard G481 selection and cultured in Dulbecco's minimum essential medium containing an antimycoplasma/antibiotic mixture and 10% fetal calf serum (PAA).
Autophagy Induction
Nutrient deprivation was performed after three washes with PBS by incubation in Earle's balanced salt solution for 2–4 h at 37 °C as indicated. When appropriate, cells were pretreated for 1 h before autophagy induction, with 10 μm E64 (Calbiochem) and 1 μg/ml pepstatin A to inhibit lysosomal enzymes. Autophagy induction was visualized by the accumulation of endogenous LC3-II by Western blot. In fluorescence microscopy, the proportions of cells with fluorescent LC3 dots among GFP-positive cells were measured in controls and after starvation. Autophagy induction is the ratio between these proportions. Routinely, basal autophagy was about 20% and reached ∼60% after starvation. For each set of experiments, the ratios shown are normalized relative to the percentage of mock-treated cells exhibiting LC3 puncta in basal conditions.
Co-immunoprecipitation
Cells were rinsed and scraped in ice-cold PBS and then centrifuged for 5 min at 4 °C. The pellet was lysed in CHAPS lysis buffer (20 mm Tris, pH 7.4, 137 mm NaCl, 2 mm EDTA, 10% glycerol, and 2% CHAPS) and incubated for 3 h at 4 °C with gentle agitation. After overnight incubation with anti-Beclin 1 antibody (1:80, 4 °C, agitation), beads of protein A-Sepharose (Amersham Biosciences) were added (agitation for 2 h at 4 °C). The beads were washed (5 min, 4 °C) once with 20 mm Tris, pH 7.4, 2 mm EDTA, 10% glycerol, 137 mm NaCl, and 0.5% CHAPS and then twice with 20 mm Tris, pH 7.4, 2 mm EDTA, 10% glycerol, 274 mm NaCl, and 0.5% CHAPS. Beads were boiled in Laemmli sample buffer prior to Bcl-2 Western blot analysis.
Cytosol Extraction and Purification of Microtubule Fractions
Labile and stable MT fractions were prepared as described previously (36) with a few modifications. Briefly, 104 cells/cm2 were plated 48 h before the experiment in flasks previously coated with 0.2% gelatin. After two rinses in MT-stabilizing PEM buffer (100 mm PIPES, 1 mm EGTA, 1 mm MgCl2, pH 6.9), cytosol was extracted using PEM supplemented with 0.02% saponin and 10% glycerol (3 min, 37 °C). Labile MTs were then allowed to depolymerize for 30 min in PEM at 37 °C. After one rinse with warm PEM, cells were subjected to an additional 3 h of incubation in PEM to collect a few residual labile MT proteins and minor contaminants from the stable MT fraction. After checking, it only contained very low protein concentration compared with labile and stable MT fractions, and this intermediate fraction was discarded. To collect the stable MT fraction, cells were incubated for 30 min on ice in PM buffer supplemented with 1 mm CaCl2 and with or without 0.05% Nonidet P-40. The labile and stable MT fractions were concentrated using Centricon 10 filtration devices (Millipore) and then subjected to MT repolymerization in the presence of 10% glycerol and 10 μm Taxol prior to SDS-PAGE and Western blot analysis on polyvinylidene difluoride membranes along with total cell lysates, cytosols, and total MT fractions. The latter was collected in parallel on cells subjected to prior cytosol extraction (as described above) and then scraped in warm Laemmli buffer.
Kinesin-1 and Dynein Inhibition
The KHC RNAi experiments were performed using two conditions of inhibition as described previously (37). Cells were either transfected with a duplex RNA oligonucleotide that targets the ubiquitous kinesin-1 heavy chain DNA in positions 2973–2991 downstream of the open reading frame (sense, 5′-AUGCAUCUCGUGAUCGCAA-dTdT-3′; antisense, 3′-dTdTUACGUAGAGCACUAGCGUU-5′) or transfected with a combination of two other double strand RNAs against KHC (38). In the first case, transfections were performed twice using HiPerfect reagent (Qiagen) according to the manufacturer's instructions with an RNA concentration of 5 nmol/liter at days 0 and 4. In the second case, transfections were performed once, 72 h prior to analysis. A nonsilencer siRNA (Qiagen) was used as a negative control in all RNAi experiments. To check the effectiveness of KHC siRNA, siRNA-transfected cells were solubilized directly in Laemmli sample buffer prior to SDS-PAGE analysis in a 6% gel, electrotransfer on polyvinylidene difluoride membranes, and Western blot analysis. Blots were probed using the KHC anti-kinesin heavy chain and anti-α-tubulin (DM1A) and then imaged using enhanced chemiluminescence assay (ECL, PerkinElmer Life Sciences) on Kodak Biomax MR films. KHC RNAi routinely yielded 65–70% KHC inhibition. As an alternative to KHC RNAi (or in combination with it), cells were transfected using a cDNA that encoded a GFP-tagged KLC-TPR domain that behaves as a dominant-negative of kinesin light chain and prevents cargo loading on kinesin-1 (32). Dynein was inhibited by overexpressing p50-dynamitin 24 or 48 h prior to analysis. The effectiveness of p50 transfection was checked retrospectively by visualizing Golgi dispersion after cell fixation in cold methanol and immunofluorescence labeling of GM-130.
Western Blot Analysis
Whole cells were lysed directly in Laemmli buffer without 2-mercaptoethanol and bromophenol blue to allow protein assay. For Western blotting, 40 μg of proteins were loaded for every sample.
Immunofluorescence, Microscopy, and Image Analysis
Cells cultured on glass coverslips were rinsed twice in 0.1 m PBS, pH 7.4, then fixed, and permeabilized with methanol at −20 °C for 5 min. Primary and secondary antibodies were diluted in PBS supplemented with 10% bovine serum albumin. Washes between incubations were made with PBS. Coverslips were mounted in Tris-buffered (80 mm, pH 8.5) 8% Mowiol and 20% glycerol mixture supplemented with diazabicyclo-octane as an anti-fading agent. Lysosomes were visualized in living cells after incubation for 1 h at 37 °C with 50 nm Dextran Cascade Blue Lysotracker® (Invitrogen). Time-lapse sequences were acquired at 37 °C, either on a Zeiss Axiovert-2 microscope through a 63× 1.4NA objective and a Zeiss mRm camera or on a Leica DM-LB microscope, through a 100× 1.3 NA objective and a Scion CFW1312M CCD camera. GFP-WIPI-1 video microscopy was conducted as described previously (39). Data were quantified using the Imaris software (Bitplane, Zürich, Switzerland) and ImageJ (W. Rasband, National Institutes of Health, Bethesda, rsb.info.nih.gov) with the kymograph plugin suite obtained on line.
