Abstract
The signal transducer and activator of transcription 3 (STAT3) is a transcription factor and downstream product of cytokine and growth factor pathways. Among members of the STAT family, STAT3 has garnered particular interest due to its role in cancer and development. Recently, it was proposed that STAT3 regulates cardiac ATP generation in vivo through protein interaction with the mitochondrial complexes of oxidative phosphorylation, specifically Complexes I/II. For this mechanism to work effectively, the cellular ratio of Complexes I/II and STAT3 must approach one. However, using three different proteomic approaches in cardiac tissue, we determined the ratio of Complexes I/II and STAT3 to be ∼105. This finding suggests that direct protein interaction between Complexes I/II and STAT3 cannot be required for optimal ATP production, nor can it dramatically modulate oxidative phosphorylation in vivo. Thus, STAT3 is likely altering mitochondrial function via transcriptional regulation or indirect signaling pathways that warrant further investigation.
Keywords: Energy Metabolism, Mitochondrial Metabolism, Protein-Protein Interactions, Proteomics, Transcription Factors
Introduction
Mitochondria are dynamic organelles that are essential for cellular ATP production, redox regulation, and apoptosis. Given these critical cell functions, it is important to identify the molecular mechanisms governing mitochondrial function in normal and pathological states. Covalent post-translational modifications, such as protein phosphorylation, acetylation, and s-nitrosylation, have increasingly been shown to modulate mitochondrial function. Similarly, mitochondrial protein-protein interactions (PPIs),2 such as the association of the inhibitory protein (IF1) with the F1F0-ATP synthase, have been shown to regulate cellular energy conversion. Importantly, like virtually all functional PPIs, IF1 combines with the F1-ATP synthase in a molar ratio of 1:1 (1). Recently, it was proposed that a nuclear encoded transcription factor, the signal transducer and activator of transcription 3 (STAT3), enters mitochondria and regulates oxidative phosphorylation at the level of Complexes I, II, and V (2–4). These studies revealed that the loss of STAT3 led to a 50–70% decrease in cellular ATP levels in murine heart tissue in vivo (4) and cells (2, 4) and provided compelling support for modulation of oxidative phosphorylation by STAT3. Toward a mechanism of this regulation, it was reported that STAT3 binds to mitochondrial Complexes I/II in vitro (3, 4). Although no direct activity measurements were made, it was suggested that the association of STAT3 with Complexes I/II is required for mitochondrial activation (3, 4). By definition, if a STAT3-Complex I/II PPI is necessary for optimal energy conversion in the cell, near equimolar amounts of STAT3 and Complexes I/II must be present in normally functioning tissue mitochondria. Following this logic, STAT3 must be expressed at a much higher concentration than most transcription factors to approach the concentration of mitochondrial proteins in heart cells. To address this issue, the current study (i) examined the stoichiometric relationship between STAT3 and Complexes I/II in purified heart mitochondria using two-dimensional gel electrophoresis and (ii) obtained an absolute concentration of STAT3 in total heart tissue using quantitative Western blotting and targeted mass spectrometry. These data reveal that the cellular ratio of Complexes I/II to STAT3 is ∼105, which implies that STAT3 cannot regulate cardiac ATP production in vivo by direct PPI. Our findings underline the importance of reporting cellular protein concentrations and biochemical mechanisms in quantitative terms when extrapolating toward functional modifications.
EXPERIMENTAL PROCEDURES
Mitochondrial Isolation
All procedures performed were in accordance with the guidelines described in the Animal Care and Welfare Act (7 United States Code 2142 Section 13) and approved by the NHLBI Animal Care and Use Committee. Porcine and murine heart and liver mitochondria were isolated from tissue that was cold-perfused in situ to remove blood and extracellular calcium as well as to prevent any warm ischemia, as described previously (5). Mitochondrial preparations were tested for viability by measuring both the respiratory control ratio and the ability to maintain matrix ATP content, as described previously (5).
