Abstract
BACE1 (β-site amyloid precursor protein-cleaving enzyme 1) is a membrane-tethered member of the aspartyl proteases, essential for the production of β-amyloid, a toxic peptide that accumulates in the brain of subjects affected by Alzheimer disease. The BACE1 C-terminal fragment contains a DXXLL motif that has been shown to bind the VHS (VPS27, Hrs, and STAM) domain of GGA1–3 (Golgi-localized γ-ear-containing ARF-binding proteins). GGAs are trafficking molecules involved in the transport of proteins containing the DXXLL signal from the Golgi complex to endosomes. Moreover, GGAs bind ubiquitin and traffic synthetic and endosomal ubiquitinated cargoes to lysosomes. We have previously shown that depletion of GGA3 results in increased BACE1 levels and activity because of impaired lysosomal degradation. Here, we report that the accumulation of BACE1 is rescued by the ectopic expression of GGA3 in H4 neuroglioma cells depleted of GGA3. Accordingly, the overexpression of GGA3 reduces the levels of BACE1 and β-amyloid. We then established that mutations in the GGA3 VPS27, Hrs, and STAM domain (N91A) or in BACE1 di-leucine motif (L499A/L500A), able to abrogate their binding, did not affect the ability of ectopically expressed GGA3 to rescue BACE1 accumulation in cells depleted of GGA3. Instead, we found that BACE1 is ubiquitinated at lysine 501 and is mainly monoubiquitinated and Lys-63-linked polyubiquitinated. Finally, a GGA3 mutant with reduced ability to bind ubiquitin (GGA3L276A) was unable to regulate BACE1 levels both in rescue and overexpression experiments. These findings indicate that levels of GGA3 tightly and inversely regulate BACE1 levels via interaction with ubiquitin sorting machinery.
Keywords: Alzheimer Disease, Amyloid, Intracellular Trafficking, Protein Degradation, Secretases, BACE1, GGA
Introduction
Alzheimer disease (AD)3 is a devastating neurodegenerative disorder that results in loss of memory and cognitive function, eventually leading to dementia. A key neuropathological event in AD is the cerebral accumulation of an ∼4-kDa peptide termed Aβ, the principal component of senile plaques. Amyloid plaques are formed by aggregates of amyloid-β-peptides, 37–43-amino acid fragments (predominantly Aβ40 and Aβ42) derived by serial proteolysis of the amyloid precursor protein (APP) by β- and γ-secretase (1).
The β-site APP-cleaving enzyme (BACE1) is a membrane-tethered member of the aspartyl proteases that has been identified as β-secretase (2–4). APP proteolysis by β-secretase results in the production of secreted β-APP polypeptide (βAPPs) along with a membrane-associated APP C-terminal fragment of 99 amino acids, which then serves as substrate for γ-secretase resulting in the production of Aβ. BACE1 is an N-glycosylated type 1 transmembrane protein that undergoes constitutive N-terminal processing in the Golgi apparatus. The ectodomain contains four glycosylation sites and two signature sequences typically associated with aspartyl proteases (D(T/S)G(T/S)). BACE1 is targeted through the secretory pathway to the plasma membrane where it can be internalized to endosomes (5). The BACE1 C-terminal fragment contains a specific di-leucine (DXXLL) sorting signal that is present in several transmembrane proteins (e.g. cation-dependent and cation-independent mannose-6-phosphate receptor) and that regulates endocytosis and lysosomal targeting (6). Mutagenesis of L499A/L500A results in retention of BACE1 at the plasma membrane (7–9). The BACE1 acidic di-leucine motif has been shown to bind GGA1–3 (Golgi-localized γ-ear-containing ARF-binding proteins), and phosphorylation of BACE1-Ser-498 appears to increase the binding (10–14).
GGA1–3 are monomeric adaptors that are recruited to the trans-Golgi network (TGN) by the Arf1-GTPase. They consist of four distinct segments as follows: a VHS (VPS27, Hrs, and STAM) domain that binds the acidic di-leucine sorting signal, DXXLL; a GAT (GGA and Tom1) domain that binds Arf:GTP; a hinge region which recruits clathrin; and a γ-adaptin ear homology domain that exhibits sequence similarity to the ear region of γ-adaptin and recruits a number of accessory proteins. GGAs are necessary for the sorting of acid hydrolases to the lysosomes. Newly synthesized acid hydrolases modified with mannose 6-phosphate groups bind to mannose 6-phosphate receptors (MPRs). MPRs bind to the VHS domain of GGAs via the DXXLL motif (15). GGAs are likely involved in the transport of proteins containing the DXXLL signal from the Golgi complex to the endosomes. However, several studies have shown that cargo proteins can be recruited in the GGA pathway not only by the di-leucine sorting motif but also by ubiquitin (16–20). Puertollano and Bonifacino (16) have reported that RNAi silencing of GGA3, but not GGA1 or GGA2, resulted in the accumulation of epithelial growth factor receptor (EGFR) in enlarged early endosomes and partially blocked its delivery to lysosomes where it is normally degraded. The sorting and lysosomal degradation of EGFR are regulated by ubiquitination of critical lysines in its cytoplasmic domain (21, 22). Ubiquitin (Ub) is attached to the target protein as a monomer or in the form of isopeptide-linked polymers termed polyubiquitin chains. Ubiquitination at one (monoubiquitination) or multiple lysines (multiubiquitination) of a target protein, e.g. EGFR, regulates its endocytosis and sorting to the lysosomes for degradation (23). Given that Ub contains seven lysine residues, once a molecule of Ub is attached to the target protein, additional Ub molecules can be linked resulting in the formation of polyubiquitin chains. Elongation in polyUb chains can occur at any of the seven lysine residues present in Ub. Lys-48-linked ubiquitination mainly targets proteins for proteasomal degradation. In contrast, Lys-63-linked polyubiquitination plays a role in endocytosis and signaling functions in a proteasome-independent fashion (24). Increasing evidence is accumulating that Lys-63-linked Ub chains are a specific signal for protein sorting into the multivesicular body (MVB) pathway (25). The endosomal machinery responsible for the delivery of EGFR and other receptors to the lumen of MVB consists of proteins containing ubiquitin-binding domains (e.g. hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), endosomal sorting complex required for transport complexes (ESCRT) I–III) (26). Depletion of the endosomal proteins HRS, STAM1, STAM2, or the tumor susceptibility gene (TSG101), a ubiquitin-binding subunit of ESCRT I, produces accumulation of EGFR in early endosomes similar to that resulting from depletion of GGA3 (27–30). The GGA3 GAT domain has also been shown to bind ubiquitin at two different sites (31–33). One of these, site 2, centers on Leu-276. Substitution L276A in GGA3 GAT domain abrogates binding to ubiquitin and to TSG101 in a yeast two-hybrid system and results in the accumulation of EGFR in early endosomes upon expression in mammalian cells (16). It is important to underscore that the short isoform of human GGA3 lacking amino acid residues 68–101 and unable to bind the di-leucine motif trafficked EGFR to lysosomes normally (16). Thus, the GGA3-mediated regulation of EGFR degradation is independent of the interaction with the GGA3 VHS domain. We have recently shown that depletion of GGA3 results in increased BACE1 levels and activity because of impaired degradation (34). Here, we report that GGA3 regulates BACE1 degradation independently of VHS/di-leucine motif interaction but via binding to the ubiquitin sorting machinery. Moreover, we have determined that BACE1 is ubiquitinated at Lys-501 and that it is mainly monoubiquitinated and Lys-63-linked polyubiquitinated.
EXPERIMENTAL PROCEDURES
Antibodies and Expression Vectors
Anti-HA and anti-Myc (mono- and polyclonal) antibodies were purchased from Cell Signaling, Danvers, MA. The anti-V5 antibody was purchased from Invitrogen. The monoclonal antibody anti-GGA3 was purchased from BD Biosciences. The GAPDH antibody was purchased from Chemicon, Temecula, CA. The anti-EEA1 and anti-LAMP2 antibodies were purchased from BD Biosciences. The anti-BACE1 polyclonal antibody PA1-757 was purchased from Affinity Bioreagents, Golden, CO. The monoclonal anti-BACE1 antibody, 3D5, was a kind gift from Dr. Robert Vassar. BACE1-Myc and HA-GGA3 (long isoform) expression vectors have been described previously (34). BACE1-V5 expression vector was a kind gift from Dr. B. T. Hyman (13). HA-ubiquitin expression vector was a kind gift from Dr. H. G. Gottlinger, Dana Farber Cancer Institute, Boston. The pRK5-HA-ubiquitin wild type, KO, Lys-63, and K48R mutant were purchased from Addgene, Cambridge MA (35).