Statistical Analysis
Quantitative data are the means ± S.E. of at least three independent experiments. Cumulative velocity distribution curves were compared using Mann-Whitney's U test. Other comparisons used Student's t test. The following were used: * means p < 0.05, ** means p < 0.01, and *** means p < 10−3. Autophagy induction was measured from at least 300 cells in each condition. Autophagosome mobility and velocity measurements were performed from at least 500 puncta in each condition.
RESULTS
Starvation-induced Autophagy Requires Microtubule Dynamics
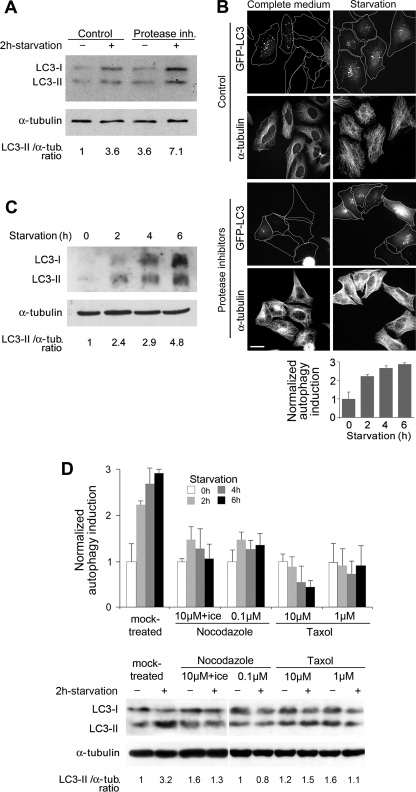
Starvation increased the amount of LC3-II (Fig. 1A) and the number of GFP-LC3 cells exhibiting puncta, without causing an apparent alteration of the MT network (Fig. 1B). Moreover, LC3-II accumulated in higher amounts in the presence of lysosomal inhibitors in starved cells compared with controls (Fig. 1A), indicating a build up of autophagosomes during starvation and an increase in the autophagic flux. Stimulation of autophagy also lasted for several hours in starved cells (Fig. 1C). To determine whether MT dynamics and/or an MT subset are specifically involved in autophagosome formation, we combined two approaches. First, the whole MT network was disassembled using nocodazole and exposure to cold. Second, to remove labile MTs and preserve stable MTs over the whole autophagy induction period, we treated cells with nanomolar nocodazole, which causes an overall inhibition of MT dynamics (40), prior to and during starvation. This treatment enriched cells in stabilized, acetylated MTs similar to those resisting a brief nocodazole treatment (supplemental Fig. S1). Because MTs were completely depolymerized or stabilized, the build up of autophagosomes was severely impaired as measured by the decreased number of cells exhibiting LC3 dots and the lack of accumulation of LC3-II (Fig. 1D), indicating that MT dynamics and/or the labile MT subset are important for autophagosome formation. Accordingly, overall MT stabilization using Taxol also prevented the build up of autophagosomes (Fig. 1D).
FIGURE 1.
Starvation-induced autophagy in HeLa cells requires MT dynamics. A, buildup of LC3-II in HeLa cells subjected or not to starvation (2 h in Earle's balanced salt solution), without or with protease inhibitors (inh.). B, HeLa cells transiently expressing GFP-LC3 were observed after immunolabeling of α-tubulin (α-tub.). The percentage of LC3-positive cells that exhibited fluorescent puncta was normalized and increased as a function of time during starvation. Values are the mean ± S.E. of data obtained from 300 cells at each time point. C, kinetics of LC3 accumulation during the course of nutrient deprivation in the presence of protease inhibitors. D, top panel histogram, accumulation of fluorescent LC3 puncta was quantified as in B after treatment with the indicated drugs to trigger partial or total MT disassembly or to stabilize MTs. Values are the mean ± S.E. obtained from 300 cells in each condition. The bottom panel shows a sample experiment demonstrating LC3-II accumulation in cells treated with protease inhibitors after the indicated treatments.
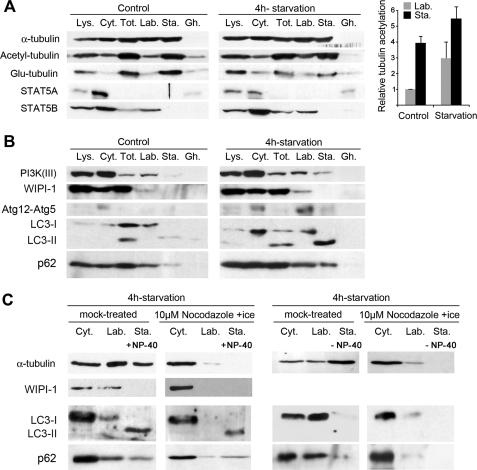
Markers of Autophagosome Formation Are Specifically Recruited on Labile MTs although Mature Autophagosomes Can Move along Stable MTs
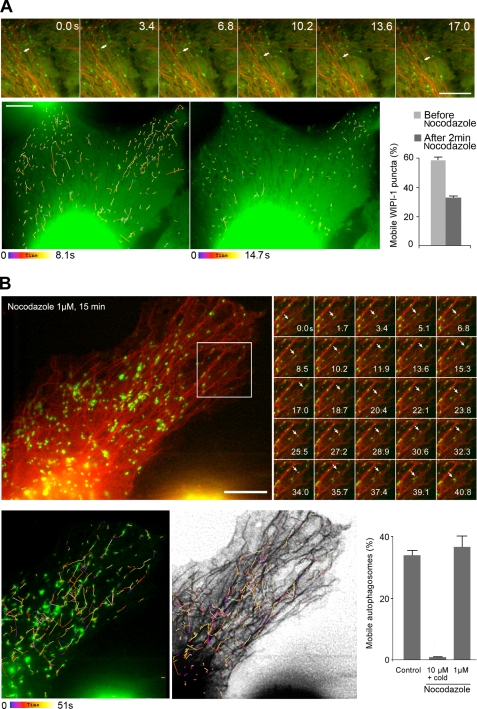
Because MT dynamics were required to allow for autophagy induction, the most labile MT subset might more specifically recruit markers of autophagosome formation. To test this hypothesis, we performed cell fractionation experiments in which we purified the proteins associated with labile MTs and with stable MTs (36). Cells were permeabilized using low saponin concentrations to avoid artifactual LC3 aggregation (supplemental Fig. S2) (41). MT protein fractionation was then validated by checking that the signaling and transcription factor STAT5B bound specifically to the labile MT fraction (Fig. 2A) and that the latter was not contaminated with soluble proteins, as STAT5A remained exclusively in the cytosol (36). The stable MT fraction was enriched in modified tubulin as shown by the distribution of the acetylated and detyrosinated tubulins. We noticed, however, that at variance with tubulin detyrosination, which seemed to decrease, tubulin acetylation increased between basal and starvation conditions, both in the labile and stable MT fractions (Fig. 2A). Consistent with the above results (Fig. 1D), the labile MT fraction contained markers of the isolation membrane like PI3K(III), WIPI-1, and the Atg12-Atg5 conjugate (Fig. 2B). Note that in contrast with the absence of WIPI-1 on labile MTs in basal conditions, PI3K(III) was readily detected on this MT subset where it might reflect the binding of early endosomes (42) to the cytoskeleton. Strikingly, LC3 was recovered differentially in the two MT fractions according to its lipidation. Although LC3-I was found in the labile MT fraction, LC3-II was extracted with stable MTs (Fig. 2B). Likewise, the polyubiquitin-binding protein p62, which is a substrate of autophagy (43), was also extracted with MTs and increased in the stable MT fraction after starvation (Fig. 2B). p62 also associated in part with labile MTs (Fig. 2, B and C) in a way that did not require autophagy induction. We further checked that the occurrence of WIPI-1, LC3-I, and p62 in the labile MT fraction actually reflected their binding to MTs. Indeed, upon complete MT disassembly, both proteins and tubulin were lost from the fraction that would otherwise have