Abstract
The transcription factor Stat3 (signal transducer and activator of transcription 3) mediates many physiological processes, including embryogenesis, stem cell self-renewal, and postnatal survival. In response to gp130 receptor activation, Stat3 becomes phosphorylated by the receptor-associated Janus kinase, forms dimers, and enters the nucleus where it binds to Stat3 target genes and regulates their expression. In this report, we demonstrate that Stat3 binds directly to the promoters and regulates the expression of three genes that are essential for cardiac differentiation: Tbx5, Nkx2.5, and GATA4. We further demonstrate that Tbx5, Nkx2.5, and GATA4 expression is dependent on Stat3 in response to ligand treatment and during ligand-independent differentiation of P19CL6 cells into cardiomyocytes. Finally, we show that Stat3 is necessary for the differentiation of P19CL6 cells into beating cardiomyocytes. All together, these results demonstrate that Stat3 is required for the differentiation of cardiomyocytes through direct transcriptional regulation of Tbx5, Nkx2.5, and GATA4.
Keywords: Cardiac Muscle, Differentiation, Gene Expression, STAT Transcription Factor, Transcription Regulation, GATA4, P19CL6, Stat3, Tbx5
Introduction
The signal transducer and activator of transcription (STAT)2 proteins are a family of transcriptional regulators that mediate a wide range of biological functions primarily in response to extracellular signaling molecules such as cytokines and growth factors (1–4). Upon ligand binding to receptors, receptor-associated Janus kinases become activated and phosphorylate tyrosine residues on the intracellular tail of the receptor, which function as docking sites for STAT molecules. The receptor-bound STAT proteins are subsequently phosphorylated on a single tyrosine residue, translocate into the nucleus as dimers, and bind to specific DNA sequences in promoters of their target genes for transcription regulation (2, 3, 5).
Stat3 is one member of the STAT family that is activated in response to cytokines that bind the gp130 receptor chain including interleukin-6, leukemia inhibitory factor (LIF), and oncostatin M (6, 7). In response to gp130 receptor activation, Stat3 becomes phosphorylated on Tyr-705 to form dimers that enter the nucleus to regulate transcription of Stat3 target genes (8, 9). Stat3 regulates transcription by recruiting other transcription factors and co-activators to form enhancersomes at the promoters of Stat3 target genes (10, 11). Stat3 is involved in many physiological and pathological processes, including embryogenesis, lymphocyte growth, postnatal survival, oncogenesis, tumor metastasis, and LIF-mediated self-renewal of murine embryonic stem cells (12–17). In addition to these processes, Stat3 has been shown to play a role in cardiac cell differentiation and development. Both Stat3 and JAK2 are involved in the differentiation of mouse embryonic stem cells into beating embryoid bodies and cardiomyocytes (18, 19).
These observations strongly support a role for Stat3 in cardiac differentiation and development. However, it is unclear how Stat3 mediates these effects during the process of cardiac differentiation. One approach to address this issue is through analysis of Stat3 target gene regulation during cardiac differentiation. We have previously reported the utilization of a genome-wide ChIP assay to identify potential Stat3 target genes (20). Among these Stat3 target genes, we identified three genes that are cardiac differentiation markers: Tbx5, Nkx2.5, and GATA4 (18, 21, 22). These genes are required for cardiac differentiation and development, and mutations in any one of the three genes can lead to congenital heart malformation (23–29). Tbx5 and Nkx2.5 interact to promote cardiomyocyte differentiation through regulation of the cardiac-specific natriuretic peptide precursor type A (Nppa) gene (30). Both Tbx5 and Nkx2.5 interact with GATA4 (31, 32). Also, overexpression of Tbx5 caused P19CL6 cells to express cardiac differentiation specific genes and beat early (30). Studies with P19CL6 cells also showed that RNAi-mediated knockdown of Nkx2.5 prevented efficient differentiation into beating cardiomyocytes (33). Overexpression of GATA4 in P19 cells enhances cardiac differentiation whereas inhibition of GATA4 blocks cardiac differentiation (34, 35). Moreover, as mouse embryonic stem cells differentiate into cardiomyocytes, there is an increase in cardiac differentiation markers including Nkx2.5, Tbx5, and GATA4; and inhibition of the Janus kinase/STAT3 pathway blocks their expression (18). However, it has not been shown how activated Stat3 regulates their expression during differentiation.
In this report, we set out to test the hypothesis that Stat3 directly regulates the expression of Nkx2.5, Tbx5, and GATA4 and is necessary for cardiomyocyte differentiation. We first demonstrate that Stat3 is phosphorylated in response to LIF treatment of P19CL6 cells, and activated Stat3 binds directly to the promoters of Tbx5, Nkx2.5, and GATA4 to induce their expression. Furthermore, Stat3 becomes phosphorylated when cells are cultured in differentiation medium independent of exogenous ligand treatment, and there is an increase in transcriptional expression of Tbx5, Nkx2.5, GATA4, and other cardiac differentiation markers. Using an RNAi knockdown approach in both ligand-induced and ligand-independent differentiation conditions, we show that Tbx5, Nkx2.5, and GATA4 expression is Stat3-dependent. Finally, we show that RNAi-mediated Stat3 knockdown decreases the number of beating regions when P19CL6 cells are cultured in differentiation conditions. Together, these results demonstrate that Stat3 is required for the differentiation of cardiomyocytes through direct transcriptional regulation of the expression of Tbx5, Nkx2.5, and GATA4.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
P19CL6 cells were obtained from Dr. Richard Kitsis (Albert Einstein College of Medicine, Bronx, NY). Cells were cultured in growth conditions as described previously (36) in α-minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 units/ml penicillin. For differentiation conditions, growth medium was supplemented with 1% dimethyl sulfoxide. The antibodies used for Western analysis were anti-phosphotyrosine Stat3 (Cell Signaling Technology) and anti-Stat3 (BD Transduction Laboratories). Transfections were performed using Lipofectamine 2000 (Invitrogen) with the plasmid RcCMV (pCMV) from Invitrogen or RcCMV expressing wild-type Stat3 (pCMV-STAT3) which was constructed previously (37). The Stat3 antibody used for ChIP analysis was from Santa Cruz Biotechnology. Recombinant mouse LIF was from Chemicon International. An Nkx2.5 antibody was from Santa Cruz Biotechnology, anti-LIF was from Millipore, and anti-Tbx5 was provided by Dr. Craig T. Basson (Weill Cornell Medical College, New York, NY).
