Abstract
The formation of primitive adipose tissue is the initial process in adipose tissue development followed by the migration of preadipocytes into adipocyte clusters. Comparatively little is known about the molecular mechanism controlling preadipocyte migration. Here, we show that cytohesin-2, the guanine-nucleotide exchange factor for the Arf family GTP-binding proteins, regulates migration of mouse preadipocyte 3T3-L1 cells through Arf6. SecinH3, a specific inhibitor of the cytohesin family, markedly inhibits migration of 3T3-L1 cells. 3T3-L1 cells express cytohesin-2 and cytohesin-3, and knockdown of cytohesin-2 with its small interfering RNA effectively decreases cell migration. Cytohesin-2 preferentially acts upstream of Arf6 in this signaling pathway. Furthermore, we find that the focal adhesion protein paxillin forms a complex with cytohesin-2. Paxillin colocalizes with cytohesin-2 at the leading edges of migrating cells. This interaction is mediated by the LIM2 domain of paxillin and the isolated polybasic region of cytohesin-2. Importantly, migration is inhibited by expression of the constructs containing these regions. These results suggest that cytohesin-2, through a previously unexplored complex formation with paxillin, regulates preadipocyte migration and that paxillin plays a previously unknown role as a scaffold protein of Arf guanine-nucleotide exchange factor.
Keywords: Adipocyte, Cell Migration, Cytoskeleton, Exchangers, Protein-Protein Interactions, Cytohesin, Paxillin
Introduction
Adipose tissue plays a key role in energy storage and in the secretion of regulatory lipid hormones such as leptin, adiponectin, and tumor necrosis factor-α (1–3). The initial stage of adipocyte differentiation begins with the proliferation of multipotent mesenchymal stem cells, which then enter the first stage of differentiation into preadipocytes. These fibroblast-like immature adipocytes still have proliferative activity; they migrate to various sites to undergo further adipogenesis. They eventually enter into the second differentiation stage, during which they become spherical mature adipocytes containing droplets of fat. In contrast to the normal development of adipose tissue, chronic overnutrition of the adipose tissue leads to macrophage infiltration, resulting in local inflammation that potentiates insulin resistance (4). Many signaling molecules, especially those for transcriptional regulatory proteins, participate in the process of adipocyte differentiation and in the formation of pathological conditions (5), but the intracellular signals responsible for cellular morphological changes and for the migration of preadipocytes before initiation of their differentiation into adipocytes remain largely unknown.
Cell migration is essential for many aspects of developmental processes and requires several spatially and temporally coordinated changes in cytoskeletons and cellular membranes. Dynamic structures critical to cell migration include filopodia, lamellipodia, and actin stress fibers. The cellular morphological changes involved in cell migration are regulated by Rho family GTPases (6), the ADP-ribosylation factors (Arfs), and the other small GTP-binding proteins. Arf proteins primarily regulate intracellular vesicular transport to control endocytic and secretory pathways and may be involved in morphological changes. In mammalian cells the Arf family consists of five or six isoforms (7, 8) that are grouped into three classes: class I (Arf1 and Arf2 and/or Arf3), class II (Arf4 and Arf5), and class III (Arf6). As this classification suggests, Arf6 is unique in important ways. It can regulate peripheral actin cytoskeleton as well as vesicle transport by controlling lipid-modifying enzymes such as phosphatidylinositol-4-phosphate 5-kinases and phospholipase D isozymes and other downstream effectors (8–10).
Like other small GTP-binding proteins, Arfs act as molecular switches and are active when bound to GTP and inactive when bound to GDP. Guanine-nucleotide exchange factors (GEFs)3 catalyze the intrinsically slow exchange reaction of GDP with free cytoplasmic GTP to generate active conformations, whereas GTPase-activating proteins (GAPs) inactivate GTP-binding proteins. The former reaction is a very important rate-limiting step, as GEFs define the specificity of small GTP-binding protein activation by integrating intracellular signals. Arf GEFs are divided into five families based on overall structure and domain organization (11, 12): Golgi brefeldin A (BFA)-resistance factor 1/BFA-inhibited GEF (GBF/BIG) (13), Arf nucleotide binding site opener (ARNO)/cytohesin, exchange factor for Arf6 (EFA6) (14, 15), BFA-resistant Arf GEF (BRAG) (16), and F-box only protein 8 (FBX8) (17). The large number of GEFs relative to the number of Arf proteins suggests that GEF activities are controlled under extensive regulatory conditions in various types of cells.
Paxillin is cytoskeletal adaptor/scaffold protein involved in the dissemination of signals from extracellular cell adhesion proteins such as integrins and growth factor receptors (18, 19). Paxillin contains only protein-protein interaction domains: five leucine-rich repeat (LD) domains and four lin-11, isl-1, mec-3 (LIM) domains. Through these, paxillin intracellularly mediates interactions with actin cytoskeleton regulatory proteins. Thus, paxillin is crucial in changing the morphology of cells and promoting cell migration (18, 20–23); for example, it binds to regulator and effector complexes of Rho GTPases and regulates their spatiotemporal actions. These facts suggest that there may be additional links between paxillin and small GTP-binding proteins and/or their regulatory proteins.
In this study we report that the cytohesin-2 and Arf6 signaling unit mediates migration of mouse preadipocyte 3T3-L1 cells. Furthermore, we find that paxillin directly involves cytohesin-2 and Arf6 in cell migration before the initiation of adipocyte differentiation, adding a unique binding partner to the components of the cellular paxillin complex.
EXPERIMENTAL PROCEDURES
Antibodies
The antibodies used were as follows: mouse monoclonal anti-Arf1 and anti-Arf6 (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-paxillin and mouse monoclonal anti-actin (1:1,000; BD Biosciences), mouse monoclonal anti-GFP (1:1,000; MBL, Nagoya, Japan), rabbit polyclonal anti-RFP (1:1,000; Evrogen, Moscow, Russia), mouse monoclonal anti-FLAG and rabbit polyclonal anti-DDDDK (1:1,000; MBL), and goat anti-mouse or rabbit IgG antibody conjugated with horseradish peroxidase (1:10,000; GE Healthcare).
Plasmids
The regions encoding full-length mouse cytohesin-1 (GenBankTM accession number NM 001112699) and mouse cytohesin-2 (GenBankTM accession number NM 001112701) were amplified from cytohesin-1 cDNA (24, 25) and 3T3-L1 cell cDNA, respectively, by the PCR method. Cytohesin-3 (GenBankTM accession number NM 011182) cDNA was obtained from the Biological Resource Center of the National Institute of Technology and Evaluation (Chiba, Japan). The PCR products were ligated into the mammalian FLAG-tagged expression vector p3×FLAG-myc-CMVTM-24 (Sigma). Each domain (coiled-coil (CC), amino acids 1–59; Sec7 domain, amino acids 60–254; pleckstrin homology (PH) domain plus C-terminal polybasic region (PH+b), amino acids 255–400; isolated PH domain, amino acids 255–386; C-terminal polybasic region, amino acids 387–400) was inserted into the p3×FLAG and pEGFP vectors. The pEGFP-paxillin and the pTurboRFP-paxillin LD1, LD2, LD3, LD4/5, LIM1, LIM2 (amino acids 381–440) and LIM3/4 plasmids were constructed as described previously (26). The ΔLIM2 was produced by the overlapping PCR method and subcloned into pTurboRFP-C1 vectors. The region encoding Golgi-localizing γ-adaptin ear homology domain (GGA) 3 (amino acids 1–343), which specifically binds to the GTP-bound, active form of Arf6 was amplified by the RT-PCR method from total RNA of human brain cells and subcloned into the Escherichia coli GST-tagged expression vector pET42a (Merck). All nucleotide sequences were confirmed by the Fasmac sequencing service (Kanagawa, Japan).
Recombinant Proteins
Recombinant GST-GGA3 and GST-paxillin LIM2 were purified using E. coli BL21 (DE3) pLysS (Takara Bio Inc., Otsu, Japan) according to the manufacturer's protocols. The transformed E. coli cells were treated with 0.4 mm isopropyl-1-thio-β-d-galactopyranoside at 30 °C for 2.5 h and were harvested through centrifugation. The precipitates were extracted with buffer A (50 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, 1 mm EDTA, and 0.5% Nonidet P-40) containing 500 μg/ml lysozyme and 100 μg/ml DNase on ice. All purification steps were performed at 4 °C. The cell lysate was centrifuged at 150,000 × g for 30 min. The supernatant was applied to glutathione-Sepharose 4B (GE Healthcare), and the resin was washed with buffer B (100 mm Tris-HCl (pH 8.0), 2 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, and 1 μg/ml leupeptin). GST-GGA3 and GST-paxillin LIM2 were eluted with column buffer B containing 20 mm glutathione (Nacalai Tesque, Kyoto, Japan). The eluted fraction was dialyzed against buffer C (10 mm HEPES-NaOH (pH 7.5), 1 mm dithiothreitol, 2 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, and 150 mm NaCl) and stored at −80 °C.