Generation of H4 Neuroglioma Stable Cell Line Expressing GGA3 or Negative Control shRNA
Lentiviral vectors (pLKO.1) carrying expression cassettes that express shRNAs targeting the human GGA3 gene were purchased from Sigma and have been described previously (34). A lentiviral vector (pLKO.1) expressing a short hairpin sequence containing 4-bp mismatches to any known human or mouse gene was used as nontarget negative control (Sigma). The packaging of the virus was performed as described previously (36). H4 human neuroglioma cells expressing APP751 (H4-APP751) have been described previously (34). H4-APP751 cells were infected with lentiviral particles expressing either negative control or GGA3 shRNA at an approximate multiplicity of infection of 100. The day after infection, the infected cells were harvested and transferred to a 150-mm dish containing selection media (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 200 μg/ml G418, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1 μg/ml puromycin). Isolated clones were screened for levels of GGA3 by Western blot (WB) analysis with monoclonal GGA3 antibody (Cell Signaling Technology, Danvers, MA) as described previously (34). The H4-APP751 cell lines expressing GGA3shRNA or negative control shRNA are referred to as H4-shGGA3 or H4-NC, respectively.
Immunofluorescence, Confocal Microscopy, and Colocalization Analysis
H4-shGGA3 or H4-NC cells were seeded at a density of 250,000 cells per well in 6-well plates. The following day, cells were transfected with 2 μg of BACE1-Myc expression vector using Superfect transfection reagent (Qiagen, Valencia, CA). Three days post-transfection, cells were trypsinized and re-plated onto 12-mm glass coverslips. Six days post-transfection, cells were fixed with 4% paraformaldehyde for 10 min at room temperature and permeabilized with 0.1% Triton X-100 for 5 min. Cells were incubated in 5% normal goat sera for 1 h at room temperature to block nonspecific binding. Primary antibodies (polyclonal anti-Myc 1:400 and monoclonal anti-EEA1 1:400 or monoclonal anti-LAMP2 1:600 in 5% goat sera) were incubated overnight at 4 °C followed by washing in phosphate-buffered saline and incubation with goat anti-mouse Alexa Fluor® 594 conjugate and rabbit Alexa Fluor® 488 conjugate antibodies (Invitrogen) for 1 h at room temperature (1:200 in 5% goat sera). Nuclei were counterstained by incubation with diamidino-2-phenylindole (Sigma) for 10 min. Coverslips were washed with phosphate-buffered saline and mounted using FluorSave (EMD Chemicals, Gibbstown, NJ). Between 14 and 25 cells expressing various levels of transfected BACE1 were imaged for colocalization studies. To ensure no biasing of data, only the channel corresponding to BACE1 staining (not the subcellular organelle) was viewed prior to image capture. Confocal microscopy was performed on a Nikon Eclipse Ti confocal microscope with a 60× oil Plan Apo VC objective in 0.130-μm steps. Pinhole was set to Airy (40.9 μm) resulting in an optical section thickness of 0.13 μm. Images were captured at 1024 × 1024 resolution with a pixel dwell of 3.8 μs. Laser intensity and gain were set using a saturation indicator to prevent saturation of pixels in any channel. Laser channels were captured sequentially to prevent bleed through. Colocalization analysis of BACE1 and subcellular organelles was performed using Metamorph version 6.1 software on a representative z-section from each cell expressing maximal fluorescence in both channels. Background correction prior to thresholding for colocalization was performed using a 32 × 32 median filter. Colocalization of BACE1 and the subcellular organelle markers EEA1 and LAMP2 was expressed as a percentage area of overlap of BACE1 with the subcellular marker.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The primers used to produce each mutation are as follows: GGA3RNAiRes (G1959A/A1962G) (along with reverse complement primer), 5′-CAG GCT GCA GTG CCC AAA TCC ATG AAA GTG AAG-3′; L276A mutation (along with reverse complement primer), 5′-GAGGACAATGATAACAGTGCGGGGGACATCCTGCAAGC-3′; N91A mutation (along with reverse complement primer), 5′-GTTCCGCTTTTTGGCTGAGTTAATCAAAGTCGTC-3′; BACE-Myc L499A/L500A (along with reverse complement primer), 5′-GATGACATCTCCGCGGCGAAGCTCGAGTC-3′; and BACE-V5 K501R (along with reverse complement primer), 5′-GACATCTCCCTGCTGAGAAAGGGCAATTCTGCAG-3′. The resulting cDNA expression vectors were verified by sequencing.
Rescue and Overexpression Experiments
H4-shGGA3 or H4-NC cells were seeded at the density of 250,000 per well in a 6-well plate. The following day, cells were cotransfected with 0.3 μg of BACE1-Myc or BACE1-Myc LL/AA expression vector and 0.3 μg of empty vector, HA-GGA3, HA-GGA3siRNARes, HA-GGA3N91AsiRNARes, or HA-GGA3L276AsiRNARes expression vectors using Superfect transfection reagent according to the manufacturer's instructions (Qiagen, Valencia, CA). Cells were harvested 6 days post-transfection and lysed as described previously (34). Equal amounts (30 μg) of each sample were separated by SDS-PAGE using 4–12% BisTris gels (Invitrogen), and WB analysis was performed using anti-Myc or anti-HA antibody to detect BACE1 or GGA3, respectively, as described previously (34). GAPDH levels were detected by WB analysis with anti-GAPDH antibody and used as a loading control.
Aβ ELISA
H4-NC cells stably expressing vector or GGA3 were transiently transfected with BACE1 expression vector. Six days post-transfection media was conditioned for ∼12 h. Secreted Aβ1–40 was measured in the conditioned media using a human Aβ1–40-specific sandwich ELISA according to manufacturer's instructions (Invitrogen). Aβ concentration (pg/μl) was normalized against the protein concentration (μg/ml) in the cell lysates.
Ubiquitination Assay
Two million H4-APP751 cells or 4 million mouse neuroglioma cells, N2A, were seeded in a 100-mm dish. The following day, the cells were cotransfected with 3 μg of BACE1-V5 or BACE1-V5 K501R and 3 μg of HA-ubiquitin using Superfect transfection reagent (Qiagen, Valencia, CA). In another set of experiments, H4-APP751 cells were cotransfected with 3 μg of BACE1-V5 and Ub WT or mutant expression vectors. Cells were harvested 24 h later and lysed with TNET buffer (25 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, 0.5% Triton X-100) containing protease inhibitors (Fisher). Approximately 50 μg of protein from each sample set was incubated with protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at 4 °C for pre-clearing. The supernatant was incubated overnight at 4 °C with protein A/G-agarose beads and anti-V5 monoclonal antibody at a 1:100 dilution (Invitrogen). The beads were washed three times with RIPA buffer (1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 5 mm EDTA, 50 mm Tris, pH 8, 150 mm NaCl), containing protease inhibitors. Immunocomplexes were eluted by adding 2× WB Sample Buffer (Invitrogen) followed by incubation at 55 °C for 10 min. The samples were separated by SDS-PAGE using 4–12% BisTris gels (Invitrogen). Western blot analysis was performed with anti-HA antibody to detect ubiquitin (Cell Signaling Technology, Danvers, MA) in conjunction with a mouse IgG TrueBlot immunoblotting reagent (eBioscience, San Diego) in lieu of the traditional anti-mouse secondary to avoid the detection of mouse immunoglobulin. This was followed by WB analysis with a monoclonal anti-V5 antibody to detect BACE1 (Invitrogen) and followed by incubation with mouse IgG TrueBlot immunoblotting reagent. Ubiquitination assay of endogenous BACE1 was performed by transfecting N2A with 6 μg of empty vector, pRK5-HA-ubiquitin WT, pRK5-HA-ubiquitin K48R, pRK5-HA-ubiquitin Lys-63, or pRK5-HA-ubiquitin KO. Cells were harvested 24 h later and lysed with RIPA buffer (1× working stock: 10 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% cholic acid, 0.1% SDS, 5 mm EDTA). Approximately 700 μg of protein from each sample set was incubated with protein A/G-agarose beads (Santa Cruz Biotechnology) and a polyclonal BACE1 antibody at a concentration of 1 μg/ml (Affinity Bioreagents, Golden, CO). Western blot analysis was performed with anti-HA or 3D5 antibody to detect ubiquitin or endogenous BACE1, respectively.
In Vitro Ubiquitin Binding Assay
Two millions H4-APP751 cells were seeded in a 100-mm dish. The following day, the cells were transfected with 10 μg of GGA3 or GGA3L276A expression vector using Superfect transfection reagent (Qiagen, Valencia, CA). Cells were harvested 72 h later and lysed with TNET buffer containing protease inhibitors. Approximately 500 μg of protein of each sample were pre-cleared by incubation with protein A-agarose beads (Sigma) for 2 h at 4 °C. The supernatant from GGA3 or GGA3L276A-expressing cells was then incubated overnight at 4 °C with protein A-agarose beads (as negative control) or with ubiquitin-agarose beads (Sigma). The beads were washed four times with RIPA buffer containing protease inhibitors. Proteins bound to the beads were eluted as described above. Western blot analysis was performed with monoclonal GGA3 antibody (Cell Signaling Technology, Danvers, MA) and mouse IgG TrueBlot immunoblotting reagent (eBioscience, San Diego).