ChIP Analysis
ChIP analysis was performed as described previously (20, 37, 38). P19CL6 cells were cultured in growth medium and treated with 10 ng/ml LIF, and immunoprecipitations were performed with 2.5 μg of Stat3 antibody. Primers were designed against the mouse Tbx5, Nkx2.5, and GATA4 promoter regions. Primers for Tbx5 were 5′-GAAGCATTTTCTATACTTTGTGAGCA-3′ and 5′-TCAGCCAGCTGTTTTCAGAG-3′; primers for GATA4 were 5′-ACTCCCTTAGGCCAGTCAGC-3′ and 5′GGAAAAGAGCAGGGACTCG3′; and primers for Nkx2.5 were 5′-AGGCAAAGAAATCACTCCACA-3′ and 5′-TGTTACAATGGCTGGGAAGG-3′.
Quantitative Real Time Reverse Transcription (RT)-PCR
P19CL6 cells were cultured in growth conditions in 6-well plates to 80–90% confluence. Cells were then treated for the indicated times with 10 ng/ml LIF. For differentiation conditions, cells were cultured for the indicated time points. RNA was extracted using the TRIzol method (Invitrogen). RNA was reverse-transcribed using either Superscript II reverse transcriptase (Invitrogen) or qScript cDNA SuperMix (Quanta Biosciences). Quantitative real-time PCR was performed using either the SYBR Green Core PCR Reagent kit (Applied Biosciences) or PerfeCTa SYBR Green FastMix, ROX (Quanta Biosciences). RT-PCR primers were as follows: Tbx5, 5′-TGACTGGCCTTAATCCCAAA-3′ and 5′-ACAAGTTGTCGCATCCAGTG-3′; Nkx2.5, 5′-GACAGGTACCGCTGTTGCTT-3′ and 5′-AGCCTACGGTGACCCTGAC-3′; GATA4, 5′-CGAGGGTGAGCCTGTATGTAA-3′ and 5′-GCTAGTGGCATTGCTGGAGT-3′; α-MHC, 5′-CAAGACTGTCCGGAATGACA-3′ and 5′-GGCTTCTTGTTGGACAGGAT-3′; GAPDH, 5′-ACGACCCCTTCATTGACC-3′ and 5′-AGACACCAGTAGACTCCACG-3′.
Stat3 siRNAs
Two sets of Stat3 siRNAs targeting two different regions of Stat3 were obtained from Qiagen (catalogue numbers SI01435294 and SI01435287) and described previously (20). A negative control siRNA (catalogue number 1022563) was also obtained from Qiagen. P19CL6 cells were cultured in either growth or differentiation conditions to a density of ∼1 × 105 cells. Cells were transfected with the siRNAs using HiPerFect transfection reagent (Qiagen).
Western Blot Analysis
Whole cell lysates were separated by SDS-PAGE and blotted with the indicated antibodies followed by chemiluminescence (PerkinElmer Life Sciences).
Quantification of Beating Regions during Cardiomyocyte Differentiation
P19CL6 cells were cultured in differentiation medium in 6-well dishes. Beating regions were observed by day 12 and counted microscopically using a grid for orientation. For each day, beating regions were counted at least three separate times to calculate the average number of beating regions and standard deviations.
RESULTS
Stat3 Binds to the Promoters of Nkx2.5, Tbx5, and GATA4 in Response to LIF
We had previously identified Tbx5, Nkx2.5, and GATA4 as potential Stat3 target genes using a genome-wide ChIP screen (20). To show that these genes are specifically and directly regulated by Stat3 during cardiac differentiation, we performed gene-specific ChIP analysis using P19CL6 cells. P19CL6 cells proliferate in growth medium and differentiate into beating cardiomyocytes in growth medium supplemented with 1% dimethyl sulfoxide, thus providing an excellent model system for the study of transcriptional regulation during cardiac development (36).
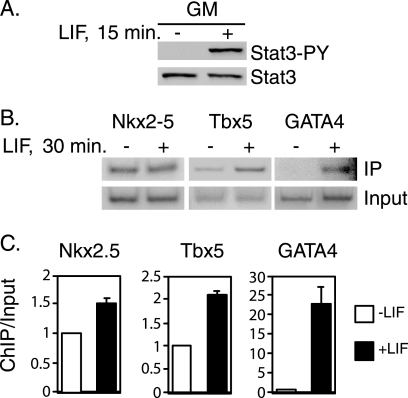
P19CL6 cells were cultured in growth medium and treated with LIF, a ligand that rapidly induces Stat3 phosphorylation. Treatment with LIF caused Stat3 to become phosphorylated, whereas there was no detectable phosphorylated Stat3 in untreated cells (Fig. 1A). Stat3 is rapidly activated by LIF within 15 min, and this activation is transient and starts to decrease after 30 min in P19CL6 cells (data not shown). This result is similar to previous work demonstrating rapid phosphorylation of STAT molecules (39). Gene-specific ChIP assays were then utilized to determine whether Stat3 binds to the promoters of Nkx2.5, Tbx5, and GATA4 in response to LIF. Cells were treated with LIF, and ChIP analysis was performed with Stat3 antibodies and primers designed to amplify potential Stat3 binding sites in the gene promoters (Fig. 1, B and C). Stat3 was bound to the Nkx2.5 promoter in untreated cells, and there was a slight increase in bound Stat3 following treatment (Fig. 1, B and C). There was a 2-fold increase in Stat3 bound to the Tbx5 promoter in response to LIF treatment compared with untreated cells (Fig. 1, B and C). There was a 20-fold increase in the amount of Stat3 bound to the GATA4 promoter in response to LIF (Fig. 1, B and C). These results show that Stat3 binds directly to the promoters of these genes, constitutively for Nkx2.5 and inducibly for Tbx5 and GATA4, in response to LIF treatment in P19CL6 cells.