FLAG-tagged cytohesin-2 PH+b protein was purified from 293T cells transiently transfected with p3×FLAG-cytohesin-2 PH+b using the CalPhos transfection reagent (Clontech, Mountain View, CA) according to the manufacturer's protocols (23). In brief, cells were lysed in lysis buffer A and centrifuged. The supernatant was mixed with protein G resin (GE Healthcare) that was preadsorbed with an anti-FLAG antibody. Bound FLAG-tagged PH+b protein was extensively washed with lysis buffer A containing 500 mm NaCl and subsequently with lysis buffer A containing 500 mm NaCl and 50 mm EDTA and eluted with lysis buffer A containing 20 mm FLAG peptide (Sigma) according to the manufacturer's protocols. The buffer contained in elution fractions was exchanged with reaction buffer (20 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, and 1 mm EDTA). The aliquot was stored at −80 °C until use.
siRNA Oligonucleotides
The 21-nucleotide siRNA duplexes were synthesized by Nippon EGT (Toyama, Japan). The specific target sequences were as follows: 5′-AAGAGCTAAGTGAGCTATGA-3′ for mouse cytohesin-2 siRNA, 5′-AAGAAAAAAGGAACTTATTGA-3′ for mouse cytohesin-3 siRNA, 5′-AAGAATATCAGCTTCACCGTG-3′ for mouse Arf1 siRNA, 5′-AAGTTCAACGTGTGGGATGTG-3′ for mouse Arf6 siRNA, and 5′-AAGAGCACGTCTACAGCTTCC-3′ for mouse paxillin siRNA. The target sequence of the control Photinus pyralis luciferase siRNA was 5′-AAGCCATTCTATCCTCTAGAG-3′, which does not have significant homology to any mammalian gene sequences.
PCR Primers
The DNA primers were synthesized by the Fasmac oligonucleotide service (Kanagawa, Japan). The primers used were as follows: 5′-ATGGAGGACGATGACAGCTATGTC-3′ (sense) and 5′-TCAGTGTCTCTTTGTGGAGGAGAC-3′ (antisense) for mouse cytohesin-1; 5′-ATGGAGGACGGTGTCTACGAG-3′ (sense) and 5′-TCAGGGTTGTTCTTGCTTCTTCTTCAC-3′ (antisense) for mouse cytohesin-2; 5′-ATGGACGAAGGCGGTGGCGGTG-3′ (sense) and 5′-CTATTTATTGGCAATCCTCCTTTTCCTCGTGGCCAAC-3′ (antisense) for mouse cytohesin-3; 5′-ATGGATGTGTGTCACACAGATCCAG-3′ (sense) and 5′-CTACTTGCCGACAATCTTCTTTTTCCGA-3′ (antisense) for mouse cytohesin-4; 5′-CTGGATGCTGCAGGGAAGACAAC-3′ (sense) and 5′-CTGAATGTACCAGTTCCTGTGGCGT3′ (antisense) for mouse Arf1; 5′-ATGGGCAATATCTTTGGGAACCTTCTGAAG-3′ (sense) and 5′-TAGCATTCGGCAAATCCTGTTTGTTTGCAAAC-3′ (antisense) for mouse Arf3; 5′-TCGGCAAGAAGCAGATGCGCATTTTG-3′ (sense) and 5′-CTGCAGACCTAGCTTGTCTGTCATCT-3′ (antisense) for mouse Arf4; 5′-GAAGTTGGGGGAGATTGTCACCAC-3′ (sense) and 5′-ACAGCCAGTCCAGCCCATCATACA-3′ (antisense) for mouse Arf5; 5′-AGTGCTATCCAAGATCTTCGGGAACAAG-3′ (sense) and 5′-CTGGATCTCATGGGGTTTCATGGCA-3′ (antisense) for mouse Arf6. The control primers for mouse β-actin were 5′-ATGGATGACGATATCGCTGCGCTC-3′ (sense) and 5′-CTAGAAGCATTTGCGGTGCACGATG-3′ (antisense).
RT-PCR
Total RNA was extracted from 3T3-L1 cells using a Trizol reagent (Invitrogen). The cDNAs were prepared from 1 μg of total RNA with Superscript III (Invitrogen) according to the manufacturer's instructions. PCR amplification was performed with ExTaq polymerase (Takara Bio) in 30 cycles, each cycle consisting of denaturation at 94 °C for 1 min, annealing at 58.5–61.5 °C (depending on the primer pair's Tm value) for 1 min, and extension at 72 °C for 1 min.
Cell Culture
Unless otherwise described, mouse preadipocyte 3T3-L1 cells and human embryonic kidney 293T cells were cultured at 37 °C in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, 50 units/ml penicillin, and 50 mg/ml streptomycin. For the migrating assay, cells were preincubated with or without 10 μm cytohesin inhibitor SecinH3 (Compound ID 1029232; CAS no. 853625-60-2; Merck) in the presence of 10 μm AraC. Less than 5% of cells incorporated trypan blue after at least 10 h of culture in the presence of 10 μm SecinH3 and/or 10 μm AraC.
Plasmid Transfection
For 3T3-L1 cells, the plasmids encoding fluorescence proteins were transfected using the Lipofectamine 2000 reagent (Invitrogen) or the cell line Amaxa Nucleofector Kit (Lonza Group Ltd, Basel, Switzerland), each according to its manufacturer's protocols. For lipofection and electroporation, the medium was replaced 4 and 24 h after transfection, respectively. Unless otherwise indicated, cells were cultured in medium containing fetal bovine serum for an additional 24 or 44 h. For 293T cells, plasmid DNAs were transfected into cells using the CalPhos transfection reagent (Clontech). The fluorescent images were captured using an Eclipse TE-300 microscope system (Nikon, Tokyo, Japan) and analyzed with AxioVision software (Carl Zeiss, Oberkochen, Germany).
siRNA Transfection
3T3-L1 cells were transfected with siRNA oligonucleotides using a CrediaTF reagent (Credia, Kyoto, Japan) according to the manufacturer's protocols. The medium was replaced 4 h after transfection, and cells were cultured in medium containing fetal bovine serum for an additional 44 h.
Immunofluorescence
Cultured cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), blocked with 20% heat-inactivated fetal bovine serum in PBS, 0.05% Tween 20, incubated with each primary antibody for 12 h at 4 °C, and treated with fluorescence-labeled secondary antibodies and/or fluorescence-labeled phalloidin (Invitrogen) in PBS, 0.1% Tween 20 for 0.5 h at room temperature. The coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA) onto slides for confocal microscopic observation. The confocal images were collected with an IX81 microscope with a Laser-Scanning FV500 System (Olympus, Tokyo, Japan) and analyzed with FluoView software (Olympus) and Adobe Photoshop (contrast value, 50; Adobe, San Jose, CA).
Evaluation of Localization on the Plasma Membrane and the Focal Adhesion Region in the Cytoplasm
The cellular localizations of cytohesin-2, paxillin, vinculin, and actin were determined as described previously (27, 28). The localization of each of these molecules was evaluated by means of fluorescence intensity profiling of a typical line scan using ImageJ 1.42q software.
Immunoblotting
Cells were lysed in lysis buffer B (50 mm HEPES-NaOH (pH 7.5), 20 mm MgCl2, 150 mm NaCl, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, 1 mm EDTA, 1 mm Na3VO4, 10 mm NaF, and 0.5% Nonidet P-40), and the lysates were centrifuged at 14,000 rpm for 10 min at 4 °C. The proteins in the supernatants were denatured in Laemmli buffer (0.4 m Tris-HCl (pH 6.8), 0.2 m dithiothreitol, 0.2% bromphenol blue, 4% SDS) and then subjected to SDS-PAGE. The electrophoretically separated proteins were transferred to polyvinylidene difluoride membranes, blocked with a Blocking One reagent (Nacalai Tesque), and immunoblotted first with each primary antibody and then with peroxidase-conjugated secondary antibodies. The bound antibodies were detected using ECL or ECL-Plus reagent (GE Healthcare).
Immunoprecipitation
Cells were lysed with lysis buffer B. The cell extracts were mixed with protein G-Sepharose CL-4B (GE Healthcare) absorbed with the primary antibody. The immune complexes were precipitated by means of centrifugation and washed three times with lysis buffer B. The immunoprecipitates were boiled in sample buffer and then separated on SDS-polyacrylamide gels. The bound proteins were detected through immunoblotting.
In Vitro Arf1 and Arf6 Activity
3T3-L1 cells were plated on 60-mm dishes and stimulated after being scratched. The Arf1 and Arf6 activities were measured as described (28, 29). Briefly, cells were lysed in ice-cold lysis buffer. Samples were incubated for 10 min at 4 °C and spun at 14,000 rpm for 10 min at 4 °C. GST-GGA3 protein coupled to glutathione-Sepharose 4B was added to each supernatant, and samples were rotated at 4 °C for 1 h. Proteins were eluted by being heated to 95 °C in Laemmli buffer (0.4 m Tris-HCl (pH 6.8), 0.2 m dithiothreitol, 0.2% bromphenol blue, 4% SDS) for 5 min, subjected to SDS-PAGE, and immunoblotted.