Densitometry and Statistical Analysis
Densitometry analysis was performed on a Macintosh computer using a Versadoc Imager and Quantity One software (Bio-Rad). Intensity values for BACE1 or GGA3 were normalized against intensity value of the loading control, GAPDH. Statistical analysis was performed using InStat3 software. Unpaired t test was employed for data sets that passed a normality test. Unpaired t test with Welch correction was employed for data sets that passed normality test but had different standard deviations.
RESULTS
Depletion and Overexpression of GGA3 Inversely Regulates BACE1 Levels
We have previously shown that both ectopically expressed and endogenous BACE1 levels increased when GGA3 was depleted by transient transfection of synthetic RNAi duplex or by infection of lentivirus expressing short hairpin RNA (shRNA) targeting different regions of the GGA3 gene in both human and mouse cell lines (human H4 and mouse N2A neuroglioma cells) as well as in primary neuronal cultures (34). We have also shown that depletion of GGA3 enhances β-secretase activity as evidenced by increased levels of the APP C-terminal fragment of 99 amino acids and Aβ. These data support the hypothesis that GGA3 plays a role in modulating BACE1 turnover and stability most likely by sorting BACE1 to late endocytic compartments/lysosomes (based on our earlier findings (9)).
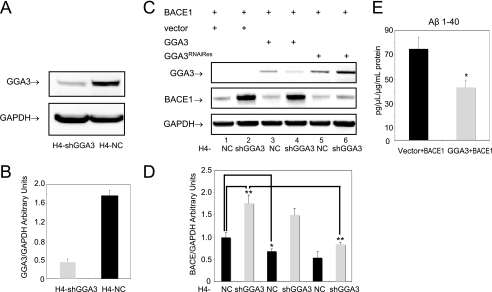
Next, we established H4 cell lines that stably express human GGA3 shRNA (H4-shGGA3) or negative control shRNA (H4-NC) in lentiviral vectors. The stable down-regulation of GGA3 was performed in H4 cell lines stably expressing APP751 (H4APP751) to be able to measure secreted Aβ by ELISA. The levels of endogenous GGA3 were decreased by 80% in the H4-shGGA3 cell line compared with control cells (H4-NC cell line) (Fig. 1, A and B).
FIGURE 1.
Depletion and overexpression of GGA3 inversely regulates BACE1 levels. A, levels of endogenous GGA3 were analyzed in H4 neuroglioma cells stably expressing GGA3 (H4-shGGA3) or negative control (H4-NC) shRNAs. Western blot analysis was performed using anti-GGA3 antibody. GAPDH was used as loading control. B, graph represents mean ± S.E. of at least six GGA3 level measurements. Densitometry was performed using a Versadoc Imager and Quantity One software (Bio-Rad). GGA3 densitometry values were normalized against GAPDH values. C, H4-NC and H4-shGGA3 cells were transiently transfected with the indicated expression vectors. Cells were collected at 6 days post-transfection. Levels of BACE1, GGA3, and GAPDH were analyzed by WB using anti-Myc, anti-HA, and anti-GAPDH antibody, respectively. D, graph represents mean ± S.E. of at least eight BACE1 level measurements. Densitometry was performed as described above. BACE1 accumulates by ∼2-fold in H4-shGGA3 cells compared with H4-NC (vector H4shGGA3 versus vector H4-NC unpaired t test with Welch correction **, p = 0.004). BACE1 accumulation was rescued by the expression of RNAi-resistant GGA3 mutant in H4-shGGA3 cells (vector versus HA-GGA3RNAiRes unpaired t test with Welch correction **, p = 0.001). Moreover, the overexpression of GGA3 in H4-NC cells reduced the levels of BACE1 (vector versus HA-GGA3 unpaired t test with Welch correction *, p = 0.03). E, H4-NC cells stably expressing vector or GGA3 were transiently transfected with BACE1 expression vector. Media were conditioned for ∼12 h at 6 days post-transfection. The graph represents mean ± S.E. of at least six Aβ1–40 measurements by ELISA. Aβ concentration (pg/μl) was normalized against the protein concentration (μg/ml) in the cell lysates. Levels of Aβ1–40 were significantly decreased in the conditioned media from cells expressing GGA3 and BACE1 compared with that from cells expressing empty vector and BACE1 (vector versus HA-GGA3 unpaired t test with Welch correction *, p = 0.02).
We next tested the effect of ectopic expression of GGA3 on BACE1 levels in H4-shGGA3 and H4-NC cells. To restore GGA3 expression in H4-shGGA3 cells, we generated a GGA3 expression vector resistant to RNAi by introducing two silent mutations at the site recognized by the GGA3shRNA (HA-GGA3RNAiRes). BACE1-Myc expression vector was cotransfected with empty vector, HA-GGA3, or HA-GGA3RNAiRes in both H4-shGGA3 and H4-NC cells. Cells were collected 6 days post-transfection. WB analysis with anti-HA tag antibody revealed that levels of ectopically expressed GGA3 were decreased in H4-shRNA cells compared with H4-NC because of RNAi. Instead, the ectopic expression of HA-GGA3RNAiRes resulted in comparable levels of GGA3 protein in both H4-shGGA3 and H4-NC cell lines demonstrating that the mutant is protected from RNAi. We confirmed that BACE1 levels increase by ∼2-fold in H4-shGGA3 cells compared with H4-NC (vector H4-shGGA3 versus vector H4-NC unpaired t test with Welch correction **, p = 0.004). However, BACE1 accumulation was rescued by the expression of the RNAi-resistant GGA3 mutant in H4-shGGA3 cells (vector versus HA-GGA3RNAiRes unpaired t test with Welch correction **, p = 0.001). Moreover, the overexpression of GGA3 in H4-NC cells reduced the levels of BACE1 (vector versus HA-GGA3 unpaired t test with Welch correction *, p = 0.03) (Fig. 1, C and D).
We next assessed whether overexpression of GGA3 reduced levels of secreted Aβ1–40, as a measure of BACE1 activity. H4-NC cells stably expressing vector or GGA3 were transiently transfected with a BACE1 expression vector. As expected, we found that levels of Aβ1–40 were significantly decreased in conditioned media from cells expressing GGA3 and BACE1 compared with that from cells expressing empty vector and BACE1 (vector versus HA-GGA3 unpaired t test with Welch correction *, p = 0.02) (Fig. 1E). These findings indicate that levels of GGA3 tightly and inversely regulate levels and activity of BACE1.
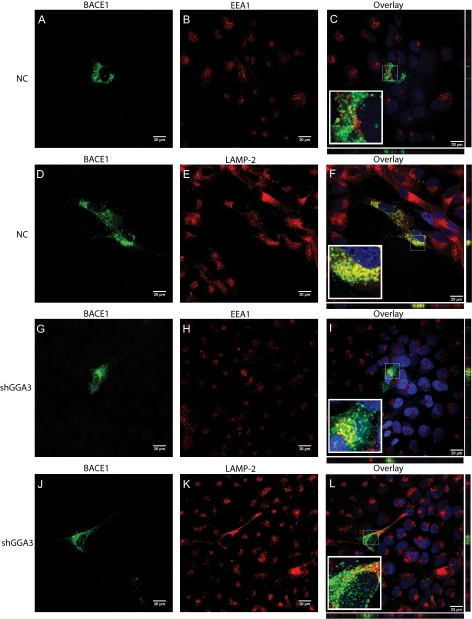
Depletion of GGA3 Results in BACE1 Accumulation in Early Endosomes Because of Impaired Trafficking of BACE1 to Late Endosomes/Lysosomes
We have previously reported that BACE1 is degraded in the lysosomes (9). Thus, we determined whether GGA3 regulates BACE1 degradation by sorting it to the lysosomes. BACE1-Myc-tagged expression vector was transiently transfected in H4-NC and H4-shGGA3 cell lines. Subcellular localization of BACE1 was assessed 6 days post-transfection. Immunofluorescent staining of EEA1 was used as a marker of early endosomes, whereas LAMP2 staining was used as a marker of late endosomes/lysosomes. Confocal microscopy showed that BACE1 was enriched in LAMP2-positive compartments in H4-NC cells (Fig. 2, A–F). Instead, BACE1 accumulates in EEA1-positive compartments in H4-shGGA3 cells most likely because of its failure to reach the lysosomes (Fig. 2, G–L). Orthogonal sections were acquired and demonstrated the authenticity of the colocalization. Quantification analysis using Metamorph software confirmed that the percentage of BACE1 staining that colocalizes with LAMP2 staining was significantly higher in H4-NC compared with H4-shGGA3 cells (unpaired t test with Welch correction ***, p < 0.0001). Instead, the percentage of BACE1 staining that colocalizes with EEA1 staining was significantly higher in H4-shGGA3 cells compared with H4-NC (unpaired t test with Welch correction ***, p < 0.0001) (supplemental Fig. S1). Previous studies, including our own, have shown that BACE1 localizes mainly in the TGN and early endosomes when analyzed between 48 and 56 h post-transfection (9, 37). It is possible that at these time points, the rate of synthesis of ectopically expressed BACE1 prevails on the rate of degradation, and as a consequence, the amount of BACE1 trafficked to the lysosomes is most likely under the detection limit of immunofluorescence microscopy. However, in this study, the subcellular localization of transiently expressed BACE1 was assessed at 6 days post-transfection when the synthesis of BACE1 by the expression vector was greatly reduced and the majority of BACE1 was targeted for degradation.