FIGURE 1.
Stat3 binds to the Nkx2.5, Tbx5, and GATA4 promoters in response to LIF treatment. A, P19CL6 cells were cultured in growth medium (GM) until cells reached 80–90% confluence. Cells in growth medium were then either treated with LIF for 15 min or left untreated. Whole cell extracts were prepared, and Western blot analyses were performed with antibodies against Stat3 phosphorylated on tyrosine 705 (Stat3-PY) or total Stat3 (Stat3). B, P19CL6 cells were cultured in growth medium and treated with LIF for 30 min or left untreated. ChIP analyses were performed with Stat3 antibody, and primers were designed to amplify potential Stat3 binding sites in the Tbx5, Nkx2.5, and GATA4 promoters. ChIP assays were repeated at least twice; one representative result is shown for each promoter. C, results from B were quantitated with a PhosphorImager. Averages ± S.D. (error bars) represent at least two independent experiments for each promoter.
LIF Induces the Expression of Nkx2.5, Tbx5, and GATA4 in P19CL6 Cells
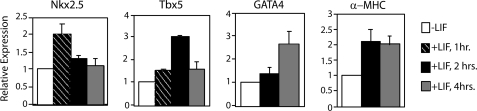
To determine whether there is activation of the expression of Nkx2.5, Tbx5, and GATA4 in response to LIF treatment, we performed quantitative real time PCR analysis with P19CL6 cells (Fig. 2). Cells were cultured in growth medium and treated with LIF for 1, 2, and 4 h. There was a 2-fold increase in Nkx2.5 expression peaking after 1 h of treatment with LIF (Fig. 2). There was a slight increase in Tbx5 after 1 h of treatment followed by a 3-fold increase at 2 h (Fig. 2). The expression of GATA4 increased 3-fold after 4 h of treatment (Fig. 2). These results suggest that, in response to LIF treatment, the expression levels of Nkx2.5, Tbx5, and GATA4 mRNA were induced. These results are similar to that of other STAT target genes which often have different kinetics in transcription activation in response to cytokine treatment (37, 40–42). As a positive control for cardiac cell differentiation, the expression of the differentiation marker α-MHC was also analyzed. The mRNA level of this marker increased over time with LIF treatment (Fig. 2). These results indicate that LIF treatment of P19CL6 cells induces expression of Tbx5, Nkx2.5, and GATA4.
FIGURE 2.
LIF induces expression of Nkx2.5, Tbx5, and GATA4. P19CL6 cells were cultured in growth medium and treated with LIF for 1, 2, or 4 h. RNA was extracted, and quantitative RT-PCR analyses were performed for Nkx2.5, Tbx5, GATA4, or α-MHC. Results are standardized to GAPDH. Values represent the averages ± S.D. (error bars) of at least three independent experiments.
LIF-mediated Induction of Nkx2.5, Tbx5, and GATA4 Is Dependent on Stat3
The results from these initial experiments suggested that in P19CL6 cells, Stat3 is activated by LIF treatment and binds to the Nkx2.5, Tbx5, and GATA4 promoters leading to the activation of these genes. To demonstrate that Stat3 is required for the activation of these genes in response to LIF, we utilized an RNAi technique to knock down Stat3 in these cells and analyze the expression of Nkx2.5, Tbx5, and GATA4.
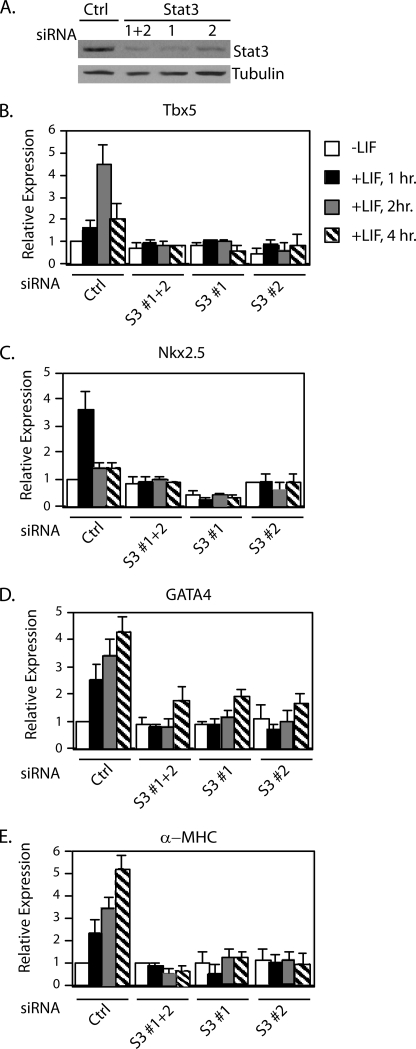
Cells were cultured in growth medium and transfected with either a control siRNA or Stat3 siRNAs. Two sets of Stat3 siRNAs specifically targeting two different regions of Stat3 were described previously (20). After 4 days, whole cell extracts were prepared, and Western blot analysis was performed with antibodies against total Stat3 or tubulin as a control. As shown in Fig. 3A, there was a significant reduction in Stat3 protein levels following transfection with either set of Stat3 siRNAs (set 1 or set 2) or both Stat3 siRNA sets combined (set 1+ set 2) compared with the control. RNA expression was analyzed with quantitative real time RT-PCR with cells treated with LIF for 1, 2, or 4 h. In cells transfected with control siRNA, the mRNA levels of Nkx2.5, Tbx5, GATA4, and the positive control differentiation marker α-MHC increased in response to LIF treatment (Fig. 3, B–E). In contrast, in Stat3 knockdown cells, the expression of Tbx5 and Nkx2.5 was not induced by LIF, whereas GATA4 was only partially induced to ∼40% of control cells. (Fig. 3, B–E). These results demonstrate that, in growth conditions, the LIF-induced expression of Nkx2.5, Tbx5, and GATA4 is dependent on Stat3.