Two-dimensional Gel Electrophoresis Studies
Mitochondrial proteins were resuspended in lysis buffer (15 mm Tris-HCl, 7 m urea, 2 m thiourea, and 4% CHAPS (w/v)) to a final concentration of ∼10 mg/ml. Samples were incubated on ice for 2 h and centrifuged at 13,000 × g for 30 min at 4 °C. The supernatant was collected, and centrifugation was repeated twice. Protein concentration was determined using a Bradford assay (USB protein determination reagent, USB Corp., Cleveland, OH) against a standard curve of bovine serum albumin. Fifty micrograms of sample or recombinant STAT3 (Active Motif, Carlsbad, CA, catalogue number 31140) matched to the concentration of Complex I were labeled with CyDyes (6) and then mixed with rehydration solution (7 m urea, 2 m thiourea, 4% CHAPS (w/v), 13 mm dithiothreitol, 1% (pH 3–11) NL Pharmalyte (v/v), and 2 μl of Destreak reagent) to a final volume of 210 μl for difference in-gel electrophoresis (DIGE). To achieve optimal focusing of STAT3, it was found necessary to incubate it with 5 mm ATP for 5 min at room temperature. Isoelectric focusing (7), electrophoresis (7), and imaging (6) were then performed using previously described methods.
Western Blot Analysis
To screen for STAT3 in mitochondria, 150 μg of porcine heart mitochondria and recombinant STAT3 were resolved by two-dimensional gel electrophoresis, as described above. Proteins were then blotted to a nitrocellulose membrane, and STAT3 was detected with anti-STAT3 (Cell Signaling Technology, Danvers, MA, catalogue number 9139) as the primary antibody and Alexa Fluor® 488 anti-mouse IgG2a (Invitrogen) as the secondary antibody. Blots were visualized using a Typhoon 9400 imager (GE Healthcare), and images were processed using Photoshop (Adobe Systems, San Jose, CA).
For quantitative Western blot analyses, porcine heart tissue was homogenized in 137 mm KCl, 10 mm HEPES, 2.5 mm MgCl2, 0.5 mm EDTA, and 0.5% maltoside on ice and mixed with Laemmli buffer, containing 15 mm dithiothreitol, to the desired concentration. Samples were run immediately on a 12.5% Tris-HCl gel in 1× Tris, glycine, and SDS buffer (Bio-Rad) for ∼200 V-h. Gels were blotted and subsequently processed as described above.
Cytochrome c Oxidase and Protein Content Determination
Using a well established method (8), the content of cytochrome c oxidase (Complex IV) in porcine heart tissue homogenates was determined to be 31 ± 0.5 nmol/g of wet weight across four measurements. Each gram of wet weight was found to contain 137 ± 9 mg of protein.
Mass Spectrometry
Porcine and murine mitochondrial proteins were prepared and identified by mass spectrometry (see Table 1), as described previously (9). To specifically screen for STAT3 using the targeted approach described here, porcine heart proteins and recombinant human STAT3 were used. It is important to note that human and STAT3 share 99.7% sequence homology across their 770 amino acid residues. Because STAT3 had not been previously identified in complex protein mixtures, preliminary experiments were performed using multiple reaction monitoring methods to obtain the best sensitivity and selectivity of the recombinant STAT3 protein. Following trypsin digestion of recombinant STAT3, IVELFR was found to be the most abundant and reproducible peptide via MRMPilot software (Applied Biosystems, Carlsbad, CA). This signature peptide was unique to STAT3, containing no Met or Cys amino acids.
TABLE 1.
Protein identification in large scale mass spectroscopy proteomic screens
Protein subunits of mitochondrial Complexes I/II were identified by mass spectrometry. Data from our laboratory (Boja) screened heart and liver mitochondria from pig and mouse. Johnson et al. (16) examined heart, liver, brain, and kidney mitochondria from rat. MitoCarta (14) and MitoP2 (15) are publically available datasets for human and mouse proteins. When mitochondria were screened from multiple tissues, we focused on heart and liver. All screens identified all or most of the Complex I/II subunits, but none identified STAT3. ●, identified.