Wound Closure Migration Assay
3T3-L1 cells were cultured in 35- or 60-mm dishes and incubated at 37 °C for 0, 6, or 10 h with or without SecinH3 in the presence of 10 μm AraC. The monolayers of confluent cells were scraped with the narrow end of a micropipette tip to generate wounds ∼50 μm in width (30). The rate of wound healing was quantified as follows: rate of healing (%) = (width of wound at 0 h minus width of wound at 0, 6, or 10 h/width of wound at 0 h) × 100.
In Vitro Binding Assay
Immobilized recombinant cytohesin-2 PH+b protein was mixed with recombinant GST-paxillin LIM2 for 2 h at 4 °C. The complex was washed three times with lysis buffer B, subjected to SDS-PAGE, and immunoblotted with an anti-GST antibody. The Kd value was calculated according to a Scatchard analysis.
Statistical Analysis
Values shown represent the means ± S.D. of data from separate experiments. ANOVA was followed by Fisher's protected least significant difference post hoc comparison (*, p < 0.01).
RESULTS
Involvement of the Cytohesin Family in Migration of Mouse Preadipocyte 3T3-L1 Cells
Preadipocytic cell lines such as the 3T3-L1 line are very useful in vitro models for studying hormonal regulation of adipocyte differentiation and metabolism (31). The signaling mechanisms underlying preadipocyte migration, however, remain largely unknown. In an attempt to clarify these, we searched for endogenous intracellular proteins that can regulate actin cytoskeleton/focal adhesion assembly and turnover. In this search we used SecinH3, a newly identified inhibitor specific to the cytohesin family of GEFs for Arf proteins, on 3T3-L1 cells (32, 33).
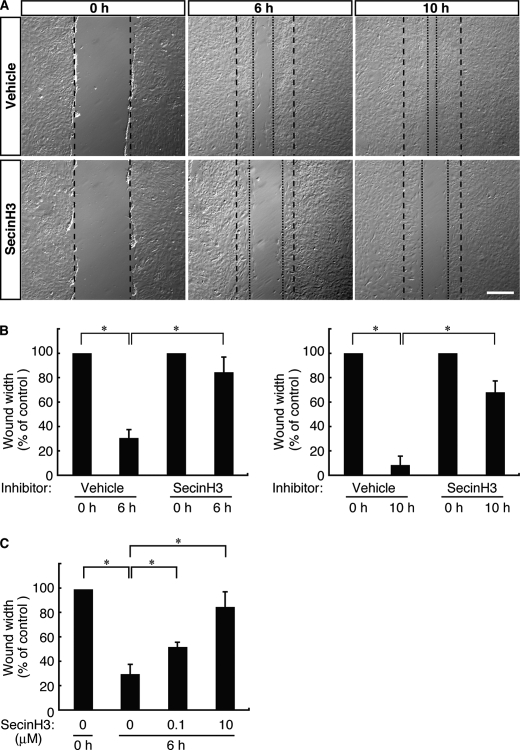
In our wound healing assay the movement of 3T3-L1 cells into artificial wounds was measured at 0, 6, and 10 h in the presence or absence of SecinH3 (10 μm (32, 33)). As shown in Fig. 1, A and B, the migration distances traveled by control cells over periods of 6 and 10 h were 43 ± 3 and 57 ± 5 μm, respectively, whereas those of SecinH3-treated cells were 16 ± 4 and 23 ± 2 μm. At both 6 and 10 h, SecinH3-treated cells had traveled approximately half as far as control cells had. This phenomenon is likely due to changes in the rate of cell migration rather than to alterations in cell proliferation, considering the fact that the experimental period was only 12 h. The experiment was deliberately kept short to minimize the potential effects of proliferation in the presence of AraC, an anti-mitotic reagent. AraC treatment did not exert a significant effect on the incorporation of trypan blue (less than 5% of treated cells incorporated trypan blue).
FIGURE 1.
SecinH3, a cytohesin family inhibitor, inhibits migration of 3T3-L1 cells. A, in the wound healing assay, 3T3-L1 cells were pretreated with or without SecinH3 (10 μm). 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 0, 6, and 10 h. The position of the representative healing edge of migrating cells at 0 h is indicated by a dashed line, and those at 6 and 10 h are indicated by dotted lines. The scale bar indicates 100 μm. B, the rate of healing was calculated based on the width of the wound at 6 h (left panel) and 10 h (right panel). C, 3T3-L1 cells were pretreated with or without SecinH3 (0.1 and 10 μm) and cultured for 10 h. Data were evaluated with one-way ANOVA (*, p < 0.01).
The effect of SecinH3 on cell migration was dependent on its concentration (Fig. 1C), suggesting that this cytohesin family GEF regulates 3T3-L1 cell migration through certain signaling pathways. Less than 5% of cells incorporated trypan blue under treatment with 0, 0.1, and 10 μm SecinH3. These results suggest that the cytohesin-Arf protein signaling unit may regulate 3T3-L1 migration.
The cytohesin family is composed of cytohesin-1, cytohesin-2/ARNO, cytohesin-3/Grp1/ARNO3, and cytohesin-4 (11). To determine which of these cytohesins is responsible for the inhibitory effect of SecinH3, we performed RT-PCR using total RNA of 3T3-L1 cells. RT-PCR analysis showed that 3T3-L1 cells express cytohesin-2, cytohesin-3, and, weakly, cytohesin-1. Any expression of cytohesin-4 was below the detection level (supplemental Fig. S1). Because the cytohesin family is the GEF for Arf proteins, we next sought to determine which Arf protein is expressed in 3T3-L1 cells. RT-PCR analysis showed that all Arf proteins exist in 3T3-L1 cells (supplemental Fig. S2), indicating that 3T3-L1 cells possess the cytohesin-2/3-Arf protein signaling unit and that this unit may be responsible for the ability of SecinH3 to inhibit migration.
Cytohesin-2 Regulates Cell Migration through Arf6 in 3T3-L1 Cells
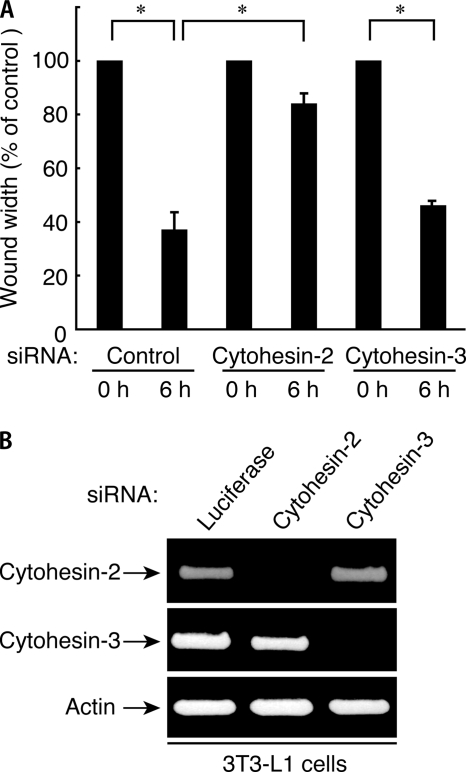
Because cytohesin-2 and cytohesin-3 are primarily expressed in 3T3-L1 cells, both or either may regulate cell migration. Accordingly, we transfected siRNA oligonucleotides specific for cytohesin-2, cytohesin-3, and control luciferase into cells. The expression levels of cytohesin-2 and cytohesin-3 were specifically and markedly reduced by transfection with siRNAs for cytohesin-2 and cytohesin-3, respectively, whereas the expression level of actin was unaffected, as revealed by RT-PCR (Fig. 2B). In the wound healing assay, we observed that knockdown of cytohesin-2 significantly inhibited migration after 6 h. In contrast, knockdown of cytohesin-3 did not have an obvious inhibitory effect on migration (Fig. 2A). These results suggest that cytohesin-2 is the primary mediator of the migration of 3T3-L1 cells.
FIGURE 2.
Suppression of cytohesin-2, but not cytohesin-3, inhibits migration of 3T3-L1 cells. A, in the wound healing assay 3T3-L1 cells were transfected with an siRNA for control (luciferase), cytohesin-2, or cytohesin-3. After siRNA treatment, 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 0 and 6 h. The rate of healing was calculated based on the width of the wound. Data were evaluated with one-way ANOVA (*, p < 0.01). B, RT-PCR of mouse cytohesin-2, cytohesin-3, and actin (as the control) was performed using total RNA of 3T3-L1 cells treated with each type of siRNA.