FIGURE 3.
Stat3 is required for the expression of Nkx2.5, Tbx5, and GATA4. A, P19CL6 cells were transfected with either a negative control siRNA or Stat3 siRNA oligonucleotide 1, 2, or 1 and 2 combined. Cells were cultured in growth medium for 4 days, and some of them were subjected to Western blot analyses. B, the remaining cells were left untreated (white bars) or treated with LIF for 1 h (black bars), 2 h (gray bars), or 4 h (striped bars) followed by quantitative real time PCR analyses. B–E, shown are Tbx5 (B), Nkx2.5 (C), GATA4 (D), and α-MHC (E). Results are standardized to GAPDH. Values represent the averages ± S.D. (error bars) of at least two independent experiments.
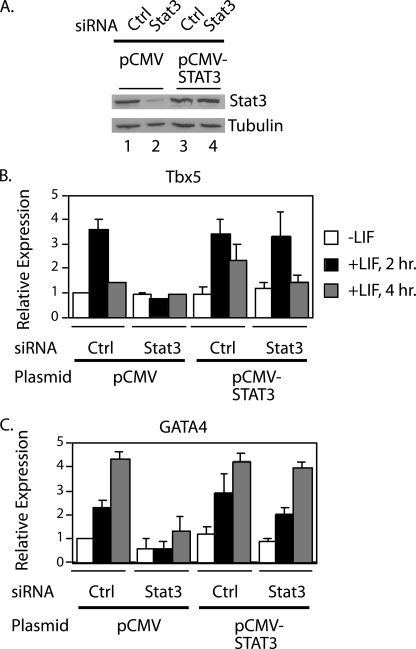
To demonstrate that the RNAi knockdown was specific for Stat3, we performed a rescue experiment to restore the levels of Stat3 protein in the Stat3 RNAi knockdown cells. Cells were transfected with either a control or combined Stat3 siRNAs as described in Fig. 3 and cultured for 3 days. Cells were then transfected with either the vector RcCMV (pCMV) or Stat3 expressed under the control of the CMV promoter (pCMV-STAT3) (37) and cultured for an additional 24 h. Western blot analysis demonstrated that cells transfected with Stat3 siRNAs and pCMV showed a decrease in Stat3 protein levels compared with control siRNA cells transfected with pCMV (Fig. 4A, lanes 1 and 2). The Stat3 direct target genes Tbx5 and GATA4 were used as examples, and their expression decreased to background levels in cells transfected with Stat3 siRNAs and pCMV in response to LIF treatment (Fig. 4, B and C). In contrast, transfection of pCMV-STAT3 restored the protein levels of Stat3 in the Stat3 knockdown cells to the level of Stat3 in control cells (Fig. 4A, lanes 3 and 4) because the residual Stat3 siRNA in cells 3 days after transfection was not effective in blocking the high level of Stat3 mRNA transcribed from the pCMV vector as described previously (43). In these Stat3-restored cells, the expression of Tbx5 and Gata4 was induced by LIF to levels similar to that in control cells (Fig. 4, B and C). These results demonstrate that the Stat3 siRNA oligonucleotides specifically knocked down Stat3 in P19CL6 cells.
FIGURE 4.
Expression of exogenous Stat3 rescues the expression of Stat3 target genes in Stat3 RNAi knockdown cells. A, P19CL6 cells were cultured in growth medium and transfected with either a control or Stat3 siRNA oligonucleotides 1 and 2 combined. After 3 days, cells were transfected with either pCMV or pCMV-STAT3 and cultured in growth medium for an additional 24 h. Whole cell extracts were prepared, and Western blot analyses were performed with Stat3 or tubulin antibodies. B and C, portions of the cells for each sample were treated with LIF for 1, 2, or 4 h. RNA was extracted, and quantitative real time RT-PCR analyses were performed for Tbx5 (B) or GATA4 (C). Results are standardized to GAPDH, and values represent the averages ± S.D. (error bars) of at least two independent experiments.