contained labile MTs (Fig. 2C). Regarding LC3-II, the situation was more complex. LC3-II was still recovered in the stable MT fraction extracted from cells subjected to complete MT disassembly (i.e. in the presence of calcium and with a nonionic detergent). In contrast, LC3-II was lost from this stable fraction when MTs were extracted in the absence of detergent (Fig. 2C). Consistently, p62 was recovered in a similar way (Fig. 2C). At this stage, detergent-dependent extraction of LC3-II and p62 did not reflect their actual compartmentalization on stable MTs but most likely indicated p62 sequestration into mature autophagosomes and LC3-II incorporation into autophagosomal membranes (which resisted saponin, see supplemental Fig. S2, not Nonidet P-40 and would thus have remained in cells although stable MTs were extracted in the absence of Nonidet P-40). From a morphological point of view, we also verified that early markers of autophagosome formation like WIPI-1 associated with MTs in living cells. After starvation, GFP-WIPI-1 occurred as puncta that were closely juxtaposed to MTs and that moved along them. A nocodazole treatment as short as 2 min inhibited puncta mobility by ∼45% (Fig. 3A) confirming that autophagosome formation involved the labile, nocodazole-sensitive MT subset. To evaluate the interaction between mature autophagosomes and MTs, we observed the displacements of GFP-LC3 puncta in cells that co-expressed mCherry-tubulin (Fig. 3B). This procedure was appropriate to follow autophagosomes as the fraction of mobile GFP-LC3 puncta and their displacement rate spectra were identical to those measured by following tandem-LC3 (30) (supplemental Fig. S3). In this construct, LC3 was fused both with GFP and monomeric red fluorescent protein. Unlike monomeric red fluorescent protein fluorescence, which persists in the late endosome/lysosome compartment after fusion with autophagosomes, GFP fluorescence is quenched upon such fusion (30), all because of distinguishable autophagosomes (green) from autolysosomes (red), which co-localized with Lysotracker® signal (blue) (supplemental Fig. S3A). Here, labile MTs were depolymerized with a mild nocodazole treatment (supplemental Fig. S1) prior to image acquisition, and we found that the trajectories of LC3 puncta always followed the tracks of nocodazole-resistant MTs (Fig. 3B). Contrasting with the WIPI-1 mobility drop upon labile MT disassembly, such treatment did not alter the percentage of mobile autophagosomes, although only a complete MT removal caused autophagosome immobilization (Fig. 3B). More accurately than LC3-II or p62 recovery in subcellular fractions, the mobility data indicate that mature autophagosomes can interact with the stable MT subset.
FIGURE 2.
Early autophagy markers compartmentalize on labile MTs. A, subcellular fractionation of HeLa cells without (Control) or with starvation (4 h in Earle's balanced salt solution). Total cell lysates (Lys.), cytosol (Cyt.), total MT (Tot.), labile MT (Lab.), and stable MT (Sta.) fractions as well as insoluble post-MT extraction cell ghosts (Gh.) were analyzed for the presence of the indicated proteins. Stable MT extractions were performed in the presence of Nonidet P-40. The histogram shows the relative increase in tubulin acetylation measured both in the labile and stable MT fractions after starvation. B, compartmentalization and enrichment of the labile MT fraction in PI3K(III), WIPI-1, the Atg12-Atg5 complex, and LC3-I after cell starvation. LC3-II occurs in the stable MT fraction after nutrient deprivation. p62 mainly associates with labile MT in basal conditions and occurs in the stable MT fraction upon starvation. Stable MT extractions were performed in the presence of Nonidet P-40. C, extensive MT disassembly prior to cellular fractionation results in the loss of the early autophagy marker WIPI-1 as well as that of LC3-I and p62 from the labile MT fraction. LC3-II and p62 were recovered in the stable MT fraction only when it was prepared in the presence of Nonidet P-40.
FIGURE 3.
WIPI-1-positive puncta movement specifically occurs on labile MTs, whereas mature autophagosomes can move along stable MTs. A, top, U2OS cells stably expressing GFP-WIPI-1 and transiently expressing mCherry-tubulin were subjected to starvation and then recorded in time-lapse video microscopy (time is in seconds). The arrow indicates a sample WIPI-1 structure that moves along an MT. Bottom, color-coded time projection of GFP-WIPI-1 puncta movements in the same cells. Count of mobile fluorescent WIPI-1 structures after a brief nocodazole treatment (1 μm, 2 min, 37 °C) indicates that WIPI-1 movement requires MT dynamics. Data are the mean ± S.E. obtained from the examination of 237 puncta (two cells). B, top, HeLa cells transiently expressing GFP-LC3 and mCherry-tubulin were subjected to a mild nocodazole treatment (1 μm, 15 min) that depolymerized labile MTs but preserved stable MTs. Cells were time-lapse recorded (Δt = 1.7 s) to evaluate autophagosome movement and trajectories. The white arrows indicate the displacement of a sample GFP-LC3 punctum in the outlined region. Bottom, trajectories of the mobile autophagosomes were color-coded as a function of time and are shown with their corresponding punctum (left) and superimposed with nocodazole-resistant MTs (middle). The histogram on the right shows that only a complete MT disassembly, not a mild nocodazole treatment blocked autophagosome movements in starved cells. Data were obtained from at least 500 autophagosomes in each condition (10 cells). Scale bars, 10 μm.