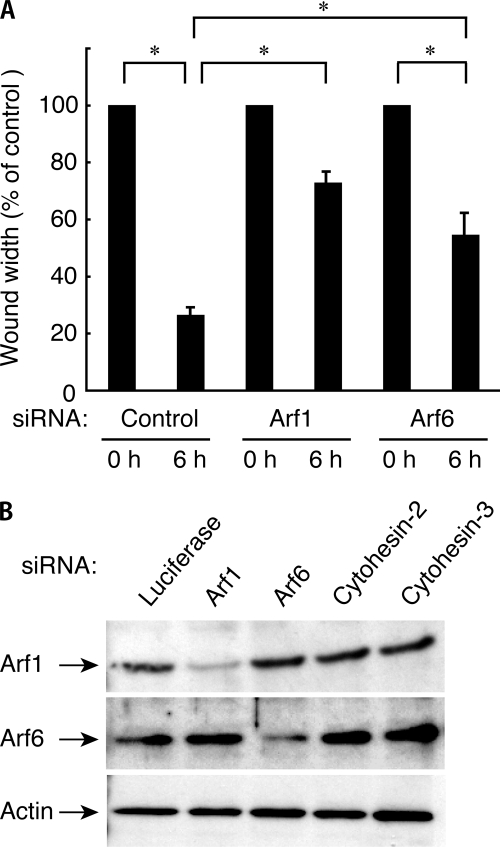
Cytohesin-2 as a GEF is preferential for Arf1 and Arf6 (11). Accordingly, we tested whether the suppression of Arf1 or Arf6 by each isoform siRNA had an effect on cell migration. The expression levels of Arf1 and Arf6 were specifically reduced by transfection with Arf1 siRNA and Arf6 siRNA, respectively, whereas the expressions of other proteins were unaffected, as revealed by immunoblotting analysis (Fig. 3B). Knockdown of Arf1 or Arf6 inhibited migration (Fig. 3A), suggesting that Arf1 and Arf6 may act downstream of cytohesin-2.
FIGURE 3.
Suppression of Arf1 or Arf6 decreases wound closure of 3T3-L1 cells. A, in the wound healing assay 3T3-L1 cells were transfected with an siRNA for control (luciferase), Arf1, or Arf6. 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 0 and 6 h. The rate of healing was calculated based on the width of the wound. Data were evaluated with one-way ANOVA (*, p < 0.01). B, cell lysates were immunoblotted with an antibody against Arf1, Afr6, or actin.
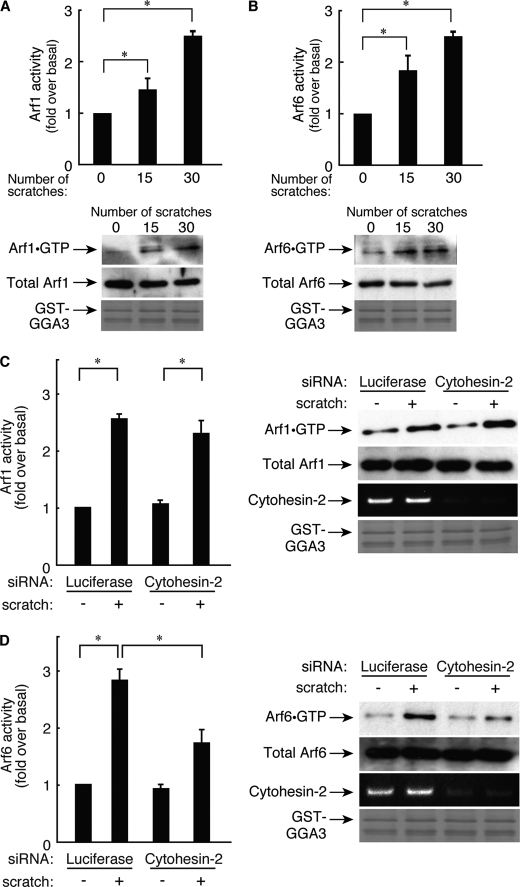
We next examined bona fide Arf protein activity in 3T3-L1 cells. We performed affinity precipitation using the recombinant GST-tagged N-terminal domain of GGA3, which binds to active GTP-bound forms of Arf1 and Arf6. Endogenous active Arf1 and Arf6 were present in a scratch-number-dependent manner 6 h after scratching (Fig. 4, A and B).
FIGURE 4.
Cytohesin-2, acting through Arf6 but not through Arf1, regulates migration of 3T3-L1 cells. A, after 0- 30 scratches, cells were stimulated for 6 h, and Arf1 activity levels were measured. Cells were lysed then affinity-precipitated with a recombinant GST-GGA3 (200 ng) and immunoblotted with an anti-Arf1 antibody. Total Arf1 was detected using an anti-Arf1 antibody. In a parallel experiment, applied GST-GGA3 was stained with Coomassie Brilliant Blue. B, after 0–30 scratches, cells were stimulated for 6 h, and Arf6 activity levels were measured. Cells were lysed and affinity-precipitated with a recombinant GST-GGA3 and immunoblotted with an anti-Arf6 antibody. Total Arf6 was detected using an anti-Arf6 antibody. In a parallel experiment, applied GST-GGA3 was stained with Coomassie Brilliant Blue. C, cells were transfected with an siRNA for control (luciferase) or for cytohesin-2. After 0 or 30 scratches, cells were incubated for 6 h, and Arf1 activity levels were measured, respectively. Cells were lysed and affinity-precipitated with a recombinant GST-GGA3 and immunoblotted with the corresponding antibody. Using aliquots of total lysates, total Arf1 was detected with anti-Arf1 antibody. To confirm the effect of cytohesin-2 siRNA on its knockdown specificity, RT-PCR of cytohesin-2 was performed using total RNA of 3T3-L1 cells that had been treated with each siRNA. In a parallel experiment, applied GST-GGA3 was stained with Coomassie Brilliant Blue. Data were evaluated with one-way ANOVA (*, p < 0.01). D, cells were transfected with an siRNA for control (luciferase) or for cytohesin-2. After 0 or 30 scratches, cells were incubated for 6 h, and Arf6 activity levels were measured, respectively. Cells were lysed and affinity-precipitated with a recombinant GST-GGA3 and immunoblotted with the corresponding antibody. Using aliquots of total lysates, total Arf6 were detected with anti-Arf6 antibody. To confirm the effect of cytohesin-2 siRNA on its knockdown specificity, RT-PCR of cytohesin-2 was performed using total RNA of 3T3-L1 cells that had been treated with each siRNA. In a parallel experiment, applied GST-GGA3 was stained with Coomassie Brilliant Blue. Data were evaluated with one-way ANOVA (*, p < 0.01).
Next, we confirmed that Arf1 and Arf6 are regulated by cytohesin-2. Knockdown of cytohesin-2 inhibited scratch-induced Arf6 activation (Fig. 4D) but not Arf1 activation (Fig. 4C). These results indicate that cytohesin-2 is the primary upstream regulator for Arf6 in this signaling pathway.
Interaction of Cytohesin-2 with Paxillin in 3T3-L1 Cells
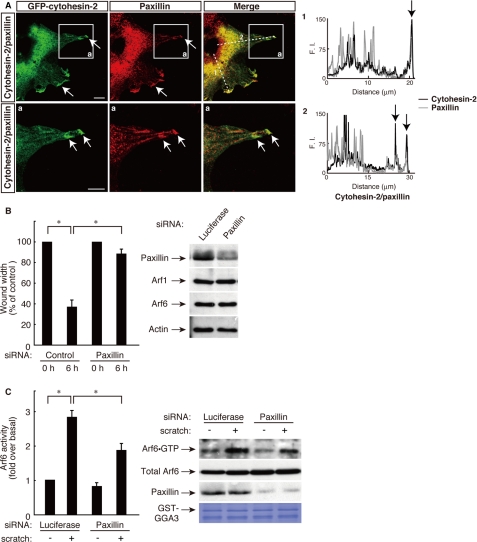
Cell migration is a complex process consisting of multiple cell signaling steps, each of which requires cytoskeletal recognition and the coordination of membrane trafficking. The migration process is controlled by the regulated activities of many proteins; one of these is paxillin, which modulates multipotent associations between various signaling molecules and cytoskeletal proteins (18, 19). Previously, we studied the relationship between paxillin and small GTP-binding protein signals in cell morphological changes (20–22, 26). These studies prompted us to investigate whether cytohesin-2 interacts with paxillin in 3T3-L1 cell migration. As shown in Fig. 5A, immunofluorescence labeling revealed that GFP-tagged cytohesin-2 was partially colocalized with paxillin in the cell peripheral regions at 6 h after scratching. These colocalization data are consistent with the finding that paxillin localizes to a greater degree in membrane-ruffling surfaces or cell peripheral regions in migrating cells (34, 35). In addition, knockdown of paxillin inhibited migration of 3T3-L1 cells (Fig. 5B) and scratch-induced Arf6 activation (Fig. 5C). The siRNA knockdown of paxillin slightly decreased the basal Arf6 activity. This reason may be because paxillin is involved in regulation of the network of signaling molecules, including small GTP-binding proteins. The expression level of paxillin was specifically reduced by transfection with siRNA for paxillin, whereas the expression levels of other proteins were unaffected, as revealed by immunoblotting. Thus, cytohesin-2, acting through paxillin, may regulate Arf6 in 3T3-L1 cells.
FIGURE 5.