Stat3 Is Activated, and the Expression of Nkx2.5, Tbx5, and GATA4 Increases in Differentiation Conditions
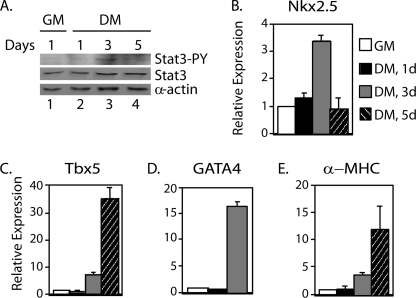
To determine further whether Stat3 is activated during the differentiation of P19CL6 cells to cardiomyocytes, cells were cultured in growth medium or differentiation medium for 1, 3, and 5 days. Western blot analyses performed with an antibody against phosphorylated Stat3 showed no detectable levels of phosphorylated Stat3 in cells cultured in growth or differentiation medium for 1 day (Fig. 5A, lanes 1 and 2). However, there was an increase in phosphorylation of Stat3 on day 3 followed by a slight decrease by day 5 (Fig. 5A, lanes 3 and 4). Total Stat3 increased at days 3 and 5 because of autoregulation of Stat3 expression by phosphorylated Stat3 (44). To see whether there is an increase in Tbx5, Nkx2.5, and GATA4 mRNA levels after 3 days in differentiation conditions, quantitative real time RT-PCR was performed. The cardiac differentiation marker α-MHC was also analyzed as a control for differentiation culture conditions. There was an increase in the expression of all of these genes after 3 days of differentiation culture (Fig. 5, B–E). These results indicate that, independent of exogenous ligand treatment, Stat3 is activated during differentiation of P19CL6 cells, and the expression of Stat3 target genes is increased.
FIGURE 5.
Stat3 is activated, and expression of Nkx2.5, Tbx5, and GATA4 increases in P19CL6 cells cultured in differentiation conditions. P19CL6 cells were cultured in growth medium (GM) (white bars) or differentiation medium (DM) for 1 day (black bars), 3 days (gray bars), or 5 days (striped bars). A, whole cell extracts were prepared, and Western blot analyses were performed with antibodies against phosphotyrosine-Stat3 (Stat3-PY), total Stat3 (Stat3), or α-actin. B–E, RNA was extracted from a portion of the cells, and quantitative real time RT-PCR analyses were performed for Nkx2.5 (B), Tbx5 (C), GATA4 (D), and α-MHC (E). Results are standardized to GAPDH. Values represent the averages ± S.D. (error bars) of at least three independent experiments.
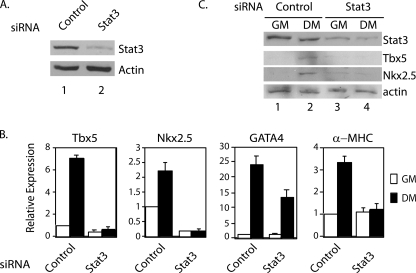
Stat3 Is Necessary for Increased Expression of Nkx2.5, Tbx5, and GATA4 in Differentiation Conditions
To determine whether Stat3 is required for the increase of expression of these genes during differentiation, P19CL6 cells were transfected with control siRNA or Stat3 siRNA in growth medium. Four days after the first round of transfection, the cells were transfected with the respective siRNA again to maintain the knockdown of Stat3 (Fig. 6A). The cells were then split, and half of them remained in growth medium, and half of them were transferred into differentiation medium. Three days after the second round of siRNA transfection, quantitative real time PCR analyses were performed. In differentiation medium, the mRNA levels of Nkx2.5, Tbx5, and GATA4 increased in cells transfected with control siRNA. In Stat3 knockdown cells, the expression of Nkx2.5 and Tbx5 could not be induced, whereas GATA4 was induced to ∼50% of the level in control cells (Fig. 6B). Five days after the second round of siRNA transfection, Western blot analyses were performed. Stat3 protein levels remained significantly decreased (Fig. 6C, lanes 3 and 4). For cells cultured in growth medium, there was no detectable level of Nkx2.5 and Tbx5 (Fig. 6C, lanes 1 and 3). When cells were cultured in differentiation medium, there was an induction of Nkx2.5 and Tbx5 proteins in cells transfected with control siRNA, but not in cells transfected with Stat3 siRNA (Fig. 6C, compare lanes 2 and 4). These results demonstrate that Stat3 is required for the full induction of Tbx5, Nkx2.5, and GATA4 when P19CL6 cells were cultured in differentiation conditions without exogenous ligand treatment.
FIGURE 6.
Stat3 is required for the expression of Tbx5, Nkx2.5, and GATA4 in P19CL6 cells in differentiation conditions. P19CL6 cells were cultured in growth medium (GM) and transfected with either control or Stat3 siRNA sets 1 and 2 combined for 4 days. Cells were further transfected with the respective siRNA and cultured in either growth medium or differentiation medium (DM). A and B, 3 days after the second round of transfection, the cells were analyzed by Western blot analysis with antibodies against total Stat3 or actin (A) and real time RT-PCR analyses of Stat3, Tbx5, Nkx2.5, and α-MHC mRNA levels (B). C, 5 days after the second round of transfection, Western blot analyses were performed with antibodies against total Stat3, Tbx5, Nkx2.5, or actin as a loading control. Error bars, S.D.
LIF Expression Increases during Differentiation of P19CL6 Cells
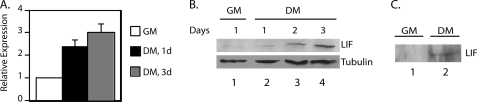
To understand how Stat3 becomes activated in differentiation medium without exogenous ligand treatment, we tested whether P19CL6 cells produce LIF when cultured in differentiation medium. Real time RT-PCR analyses showed that LIF mRNA levels increased when cells were transferred into differentiation medium (Fig. 7A). Western blot analyses of whole cell extracts showed that the protein level of LIF also increased when cells were cultured in differentiation medium (Fig. 7B, lanes 3 and 4). Furthermore, Western blot analysis was performed on the medium from cells cultured in growth medium or differentiation medium for 3 days. Secreted LIF protein was detected in the medium of cells cultured in differentiation medium for 3 days (Fig. 7C, lane 2). There was no detectable LIF protein in the medium of cells cultured in growth medium (Fig. 7C, lane 1). These results suggest that LIF expression increases in P19CL6 cells cultured in differentiation medium, which leads to Stat3 activation.
FIGURE 7.