FIGURE 2.
Depletion of GGA3 results in BACE1 accumulation in early endosomes because of impaired delivery of BACE1 to late endosomes/lysosomes. BACE1-Myc-tagged expression vector was transiently transfected in H4-NC (A–F) and H4-shGGA3 cell lines (G–L). Subcellular localization of BACE1 was assessed 6 days post-transfection. BACE1 was immunostained using polyclonal anti-Myc (1:400) followed by incubation with goat anti-rabbit Alexa Fluor® 488 conjugate antibody (A, D, G, and J). EEA1 (B and H) and LAMP2 (E and K) were used as a marker for early endosomes and late endosomes-lysosomes, respectively, using monoclonal anti-EEA1 (1:400) or monoclonal anti-LAMP2 (1:600 in 5% goat sera) followed by incubation with goat anti-mouse Alexa Fluor® 594 conjugate antibody. Nuclei were counterstained by incubation with 4′,6-diamidino-2-phenylindole. The images for BACE1 and EEA1 or LAMP2 staining were merged (Overlay) to determine the area of colocalization (C, F, I, and L), Orthogonal sections were acquired and revealed the authenticity of the colocalization (C, F, I, and L). NC, negative control.
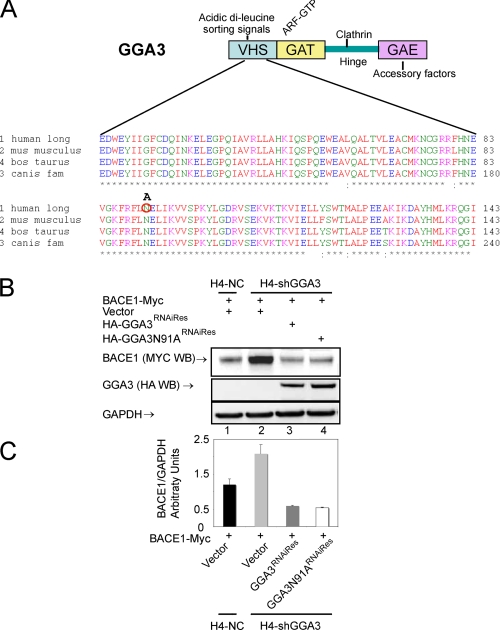
Interaction between GGA3 VHS Domain and BACE1 C-terminal Acidic Di-leucine Motif Is Not Necessary for GGA3-mediated Regulation of BACE1
It has previously been shown that the VHS domain of GGA1–3 binds to the acidic di-leucine motif in the BACE1 C terminus (10–14). Thus, we determined whether the interaction between the GGA3 VHS domain and the BACE1 di-leucine motif is required for GGA3-mediated regulation of BACE1. The asparagine at position 91 in the VHS domain of GGA3 is essential for the interaction with the acidic di-leucine motif. It has previously been demonstrated that amino acid substitution N91A in GGA3 VHS domain abrogates the interaction with the MPR di-leucine motif (Fig. 3A) (38). First, the N91A substitution was introduced into the HA-GGA3RNAiRes expression vector by site-directed mutagenesis. We next assessed whether HA-GGA3N91ARNAiRes was able to rescue BACE1 accumulation in H4-shGGA3 cells to a similar extent as that of HA-GGA3RNAiRes. H4-shGGA3 cells were transfected with BACE1-Myc and empty vector, HA-GGA3RNAiRes, or HA-GGA3N91ARNAiRes. H4-NC cells were transfected with BACE1-Myc and empty vector. BACE1 and GGA3 protein levels were analyzed by WB using an anti-Myc or anti-HA antibody, respectively (Fig. 3B). We found that HA-GGA3N91ARNAiRes was able to restore normal levels of BACE1 similarly to that of HA-GGA3RNAiRes in H4-shGGA3 cells (Fig. 3C). These data indicate that the binding between the GGA3 VHS domain and the di-leucine motif in BACE1 is not required for the GGA3-mediated regulation of BACE1.
FIGURE 3.
Expression of GGA3N91A rescues BACE1 accumulation in cells depleted of GGA3. A, schematic representation of GGA3 domains and multiple sequence alignment of human (NP_619525), Mus musculus (NP_766636), Bos taurus (XP_587687), and Canis familiaris (XP_540429) GGA3 VHS domain (amino acids 24–143). Asparagine at position 91 was substituted by alanine (A) and is indicated by the red circle. B, H4-NC and H4-shGGA3 cells were transiently transfected with the indicated expression vectors. Cells were collected at 6 days post-transfection. Levels of BACE1, GGA3, and GAPDH were analyzed by WB using anti-Myc, anti-HA, and anti-GAPDH antibody, respectively. C, graph represents mean ± S.E. of at least four BACE1 level measurements. Densitometry was performed as described in Fig. 1. HA-GGA3N91ARNAiRes was able to rescue BACE1 accumulation similarly to HA-GGA3RNAiRes in H4-shGGA3 cells.
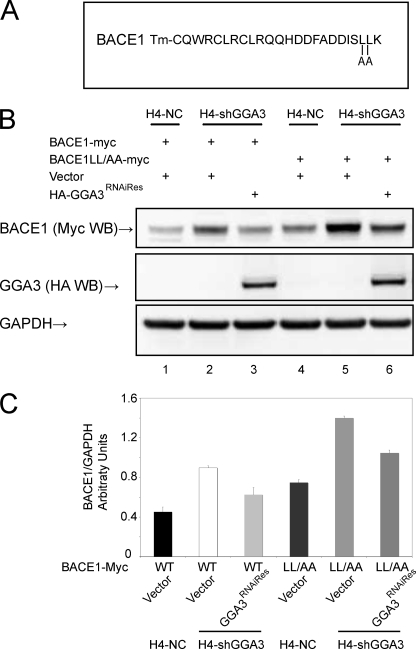
To confirm these findings, a BACE1 mutant in which Leu-499 and Leu-500 had been substituted by alanines (BACE1LL/AA-Myc) was also used (Fig. 4A). These substitutions have previously been shown to abrogate the binding between BACE1 C-terminal peptide and the GGA VHS domain (10). Thus, we set out to determine whether the accumulation of BACE1LL/AA in H4-shGGA3 was rescued by the ectopic expression of GGA3RNAiRes. We found that both BACE1 and BACE1LL/AA accumulate in shGGA3 cells when compared with H4-NC cells. Levels of BACE1LL/AA were increased in both H4-NC and H4-shGGA3 cells compared with BACE1 as reported previously (8). However, the ectopic expression of GGA3RNAiRes reduced the levels of BACE1LL/AA (by ∼30%) similarly to that of BACE1 in H4-shGGA3 cells (Fig. 4, B and C). These findings indicate that GGA3 regulates BACE1 levels independently of the interaction between its VHS domain and di-leucine motif in the BACE1 C terminus.
FIGURE 4.
Expression of GGA3 rescues the accumulation of BACE1LL/AA as well as BACE1 wild type in cells depleted of GGA3. A, amino acid sequence of BACE1 C-terminal fragment. Tm stands for transmembrane domain. Amino acid substitutions L499A and L500A are indicated. B, H4-NC and H4-shGGA3 cells were transiently transfected with the indicated expression vectors. Cells were collected at 6 days post-transfection. Levels of BACE1, GGA3, and GAPDH were analyzed by WB using anti-Myc, anti-HA, and anti-GAPDH antibody, respectively. C, graph represents mean ± S.E. of at least three BACE1 level measurements. Densitometry was performed as described in Fig. 1. HA-GGA3RNAiRes was able to rescue the accumulation of wild type BACE1 as well as BACE1LL/AA in H4-shGGA3 cells.