Cytohesin-2, acting through Arf6, cooperates with paxillin at the leading edge of migrating 3T3-L1 cells. A, the plasmid encoding GFP-tagged cytohesin-2 was electroporated into 3T3-L1 cells, and cells were immunostained using anti-GFP antibody and anti-paxillin antibody 6 h after scratching. Representative confocal images of GFP-tagged cytohesin-2 (green) and endogenous paxillin (red) were collected with a confocal microscope system. Staining intensities measured according to pixel brightness were evaluated through a fluorescence intensity (F.I.) profile of a typical line scan (lines 1 and 2). The lower panels (a) are high magnification images of the areas boxed with white lines in the corresponding upper panels. The scale bar indicates 50 μm. B and C, 3T3-L1 cells were transfected with an siRNA for control (luciferase) or paxillin. B, cell monolayers were wounded by scraping and observed microscopically at 0 and 6 h. The rate of healing was calculated based on the width of the wound. C, after 0 or 30 scratches, the Arf6 GTP form was measured with GST-GGA3, respectively. Cell lysates were immunoblotted with an antibody against paxillin, small GTP-binding protein, or actin. Data were evaluated with one-way ANOVA (*, p < 0.01).
The C-terminal Polybasic Region of Cytohesin-2 Is Required for Binding to the LIM2 Domain of Paxillin
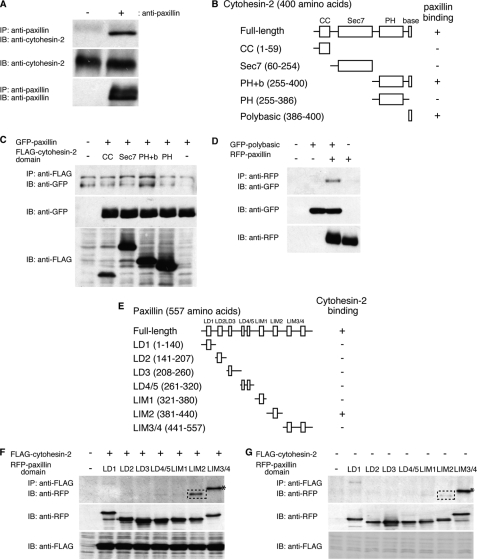
Because cytohesin-2 partially colocalizes and interacts with paxillin in migrating 3T3-L1 cells, we assessed the direct interaction between paxillin and cytohesin-2. We first performed co-immunoprecipitation of endogenous paxillin with endogenous cytohesin-2 from 3T3-L1 cell lysates and found that cytohesin-2 formed an immune complex (Fig. 6A). It is possible that in 3T3-L1 cells cytohesin-2 may be the primary partner of paxillin among the cytohesin family.
FIGURE 6.
Specific interaction of cytohesin-2 with paxillin in cells. A, endogenous paxillin was immunoprecipitated from 3T3-L1 cell lysates using anti-paxillin antibody and immunoblotted with an anti-cytohesin-2 antibody. The total lysates were also used for immunoblotting with an anti-paxillin antibody or an anti-cytohesin-2 antibody. B, the schematic structures of full-length cytohesin-2 and its domains are illustrated. C, 293T cells were transfected with plasmids coding either FLAG-tagged cytohesin-2 domain (CC, Sec7, PH+b, PH) or GFP-tagged full-length paxillin. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with an anti-GFP antibody. The total lysates were also used for immunoblotting with an anti-FLAG antibody or anti-GFP antibody. D, 293T cells were transfected with plasmids coding GFP-tagged isolated C-terminal polybasic region of cytohesin-2 and RFP-tagged full-length paxillin. After 48 h, cells were lysed, immunoprecipitated with an anti-RFP antibody, and immunoblotted with an anti-GFP antibody. The total lysates were also used for immunoblotting with an anti-GFP antibody or anti-RFP antibody. E, the schematic structures of full-length paxillin, and its domains are illustrated. F and G, 293T cells were transfected with plasmids coding both FLAG-tagged full-length cytohesin-2 and one of the RFP-tagged paxillin domains. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with an anti-RFP antibody. The total lysates were also used for immunoblotting with an anti-FLAG antibody or anti-RFP antibody. The squares with dotted lines indicate specific binding (F) compared with the negative control that does not express FLAG-cytohesin-2 (G). Asterisks indicate nonspecific binding. IP, immunoprecipitation; IB, immunoblotting.
Next, to determine which domain of cytohesin-2 interacts with paxillin, we constructed four FLAG-tagged truncated mutants of cytohesin-2: the CC domain (amino acids 1–59), the catalytic Sec7 domain (amino acids 60–254), the phosphoinositide binding PH domain plus C-terminal polybasic region (PH+b) (amino acids 255–400), and the isolated PH domain (amino acids 255–386) (Fig. 6B). We found that the FLAG-tagged PH+b domain of cytohesin-2 was responsible for interaction with GFP-paxillin (Fig. 6C). Interestingly, the cytohesin-2 PH+b domain also binds to Arl4D, the other type of Arf protein, whereas the isolated PH domain does not (28, 36); in a similar manner, the interaction between cytohesin-2 and paxillin may require the C-terminal polybasic region in addition to the PH domain. Thus, we tested the possibility that the isolated C-terminal polybasic region binds paxillin. As shown Fig. 6D, the GFP-tagged isolated polybasic region of cytohesin-2 weakly coimmunoprecipitated with RFP-tagged paxillin, suggesting that it is the minimum interaction site with paxillin.
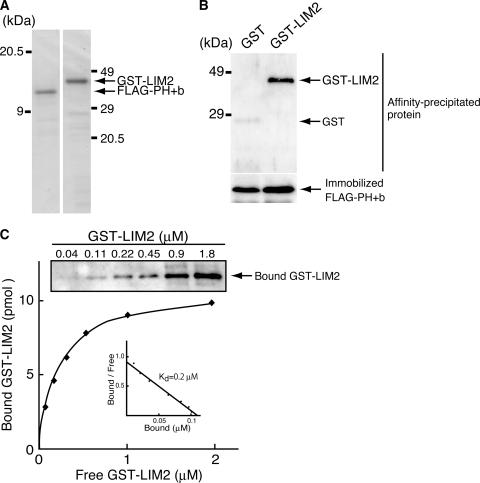
To determine which region of paxillin interacts with cytohesin-2, we performed co-immunoprecipitation of RFP-tagged LD1 (amino acids 1–140), LD2 (amino acids 141–207), LD3 (amino acids 208–260), LD4/5 (amino acids 261–320), LIM1 (amino acids 321–380), LIM2 (amino acids 381–440), or LIM3/4 (amino acids 441–557) (Fig. 6E) with FLAG-tagged cytohesin-2. The RFP-tagged LIM2 domain of paxillin co-immunoprecipitated with FLAG-tagged cytohesin-2 (Fig. 6F) as a comparison with the negative control without FLAG-tagged cytohesin-2 transfection shows (Fig. 6G). We further checked the direct interaction of paxillin with cytohesin-2 using recombinant proteins. The recombinant GST-tagged paxillin LIM2 domain was produced in E. coli, and the FLAG-tagged cytohesin-2 PH+b domain was produced in 293T cells; each protein was purified for further examination (Fig. 7A). The GST-tagged paxillin LIM2 domain with the FLAG-tagged cytohesin-2 PH+b domain exhibited a specific band, whereas the sample with GST protein and the FLAG-tagged cytohesin-2 PH+b domain exhibited no band (Fig. 7B).
FIGURE 7.
The cytohesin-2 PH+b domain directly interacts with the paxillin LIM2 domain in vitro. A, purified recombinant GST-tagged paxillin LIM2 domain (1 μg, left panel) and FLAG-tagged cytohesin-2 PH+b domain (1 μg, right panel) were stained with Coomassie Brilliant Blue. B, 1 μm FLAG-tagged cytohesin-2 PH+b domain was mixed for 2 h at 4 °C with 1 μm concentrations of either immobilized GST or GST-tagged paxillin LIM2 domain and GST. Affinity-precipitated GST or GST-tagged LIM2 domain was subjected to SDS-PAGE followed by immunoblotting with anti-GST antibody (upper panel). The LIM2 domain specifically bound to the PH+b domain. Immobilized FLAG-tagged cytohesin-2 PH+b domain was stained with Coomassie Brilliant Blue (lower panel). C, quantitative analysis of the interaction between FLAG-tagged cytohesin-2 PH+b domain and GST-tagged paxillin LIM2 domain was performed. Various concentrations (0.04–1.8 μm) of recombinant GST-tagged paxillin LIM2 domain were added to immobilized FLAG-tagged cytohesin-2 PH+b domain (1 μm), and each sample was washed and subjected to SDS-PAGE followed by immunoblotting with anti-GST antibody (upper panel). The amount of bound versus free GST-tagged paxillin LIM2 is plotted. Scatchard analysis indicated that the Kd was ∼0.2 μm (inset).
To assess the interaction between E. coli produced, GST-tagged paxillin LIM2, and the 293T cell-produced FLAG-tagged cytohesin-2 PH+b domain before further analysis, we performed an in vitro binding assay. The dissociation constant (Kd) was ∼0.2 μm (Fig. 7C), indicating that the association of paxillin with cytohesin-2 is direct and significant. This association constant is comparable with that of the GIT1 paxillin binding domain with paxillin LD2 (25.1 ± 0.2 μm) or LD4 motifs (7.0 ± 0.2 μm) (38). Together with the results shown in Figs. 6 and 7, this indicates that cytohesin-2 binds to paxillin and that the interaction occurs through the PH+b domain and the LIM2 domain.