Nutrient Deprivation Triggers a Hyper-acetylation of Microtubules That Enhances Kinesin-1-dependent JNK Activation and Subsequent Autophagy Induction
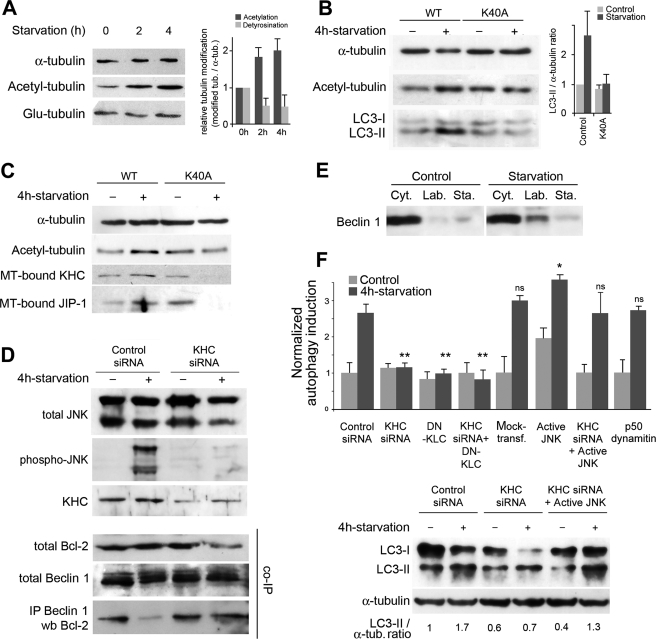
In the MT fractionation experiments, we noticed that along with the increased recruitment of PI3K(III), WIPI-1, or of the Atg12-Atg5 complex on labile MTs, tubulin underwent changes in its post-translational modifications upon starvation (see Fig. 2A). We verified this finding on a whole MT fraction in which tubulin acetylation approximately doubled by 2–4 h of starvation (Fig. 4A). As we observed previously that similar MT hyperacetylation occurs after a genotoxic stress to coordinate MT-dependent signaling (28), we tested whether starvation-induced hyperacetylation was important to allow autophagy induction. Strikingly, the expression of a nonacetylatable tubulin (the K40A mCherry-α-tubulin mutant), which prevented tubulin hyperacetylation upon starvation without an obvious MT network alteration (supplemental Fig. S4), prevented autophagy induction as shown by the impairment of LC3-II accumulation in the presence of protease inhibitors (Fig. 4B).
FIGURE 4.
Starvation-induced hyperacetylation of tubulin enhances JNK activation via a kinesin-1-dependent mechanism to allow autophagy induction. A, nutrient deprivation causes an ∼2-fold increase in tubulin acetylation of MTs. Whole MT fractions were prepared from cells subjected or not to incubation in Earle's balanced salt solution medium for the indicated times. Left, sample Western blot of total, acetylated, and detyrosinated (Glu-) tubulins. Right, histogram shows the mean ± S.E. of the acetylated-to-total and Glu-to-total tubulin ratios (n = 3 independent experiments). B, expression of a nonacetylatable mutant of mCherry-tubulin (K40A) impairs autophagy induction as measured by LC3-II accumulation. Cells pretreated with antiproteases to inhibit lysosomal degradation of autophagosome-associated proteins were subjected or not to starvation as indicated, lysed, and subjected to the analysis of tubulin and LC3 content. Left, sample of Western blots showing the effectiveness of K40A tubulin expression in preventing tubulin hyperacetylation and the inhibition of LC3-II buildup. Control cells expressed wild-type mCherry-tubulin (WT). Right, histogram showing the mean ± S.E. of cellular LC3-II-to-total tubulin ratios relative to that measured in cells expressing wild-type mCherry-tubulin (Control) without autophagy induction (n = 3 experiments). C, tubulin hyperacetylation caused by starvation increased the recruitment of KHC and JIP-1. Proteins were analyzed from total MT fractions prepared from cells expressing wild-type mCherry-tubulin (WT) or a nonacetylatable mCherry-tubulin mutant (K40A) that was subjected to starvation or not as indicated. D, JNK phosphorylation and the dissociation of the Bcl-2-Beclin 1 complex require kinesin-1 in conditions of starvation. Kinesin-1 inhibition was performed using RNAi of KHC and then cells were subjected or not to starvation as indicated. Top, whole cell lysates were analyzed for the presence of total JNK, phospho-JNK, and KHC. Bottom, the association of Beclin 1 with Bcl-2 was analyzed after co-immunoprecipitation (co-IP) of Beclin 1 and Bcl-2. The top two gels show Beclin 1 and Bcl-2 signals in the lysates before immunoprecipitation (IP). E, starvation results in increased Beclin 1 recruitment on the labile (Lab.) MT fractions and not in the stable (Sta.) fraction. Cyt., cytosol. F, autophagy induction requires kinesin-1. Top, accumulation of fluorescent LC3 puncta in cells expressing GFP-LC3 after kinesin-1 or dynein inhibition. Kinesin-1 was inhibited using KHC RNAi and the expression of a dominant-negative of kinesin light chain (DN-KLC) alone or in combination. Dynein was inhibited by overexpressing p50 dynamitin. Constitutive active JNK prevented the drop in autophagy stimulation that resulted from kinesin-1 inhibition. Data are the mean ± S.E. from at least 400 cells in each condition, except for the active JNK control as follows: 565 cells in basal autophagy and 488 cells after starvation. Comparisons were performed between starvation conditions; * means p < 0.05; ** means p < 0.01, and ns means nonsignificant. Bottom, example of LC3-II accumulation in cells treated with protease inhibitors in cells subjected or not to KHC inhibition and complementation with active JNK.