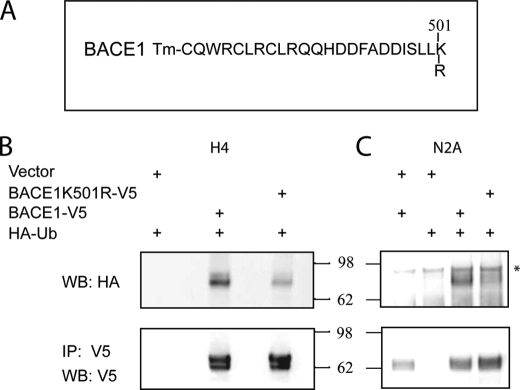
BACE1 Is Ubiquitinated at Lysine 501
Given that GGA3 regulates the degradation of ubiquitinated cargoes, we next set out to determine whether ubiquitin is the signal for BACE1 recruitment in the GGA3 pathway. Ubiquitination is a reversible post-translation modification of cellular proteins. Ub is covalently attached to the ϵ-amino group of lysine residues of the target protein. The C terminus of BACE1 contains a lysine at amino acid position 501 (Fig. 5A). To test whether lysine 501 is a site of ubiquitination, we substituted lysine 501 to arginine (K501R) in a V5-tagged BACE1 expression vector. HA-ubiquitin was cotransfected with empty vector, BACE1-V5, or BACE1K501R-V5 expression vectors in H4-APP751 cells. Cell lysates were immunoprecipitated with anti-V5 antibody. Western blot analysis of the immunoprecipitates with anti-HA antibody revealed that BACE1 is ubiquitinated, but amino substitution K501R greatly reduced BACE1 ubiquitination. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector and HA-ubiquitin as a negative control (Fig. 5B). Similar results were obtained when lysine 501 was substituted to alanine in the BACE1-Myc expression vector (supplemental Fig. S2). To confirm these findings in additional cell lines, murine neuroglioma N2A cells were cotransfected with HA-ubiquitin and BACE1-V5 or BACE1K501R-V5 expression vectors. Cell lysates were immunoprecipitated with anti-V5 antibody. Western blot analysis of the immunoprecipitates with anti-HA antibody revealed that BACE1 is ubiquitinated, but amino substitution K501R greatly reduced BACE1 ubiquitination in N2A cells as well as in H4 cells. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector and HA-ubiquitin or empty vector and BACE1-V5 as negative controls. As a further control, we showed that BACE1 was immunoprecipitated in a similar amount both in cells expressing BACE1-V5 or BACE1K501R-V5 (Fig. 5, B and C).
FIGURE 5.
BACE1 is ubiquitinated, and amino substitution K501R greatly reduces BACE1 ubiquitination. A, amino acid sequence of BACE1 C-terminal fragment. Tm stands for transmembrane domain. Amino acid substitution K501R is indicated. B and C, HA-ubiquitin was cotransfected with empty vector, BACE1-V5, or BACE1K501R-V5 expression vectors in H4-APP751 or N2A cells, respectively. Cells were collected and lysed 24 h post-transfection. BACE1 was immunoprecipitated using anti-V5 antibody. Ubiquitination of BACE1 was assessed by WB analysis of the immunoprecipitates with anti-HA antibody. Please note that ubiquitinated BACE1 has a higher molecular weight because of the presence of Ub. No ubiquitin was detected in immunoprecipitate from cells transfected with empty vector and HA-ubiquitin or empty vector and BACE1-V5. Western blot analysis with anti-V5 antibody revealed that similar amount of BACE1 was immunoprecipitated (IP). BACE1 immunoprecipitates were ubiquitinated, but amino substitution K501R greatly reduced BACE1 ubiquitination. * indicates a nonspecific band.
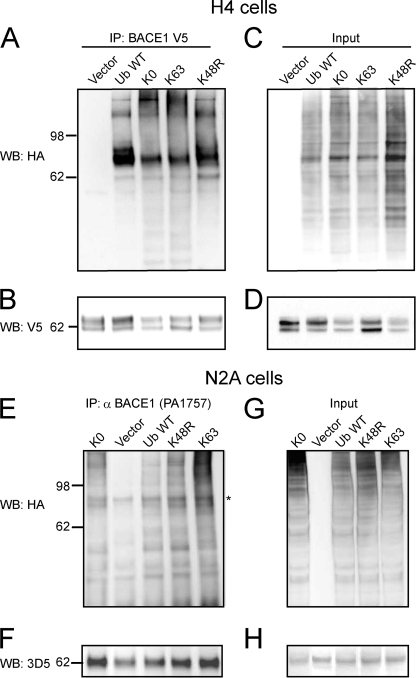
Endogenous and Ectopically Expressed BACE1 Is Mainly Monoubiquitinated and Lys-63-linked Polyubiquitinated
To determine whether BACE1 is monoubiquitinated, Lys-48- and/or Lys-63-linked polyubiquitinated, we employed mutant HA-tagged Ub expression vectors. Monoubiquitination of cellular proteins has been previously assessed by using a mutant Ub in which all seven lysine residues are substituted with arginines (Ub-KO), and thus it is unable to form any Ub chains (39). Lys-48-linked polyubiquitination mainly targets ubiquitinated proteins for proteasomal degradation and can be assessed by using a mutant Ub in which Lys-48 has been substituted to arginine and thus is unable to form polyUb chains at Lys-48 (UbK48R). The lack of ubiquitination following expression of K48R mutant indicates that the target protein is mainly Lys-48-linked ubiquitinated. In contrast, a mutant Ub having all lysines but Lys-63 substituted by arginines will allow only Lys-63-linked ubiquitination of the target protein (35). Thus, H4 cells were cotransfected with BACE1-V5 and empty vector, HA-ubiquitin WT, HA-ubiquitin Lys-63, HA-ubiquitin KO, or HA-ubiquitin K48R. Cell lysates were immunoprecipitated with anti-V5 antibody. Western blot analysis of the immunoprecipitates with anti-HA antibody revealed that BACE1 was ubiquitinated upon expression of Ub WT, Lys-63, KO, or K48R mutant. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector as a negative control (Fig. 6A). As a further control, Western blot analysis of immunoprecipitates with anti-V5 revealed that BACE1 was immunoprecipitated in each sample (Fig. 6B). Western blot analysis of cell lysates (input) with anti-HA or anti-V5 was performed to assess the levels of expressions of BACE1-V5 and HA-Ub WT or mutants (Fig. 6, C and D). Next, we set out to determine whether endogenous BACE1 is ubiquitinated in a pattern similar to ectopically expressed BACE1. N2A cells were transfected with empty vector, HA-ubiquitin WT, HA-ubiquitin Lys-63, HA-ubiquitin KO, or HA-ubiquitin K48R expression vectors. Endogenous BACE1 was immunoprecipitated from cell lysates using a polyclonal anti-BACE1 antibody (PA1-757, Affinity Bioreagents, Golden, CO). Western blot analysis of the immunoprecipitates or cell lysates (input) was performed with anti-HA or monoclonal anti-BACE1 antibody, 3D5, to detect ubiquitin or endogenous BACE1 (Fig. 6, E–G and F–H), respectively. Endogenous BACE1 was ubiquitinated upon expression of Ub WT, Lys-63, KO, or K48R mutants similarly to ectopically expressed BACE1. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector as a negative control. The data indicate that both endogenous and ectopically expressed BACE1 are ubiquitinated not only upon expression of Ub WT but also of a Ub mutant that allows only monoubiquitination (Ub KO), a Ub mutant that allows the formation of only Lys-63-linked polyUb chains, and a Ub mutant that does not allow the formation of Lys-48-linked polyUb chains (K48R). Thus, we have determined that both ectopically expressed and endogenous BACE1, in H4 and N2A cell lines, respectively, are mainly monoubiquitinated and Lys-63-linked ubiquitinated, although Lys-48-linked ubiquitination does not appear to be a major post-translation modification of BACE1.
FIGURE 6.
