Abstract
Cytoglobin (Cygb) was investigated for its capacity to function as a NO dioxygenase (NOD) in vitro and in hepatocytes. Ascorbate and cytochrome b5 were found to support a high NOD activity. Cygb-NOD activity shows respective Km values for ascorbate, cytochrome b5, NO, and O2 of 0.25 mm, 0.3 μm, 40 nm, and ∼20 μm and achieves a kcat of 0.5 s−1. Ascorbate and cytochrome b5 reduce the oxidized Cygb-NOD intermediate with apparent second order rate constants of 1000 m−1 s−1 and 3 × 106 m−1 s−1, respectively. In rat hepatocytes engineered to express human Cygb, Cygb-NOD activity shows a similar kcat of 1.2 s−1, a Km(NO) of 40 nm, and a kcat/Km(NO) (k′NOD) value of 3 × 107 m−1 s−1, demonstrating the efficiency of catalysis. NO inhibits the activity at [NO]/[O2] ratios >1:500 and limits catalytic turnover. The activity is competitively inhibited by CO, is slowly inactivated by cyanide, and is distinct from the microsomal NOD activity. Cygb-NOD provides protection to the NO-sensitive aconitase. The results define the NOD function of Cygb and demonstrate roles for ascorbate and cytochrome b5 as reductants.
Keywords: Ascorbic Acid, Enzyme Catalysis, Heme, Hemoglobin Myoglobin, Nitric Oxide, Superoxide Ion, Cytochrome b5, Cytoglobin, Neuroglobin, Nitric-oxide Dioxygenase
Introduction
The functions of cytosolic O2-binding hemoglobins in eubacteria, archaebacteria, yeast, fungi, plants, nematodes, protozoa, and mammals have remained largely enigmatic (1, 2). The discovery of a nitric-oxide dioxygenase (NOD)3 (EC 1.14.12.17) function for the inducible Escherichia coli flavohemoglobin suggested a primal and common function for hemoglobins and their associated reductases (3). Indeed, evolutionarily distant globins catalytically dioxygenate NO with high fidelity (4, 5). For a number of cytosolic hemoglobins, including the nonsymbiotic plant hemoglobin (6) and muscle Mb (7, 8), an enzymic NO scavenging function has been convincingly demonstrated in vivo and generally accepted, yet for many others, including the mammalian Cygb and Ngb, function remains largely unsettled (9–11).
Measurements of the NO reactivities of the oxy-complexes of Ngb and Cygb in vitro (12, 13) support a NOD function. However, O2 transport-storage (14, 15), peroxidase (16, 17), oxidase (18), heterotrimeric Gα protein guanine nucleotide dissociation inhibitor (19), and other functions (11, 20, 21) have also been postulated, and it remains unclear which of the proposed functions is physiologically significant and under what circumstances. For example, several NO metabolic activities and sinks have been measured in various cells and organelles (22–24) that would make a NOD activity appear either redundant or inconsequential. In addition, measurements of NO metabolism by globins with autooxidizable electron donors in the absence of SOD (13) do not allow the discernment of catalytic NO dioxygenation from NO oxidation by O2˙̄.
Understanding globin function(s) continues to demand a synthesis of knowledge of structure, ligand affinities, reactivities, autooxidation rates, and electron donors. Moreover, to assess the NOD function of a globin, knowledge of its interactions with NO and O2 within the cellular milieu is essential. Recently, Cygb-dependent NO dioxygenation and protection of NO-sensitive respiration within mouse NIH3T3 fibroblasts was reported (25), yet the relative capacity of Cygb for a NOD function with NO, O2, and cellular reductant(s), as well as its relation to other cellular NO sinks, remains to be defined.
Here we describe interactions of Cygb with NO, O2, and reductants that govern its capacity to function as a NOD in vitro and in rat hepatocytes. Investigations of the NOD activities of Cygb, Ngb, and Mb with ascorbate reveal a uniquely efficient redox coupling with Cygb that may be attributed to a unique Cygb structure. In addition, we report that the Cygb-NOD activity is physically and kinetically distinct from the microsomal NOD activity (23, 26, 27).