Tubulin acetylation has been shown previously to enhance the binding of molecular motors to MTs (25, 26) as well as that of Hsp90 and some of its client proteins (28, 44). This together with the fact that JNK regulates autophagy (45) and that kinesin-1 controls JNK activation on MTs (37) prompted us to determine whether the regulation of autophagy induction by tubulin acetylation involved kinesin-1 and JNK. To this end, we first checked that the increase in tubulin acetylation we observed after nutrient deprivation actually triggered an increased recruitment of KHC on MTs. This recruitment indeed required the hyperacetylation of tubulin as it was inhibited when cells expressed the K40A mutant (Fig. 4C). Next, if this kinesin-1 increase on MTs was not linked to increased organelle or vesicle trafficking but to JNK activation, then we should observe a similar behavior of JIP-1. JIP-1 is a molecular scaffolding protein that is transported by kinesin-1 (46) and binds a few signaling enzymes like JNK or its upstream protein kinases (MKK4 and MKK7) (47) to allow JNK phosphorylation and activation (48). Tubulin hyperacetylation was also responsible for the binding of JIP-1 to MTs (Fig. 4C), suggesting that enhanced kinesin-1 recruitment on MTs was specifically involved in JNK activation. Note that as in the experiments in which we measured LC3-II accumulation, the expression of nonacetylatable tubulin did not cause the entire loss of acetyl-tubulin from cells (as expected if some endogenous tubulin is still present) but only prevented its hyperacetylation. In the presence of nonacetylatable tubulin, the levels of MT-bound KHC and JIP-1 were regularly lower than in untreated controls, suggesting that it is not the static marking of specific MT tracks (stable ones) with acetyl-tubulin that is important to allow motor recruitment and autophagy induction but is most probably the dynamics of tubulin acetylation itself. To demonstrate that the acetylation-dependent increase in microtubular kinesin-1 and JIP-1 was actually responsible for the induction of autophagy, we next tested whether JNK activation required kinesin-1 upon nutrient deprivation. As shown in Fig. 4D (top panel), we knocked down KHC expression using RNAi, then subjected cells to starvation, and analyzed JNK phosphorylation by Western blot. Although nutrient deprivation dramatically increased JNK phosphorylation in controls, a reduction of cellular KHC levels by 65–70% prevented such phosphorylation, confirming that JNK activation was actually the consequence of kinesin recruitment on MTs. Then, as JNK regulates autophagy by phosphorylating Bcl-2 (45) and to assess that the kinesin-1-dependent JNK activation we observed was actually responsible for the onset of autophagy, we examined the status of the Bcl-2-Beclin 1 complex in co-immunoprecipitation experiments (Fig. 4D, bottom panel). Although the complex normally dissociated upon nutrient deprivation yielding free Beclin 1 that could associate with labile MTs (Fig. 4E), dissociation of Bcl-2 and Beclin 1 was blocked in cells that underwent kinesin-1 inhibition (Fig. 4D, bottom panel). Finally, we confirmed the involvement of kinesin-1-mediated JNK activation in regulating starvation-induced autophagy by studying the effects of molecular motor inhibition on the proportion of cells exhibiting LC3 puncta and on LC3-II accumulation (Fig. 4F). As expected from the above experiments, kinesin-1 inhibition (using either RNAi and/or using a dominant-negative of the light chain) completely blocked starvation-induced autophagy. As a control, we checked that dynein, the recruitment of which also increased on MTs upon cell starvation (data not shown), was not involved at all in autophagosome formation. In addition, we found that kinesin-1 inhibition could be compensated for by the expression of a constitutively active JNK, confirming that the JNK fraction that regulates autophagy requires a kinesin-1-dependent and hence a tubulin hyperacetylation-dependent activation.
Kinesin-1-dependent Autophagosome Traffic Is Switched Off upon Starvation
The most abundant fraction of cellular kinesin-1 is likely to be kept in an auto-inhibited form bound to Golgi membranes (49). Meanwhile, two motor subsets function in various membrane trafficking steps, including in autophagosome transport (50) and in basal JNK activation that is essential to sustain MT growth (37). It is clear from our above data that tubulin hyperacetylation is required to allow autophagy induction by making the stimulation of the kinesin-1/JNK pathway possible. As the mechanisms that regulate kinesin-1 commitment into each pathway are still obscure, we next asked whether kinesin-1 involvement in JNK activation operates independently from its involvement in membrane traffic, especially in that of autophagosomes (50).
To answer this question, we first determined whether kinesin-1 actually functioned in autophagosome traffic and whether its involvement changed between basal autophagy and nutrient deprivation conditions. Having verified above that the presence of MTs was required to allow autophagosome movements (Fig. 3B) (15, 16, 51), we first checked that autophagosomes actually underwent centrifugal movements (Fig. 5A). Next, we measured autophagosome mobility in cells that were subjected or not to molecular motor inhibition. Note that although the kinesin-1/JNK pathway functions to stimulate autophagy, it did not affect basal autophagy levels (see Fig. 4). Thus, we measured autophagosome mobility on the fraction (∼20%) of the LC3-positive cells that exhibited LC3 puncta. Strikingly, in control conditions, the fraction of mobile autophagosomes dropped by ∼50% upon starvation (Fig. 5B and supplemental Fig. S5). In contrast, all the means of kinesin-1 inhibition we used affected only basal autophagosome mobility and not that measured after starvation (Fig. 5B). This observation confirms that kinesin-1 is involved in autophagosome traffic in basal conditions and indicates that, in parallel with its increased recruitment on MTs to activate JNK, it is no longer involved in motoring autophagosomes upon nutrient deprivation. As a control, we also checked that autophagosome mobility involved dynein, which participated in motoring autophagosomes both in basal and in starvation conditions (Fig. 5B) as described previously (17, 51, 52).
FIGURE 5.