LIF expression increases during cardiac differentiation. A, P19CL6 cells were cultured in growth medium (GM) or differentiation medium (DM) for 1, 2, or 3 days. RNA was extracted using the TRIzol method, and quantitative real time RT-PCR analyses were performed for LIF expression. Results were standardized to GAPDH levels with the value for growth medium set at 1. Results represent the average ± S.D. (error bars) of three independent experiments. B, cells were cultured as described in A, and whole cell extracts were prepared. Western blot analyses were performed with antibodies against tubulin and LIF. C, cells were cultured in growth medium or differentiation medium for 3 days. A total of 200 μl of medium from each set of cells was subjected to Western blot analyses and blotted with an antibody to LIF.
Stat3 Is Required for Differentiation of P19CL6 Cells into Cardiomyocytes
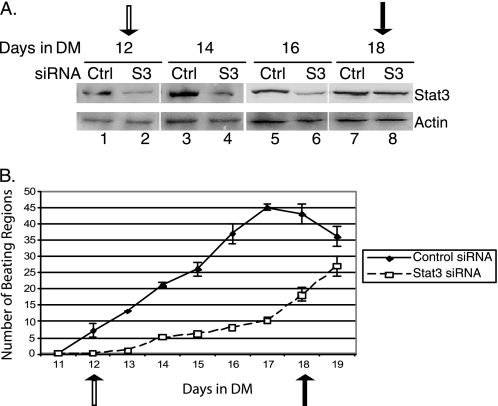
Nkx2.5, Tbx5, and GATA4 have previously been shown to promote cardiac differentiation and development (30, 35). RNAi-mediated knockdown of Nkx2.5 caused a decrease in beating efficiency of P19CL6 cells during cardiomyocyte differentiation, suggesting that Nkx2.5 is required for differentiation (33). To determine whether Stat3 plays a direct physiological role in the process of P19CL6 differentiation into beating cardiomyocytes, P19CL6 cells were transfected with Stat3 or control siRNAs in growth medium. Four days after the initial round of siRNA transfection, Stat3 was significantly knocked down (Fig. 3). The cells were transferred to differentiation medium and were further transfected with the respective siRNA every 4 days until day 12 in differentiation medium. Stat3 knockdown was maintained throughout the course of differentiation procedure until day 16 (Fig. 8A). Because these experiments utilize transfected Stat3 siRNA oligonucleotides, the Stat3 knockdown is transient. Stat3 protein levels began to increase back to normal levels on day 18 (Fig. 8A, lanes 7 and 8). The transient Stat3 RNAi knockdown therefore provides an internal “rescue” experiment when the Stat3 protein returns to normal levels in the same cells. Starting from day 11, the cultured cells were examined daily under microscope, and the number of beating regions were counted. The number of beating regions was significantly lower in Stat3 knockdown cells compared with that in the cells transfected with control siRNA (Fig. 8B). However, once Stat3 proteins became normal after Stat3 siRNA transfection was stopped, the number of beating regions in these same cells began to increase (Fig. 8B). The cells were growing normally in both control and Stat3 siRNA-transfected samples (data not shown). These results strongly demonstrate that Stat3 is required for the differentiation of P19CL6 cells into cardiomyocytes.
FIGURE 8.
Stat3 is necessary for the differentiation of P19CL6 cells into beating cardiomyocytes. P19CL6 cells were transfected with either control or Stat3 siRNA sets 1 and 2 combined for 4 days. After transfer to differentiation medium (DM), cells were transfected every 4 days until day 12 in differentiation medium. A, Western blot analyses were performed with Stat3 and actin antibodies on whole cell extracts on the days indicated. B, time course of the generation of beating regions in control and Stat3 knockdown cells from days 11 to 21. Beating regions were observed microscopically and counted using a grid for orientation. For each day, beating regions were counted at least three times to obtain an average ± S.D. (error bars). Four experiments were performed. The time course and Western blot analysis from one representative experiment are shown. Open arrows correspond to the last day of Stat3 siRNA transfection (day 12), and closed arrows correspond to the point at which Stat3 protein levels recovered (day 18).
DISCUSSION
It has been suggested previously that Stat3 plays an important role during differentiation of mouse embryonic stem cells into beating cardiomyocytes (18, 19). However, it was not known how Stat3 is involved in the process of cardiac differentiation. In a genome-wide ChIP screen designed to identify potential direct Stat3 target genes (20), we identified three cardiac differentiation markers, Nkx2.5, Tbx5, and GATA4, suggesting that Stat3 directly regulates the expression of these genes critical for cardiomyocyte differentiation. In this report, we demonstrate that Tbx5, Nkx2.5, and GATA4 are direct target genes of Stat3, and activated Stat3 increases expression of these genes (Figs. 1 and 2). Furthermore, we showed that Stat3 is required for the expression of these genes during cardiomyocyte differentiation (Fig. 3). Tbx5 and Nkx2.5 interact with each other, and they genetically and physically interact with GATA4 to activate genes necessary for cardiac differentiation (30–32, 45). Therefore, it is likely that Stat3 is at the upstream of the differentiation cascade by controlling the expression of a group of genes essential for cardiac muscle differentiation.
It is interesting that our ChIP results suggest that some levels of Stat3 are bound to the Nkx2.5 and Tbx5 promoters in untreated P19CL6 cells (Fig. 1). This level of bound Stat3 increases following LIF treatment. Stat3 has been shown to shuttle between the cytoplasm and nucleus regardless of tyrosine phosphorylation, and several reports showed a role for unphosphorylated Stat3 in transcription including recruitment of Stat3 to promoters through interactions with NFκB (46–49). Our laboratory has also observed Stat3 bound to promoters in the absence of tyrosine phosphorylation (20). Unphosphorylated Stat3 bound to these promoters could partially clear the promoter and in this case could explain the more rapid induction of Nkx2.5 transcription (Fig. 2A). In addition, Nkx2.5 has been shown to bind to the promoter of GATA4 and is partially necessary for its expression (50). Therefore, it is likely that Stat3 first induces the expression of Nkx2.5 and then together with Nkx2.5 induces the expression of GATA4. Hence, the expression of Nkx2.5 peaks at 1 h whereas GATA4 expression continues to increase after 4 h due to the presence of Nkx2.5 (Fig. 2). This could also explain why GATA4 expression is only partially dependent on Stat3 (Figs. 3 and 6).