| Boja | Johnson | MitoCarta | MitoP2 | |
|---|---|---|---|---|
| Complex I | ||||
| NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, 51 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 2, 8 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 4, 9 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 5, 13 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 6, 14 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 8, 19 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 9, 39 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 10, 42 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 11, 14.7 kDa | ● | ● | ||
| NADH dehydrogenase (ubiquinone) 1 α subcomplex, 13, 16.7 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 3, 12 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 4, 15 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 6, 17 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 7, 18 kDa | ● | ● | ||
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 8, 19 kDa | ● | ● | ● | ● |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 9, 22 kDa | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) 1 β subcomplex, 10, 22 kDa | ● | |||
| NADH dehydrogenase (ubiquinone) 1 α/β subcomplex, 1, 8 kDa | ● | ● | ||
| NADH dehydrogenase (ubiquinone) subunit 1 | ● | ● | ● | |
| NADH dehydrogenase (ubiquinone) subunit 5 | ● | ● | ● | |
| Complex II | ||||
| Succinate dehydrogenase complex, subunit A | ● | ● | ● | ● |
| Succinate dehydrogenase complex, subunit B | ● | ● | ● | ● |
| Succinate dehydrogenase complex, subunit C | ● | ● | ● | |
| Succinate dehydrogenase complex, subunit D | ● | ● | ● | |
| STAT3 | No | No | No | No |
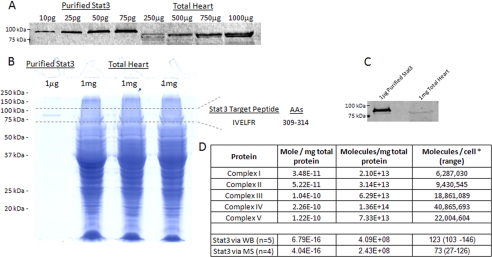
Initial attempts to detect and quantify STAT3 in total heart homogenates were unsuccessful. To reduce the complexity, 1 μg of recombinant STAT3 and three lanes with 1 mg of total heart protein were loaded onto 12.5% Tris-HCl gels as described above. The 88-kDa molecular mass region was excised and digested with trypsin. Forty percent of the total heart digest (from the three 1-mg bands) were used for analysis. Liquid chromatography-tandem mass spectrometry was then performed using an Eksigent nanoLC-Ultra 1D Plus system (Dublin, CA) coupled to an LTQ orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Peptides were first loaded onto a C18 trap (Agilent, Palo Alto, CA) at a flow rate of 6 μl/min for 6 min and then separated on a reversed-phase PicoFrit analytical column (New Objective, Woburn, MA) using a 40-min linear gradient of 5–35% acetonitrile in 0.1% formic acid at a flow rate of 250 nl/min. The monoisotopic masses of the STAT3 peptides were entered into a parent mass list, and survey scans (MS1) were acquired in the orbitrap at resolution 60,000. A precursor peak above 500 counts in the parent mass list (±50 ppm) triggered an MS/MS scan in the ion trap. Peptide IVELFR was followed throughout the analysis, and the measured intensities of fragment ion 564.4 obtained in the MS/MS spectra of 388.74 m/z were used for quantification. Quantification of STAT3 in the heart homogenate was based on the intensity of the signature MS/MS fragment ion 564.4 relative to its intensity in the 1 μg of recombinant STAT3 digest, which revealed 150 and 183 amol of STAT3/mg of heart protein in two measurements. For confirmation purposes, quantification was also performed against a known concentration (10 fmol) of 13C-labeled IVELFR, which revealed 583 and 700 amol STAT3/mg of heart protein in two measurements. Fig. 2D converts these values into molecules per cell.
FIGURE 2.
Absolute quantification of STAT3 in total heart tissue. A, a representative Western analysis of purified STAT3 and total porcine heart protein homogenate. B shows the Coomassie Blue-stained SDS gel of purified STAT3 and total porcine heart protein, with the excised region used for absolute mass spectrometry quantification, based on the target tryptic peptide, IVELFR. AAs, amino acids. C, a Western analysis confirming the presence of STAT3 in the gel used for mass spectrometry. The absolute quantity of mitochondrial proteins and STAT3 in moles per mg of protein, molecules per mg of protein, and molecules per cell are shown in D. The * in D denotes that 300 pg per cell was used as a conversion factor (12). WB, Western blot; MS, mass spectrometry.