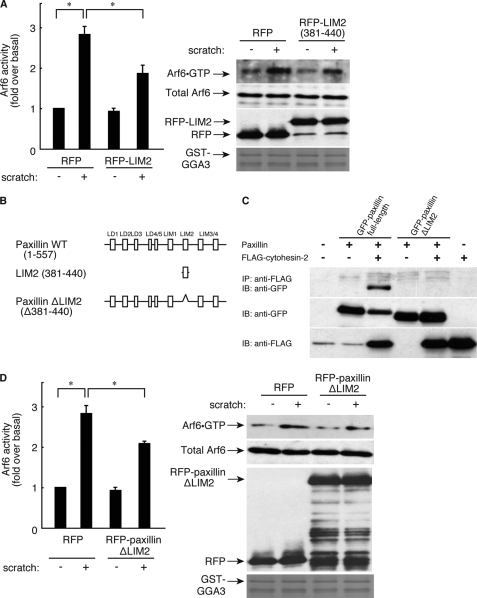
The LIM2 Domain of Paxillin Plays an Important Role in Arf6 Activation
We next checked whether the LIM2 domain of paxillin is required for Arf6 activation. Transfection with the plasmid encoding the isolated LIM2 domain (381–440) (Fig. 8B) into 3T3-L1 cells inhibited scratch-induced Arf6 activation (Fig. 8A), as did transfection with the LIM2 domain-deficient paxillin mutant (Fig. 8, B and D), which has no ability to bind to cytohesin-2 (Fig. 8C). These results further support the idea that the LIM2 domain structurally participates in the binding of paxillin to cytohesin-2 and the regulation of Arf6 GEF activity in cells.
FIGURE 8.
Role of paxillin LIM2 domain in interaction with cytohesin-2. A, 3T3-L1 cells were electroporated with RFP and RFP-paxillin LIM2 domain. After 0 or 30 scratches, cells were incubated for 6 h, and Arf6 activity was measured, respectively. Cells were lysed, affinity-precipitated with GST-GGA3, and immunoblotted with the corresponding antibody. B, the schematic structures of paxillin wild type, isolated paxillin LIM2 domain, and paxillin ΔLIM2 domain are shown. C, 293T cells were transfected with various combinations of plasmids of pEGFP-paxillin, pEGFP-paxillin ΔLIM2 domain, and p3×FLAG-cytohesin-2. After 48 h, cells were lysed, and supernatants were incubated with an anti-FLAG antibody and collected with protein G-Sepharose CL-4B. After washing the resin with ice-cold lysis buffer, bound proteins as well as total lysate were separated by SDS-PAGE and detected by Western blotting with antibodies against FLAG tag and GFP-tag. IP, immunoprecipitation; IB, immunoblotting. D, 3T3-L1 cells were electroplated with RFP and RFP-paxillin ΔLIM2 domain. After 0 or 30 scratches, cells were incubated for 6 h, and Arf6 activity was measured, respectively. Cells were lysed, affinity-precipitated with GST-GGA3, and immunoblotted with the corresponding antibody.
Migration of 3T3-L1 Cells Is Meditated through the Region Containing the Polybasic Amino Acid Sequence of Cytohesin-2 and the LIM2 Domain of Paxillin
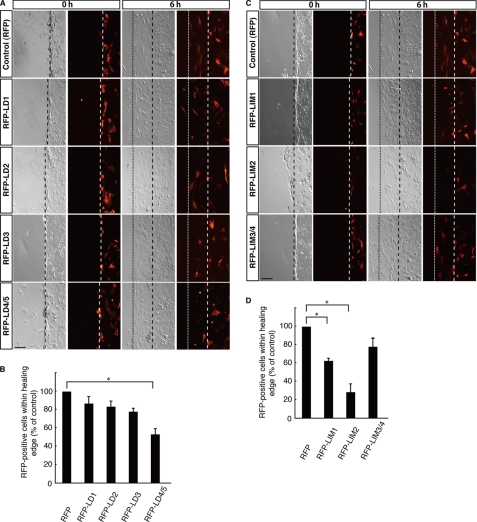
To investigate whether paxillin interacts functionally with cytohesin-2, we electroporated plasmids encoding each RFP-tagged domain of paxillin into 3T3-L1 cells. The number of migrating RFP-positive cells was counted 6 h after scratching and compared with the number of migrating cells expressing only control RFP. Cells within the healing area were inhibited by transfection with the RFP-tagged LD4/5, LIM1, and LIM2 domains of paxillin (Fig. 9, A–D) but not by transfection with the RFP-tagged LD1, LD2, or LD3 domains (Fig. 9, A and B). The RFP-tagged LIM3/4 domain modestly inhibited migration (Fig. 9, C and D). These inhibitory effects exhibited by domains other than the LIM2 domain may be caused by the existence of many other paxillin-interacting proteins (18, 19).
FIGURE 9.
Effects of paxillin domains on 3T3-L1 cell migration. A, in the wound healing assay, a plasmid encoding either RFP or one of the RFP-tagged paxillin domains (LD1-LD4/5) was electroporated into 3T3-L1 cells. 3T3-L1 cell monolayers were wounded by scraping and were observed microscopically at 6 h. Images for phase contrast (left panel) and RFP fluorescent images (right panel) were obtained at 0 and 6 h. The position of the representative healing edge of migrating cells at 0 h is indicated by a dashed line, and that at 6 h is indicated by a dotted line. The scale bar indicates 100 μm. B, based on the images described above in A, the population of RFP-positive cells within each healing area was measured. Values are expressed relative to the healing area of RFP (control)-expressing cells. C, in the wound healing assay, a plasmid encoding RFP or one of the RFP-tagged paxillin domains (LIM1-LIM3/4) was electroporated into 3T3-L1 cells. 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 6 h. Images for phase contrast (left panel) and RFP fluorescent images (right panel) were obtained at 0 and 6 h. The position of the representative healing edge of migrating cells at 0 h is indicated by a dashed line, and that at 6 h is indicated by a dotted line. The scale bar indicates 100 μm. D, based on the images described above in C, the population of RFP-positive cells within each healing area was measured. Values are expressed relative to the healing area of RFP (control)-expressing cells. Data were evaluated with one-way ANOVA (*, p < 0.01).
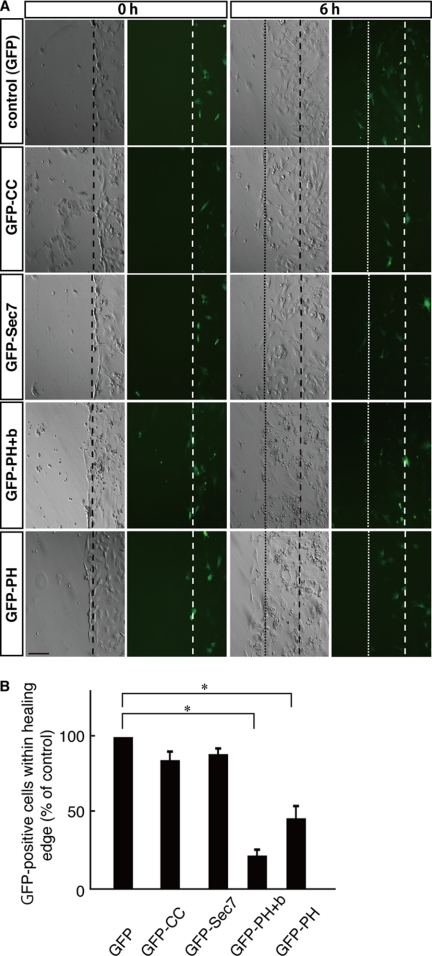
We next transfected plasmids encoding each GFP-tagged domain of cytohesin-2. The number of migrating GFP-positive cells was counted 6 h after scratching and compared with the number of migrating cells expressing only control GFP. Cells within the healing area were inhibited by transfection with the GFP-tagged PH+b domain of cytohesin-2 and modestly by transfection with the isolated PH domain of cytohesin-2 but not by transfection with the GFP-tagged CC or Sec7 domains of cytohesin-2 (Fig. 10, A and B). The PH+b domain is also known to be a minimally functional domain that participates in the interaction of cytohesin-2 with the Arf family small GTP-binding protein Arl4D and that acts upstream of cytohesin-2 in migrating cells (28, 36). Taken together, these data indicate that the paxillin-cytohesin-2 interaction is involved in the migration of 3T3-L1 cells and that this is mediated mostly by the LIM2 domain of paxillin and possibly through the region containing the polybasic amino acid sequence of cytohesin-2.
FIGURE 10.
Effects of cytohesin-2 domains on 3T3-L1 cell migration. A, in the wound healing assay, a plasmid encoding GFP or one of the GFP-tagged cytohesin-2 domains was electroporated into 3T3-L1 cells. 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 6 h. Images for phase contrast (left panel) and GFP fluorescent images (right panel) were obtained at 0 and 6 h. The position of the representative healing edge of migrating cells at 0 h is indicated by a dashed line, and that at 6 h is indicated by a dotted line. The scale bar indicates 100 μm. B, based on the images described above in A, the population of GFP-positive cells within each healing area was measured. Values are expressed relative to the healing area of GFP (control)-expressing cells. Data were evaluated with one-way ANOVA (*, p < 0.01).