Stat3 becomes activated in P19CL6 cells cultured in differentiation medium without the addition of exogenous ligand (Fig. 5A). Because LIF has been suggested to contribute to cardiac differentiation (19), we analyzed the expression of LIF in differentiation culture conditions. These experiments demonstrated that LIF expression is induced and secreted into the medium when P19CL6 cells were cultured in differentiation medium.
Stat3 has been shown to function in a number of biological processes. Often, Stat3 function is associated with the processes of proliferation and oncogenesis rather than differentiation. Stat3 activation inhibits differentiation and promotes proliferation during skeletal muscle development (20, 51–53). In contrast to this observation, results from this report along with those of others (18, 19) suggest that Stat3 promotes differentiation in cardiac cells. These observations suggest that Stat3 might be a key regulator in both cardiac and skeletal muscle development through transcriptional regulation of its target genes. It is important to note that cardiac cells continue to proliferate during differentiation. This is in contrast to skeletal muscle cells that exit the cell cycle and cease proliferation during differentiation (54). We have shown in this work that Stat3 regulates expression of GATA4, Tbx5, and Nkx2.5, three genes that function in cardiac differentiation (33). Our data and others suggest a role for Stat3 during the process of cardiac differentiation (18, 19). Although we did not observe an obvious effect of Stat3 knockdown on cell growth in differentiation conditions, we cannot rule out the possibility that Stat3 could have a separate function in cardiac cell proliferation. Further analyses of more Stat3 target genes should lead to a greater understanding of the mechanisms governing both skeletal and cardiac muscle proliferation and differentiation.
Acknowledgments
We thank all laboratory members for helpful discussions, the Department of Microbiology and Immunology at the Weill Medical College of Cornell University for the use of the Applied Biosystems PRISM 7900HT Sequence Detection Equipment, Richard Kitsis for the P19CL6 cell line, and Craig T. Basson and Cathy Hatcher for Tbx5 antibodies and technical advice.
This work was supported, in whole or in part, by National Institutes of Health Grant HL091525 (to X.-Y. H.). This work was also supported by American Heart Association Grant 0455896T (to J. J. Z.).
- STAT
- signal transducer and activator of transcription
- ChIP
- chromatin immunoprecipitation
- GAPDH
- glucose-3-phosphate dehydrogenase
- LIF
- leukemia inhibitory factor
- MHC
- myosin heavy chain
- RNAi
- RNA interference
- RT
- reverse transcription
- siRNA
- small interfering RNA.
REFERENCES
- 1.Yu H., Pardoll D., Jove R. (2009) Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 3.O'Shea J. J., Gadina M., Schreiber R. D. (2002) Cell 109, (suppl.) S121–131 [DOI] [PubMed] [Google Scholar]
- 4.Sehgal P. B. (2008) Semin. Cell Dev. Biol. 19, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akira S. (1999) Stem Cells 17, 138–146 [DOI] [PubMed] [Google Scholar]
- 6.Akira S., Nishio Y., Inoue M., Wang X. J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M., Kishimoto T. (1994) Cell 77, 63–71 [DOI] [PubMed] [Google Scholar]
- 7.Heinrich P. C., Horn F., Graeve L., Dittrich E., Kerr I., Müller-Newen G., Grötzinger J., Wollmer A. (1998) Z. Ernahrungswiss. 37, Suppl. 1, 43–49 [PubMed] [Google Scholar]
- 8.Becker S., Corthals G. L., Aebersold R., Groner B., Müller C. W. (1998) FEBS Lett. 441, 141–147 [DOI] [PubMed] [Google Scholar]
- 9.Seidel H. M., Milocco L. H., Lamb P., Darnell J. E., Jr., Stein R. B., Rosen J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3041–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X., Wrzeszczynska M. H., Horvath C. M., Darnell J. E., Jr. (1999) Mol. Cell. Biol. 19, 7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerner L., Henriksen M. A., Zhang X., Darnell J. E., Jr. (2003) Genes Dev. 17, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haura E. B., Turkson J., Jove R. (2005) Nat. Clin. Pract. Oncol. 2, 315–324 [DOI] [PubMed] [Google Scholar]
- 13.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr. (1999) Cell 98, 295–303 [DOI] [PubMed] [Google Scholar]
- 14.Shen Y., Schlessinger K., Zhu X., Meffre E., Quimby F., Levy D. E., Darnell J. E., Jr. (2004) Mol. Cell. Biol. 24, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akira S. (2000) Oncogene 19, 2607–2611 [DOI] [PubMed] [Google Scholar]
- 16.Raz R., Lee C. K., Cannizzaro L. A., d'Eustachio P., Levy D. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niwa H., Burdon T., Chambers I., Smith A. (1998) Genes Dev. 12, 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foshay K., Rodriguez G., Hoel B., Narayan J., Gallicano G. I. (2005) Stem Cells 23, 530–543 [DOI] [PubMed] [Google Scholar]