Calculations
To determine the concentrations of the mitochondrial oxidative phosphorylation complexes and STAT3 in heart tissue, the following values were used: 31 nmol of Complex IV/g of wet heart weight; 137 mg of heart protein/g of wet weight; 2.26 mol of Complex IV/mg of protein; 88058 g/mol (molecular weight of STAT3); CI1CII1.5CIII3CIV6.5CV3.5 (predetermined ratio of oxidative phosphorylation complexes (10, 11)); and 300 pg of protein/cell (predetermined conversion factor (12)).
RESULTS AND DISCUSSION
To determine whether STAT3 regulates mitochondrial ATP generation via direct PPI with the complexes of oxidative phosphorylation, we first used two-dimensional gel electrophoresis to examine the stoichiometric relationship between STAT3 and Complexes I/II in purified heart mitochondria. Subsequently, we applied quantitative Western analyses and targeted mass spectrometry to obtain an absolute cellular concentration of STAT3 in total heart tissue.
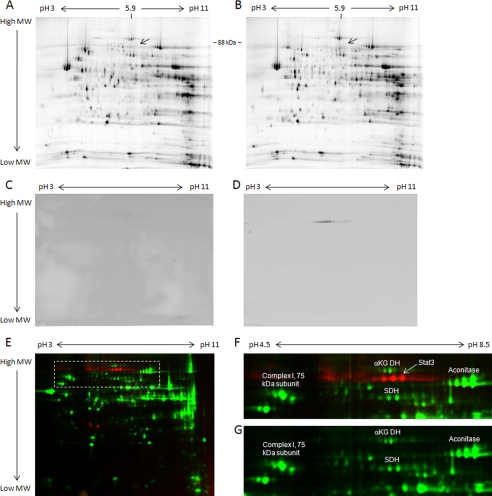
In our experience, the complexes of oxidative phosphorylation are readily observed when heart mitochondrial protein extracts are resolved with two-dimensional gel techniques (5, 13). If STAT3 were stoichiometric with Complexes I/II, it would be detected at a molecular mass of 88 kDa and an isoelectric focusing point (pI) of 5.9. However, we could not detect any protein in this region of the gel (Fig. 1, A and B). A two-dimensional Western analysis was then performed on heart mitochondria to eliminate the possibility that proteolysis or post-translational modifications altered the molecular weight or pI of STAT3 upon localization to the mitochondria. Again, STAT3 was not detected (Fig. 1, C and D). To further establish the position of STAT3 in a complex mitochondrial protein mixture, porcine heart mitochondria were spiked with recombinant STAT3 at a 1:1 ratio with mitochondrial Complex I and visualized by two-dimensional DIGE (Fig. 1E). Although the purified STAT3 was easily visualized at its expected molecular weight and pI (Fig. 1F), again no STAT3 was detected in the unspiked mitochondrial sample (Fig. 1G). Based on these experiments, we estimate that the ratio of Complex I to STAT3 must exceed several orders of magnitude.
FIGURE 1.
Relationship of heart mitochondrial proteins and STAT3. A and B, representative two-dimensional gels of porcine (A) and murine (B) heart mitochondrial proteins, with an arrow indicating the expected position of STAT3. C and D, representative two-dimensional Western analyses of porcine heart mitochondrial proteins, probing for STAT3 (C), and of purified STAT3 (D). E–G, a two-dimensional DIGE gel of porcine heart mitochondrial proteins (labeled with Cy3, green) and a stoichiometric amount (relative to Complex I) of purified STAT3 (labeled with Cy5, red) (E), with enlargement of the highlighted region with (F) and without (G) purified STAT3. For all gels, proteins were separated in the first dimension by isoelectric focusing point over a nonlinear pH range of 3–11 and in the second dimension by molecular mass, ranging from ∼150 to 10 kDa. aKG DH, α-ketoglutarate dehydrogenase; SDH, succinate dehydrogenase.