DISCUSSION
Preadipocytes differentiate from multipotent mesenchymal stem cells, then migrate to various sites where they undergo adipogenesis into mature adipocytes (3, 39). Although an array of transcription factors are known to control adipocyte differentiation, the mechanism underlying preadipocyte migration remains poorly understood. Here, using the preadipocyte 3T3-L1 cell line as a model, we have discovered a key signaling pathway through Arf protein, the small GTP-binding protein involved in cellular morphological changes essential for cell migration.
A large part of the signaling mechanism underlying 3T3-L1 cell migration is mediated by the Arf GEF cytohesin-2 and Arf6. This conclusion is supported by our observation that inhibition of cytohesin-2 or Arf6 with a pharmacological inhibitor or knockdown of the specific molecule through RNA interference attenuates migration and by our observation that cytohesin-2-dependent Arf6 activity is stimulated in a scratch-dependent manner. Importantly, 3T3-L1 cell migration requires the interaction of paxillin with cytohesin-2. This interaction occurs between the LIM2 domain of paxillin and the C-terminal PH domain in tandem with a short polybasic amino acid sequence of cytohesin-2. Expression of the LIM domain or C-terminal region containing the PH domain in cells results in an inhibitory effect on migration. These results demonstrate that migration of 3T3-L1 cells involves a previously unknown functional signaling unit, namely, the cytoskeletal scaffold protein paxillin-Arf GEF cytohesin-2 complex.
It is clear that cytohesin-2, acting through Arf6, regulates migration of 3T3-L1 cells, but other GEFs for Arf6 may also participate in migration. EFA6 and BRAG family proteins as well as cytohesin family proteins are also GEFs for Arf6 (11). All of these GEFs consist of similar domains organized in a similar manner; the catalytic Sec7 domain is in tandem with the PH domain and the coiled-coil region. The EFA6 family of proteins is composed of four members: EFA6A, EFA6B, EFA6C, and EFA6D. EFA6B and EFA6D are widely expressed in tissues, whereas EFA6A and EFA6C are predominantly expressed in brain tissues (14, 15). The BRAGs include three members, among which BRAG2 shows ubiquitous expression; in contrast, BRAG1 and BRAG3 display primal expression in the brain (16). It is conceivable that EFA6A, EFA6E, and/or BRAG2 could act together with cytohesin-2 in 3T3-L1 cell migration.
It is worth noting that cytohesin family proteins also act as GEFs for Arf1, as migration also involves Arf1. Moreover, 3T3-L1 cells weakly express other cytohesin family proteins in addition to cytohesin-2. These GEFs may act upstream of Arf1 and control migration, at least in part. Alternatively, the well characterized brefeldin A-sensitive GEFs may be upstream regulators of Arf1 in this signaling pathway. In fact, BIG1 mediates migration of HepG2 cells (40), and BIG2 promotes neural progenitor cell migration in the cerebral cortex (41). It will be interesting to study how the GEFs for Arf1 and/or Arf6 cooperatively regulate 3T3-L1 cell migration.
Paxillin plays crucial roles in cytoskeletal organization, cell adhesion, migration, and cellular morphological changes (18, 19). It is a multidomain adaptor or scaffold protein that links extracellular signals from cell adhesion proteins and growth factor receptors to intracellular signaling proteins through direct interaction and/or phosphorylation-dependent association. Herein, we identify the Arf GEF cytohesin-2 as a novel binding partner of paxillin. Paxillin is already known to form a complex with some Arf GAPs, including G protein-coupled receptor kinase-interacting protein 1 (GIT1; also called Cat-1/p95-APP1), GIT2 (also called Cat-2/p95-APP2/PKL), Arf GAP with Src homology 3 domain, ankyrin repeats, and PH domains 2 (ASAP2; also called PAP/AMAP2/DEF1/PAG3) (42, 43). GIT2 displays nearly ubiquitous expression in tissues, whereas GIT1 appears to be expressed only in endothelial cells. GIT1/2 binds to the LD4 domain of paxillin (42). ASAP2, which ubiquitously expresses in tissues, binds to the N-terminal region involving all LD domains (44). Because the paxillin-binding sites of Arf GAPs GIT1/2 and ASAP2 are different from that of cytohesin-2, these Arf GAPs and cytohesin-2 can bind to paxillin simultaneously. Interestingly, GIT1/2 also exists as a tight physiological complex with p21-activated kinase-interacting exchange factor (PIX)/Cool proteins, the GEFs for Rac and Cdc42, members of the Rho family of small GTPases (45). In addition, CdGAP, a GAP for Rho GTPases, interacts with paxillin (46). Cell migration involves dynamic cytoskeletal changes and intracellular membrane traffic and is mediated by organized cycles of on/off switch mechanisms in signaling molecules. Paxillin may provide a platform for small GTP-binding protein regulators, including the GEFs and GAPs for the Arf and Rho proteins; this idea is consistent with our observation that expression of domains other than the LIM2 domain in cells partially inhibits migration of 3T3-L1 cells.
Cytohesin-2 may regulate 3T3-L1 cell migration not only through paxillin but also through other known binding partners. Some cytohesin family binding proteins have been identified and characterized. The coiled-coil domain provides a binding site with other coiled-coil domain-containing proteins, including Grp1 signaling partner (GRSP)1/mKIAA1013 (47), Grp1-associated scaffold protein (GRASP)/tamalin (48, 49), cytohesin-associated scaffold protein (CASP)/Cybr/cytohesin-interacting protein (CYTIP) (50–52), and interaction protein for cytohesin exchange factor (IPCEF) 1/KIAA0403 (53). Although CASP probably interacts specifically with cytohesin-1, GRSP1, GRASP, and IPCEF can bind to most cytohesin proteins. These adaptor-like proteins may be involved in this signaling pathway, although their precise roles in cell migration are not yet thoroughly understood.
It is well established that Arf6 directly activates phosphatidylinositol-4-phosphate 5-kinases and phospholipase D isoenzymes in many types of cells (6, 8, 10). Phosphatidylinositol-4-phosphate 5-kinases and phospholipase D isoenzymes participate in actin cytoskeletal changes that generate phosphoinositides and phospholipids as the products. These lipid-modifying enzymes may, therefore, act as the functional effectors downstream of Arf6 in this signaling pathway. Alternatively, cytohesin-2 and Arf6 may regulate Rac, which contributes to actin cytoskeletal changes. Arf6 activation of Rac is proposed to be mediated by the Rac-GEF Dock180-engulfment and cell motility (ELMO) complex (54). Montagnac et al. (55) have recently reported that c-Jun N-terminal kinase (JNK)-interacting protein (JIP) 3 and JIP4 are targets of Arf6. JIP3/4 may mediate migration of 3T3-L1 cells, as it forms a complex with kinesin-1 and dynactin to control the microtube-dependent endosome trafficking required for migration. Because these Arf6 effectors are involved in basic cellular functions such as phosphoinositide cycling and intracellular trafficking, it is conceivable that they also function coordinately in this signaling pathway.
In this study we show for the first time that paxillin forms a functional complex with the Arf-GEF cytohesin-2 to mediate scratch-induced migration of 3T3-L1 preadipocytes before the initiation of differentiation to adipocytes. It is known that adipogenesis is regulated by activities of small GTP-binding proteins (56, 57). For example, RhoA regulates adipogenesis (37) and maintains the cell morphology of primary mesenchymal stem cells. Inhibition of RhoA and the downstream Rho kinase leads to differentiation to adipocytes. Similarly, our preliminary data show that inhibition of cytohesin family proteins by means of the pharmacological inhibitor SecinH3 promoted differentiation of 3T3-L1 cells (data not shown). As in the case of RhoA, Arf6 activity may support the preservation of the undifferentiated phenotype as well as migration. Further studies on the role of the cytohesin-2-Arf6 signaling pathway in 3T3-L1 cells and also in primary cells will advance our understanding of the involvement of small GTP-binding protein activity in adipocyte differentiation. In addition, because inhibition of cytohesin-2 in vivo results in hepatic insulin resistance (32, 33) and insulin resistance is a hallmark of type 2 diabetes, inhibition of this signaling pathway through cytohesin-2 is closely associated with pathological changes. Conversely, if this pathway is properly regulated, it may present a means for improving type 2 diabetes. Elucidation of the details of this pathway molecular mechanism and of the cytohesin-2 role therein may contribute to the development of drug target-specific medicines for type 2 diabetes.
Supplementary Material
Acknowledgments
We thank Drs. M. Imagawa (Nagoya City University) and M. Nishizuka (Nagoya City University) for providing 3T3-L1 cells and for participation in helpful discussions.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to J. Y.) and from the Ministry of Health, Labor, and Welfare, by the Japan Health Science Foundation and the Ministry of Human Health and Welfare (to Y. M., J. Y., and A. T.) and by research grants from the Human Health Sciences Foundation (to J. Y.), the Naito Foundation (to J. Y.), the Suzuken Foundation (to J. Y.), and the Takeda Science Foundation (to J. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GEF
- guanine-nucleotide exchange factor
- siRNA
- small interfering RNA
- GAP
- GTPase-activating proteins
- LD
- leucine-rich repeat domain
- LIM domain
- lin-11, isl-1, mec-3 domain
- CC
- coiled-coil
- PH
- pleckstrin homology
- PH+b
- PH domain plus C-terminal polybasic region
- GGA
- Golgi-localizing γ-adaptin ear homology domain
- BRAG
- BFA-resistant Arf GEF
- RFP
- red fluorescent protein
- GFP
- green fluorescent protein
- RT
- reverse transcription
- GST
- glutathione S-transferase
- ANOVA
- analysis of variance
- BRAG
- BFA-resistant Arf GEF.