- 19.Rajasingh J., Bord E., Hamada H., Lambers E., Qin G., Losordo D. W., Kishore R. (2007) Circ. Res. 101, 910–918 [DOI] [PubMed] [Google Scholar]
- 20.Snyder M., Huang X. Y., Zhang J. J. (2008) J. Biol. Chem. 283, 3791–3798 [DOI] [PubMed] [Google Scholar]
- 21.Olson E. N. (2006) Science 313, 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N., Olson E. N. (2006) Curr. Opin. Cell Biol. 18, 715–722 [DOI] [PubMed] [Google Scholar]
- 23.Hatcher C. J., Diman N. Y., McDermott D. A., Basson C. T. (2003) Trends Mol. Med. 9, 512–515 [DOI] [PubMed] [Google Scholar]
- 24.Garry D. J., Olson E. N. (2006) Cell 127, 1101–1104 [DOI] [PubMed] [Google Scholar]
- 25.Hatcher C. J., Diman N. Y., Kim M. S., Pennisi D., Song Y., Goldstein M. M., Mikawa T., Basson C. T. (2004) Physiol. Genomics 18, 129–140 [DOI] [PubMed] [Google Scholar]
- 26.Horb M. E., Thomsen G. H. (1999) Development 126, 1739–1751 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M., Chen Z., Bartunkova S., Yamasaki N., Izumo S. (1999) Development 126, 1269–1280 [DOI] [PubMed] [Google Scholar]
- 28.Schott J. J., Benson D. W., Basson C. T., Pease W., Silberbach G. M., Moak J. P., Maron B. J., Seidman C. E., Seidman J. G. (1998) Science 281, 108–111 [DOI] [PubMed] [Google Scholar]
- 29.Pikkarainen S., Tokola H., Kerkelä R., Ruskoaho H. (2004) Cardiovasc. Res. 63, 196–207 [DOI] [PubMed] [Google Scholar]
- 30.Hiroi Y., Kudoh S., Monzen K., Ikeda Y., Yazaki Y., Nagai R., Komuro I. (2001) Nat. Genet. 28, 276–280 [DOI] [PubMed] [Google Scholar]
- 31.Garg V., Kathiriya I. S., Barnes R., Schluterman M. K., King I. N., Butler C. A., Rothrock C. R., Eapen R. S., Hirayama-Yamada K., Joo K., Matsuoka R., Cohen J. C., Srivastava D. (2003) Nature 424, 443–447 [DOI] [PubMed] [Google Scholar]
- 32.Durocher D., Charron F., Warren R., Schwartz R. J., Nemer M. (1997) EMBO J. 16, 5687–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H., Harris T. M., Kim H. H., Childs G. (2005) Funct. Integr. Genomics 5, 218–239 [DOI] [PubMed] [Google Scholar]
- 34.Grépin C., Robitaille L., Antakly T., Nemer M. (1995) Mol. Cell. Biol. 15, 4095–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grépin C., Nemer G., Nemer M. (1997) Development 124, 2387–2395 [DOI] [PubMed] [Google Scholar]
- 36.Habara-Ohkubo A. (1996) Cell Struct. Funct. 21, 101–110 [DOI] [PubMed] [Google Scholar]
- 37.Sun W., Snyder M., Levy D. E., Zhang J. J. (2006) FEBS Lett. 580, 5880–5884 [DOI] [PubMed] [Google Scholar]
- 38.Aparicio O., Geisberg J. V., Sekinger E., Yang A., Moqtaderi Z., Struhl K. (2005) Curr. Protoc. Mol. Biol. 21, 21.23–21.33 [DOI] [PubMed] [Google Scholar]
- 39.Haspel R. L., Salditt-Georgieff M., Darnell J. E., Jr. (1996) EMBO J. 15, 6262–6268 [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsauer K., Farlik M., Zupkovitz G., Seiser C., Kröger A., Hauser H., Decker T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2849–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder M., He W., Zhang J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W., Nair J. S., Malhotra A., Zhang J. J. (2005) J. Interferon Cytokine Res. 25, 113–124 [DOI] [PubMed] [Google Scholar]
- 43.Snyder M., Huang X. Y., Zhang J. J. (2009) J. Biol. Chem. 284, 13466–13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichiba M., Nakajima K., Yamanaka Y., Kiuchi N., Hirano T. (1998) J. Biol. Chem. 273, 6132–6138 [DOI] [PubMed] [Google Scholar]
- 45.Maitra M., Schluterman M. K., Nichols H. A., Richardson J. A., Lo C. W., Srivastava D., Garg V. (2009) Dev. Biol. 326, 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Z., Zhang W., Kone B. C. (2002) Biochem. J. 367, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagihara K., Nishikawa T., Sugamata Y., Song J., Isobe T., Taga T., Yoshizaki K. (2005) Genes Cells 10, 1051–1063 [DOI] [PubMed] [Google Scholar]
- 48.Liu L., McBride K. M., Reich N. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8150–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Stark G. R. (2008) Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 50.Riazi A. M., Takeuchi J. K., Hornberger L. K., Zaidi S. H., Amini F., Coles J., Bruneau B. G., Van Arsdell G. S. (2009) PLoS One 4, e5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kami K., Senba E. (2002) J. Histochem. Cytochem. 50, 1579–1589 [DOI] [PubMed] [Google Scholar]
- 52.Megeney L. A., Perry R. L., LeCouter J. E., Rudnicki M. A. (1996) Dev. Genet. 19, 139–145 [DOI] [PubMed] [Google Scholar]
- 53.Kataoka Y., Matsumura I., Ezoe S., Nakata S., Takigawa E., Sato Y., Kawasaki A., Yokota T., Nakajima K., Felsani A., Kanakura Y. (2003) J. Biol. Chem. 278, 44178–44187 [DOI] [PubMed] [Google Scholar]
- 54.Olson E. N. (1993) Circ. Res. 72, 1–6 [DOI] [PubMed] [Google Scholar]