To confirm these findings, we searched mass spectrometry databases of mitochondrial proteins (MitoCarta (14) and MitoP2 (15)) and data from our laboratory in porcine (5, 13), rat (16), and murine mitochondria but found no evidence of STAT3 localization to heart or liver mitochondria (Table 1). Collectively, these data are consistent with the notion that STAT3 levels are much lower than Complex I in heart mitochondria.
To determine the absolute cellular concentration of STAT3, quantitative Western blotting of total porcine heart proteins was performed, using recombinant STAT3 as a standard (Fig. 2A). These experiments revealed that there are ∼0.7 fmol of STAT3 per mg of porcine heart protein. To validate this finding, we utilized a targeted mass spectrometry approach against peptide IVELFR from STAT3 trypsin digestion (Fig. 2, B and D), which yielded ∼0.4 fmol of STAT3 per mg of porcine heart protein. Using 300 pg of total protein per cell as a conversion factor (12), these methods establish the cellular concentration of STAT3 to be 73–123 molecules/cell (Fig. 2D).
To quantitatively assess the relationship between STAT3 and Complexes I/II, we next determined the absolute concentration of the oxidative phosphorylation complexes in porcine heart mitochondria. Applying spectrophotometric techniques, we determined the concentration of Complex IV in porcine heart tissue to be 226 pmol/mg of protein, which is in good agreement with previous studies (8). We next used previously determined ratios for the oxidative phosphorylation complexes, CI1CII1.5CIII3CIV6.5CV3.5 (10, 11), and calculated the cellular concentrations of Complexes I/II to be 35 and 52 pmol/mg of protein, respectively. Assuming 300 pg of protein per cell, this translates to more than 6 million molecules of Complex I and 9 million molecules of Complex II per cell as compared with ∼100 molecules of STAT3 per cell (Fig. 2D). This result implies that the ratio of Complexes I/II to STAT3 is on the order of 105, in agreement with our two-dimensional gel electrophoresis studies in Fig. 1.
Using these various methods to detect STAT3, we determined that its absolute concentration is on the order of 0.5 fmol/mg of protein or 100 molecules/cell. This low level of protein expression is consistent with other cellular transcription factors (17–19). Indeed, most modeling efforts place transcription factors in the high picomolar to low micromolar range (20). The small number of transcription factor molecules is likely due to the limited number of DNA targets responsible for controlling protein expression. Relative to Complexes I/II, our studies reveal that STAT3 is 105-fold less abundant. This low concentration implies that there are not enough STAT3 molecules for every mitochondrion in the heart cell, let alone enough STAT3 molecules to bind to each molecule of Complexes I/II. Because only a vanishingly small fraction of Complexes I/II can be influenced by STAT3, it is highly unlikely that under normal conditions, protein-protein interaction between STAT3 and Complexes I/II could considerably alter oxidative phosphorylation. In fact, if the number of STAT3 molecules was increased to the level required for PPI with Complexes I/II (i.e. from 100 molecules/cell to more than 1million molecules/cell), it is likely that STAT3 would saturate the binding sites on DNA and lead to defective protein expression.
Given the central role of the mitochondrion in cellular energy metabolism, redox regulation, apoptosis, heme and protein processing, and various biosynthetic and anabolic reaction pathways, it would be surprising if the elimination or overexpression of any general transcription factor did not affect mitochondrial programming and regulation. In the case of STAT3, the high ratio of Complexes I/II to STAT3 implies that PPI between these molecules would be highly ineffective in regulating oxidative phosphorylation. It is possible that STAT3 activates a small fraction of oxidative phosphorylation at the level of Complexes I/II via protein association. However, previous studies demonstrate that porcine, canine, and murine heart need to acutely regulate mitochondrial ATP production near Vmax in vivo (21, 22) and, therefore, require virtually complete activation of the oxidative phosphorylation complexes. This necessitates that a near equimolar concentration of STAT3 and Complexes I/II is present in cardiac mitochondria to generate sufficient ATP to support maximum cardiac performance. The current study revealed that the cellular ratio of Complexes I/II to STAT3 is not 1:1, but rather 105, which implies that STAT3 is not modulating cardiac ATP generation via direct PPIs with the complexes of oxidative phosphorylation. We suggest that the effect of STAT3 on mitochondrial function, as demonstrated in previous studies (2, 4), may result instead from amplified indirect events such as protein expression regulation, alternate signaling pathways, or other undefined cooperative mechanisms that clearly warrant further investigation.