REFERENCES
- 1.Lazar M. A. (2005) Science 307, 373–375 [DOI] [PubMed] [Google Scholar]
- 2.Rosen E. D., Spiegelman B. M. (2006) Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gálvez B. G., San Martín N., Rodríguez C. (2009) PLoS ONE 4, e4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Nature 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDougald O. A., Lane M. D. (1995) Annu. Rev. Biochem. 64, 345–373 [DOI] [PubMed] [Google Scholar]
- 6.Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 7.Kahn R. A., Kern F. G., Clark J., Gelmann E. P., Rulka C. (1991) J. Biol. Chem. 266, 2606–2614 [PubMed] [Google Scholar]
- 8.Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson J. G., Honda A. (2005) Biochem. Soc. Trans. 33, 639–642 [DOI] [PubMed] [Google Scholar]
- 10.D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 11.Casanova J. E. (2007) Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 12.DiNitto J. P., Delprato A., Gabe Lee M. T., Cronin T. C., Huang S., Guilherme A., Czech M. P., Lambright D. G. (2007) Mol. Cell 28, 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. (2005) Mol. Biol. Cell 16, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco M., Peters P. J., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. (1999) EMBO J. 18, 1480–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luton F., Klein S., Chauvin J. P., Le Bivic A., Bourgoin S., Franco M., Chardin P. (2004) Mol. Biol. Cell 15, 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy J. A., Jensen O. N., Walikonis R. S. (2006) Brain Res. 1120, 35–45 [DOI] [PubMed] [Google Scholar]
- 17.Yano H., Kobayashi I., Onodera Y., Luton F., Franco M., Mazaki Y., Hashimoto S., Iwai K., Ronai Z., Sabe H. (2008) Mol. Biol. Cell 19, 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown M. C., Turner C. E. (2004) Physiol. Rev. 84, 1315–1339 [DOI] [PubMed] [Google Scholar]
- 19.Deakin N. O., Turner C. E. (2008) J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamauchi J., Miyamoto Y., Sanbe A., Tanoue A. (2006) Exp. Cell Res. 312, 2954–2961 [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto Y., Yamauchi J., Chan J. R., Okada A., Tomooka Y., Hisanaga S., Tanoue A. (2007) J. Cell Sci. 120, 4355–4366 [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi J., Miyamoto Y., Murabe M., Fujiwara Y., Sanbe A., Fujita Y., Murase S., Tanoue A. (2007) Exp. Cell Res. 313, 1886–1896 [DOI] [PubMed] [Google Scholar]
- 23.Yamauchi J., Miyamoto Y., Chan J. R., Tanoue A. (2008) J. Cell Biol. 181, 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goda N., Tanoue A., Kikuchi S., Tsujimoto G. (2000) Biochim. Biophys. Acta 1493, 195–199 [DOI] [PubMed] [Google Scholar]
- 25.Koga H., Yuasa S., Nagase T., Shimada K., Nagano M., Imai K., Ohara R., Nakajima D., Murakami M., Kawai M., Miki F., Magae J., Inamoto S., Okazaki N., Ohara O. (2004) DNA Res. 11, 293–304 [DOI] [PubMed] [Google Scholar]
- 26.Yamauchi J., Miyamoto Y., Kusakawa S., Torii T., Mizutani R., Sanbe A., Nakajima H., Kiyokawa N., Tanoue A. (2008) Exp. Cell Res. 314, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 27.Nagel W., Schilcher P., Zeitlmann L., Kolanus W. (1998) Mol. Biol. Cell 9, 1981–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C. C., Chiang T. C., Wu T. S., Pacheco-Rodriguez G., Moss J., Lee F. J. (2007) Mol. Biol. Cell 18, 4420–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamauchi J., Miyamoto Y., Torii T., Mizutani R., Nakamura K., Sanbe A., Koide H., Kusakawa S., Tanoue A. (2009) Exp. Cell Res. 315, 2043–2052 [DOI] [PubMed] [Google Scholar]
- 30.Tan I., Yong J., Dong J. M., Lim L., Leung T. (2008) Cell 135, 123–136 [DOI] [PubMed] [Google Scholar]
- 31.Green H., Kehinde O. (1976) Cell 7, 105–113 [DOI] [PubMed] [Google Scholar]
- 32.Fuss B., Becker T., Zinke I., Hoch M. (2006) Nature 444, 945–948 [DOI] [PubMed] [Google Scholar]
- 33.Hafner M., Schmitz A., Grüne I., Srivatsan S. G., Paul B., Kolanus W., Quast T., Kremmer E., Bauer I., Famulok M. (2006) Nature 444, 941–944 [DOI] [PubMed] [Google Scholar]
- 34.Tsubouchi A., Sakakura J., Yagi R., Mazaki Y., Schaefer E., Yano H., Sabe H. (2002) J. Cell Biol. 159, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank S. R., Adelstein M. R., Hansen S. H. (2006) EMBO J. 25, 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann I., Thompson A., Sanderson C. M., Munro S. (2007) Curr. Biol. 17, 711–716 [DOI] [PubMed] [Google Scholar]
- 37.McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S. (2004) Dev. Cell 6, 483–495 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z. M., Simmerman J. A., Guibao C. D., Zheng J. J. (2008) J. Biol. Chem. 283, 18685–18693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wasserman F. (1965) in Handbook of Physiology (Renold A. E., Cahill G. F. eds) pp. 87–100, American Physiological Society, Washington, D. C. [Google Scholar]
- 40.Shen X., Hong M. S., Moss J., Vaughan M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1230–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheen V. L., Ganesh V. S., Topcu M., Sebire G., Bodell A., Hill R. S., Grant P. E., Shugart Y. Y., Imitola J., Khoury S. J., Guerrini R., Walsh C. A. (2004) Nat. Genet. 36, 69–76 [DOI] [PubMed] [Google Scholar]
- 42.Inoue H., Randazzo P. A. (2007) Traffic 8, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 43.Kahn R. A., Bruford E., Inoue H., Logsdon J. M., Jr., Nie Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L., Cassel D. (2008) J. Cell Biol. 182, 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo A., Hashimoto S., Yano H., Nagayama K., Mazaki Y., Sabe H. (2000) Mol. Biol. Cell 11, 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoefen R. J., Berk B. C. (2006) J. Cell Sci. 119, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 46.LaLonde D. P., Grubinger M., Lamarche-Vane N., Turner C. E. (2006) Curr. Biol. 16, 1375–1385 [DOI] [PubMed] [Google Scholar]
- 47.Klarlund J. K., Holik J., Chawla A., Park J. G., Buxton J., Czech M. P. (2001) J. Biol. Chem. 276, 40065–40070 [DOI] [PubMed] [Google Scholar]
- 48.Nevrivy D. J., Peterson V. J., Avram D., Ishmael J. E., Hansen S. G., Dowell P., Hruby D. E., Dawson M. I., Leid M. (2000) J. Biol. Chem. 275, 16827–16836 [DOI] [PubMed] [Google Scholar]
- 49.Kitano J., Kimura K., Yamazaki Y., Soda T., Shigemoto R., Nakajima Y., Nakanishi S. (2002) J. Neurosci. 22, 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansour M., Lee S. Y., Pohajdak B. (2002) J. Biol. Chem. 277, 32302–32309 [DOI] [PubMed] [Google Scholar]
- 51.Boehm T., Hofer S., Winklehner P., Kellersch B., Geiger C., Trockenbacher A., Neyer S., Fiegl H., Ebner S., Ivarsson L., Schneider R., Kremmer E., Heufler C., Kolanus W. (2003) EMBO J. 22, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang P., Cheng T. P., Agnello D., Wu C. Y., Hissong B. D., Watford W. T., Ahn H. J., Galon J., Moss J., Vaughan M., O'Shea J. J., Gadina M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2625–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venkateswarlu K. (2003) J. Biol. Chem. 278, 43460–43469 [DOI] [PubMed] [Google Scholar]
- 54.Santy L. C., Ravichandran K. S., Casanova J. E. (2005) Curr. Biol. 15, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 55.Montagnac G., Sibarita J. B., Loubéry S., Daviet L., Romao M., Raposo G., Chavrier P. (2009) Curr. Biol. 19, 184–195 [DOI] [PubMed] [Google Scholar]
- 56.Liu J., DeYoung S. M., Zhang M., Zhang M., Cheng A., Saltiel A. R. (2005) Cell Metab. 2, 165–177 [DOI] [PubMed] [Google Scholar]
- 57.Bryan B. A., Mitchell D. C., Zhao L., Ma W., Stafford L. J., Teng B. B., Liu M. (2005) Mol. Cell. Biol. 25, 11089–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.