Endogenous and ectopically expressed BACE1 is mainly monoubiquitinated and Lys-63-linked polyubiquitinated. A, H4 cells were cotransfected with BACE1-V5 and empty vector, HA-ubiquitin wild type (WT), HA-ubiquitin Lys-63, HA-ubiquitin KO, or HA-ubiquitin K48R. Cell lysates were immunoprecipitated (IP) with anti-V5 antibody. Western blot analysis of the immunoprecipitates with anti-HA antibody revealed that BACE1 was ubiquitinated upon expression of Ub WT, Lys-63, KO, or K48R mutant. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector as a negative controls. B, Western blot analysis of immunoprecipitates with anti-V5 revealed that BACE1 was immunoprecipitated in each sample. C and D, Western blot analysis of cell lysates (input) with anti-HA or anti V5 was performed to assess the levels of expressions of BACE1-V5 and HA-Ub WT or mutants. E, N2A cells were transfected with empty vector, HA-ubiquitin wild type (WT), HA-ubiquitin Lys-63, HA-ubiquitin KO, or HA-ubiquitin K48R expression vectors. Endogenous BACE1 was immunoprecipitated from cell lysates using a polyclonal anti-BACE1 antibody (PA1-757, Affinity Bioreagents, Golden, CO). E, Western blot analysis of the immunoprecipitate anti-HA that endogenous BACE1 was ubiquitinated upon expression of Ub WT, Lys-63, KO, or K48R mutant similarly to ectopically expressed BACE1. No ubiquitin was detected in the immunoprecipitates from cells transfected with empty vector as a negative controls. F, Western blot analysis of immunoprecipitates with monoclonal anti-BACE1 antibody, 3D5, revealed that endogenous BACE1 was immunoprecipitated in each sample. G and H, Western blot analysis of cell lysates (input) with anti-HA or anti-3D5 antibody was performed to assess the levels HA-Ub WT or mutants and endogenous BACE1. * indicates a nonspecific band.
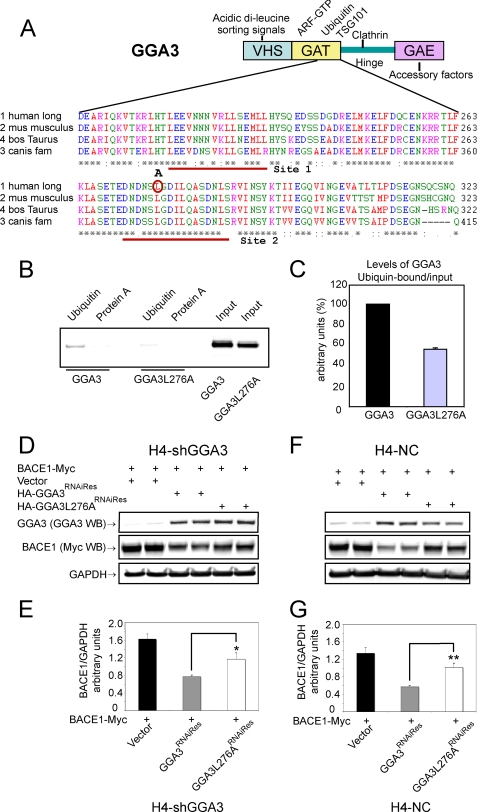
Binding of GGA3 to Ubiquitin Is Necessary to Regulate BACE1 Degradation
It has previously been shown that the amino acid substitution L276A in the GGA3 GAT domain abrogates binding to ubiquitin and TSG101 in a yeast two-hybrid system and that GGA3L276A does not rescue the accumulation of EGFR in early endosomes of mammalian cells depleted of GGA3 (Fig. 7A) (16). Thus, we set out to determine whether GGA3 binding to the ubiquitin sorting machinery is necessary to regulate BACE1 degradation. First, we generated a HA-GGA3L276A mutant by site-directed mutagenesis and tested the ability of GGA3 and GGA3L276A to bind ubiquitin in vitro. H4-APP751 cells were transfected with HA-GGA3 or HA-GGA3L276A expression vectors and cell lysates were incubated with ubiquitin-coated agarose beads or protein A-coated agarose beads as a negative control. As shown previously, GGA3 was able to bind ubiquitin, whereas the amino acid substitution L276A greatly impaired GGA3 binding to ubiquitin (Fig. 7B). GGA3L276A binding to ubiquitin was ∼50% decreased relative to GGA3 binding after normalization to the amount of GGA3 or GGA3L276A present in the input (Fig. 5C). It is important to emphasize that Puertollano and Bonifacino (16) found that this mutation abrogates the binding of GGA3 to ubiquitin and Tsg101 employing a GGA3 VHS-GAT truncated construct as bait in a yeast two-hybrid system. In contrast, we assessed the Ub binding of the endogenous and whole GGA3 protein immunopurified from mammalian cells. Our data are consistent with the presence of two distinct ubiquitin-binding sites in the GAT domain of GGA3 (31–33). Moreover, the solution of the crystal structure of the GGA3 GAT domain with ubiquitin has revealed that ubiquitin binds to a hydrophobic and acidic patch on helices α1 and α2 of the GAT three-helix bundle and that site 1 is the primary binding site for ubiquitin (32). Given that changes in conformation can strongly affect protein binding to ubiquitin, it possible that GGA3/Ub binding can be affected by the presence of the other domains of GGA3, which may produce a change in conformation of the Ub-binding hydrophobic patch both directly and/or via interaction with additional binding partners resulting in the decrease in the Ub binding that we observed.
FIGURE 7.
Binding of GGA3 to ubiquitin is necessary to regulate BACE1 degradation. A, schematic representation of GGA3 domains and multiple sequence alignment of human (NP_619525), M. musculus (NP_766636), B. taurus (XP_587687) and C. familiaris (XP_540429) GGA3 GAT domain (amino acids 204–323). Ubiquitin site 1 and 2 are underlined. Leucine at position 276 was substituted by alanine (A) and is indicated by the red circle. B, in vitro ubiquitin binding assay. H4-APP751 cells were transiently transfected with the indicated expression vectors. Cell lysates were incubated overnight at 4 °C with protein A-agarose beads (as negative control) or with ubiquitin-agarose beads. Western blot analysis with anti-HA antibody detected GGA3 protein bound to beads and in the cell lysates used as input. C, graph represents mean ± S.E. of at least three GGA3 or GGA3L276A level measurements. Densitometry was performed as described in Fig. 1. Levels of ubiquitin-bound GGA3 were normalized against levels of GGA3 in input. D–F, H4-shGGA3 and H4-NC cells were transiently transfected with the indicated expression vectors. Cells were collected at 6 days post-transfection. Duplicate samples were analyzed. Levels of BACE1, GGA3, and GAPDH were analyzed by WB using anti-Myc, anti-GGA3, and anti-GAPDH antibody, respectively. E, graph represents mean ± S.E. of at least 14, 17, and 12 BACE1 level measurements in H4-shGGA3 cells expressing empty vector, HA-GGA3RNAiRes, or HA-GGA3L276ARNAiRes, respectively. Densitometry was performed as described in Fig. 1. BACE1 accumulation was reverted by the expression of HA-GGA3RNAiRes but not by the expression of HA-GGA3L276ARNAiRes in H4-shGGA3 cells (HA-GGA3RNAiRes versus HA-GGA3L276ARNAiRes unpaired t test with Welch correction *, p = 0.02). G, graph represents mean ± S.E. of at least 16, 20, and 11 BACE1 levels measurements in H4-NC cells expressing empty vector, HA-GGA3RNAiRes, or HA-GGA3L276ARNAiRes, respectively. The ectopic expression of HA-GGA3RNAiRes in H4-NC cells resulted in GGA3 overexpression and reduced levels of BACE1 compared with vector alone (E). However, the overexpression of HA-GGA3L276ARNAiRes failed to down-regulate BACE1 in H4-NC cells (HA-GGA3RNAiRes versus HA-GGA3L76ARNAiRes unpaired t test with Welch correction **, p = 0.001).
Next, we tested the ability of GGA3L276A to rescue the accumulation of BACE1 in H4-shGGA3 cells. BACE1-Myc expression vector was cotransfected with an empty vector, HA-GGA3RNAiRes, or HA-GGA3L276ARNAiRes expression vector in H4-shGGA3 and H4-NC cells. Western blotting with an anti-GGA3 antibody revealed that levels of endogenous GGA3 are decreased in H4-shGGA3 compared with H4-NC cells expressing empty vector because of RNAi-mediated down-regulation. Instead, protein levels of HA-GGA3RNAiRes and HA-GGA3L276ARNAiRes were comparable in both cell lines (Fig. 7, D and F, respectively). BACE1 accumulation was reversed by the expression of HA-GGA3RNAiRes but not by the expression of HA-GGA3L276ARNAiRes in H4-shGGA3 cells (HA-GGA3RNAiRes versus HA-GGA3L76ARNAiRes unpaired t test with Welch correction *, p = 0.02) (Fig. 7, D and E). The ectopic expression of HA-GGA3RNAiRes in H4-NC cells resulted in GGA3 overexpression and reduced levels of BACE1 compared with vector alone (Fig. 7, F and G). However, the overexpression of HA-GGA3L276ARNAiRes failed to down-regulate BACE1 in H4-shGGA3 cells (HA-GGA3RNAiRes versus HA-GGA3L276ARNAiRes unpaired t test with Welch correction **, p = 0.001). These data indicate that GGA3 binding to the ubiquitin sorting machinery is necessary for the regulation of BACE1 levels.