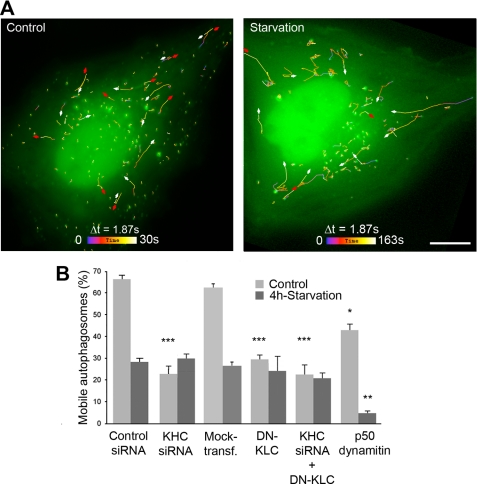
Kinesin-1 involvement in carrying autophagosomes decreases upon starvation. A, autophagosomes exhibit centrifugal movements. HeLa cells expressing tandem fluorescent LC3 were followed in the GFP channel by time-lapse microscopy with 1.87-s intervals during the indicated periods of time. Mobile autophagosomes were tracked, and their trajectories were color-coded as indicated. Red and white arrows show centrifugal and centripetal movements, respectively. Scale bar, 10 μm. B, kinesin-1 functions to carry autophagosomes in basal conditions, whereas autophagosome mobility drops by ∼50% after nutrient deprivation. The percentage of mobile autophagosomes was measured by following tandem LC3 as shown above, without and with kinesin-1 inhibition. As kinesin-1 inhibition prevents autophagy stimulation, the proportion of cells that exhibited fluorescent puncta was ∼20% after kinesin-1 inhibition, although it reached ∼60% in control conditions. Dynein inhibition caused by p50 dynamitin overexpression was used to control that autophagosome mobility could actually be inhibited in starvation conditions. Asterisks indicate the levels of significance relative to the corresponding control measured in the same condition as follows: *** means p < 0.001; ** means p < 0.01; * means p < 0.05. Data were obtained from the examination of at least 500 autophagosomes (10 cells) in each condition of kinesin-1 inhibition.
Tubulin Hyperacetylation Is Responsible for a Traffic Slowdown upon Starvation
We finally sought to explain the massive reduction in the fraction of mobile autophagosomes that occurs upon starvation. This phenomenon could either reflect the switch in kinesin-1 function shown above (from carrying autophagosomes to activating JNK) or a more global change of MTs like hyperacetylation. The latter hypothesis was supported by the fact that in conditions of starvation, other organelles and vesicles like late endosomes/lysosomes (visualized using fluorescent CD63) also exhibited similarly reduced mobility (Fig. 6A). To determine whether tubulin hyperacetylation was responsible for this change in mobility, we measured both the percentage of mobile autophagosomes and their velocities in cells that expressed or not a nonacetylatable tubulin (Fig. 6, B and C). The impairment of tubulin hyperacetylation prevented the decrease in autophagosome mobility triggered by starvation (Fig. 6B). Accordingly, a comparison of the cumulative autophagosome velocity distribution spectra revealed the slowdown expected to occur upon starvation and confirmed that such a slowdown required increased tubulin acetylation (Fig. 6C).
FIGURE 6.
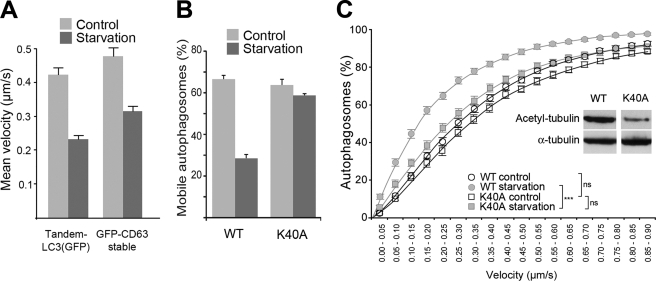
Drop in autophagosome mobility that occurs upon starvation results from tubulin hyperacetylation. A, starvation triggers a traffic slowdown that does not specifically affect autophagosomes. Autophagosome velocities were measured from kymographs computed from time-lapse sequences acquired with cells expressing tandem LC3 (in the GFP channel) or GFP-CD63. Data are the mean ± S.E. of at least 1000 autophagosomes measured from 15 cells in each condition. B, percentage of mobile autophagosomes decreases upon nutrient deprivation as result of tubulin hyperacetylation. Nonacetylatable mCherry-tubulin (K40A) or wild-type mCherry-tubulin (WT) was co-transfected with tandem LC3 48 h prior to time-lapse observation. Data are the mean ± S.E. of at least 1000 puncta observed from 15 cells per condition. C, tubulin hyperacetylation is responsible for the drop in autophagosome velocities observed upon starvation. The velocity distribution spectra measured from kymographs were transformed into cumulative curves for easier comparison. The curves show the mean ± S.E. of the velocities of at least 1000 LC3 puncta (15 cells) per condition. The inset shows the effectiveness of K40A-mCherry-tubulin expression on overall tubulin acetylation.
DISCUSSION
As shown previously in vesicular trafficking and signaling situations (36, 53), labile MTs may exhibit specialized functions. The molecular basis of such functional specialization is not yet clearly established. However, tubulin acetylation on lysine 40 may change tubulin conformation and allow stable MTs to modulate their protein environment, as shown for the molecular motors kinesin-1 and dynein (25, 26), and for the chaperone Hsp90 and some of its client proteins (28, 44). Such changes in the pattern of MT-bound proteins might cause the global decrease in the effectiveness of vesicle trafficking we observed. Like after genotoxic stress (28), we show now that nutritional stress also triggers hyperacetylation of tubulin, not only on stable but also on labile MTs, to coordinate cell response by enhancing kinesin-1 and JIP-1 recruitment on MTs. As we found that kinesin-1 inhibition almost completely abolished JNK phosphorylation in starvation conditions, and as JNK is a major cargo of MT-bound kinesin-1 (37), we may conclude that tubulin hyperacetylation is necessary to allow kinesin-1-dependent JNK activation and hence stimulation of autophagosome formation after nutrient deprivation. Although the signaling mechanism that causes tubulin hyperacetylation is unknown, it may change the balance between a tubulin acetyltransferase activity (like the Elongator complex ELP3 subunit (54)) and the deacetylase activities of HDAC6 (55) and SIRT2 (56). Interestingly, HDAC6 was shown to participate in autophagic responses in conditions of cell stress such as after the ubiquitin-proteasome inhibition or in cells with aggregates of misfolded proteins (57, 58). Unlike what happens with tubulin, Atg proteins, including Atg5, Atg7, and Atg8, are deacetylated to stimulate autophagy upon starvation, in a way that involves p300 and SIRT1 (59, 60). Contrasting with a still incomplete view in basal autophagy conditions (Fig. 7A), we may now propose a more comprehensive picture of how MTs participate in starvation-induced autophagy (Fig. 7B). Hyperacetylation of tubulin during starvation increases the recruitment of dynein (data not shown) and kinesin-1. This MT change is important to enhance JNK activation, but it is not sufficient to do so as histone deacetylase inhibition alone did not stimulate autophagy (data not shown). Following JNK activation, the upstream MAPK kinase and MAPK kinase kinase might cause the release of JIP from kinesin (61) and hence inactivate the motor. Kinesin-1 is thus required to allow JNK activation and consecutive Bcl-2 phosphorylation to release Beclin 1 (45). Beclin 1 would then associate with the labile MT subset where, together with PI3K(III), it might trigger phosphatidylinositol 3-phosphate synthesis, the subsequent recruitment of the phosphatidylinositol 3-phosphate-binding protein WIPI-1, and the assembly of autophagosome-forming scaffold as suggested by the recruitment of the Atg12-Atg5 conjugate. As mature autophagosomes can interact with stable MTs, the two MT subsets may cooperate to organize autophagosome trafficking and subcellular localization in starved cells (Fig. 7B). The exact step and the molecular mechanism(s) by which autophagosomes or immature autophagosomes (i.e. unsealed autophagosomal membrane) may switch from one MT subset to the other remain to be determined precisely. Whether such a switch is the rate-limiting step identified in autophagosome clustering and fusion with lysosomes (16) is also unknown. If it were the case, then the switch from dynamic to stable MTs would look like crossing the functional barrier proposed to isolate pre-autophagosomal structures from mature autophagosomes and thus from lysosomes (16). If the latter actually traffic on stable MTs, this would ensure that immature autophagosomes might not prematurely fuse and/or possibly prevent harmful lysosomal enzyme release in the cytoplasm.