Acknowledgments
We thank David Chess and Edward Neufeld for insightful discussions and editorial advice.
This work was supported, in whole or in part, by the National Institutes of Health Division of Intramural Research.
- PPI
- protein-protein interaction
- STAT3
- signal transducer and activator of transcription 3
- DIGE
- difference in-gel electrophoresis
- MS/MS
- tandem mass spectrometry
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Gomez-Fernandez J. C., Harris D. A. (1978) Biochem. J. 176, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., Levy D. E. (2009) Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers M. G., Jr. (2009) Science 323, 723–724 [DOI] [PubMed] [Google Scholar]
- 4.Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aponte A. M., Phillips D., Hopper R. K., Johnson D. T., Harris R. A., Blinova K., Boja E. S., French S., Balaban R. S. (2009) J. Proteome. Res. 8, 2679–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips D., Ten Hove M., Schneider J. E., Wu C. O., Sebag-Montefiore L., Aponte A. M., Lygate C. A., Wallis J., Clarke K., Watkins H., Balaban R. S., Neubauer S. (2010) J. Mol. Cell Cardiol. 48, 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips D., Aponte A. M., French S. A., Chess D. J., Balaban R. S. (2009) Biochemistry 48, 7140–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaban R. S., Mootha V. K., Arai A. (1996) Anal. Biochem. 237, 274–278 [DOI] [PubMed] [Google Scholar]
- 9.Boja E. S., Phillips D., French S. A., Harris R. A., Balaban R. S. (2009) J. Proteome Res. 8, 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatefi Y. (1985) Annu. Rev. Biochem. 54, 1015–1069 [DOI] [PubMed] [Google Scholar]
- 11.Schägger H., Pfeiffer K. (2001) J. Biol. Chem. 276, 37861–37867 [DOI] [PubMed] [Google Scholar]
- 12.Volpe P., Eremenko-Volpe T. (1970) Eur. J. Biochem. 12, 195–200 [DOI] [PubMed] [Google Scholar]
- 13.Hopper R. K., Carroll S., Aponte A. M., Johnson D. T., French S., Shen R. F., Witzmann F. A., Harris R. A., Balaban R. S. (2006) Biochemistry 45, 2524–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prokisch H., Andreoli C., Ahting U., Heiss K., Ruepp A., Scharfe C., Meitinger T. (2006) Nucleic Acids Res. 34, D705–D711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson D. T., Harris R. A., French S., Blair P. V., You J., Bemis K. G., Wang M., Balaban R. S. (2007) Am. J. Physiol. Cell Physiol. 292, C689–C697 [DOI] [PubMed] [Google Scholar]
- 17.Elf J., Li G. W., Xie X. S. (2007) Science 316, 1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert W., Müller-Hill B. (1966) Proc. Natl. Acad. Sci. U.S.A. 56, 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripperger J. A., Fritz S., Richter K., Hocke G. M., Lottspeich F., Fey G. H. (1995) J. Biol. Chem. 270, 29998–30006 [DOI] [PubMed] [Google Scholar]
- 20.Zeiser S., Liebscher H. V., Tiedemann H., Rubio-Aliaga I., Przemeck G. K., de Angelis M. H., Winkler G. (2006) Theor. Biol. Med. Model. 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaban R. S. (2006) Ann. N.Y. Acad. Sci. 1080, 140–153 [DOI] [PubMed] [Google Scholar]
- 22.Mootha V. K., Arai A. E., Balaban R. S. (1997) Am. J. Physiol. 272, H769–H775 [DOI] [PubMed] [Google Scholar]