DISCUSSION
Our major finding is the identification of a novel post-translational mechanism by which GGA3 level tightly and inversely regulates levels and activity of BACE1. We demonstrate here that, unexpectedly, direct binding of GGA3 VHS domain to BACE1 via the di-leucine motif is not necessary for this regulation. Instead, GGA3 interaction with ubiquitin is essential for regulating BACE1 levels.
Our previous studies demonstrated that RNAi-mediated depletion of GGA3 results in increased levels of BACE1 and Aβ in vitro (34). Furthermore, we determined that depletion of GGA3 naturally occurs following caspase activation both in cellular models of apoptosis and in a rodent model of stroke. More importantly, we discovered that GGA3 protein levels were significantly decreased in the temporal cortex of AD brains samples compared with nondemented control. In contrast, BACE1 levels were significantly increased and inversely correlated with GGA3 levels in the AD group but not in the nondemented control group (34). Accordingly, we report here that although GGA3 depletion increases BACE1 levels, the overexpression of GGA3 reduces levels of BACE1 and Aβ1–40 in vitro.
We then determined that GGA3 is necessary for the trafficking of BACE1 to the lysosomes where it is normally degraded (9). We have shown that BACE1 accumulates in early endosomes in H4 cells depleted of GGA3, whereas BACE1 is transported to lysosomes in control cells.
Because GGAs have been shown to bind the acidic di-leucine motif in the BACE1 C terminus, we assessed whether this binding is necessary for GGA3-mediated regulation of BACE1. Surprisingly, we found that mutations in the GGA3 VHS domain or in the BACE1 di-leucine motif, which are able to abrogate their binding, did not affect the ability of ectopically expressed GGA3 to rescue BACE1 accumulation in H4 cells depleted of GGA3.
Having ruled out that the VHS/di-leucine interaction is required for GGA3-mediated regulation of BACE1 and given that GGA3 regulates the degradation of ubiquitinated cargoes, we next set out to determine whether ubiquitin is the signal for BACE1 recruitment in the GGA3 pathway. We found that BACE1 is ubiquitinated at lysine 501, indicating that BACE1 can bind the GGA3 GAT domain via ubiquitin. Moreover, we have determined that both endogenous and ectopically expressed BACE1 is mainly monoubiquitinated and Lys-63-linked polyubiquitinated. Finally, we determined that the binding of GGA3 to ubiquitin is necessary to regulate BACE1 levels by demonstrating that a GGA3 mutant with reduced ability to bind ubiquitin (GGA3L276A) is unable to regulate BACE1 levels both in rescue and overexpression experiments.
Our findings are in agreement with increasing evidence showing that GGAs bind ubiquitin and traffic both synthetic and endosomal ubiquitinated cargoes to lysosomes (16–20). In further support of a role for ubiquitin versus VHS/di-leucine motif in the recruitment of cargoes in the GGA pathway, the amino acid residues in the GGA VHS domain necessary to bind the di-leucine motif are not conserved in Saccharomyces cerevisiae (38). Moreover, yeast GGAs traffic cargo proteins that do not contain the acidic di-leucine motif (15). Ubiquitin is a sorting signal for membrane proteins at the TGN, plasma membrane, and endosomes to be delivered to lysosomes (6, 40). MVB cargoes are monoubiquitinated, multiubiquitinated, and Lys-63-linked polyubiquitinated. Lysine 48-linked chains can also lead to lysosomal delivery but less efficiently than Lys-63-linked chains (23). Increasing evidence is accumulating that Lys-63-linked ubiquitin chains are a specific signal for protein sorting into the MVB pathway (25). More importantly, it has recently been shown that trafficking of the yeast membrane protein Gap1 to MVB required both its Lys-63-linked ubiquitination and the yeast Gga GAT domain suggesting that Ggas recognize Lys-63-linked Ub chains (19). While this manuscript was under review, Ren and Hurley (41) reported that the VHS domain of Hrs, STAM, GGAs and other trafficking molecules binds Lys-63-linked tetra-Ub chains 50-fold more tightly than monoubiquitin, although only 2-fold more than Lys-48-linked tetraubiquitin.
Our present findings showing that BACE1 is monoubiquitinated and polyubiquitinated via Lys-63-linked Ub chains further support our previous observations that BACE1 is degraded via the lysosomal pathway (9) and a role for GGA3 in the delivery of BACE1 to the MVB/lysosomes pathway. In contrast, Lys-48-linked ubiquitination, the main signal for proteasomal degradation, is negligible in BACE1 supporting our previous observation that inhibition of lysosomal hydrolases but not of the proteasome results in accumulation of endogenous levels of BACE1 in a variety of cell lines and in primary neuronal cultures (9). A previous study reported that ectopically expressed BACE1 is ubiquitinated and that BACE1 accumulates following inhibition of the proteasome (42). Although we confirm and extend the observation by Qing et al. (42) regarding BACE1 ubiquitination, our previous (9) and current data do not support a major role for the proteasome in the degradation of BACE1. Given that caspase activation results in BACE1 stabilization (34), the accumulation of BACE1 observed by Qing et al. (42) following treatment with proteasome inhibitors is most likely due to the induction of apoptosis associated with proteasome inhibition (43). More recently, Zhang et al. (44) have reported that UBB+1, a mutant ubiquitin that consists of a ubiquitin moiety and a 19-amino acid C-terminal extension, accumulates following cerebral ischemia and binds to BACE1 in a caspase-dependent fashion. It has been shown that excessive UBB+1 becomes an endogenous inhibitor of the proteasome, and once a protein is tagged by UBB+1, it becomes resistant to proteasomal degradation (45). We have previously reported that caspase activation results in GGA3 cleavage and depletion resulting in elevation of BACE1 levels following experimental cerebral ischemia (34). Although we cannot rule out that the inhibition of the proteasome by UBB+1 may play a role in BACE1 stabilization in stress conditions, an alternative explanation could be that under stress conditions UBB+1 binds to BACE1 preventing its normal ubiquitination, and as a result, BACE1 trafficking to MVBs and lysosomes is impaired.
Our findings that GGA3-mediated regulation of BACE1 is independent of VHS/di-leucine motif interaction can help to explain the differential effects that mutation of the di-leucine sorting motif and depletion that GGAs has on BACE1 trafficking and activity. He et al. (37) have demonstrated that depletion of GGA1–3 results in BACE1 accumulation in early endosomes but not at the cell membrane, excluding a role for GGAs in BACE1 internalization. Instead mutagenesis of the di-leucine motif impairs BACE1 internalization and results in retention of BACE1 at the cell membrane (7–9, 37). Interestingly, He et al. (37) have observed that GGA3 but not GGA1 partially colocalizes with BACE1LL/AA suggesting the presence of alternative binding sites for GGA3 and BACE1. Moreover, they have also shown that mutation of the BACE1 di-leucine motif has no effect on Aβ production, although we have reported that depletion of GGA3 increases Aβ levels (34).
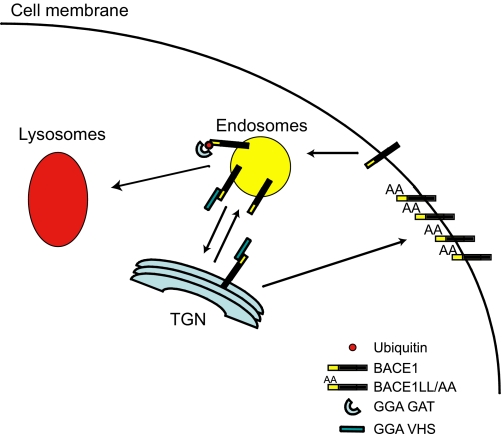
Taking all these findings together, we can hypothesize that GGAs may play a dual role in the trafficking of BACE1. It is possible that GGAs traffic BACE1 as well as MPRs from the TGN to early endosomes (37). Then BACE1 is transported from the endosomes to the cell membrane, where it can be internalized to the endosomes again. At this point, GGAs can transport BACE1 back to the TGN (12) or sort it to the lysosomes for degradation (Fig. 8). Thus, depletion of GGAs interrupts both routes resulting in the accumulation of BACE1 in early endosomes. Given that a low pH is necessary for BACE1 activity, the accumulation of BACE1 in these acidic organelles results in increased β-secretase activity and Aβ production in cells depleted of GGA3 (34). When the VHS/di-leucine motif binding is abrogated because of the LL/AA mutation, BACE1 is transported directly to the cell membrane escaping the endosomes. Because the internalization of the LL/AA mutant is impaired, BACE1 accumulates at the membrane where β-secretase activity may be impaired and as a consequence Aβ levels do not increase (37).
FIGURE 8.
Role of GGAs in BACE1 trafficking. Schematic representation of BACE1 intracellular trafficking mediated by GGAs.