FIGURE 7.
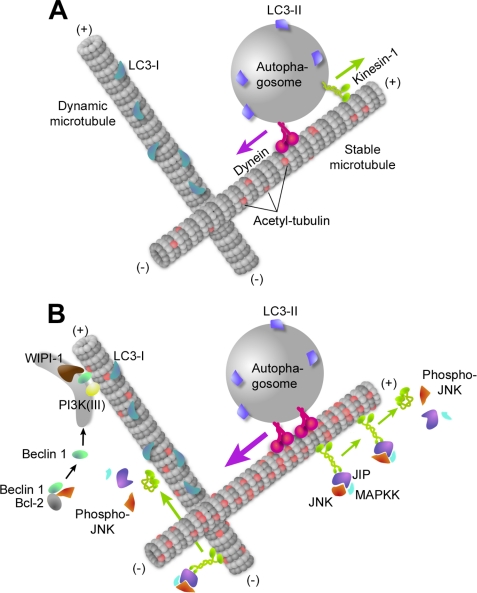
Model of microtubule participation in basal and starvation-induced autophagy. A, in normal nutritional conditions, LC3-I localizes on labile MTs. When autophagosomes form, they are loaded with LC3-II and would be recruited to stable, acetylated MTs. Then they rapidly move toward the MT minus and plus ends using dynein and kinesin-1. B, upon starvation, tubulin hyperacetylation results in an increased motor recruitment. Kinesin-1 is diverted from its autophagosome-carrying function to transport the scaffolding protein JIP and allow JNK activation by upstream MAPK kinases (MAPKK), which may then deactivate kinesin-1. Activated JNK phosphorylates Bcl-2, allowing the release of Beclin 1. Beclin 1 is then recruited on isolation membranes together with PI3K(III) to initiate phosphatidylinositol 1,4,5-trisphosphate-dependent WIPI-1 recruitment to mobile membrane structures. Once mature, autophagosomes might switch from labile to stable MTs to allow dynein-dependent, long distance transport toward MT minus ends prior to fusion with lysosomes.
Supplementary Material
Acknowledgments
We are grateful to Valérie Nicolas for help in operating the video microscope at the imaging facility of IFR 141. We thank Drs. R. Davis, S. Etienne-Manneville, L. Lafanechère, F. Saudou, R. Y. Tsien, M. Way, and T. Yoshimori for sharing materials and Dr. A. J. Meijer for critical reading of the manuscript.
This work was supported in part by institutional funding from INSERM (to P. C.) and a grant from the Association pour la Recherche sur le Cancer (to S. G. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PI3K
- phosphatidylinositol 3-kinase
- PBS
- phosphate-buffered saline
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- PIPES
- 1,4-piperazinediethanesulfonic acid
- JNK
- c-Jun N-terminal kinase
- TRITC
- tetramethylrhodamine isothiocyanate
- MT
- microtubule
- GFP
- green fluorescent protein
- KHC
- kinesin heavy chain
- MAPK
- mitogen-activated protein kinase
- RNAi
- RNA interference
- siRNA
- small interfering RNA.
REFERENCES
- 1.Levine B., Kroemer G. (2008) Cell 132, 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehrpour M., Esclatine A., Beau I., Codogno P. (2010) Am. J. Physiol. Cell Physiol 298, C776–C785 [DOI] [PubMed] [Google Scholar]
- 4.Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 5.Proikas-Cezanne T., Ruckerbauer S., Stierhof Y. D., Berg C., Nordheim A. (2007) FEBS Lett. 581, 3396–3404 [DOI] [PubMed] [Google Scholar]
- 6.Proikas-Cezanne T., Waddell S., Gaugel A., Frickey T., Lupas A., Nordheim A. (2004) Oncogene 23, 9314–9325 [DOI] [PubMed] [Google Scholar]
- 7.Longatti A., Tooze S. A. (2009) Cell Death Differ. 16, 956–965 [DOI] [PubMed] [Google Scholar]
- 8.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 9.Kuznetsov S. A., Gelfand V. I. (1987) FEBS Lett. 212, 145–148 [DOI] [PubMed] [Google Scholar]
- 10.Mann S. S., Hammarback J. A. (1994) J. Biol. Chem. 269, 11492–11497 [PubMed] [Google Scholar]
- 11.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aplin A., Jasionowski T., Tuttle D. L., Lenk S. E., Dunn W. A., Jr. (1992) J. Cell. Physiol. 152, 458–466 [DOI] [PubMed] [Google Scholar]
- 13.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouno T., Mizuguchi M., Tanida I., Ueno T., Kanematsu T., Mori Y., Shinoda H., Hirata M., Kominami E., Kawano K. (2005) J. Biol. Chem. 280, 24610–24617 [DOI] [PubMed] [Google Scholar]
- 15.Köchl R., Hu X. W., Chan E. Y., Tooze S. A. (2006) Traffic 7, 129–145 [DOI] [PubMed] [Google Scholar]
- 16.Fass E., Shvets E., Degani I., Hirschberg K., Elazar Z. (2006) J. Biol. Chem. 281, 36303–36316 [DOI] [PubMed] [Google Scholar]
- 17.Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O'Kane C. J., Brown S. D., Rubinsztein D. C. (2005) Nat. Genet. 37, 771–776 [DOI] [PubMed] [Google Scholar]
- 18.Rubinsztein D. C., Ravikumar B., Acevedo-Arozena A., Imarisio S., O'Kane C. J., Brown S. D. (2005) Autophagy 1, 177–178 [DOI] [PubMed] [Google Scholar]
- 19.Jahreiss L., Menzies F., Rubinsztein D. (2008) Traffic 9, 574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura S., Noda T., Yoshimori T. (2008) Cell Struct. Funct. 33, 109–122 [DOI] [PubMed] [Google Scholar]
- 21.vom Dahl S., Dombrowski F., Schmitt M., Schliess F., Pfeifer U., Häussinger D. (2001) Biochem. J. 354, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster D. R., Gundersen G. G., Bulinski J. C., Borisy G. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 9040–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhey K. J., Gaertig J. (2007) Cell Cycle 6, 2152–2160 [DOI] [PubMed] [Google Scholar]
- 24.Hammond J. W., Cai D., Verhey K. J. (2008) Curr. Opin. Cell Biol. 20, 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed N. A., Cai D., Blasius T. L., Jih G. T., Meyhofer E., Gaertig J., Verhey K. J. (2006) Curr. Biol. 16, 2166–2172 [DOI] [PubMed] [Google Scholar]
- 26.Dompierre J. P., Godin J. D., Charrin B. C., Cordelières F. P., King S. J., Humbert S., Saudou F. (2007) J. Neurosci. 27, 3571–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreitzer G., Liao G., Gundersen G. G. (1999) Mol. Biol. Cell 10, 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giustiniani J., Daire V., Cantaloube I., Durand G., Poüs C., Perdiz D., Baillet A. (2009) Cell. Signal. 21, 529–539 [DOI] [PubMed] [Google Scholar]
- 29.Chaumorcel M., Souquère S., Pierron G., Codogno P., Esclatine A. (2008) Autophagy 4, 46–53 [DOI] [PubMed] [Google Scholar]
- 30.Kimura S., Noda T., Yoshimori T. (2007) Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 31.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 32.Rietdorf J., Ploubidou A., Reckmann I., Holmström A., Frischknecht F., Zettl M., Zimmermann T., Way M. (2001) Nat. Cell Biol. 3, 992–1000 [DOI] [PubMed] [Google Scholar]
- 33.Etienne-Manneville S., Hall A. (2001) Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]
- 34.Lei K., Nimnual A., Zong W. X., Kennedy N. J., Flavell R. A., Thompson C. B., Bar-Sagi D., Davis R. J. (2002) Mol. Cell. Biol. 22, 4929–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bampton E. T., Goemans C. G., Niranjan D., Mizushima N., Tolkovsky A. M. (2005) Autophagy 1, 23–36 [DOI] [PubMed] [Google Scholar]
- 36.Phung-Koskas T., Pilon A., Poüs C., Betzina C., Sturm M., Bourguet-Kondracki M. L., Durand G., Drechou A. (2005) J. Biol. Chem. 280, 1123–1131 [DOI] [PubMed] [Google Scholar]
- 37.Daire V., Giustiniani J., Leroy-Gori I., Quesnoit M., Drevensek S., Dimitrov A., Perez F., Poüs C. (2009) J. Biol. Chem. 284, 31992–32001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta V., Palmer K. J., Spence P., Hudson A., Stephens D. J. (2008) Traffic 9, 1850–1866 [DOI] [PubMed] [Google Scholar]
- 39.Proikas-Cezanne T., Pfisterer S. G. (2009) Methods Enzymol. 452, 247–260 [DOI] [PubMed] [Google Scholar]
- 40.Vasquez R. J., Howell B., Yvon A. M., Wadsworth P., Cassimeris L. (1997) Mol. Biol. Cell 8, 973–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciechomska I. A., Tolkovsky A. M. (2007) Autophagy 3, 586–590 [DOI] [PubMed] [Google Scholar]
- 42.Backer J. M. (2008) Biochem. J. 410, 1–17 [DOI] [PubMed] [Google Scholar]
- 43.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giustiniani J., Couloubaly S., Baillet A., Pourci M. L., Cantaloube I., Fourniat C., Paul J. L., Poüs C. (2009) Exp. Cell Res. 315, 3509–3520 [DOI] [PubMed] [Google Scholar]
- 45.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhey K. J., Meyer D., Deehan R., Blenis J., Schnapp B. J., Rapoport T. A., Margolis B. (2001) J. Cell Biol. 152, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhey K. J., Rapoport T. A. (2001) Trends Biochem. Sci. 26, 545–550 [DOI] [PubMed] [Google Scholar]
- 48.Bogoyevitch M. A., Kobe B. (2006) Microbiol. Mol. Biol. Rev. 70, 1061–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marks D. L., Larkin J. M., McNiven M. A. (1994) J. Cell Sci. 107, 2417–2426 [DOI] [PubMed] [Google Scholar]
- 50.Cardoso C. M., Groth-Pedersen L., Høyer-Hansen M., Kirkegaard T., Corcelle E., Andersen J. S., Jäättelä M., Nylandsted J. (2009) PLoS ONE 4, e4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jahreiss L., Menzies F. M., Rubinsztein D. C. (2008) Traffic 9, 574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura S., Noda T., Yoshimori T. (2008) Cell Struct. Funct. 33, 109–122 [DOI] [PubMed] [Google Scholar]
- 53.Poüs C., Chabin K., Drechou A., Barbot L., Phung-Koskas T., Settegrana C., Bourguet-Kondracki M. L., Maurice M., Cassio D., Guyot M., Durand G. (1998) J. Cell Biol. 142, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Creppe C., Malinouskaya L., Volvert M. L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S., Belachew S., Malgrange B., Chapelle J. P., Siebenlist U., Moonen G., Chariot A., Nguyen L. (2009) Cell 136, 551–564 [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., Matthias P. (2003) EMBO J. 22, 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003) Mol. Cell 11, 437–444 [DOI] [PubMed] [Google Scholar]
- 57.Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 58.Pandey U., Nie Z., Batlevi Y., McCray B., Ritson G. (2007) Nature 447, 860–864 [DOI] [PubMed] [Google Scholar]
- 59.Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., Tsokos M., Alt F. W., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3374–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee I. H., Finkel T. (2009) J. Biol. Chem. 284, 6322–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horiuchi D., Collins C. A., Bhat P., Barkus R. V., Diantonio A., Saxton W. M. (2007) Curr. Biol. 17, 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.