Thus, we can hypothesize that VHS/di-leucine binding regulates the cycling of BACE1 from the TGN to the endosomes and vice versa. Instead, the binding of ubiquitinated BACE1 to the GGA3 GAT domain regulates the sorting of synthetic and endosomal BACE1 to lysosomes for degradation. Further studies will be necessary to determine whether GGAs play different roles in the trafficking of BACE1, e.g. GGA3 targeting ubiquitinated BACE1 for degradation and GGA1/2 cycling BACE1 between TGN and endosomes. Several sources of evidence support a unique role for GGA3 in the regulation of ubiquitinated cargo degradation. Depletion of GGA3, but not GGA1 and -2, impairs the degradation of EGFR (16). Among three human GGAs (GGA1–3), ubiquitin binds most strongly to GGA3 (16, 17, 46). It has been shown previously that, unlike GGA1-GAT and GGA2-GAT, the GAT domain of GGA3 does not bind to Rabaptin-5, which in complex with Rabex-5, a guanine nucleotide exchange factor, participates in endosomal tethering/fusion events (47). More recently, Ren and Hurley (41) reported that among GGAs only GGA3VHS domain binds ubiquitin, supporting a specific role for GGA3 in the trafficking of ubiquinated cargoes to MVBs.
Given that the subcellular localization of BACE1 affects β-secretase activity, factors that regulate BACE1 trafficking may represent novel therapeutic targets for the treatment of AD. In addition to GGAs, the reticulon/Nogo family of proteins has also been shown to interact with BACE1 and regulate β-secretase activity. The overexpression of RTN3 results in the retention of BACE1 in the endoplasmic reticulum and the reduction of Aβ production both in vitro and in vivo (48, 49).
Because levels of BACE1 are elevated in AD brains and they are inversely correlated with GGA3 levels (34), our studies suggest that therapies able to increase GGA3 expression in the brain may represent a potential treatment for AD.
Supplementary Material
Acknowledgments
We thank Drs. Stephen Moss and Michele Jacob for helpful comments on this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01AG025952-04 and 1R01AG033016-01A1 (to G. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- AD
- Alzheimer disease
- APP
- β-amyloid precursor protein
- Aβ
- amyloid β-protein
- GGA
- Golgi-localized γ-ear-containing ARF binding proteins
- VHS
- VPS27, Hrs, and STAM domain
- GAT
- GGA and Tom1 domain
- GAE
- γ-adaptin ear homology domain
- MPR
- mannose 6-phosphate receptor
- TGN
- trans-Golgi network
- EGFR
- epithelial growth factor receptor
- shRNA
- short hairpin RNA
- ELISA
- enzyme-linked immunosorbent assay
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HA
- hemagglutinin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RNAi
- RNA interference
- WT
- wild type
- WB
- Western blot
- MVB
- multivesicular body
- NC
- negative control
- KO
- knock-out.
REFERENCES
- 1.De Strooper B., Annaert W. (2000) J. Cell Sci. 113, 1857–1870 [DOI] [PubMed] [Google Scholar]
- 2.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 3.Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 4.Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., Carter D. B., Tomasselli A. G., Parodi L. A., Heinrikson R. L., Gurney M. E. (1999) Nature 402, 533–537 [DOI] [PubMed] [Google Scholar]
- 5.Vassar R., Kovacs D. M., Yan R., Wong P. C. (2009) J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino J. S., Traub L. M. (2003) Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 7.Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 8.Pastorino L., Ikin A. F., Nairn A. C., Pursnani A., Buxbaum J. D. (2002) Mol. Cell. Neurosci. 19, 175–185 [DOI] [PubMed] [Google Scholar]
- 9.Koh Y. H., von Arnim C. A., Hyman B. T., Tanzi R. E., Tesco G. (2005) J. Biol. Chem. 280, 32499–32504 [DOI] [PubMed] [Google Scholar]
- 10.He X., Chang W. P., Koelsch G., Tang J. (2002) FEBS Lett. 524, 183–187 [DOI] [PubMed] [Google Scholar]
- 11.He X., Zhu G., Koelsch G., Rodgers K. K., Zhang X. C., Tang J. (2003) Biochemistry 42, 12174–12180 [DOI] [PubMed] [Google Scholar]
- 12.Wahle T., Prager K., Raffler N., Haass C., Famulok M., Walter J. (2005) Mol. Cell. Neurosci. 29, 453–461 [DOI] [PubMed] [Google Scholar]
- 13.von Arnim C. A., Tangredi M. M., Peltan I. D., Lee B. M., Irizarry M. C., Kinoshita A., Hyman B. T. (2004) J. Cell Sci. 117, 5437–5445 [DOI] [PubMed] [Google Scholar]
- 14.Shiba T., Kametaka S., Kawasaki M., Shibata M., Waguri S., Uchiyama Y., Wakatsuki S. (2004) Traffic 5, 437–448 [DOI] [PubMed] [Google Scholar]
- 15.Bonifacino J. S. (2004) Nat. Rev. Mol. Cell Biol. 5, 23–32 [DOI] [PubMed] [Google Scholar]
- 16.Puertollano R., Bonifacino J. S. (2004) Nat. Cell Biol. 6, 244–251 [DOI] [PubMed] [Google Scholar]
- 17.Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. (2004) Nat. Cell Biol. 6, 252–259 [DOI] [PubMed] [Google Scholar]
- 18.Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauwers E., Jacob C., André B. (2009) J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Y., Guo Y., Watson H., Au W. C., Shakoury-Elizeh M., Basrai M. A., Bonifacino J. S., Philpott C. C. (2009) J. Biol. Chem. 284, 23830–23841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longva K. E., Blystad F. D., Stang E., Larsen A. M., Johannessen L. E., Madshus I. H. (2002) J. Cell Biol. 156, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 23.Davies B. A., Lee J. R., Oestreich A. J., Katzmann D. J. (2009) Chem. Rev. 109, 1575–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 25.Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 26.Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 27.Bache K. G., Brech A., Mehlum A., Stenmark H. (2003) J. Cell Biol. 162, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bache K. G., Raiborg C., Mehlum A., Stenmark H. (2003) J. Biol. Chem. 278, 12513–12521 [DOI] [PubMed] [Google Scholar]
- 29.Bishop N., Horman A., Woodman P. (2002) J. Cell Biol. 157, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7626–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilodeau P. S., Winistorfer S. C., Allaman M. M., Surendhran K., Kearney W. R., Robertson A. D., Piper R. C. (2004) J. Biol. Chem. 279, 54808–54816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prag G., Lee S., Mattera R., Arighi C. N., Beach B. M., Bonifacino J. S., Hurley J. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawasaki M., Shiba T., Shiba Y., Yamaguchi Y., Matsugaki N., Igarashi N., Suzuki M., Kato R., Kato K., Nakayama K., Wakatsuki S. (2005) Genes Cells 10, 639–654 [DOI] [PubMed] [Google Scholar]
- 34.Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., Tanzi R. E. (2007) Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sena-Esteves M., Tebbets J. C., Steffens S., Crombleholme T., Flake A. W. (2004) J. Virol. Methods 122, 131–139 [DOI] [PubMed] [Google Scholar]
- 37.He X., Li F., Chang W. P., Tang J. (2005) J. Biol. Chem. 280, 11696–11703 [DOI] [PubMed] [Google Scholar]
- 38.Misra S., Puertollano R., Kato Y., Bonifacino J. S., Hurley J. H. (2002) Nature 415, 933–937 [DOI] [PubMed] [Google Scholar]
- 39.Terrell J., Shih S., Dunn R., Hicke L. (1998) Mol. Cell 1, 193–202 [DOI] [PubMed] [Google Scholar]
- 40.Piper R. C., Luzio J. P. (2007) Curr. Opin. Cell Biol. 19, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren X., Hurley J. H. (2010) EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qing H., Zhou W., Christensen M. A., Sun X., Tong Y., Song W. (2004) FASEB J. 18, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 43.Hoeller D., Dikic I. (2009) Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Xiong M., Yan R. Q., Sun F. Y. (2010) J. Cereb. Blood Flow Metab. 30, 566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Tijn P., de Vrij F. M., Schuurman K. G., Dantuma N. P., Fischer D. F., van Leeuwen F. W., Hol E. M. (2007) J. Cell Sci. 120, 1615–1623 [DOI] [PubMed] [Google Scholar]
- 46.Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., Kobayashi H., Shin H. W., Wakatsuki S., Nakayama K. (2004) J. Biol. Chem. 279, 7105–7111 [DOI] [PubMed] [Google Scholar]
- 47.Mattera R., Arighi C. N., Lodge R., Zerial M., Bonifacino J. S. (2003) EMBO J. 22, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He W., Lu Y., Qahwash I., Hu X. Y., Chang A., Yan R. (2004) Nat. Med. 10, 959–965 [DOI] [PubMed] [Google Scholar]
- 49.Shi Q., Prior M., He W., Tang X., Hu X., Yan R. (2009) J. Neurosci. 29, 9163–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.