MATERIALS AND METHODS
Reagents
l-Ascorbic acid, sodium citrate, glucose, NADP+, NADPH, DPI, Me2SO, sodium cyanide, 98.5% NO, Brilliant Blue G, recombinant human CYPOR (27 units/mg), spinach ferredoxin-NADP+ oxidoreductase (3.9 units/mg), porcine heart isocitrate dehydrogenase, horse heart cytochrome c, Aspergillus niger glucose oxidase, bovine serum albumin, EDTA, HEPES, Tris, glycine, dithiothreitol, chloramphenicol, and ampicillin were from Sigma-Aldrich. Sodium diphosphate and sodium monophosphate were obtained from Fisher. NADH, bovine liver catalase (260,000 units/ml), bovine pancreas deoxyribonuclease I (2000 units/mg), bovine erythrocyte copper- and zinc-containing SOD (5000 units/mg), and nitrate reductase (10 units/mg) were obtained from Roche Applied Science. DNA restriction and modifying enzymes were obtained from New England Biolabs, Inc. DNA primers, G418, and SeeBlueTM prestained protein molecular weight standards were purchased from Invitrogen. AG 1-X8 ion exchange resin (acetate form) was purchased from Bio-Rad. 99.993% O2, 99.999% CO, 99.998% N2, and 99.99% argon were from Praxair (Bethlehem, PA). Recombinant rat cytochrome b5 (soluble form) was expressed in E. coli and purified essentially as described (28). Recombinant human NCB5OR (29) was provided by Drs. H. Zhu and F. Bunn (Brigham and Women's Hospital, Boston, MA). Sperm whale Mb (4, 30) was obtained from Dr. J. Olson (Rice University, Houston, TX). E. coli MnSOD (2400 units/mg) was supplied by Miami Valley Biotech (Dayton, OH). Microsomes containing 10 mg/ml of protein and 70 milliunits/mg of membrane-bound CYPOR activity were isolated from Caco-2 cells (23).
Expression and Purification of Cygb and Ngb
Human Cygb cDNA Image clone 5193583 (ATCC number 7498923) was obtained from the American Type Culture Collection (Manassas, VA). Cygb cDNA was PCR-amplified with the respective sense and antisense primers GGAGCTGCATATGGAGAAAGTGCCAGGCGA and CCTCAAGCTTCCTTGGCACCCAGAAATGGA engineered with respective NdeI and HindIII sites, cloned into the pET17b vector (Novagen) polylinker, and sequenced. Cygb was expressed and isolated from E. coli BL21(DE3)pLysS. Mouse Ngb cDNA in pET3A was provided by Dr. Thorsten Burmester and Thomas Hankeln (Johannes Gutenberg University of Mainz, Mainz, Germany) and was expressed and isolated from E. coli BL21(DE3)pLysS (31). E. coli expressing isopropylthio-β-d-galactoside-inducible globin were inoculated in 2 liters of Luria-Bertani medium (32) containing 50 μg/ml ampicillin, 1 μm hemin, and 5 units/ml catalase in 2.8-liter Fernbach flasks and grown in a 37 °C gyrorotatory shaker at 175 rpm. Globin expression was induced with 0.1 mm isopropylthio-β-d-galactoside when the cell density, as measured by absorbance at 550 nm, was 1.2. The cells were treated with isopropylthio-β-d-galactoside for 16 h and were harvested by centrifugation. The cell pellets were resuspended and sonicated in a cell:buffer volume ratio of 2:3 in chilled 20 mm potassium phosphate buffer, pH 7.0, containing 1 mm EDTA, 10 μg/ml deoxyribonuclease I, and 260 units/ml catalase. The cell lysates were clarified by centrifugation, and the extracts were dialyzed extensively against 1 mm potassium phosphate, pH 7.0. Cell-free extracts were diluted to 20 mg/ml protein, heated for 10 min at 65 °C, chilled, and clarified by centrifugation. Cygb and Ngb were isolated by column chromatography on DEAE-Sepharose, Superdex 75, and hydroxyapatite. Protein was measured by the method of Lowry et al. (33) with bovine serum albumin as the standard. Protein purity was assessed by SDS-PAGE using Brilliant Blue G staining. Heme was assayed using the alkaline-pyridine method (34). The spectra were recorded with a Beckman DU® 7500 diode array spectrophotometer.
Mammalian Cell Culture
The rat hepatocyte, K9 (CRL-1439), the human lung adenocarcinoma A549 (CCL185), and the human colorectal carcinoma Caco-2 (HTB-37) were obtained from the American Type Culture Collection. The cells were grown, passaged, and harvested as previously described (27). The cells were counted with a hemacytometer, and the weights of the cell pellets were measured with an analytical balance.
Western Blot Analysis
The cells were lysed with an equal volume of detergent buffer containing 20 mm Tris-Cl, pH 7.4, 50 mm NaCl, 1 mm EDTA, and 1% Triton X-100. The extract protein was assayed (33), separated by reducing SDS-PAGE in 1.0-mm Precast 8–16% gradient gels (Invitrogen), and transferred to nitrocellulose membranes. The membranes were washed, blocked with 5% milk, probed with rabbit anti-Cygb antibodies (15, 35), and detected using peroxidase-conjugated goat anti-rabbit IgG (Pierce) and the SuperSignal® West Dura extended duration substrate kit (Thermo Scientific) on Kodak X-OmatTM film. Cygb was measured by Western analysis and densitometry with purified human Cygb standards.
Construction of Cygb-expressing Hepatocytes
The 995-bp EcoRI-XbaI fragment of cytoglobin cDNA Image clone 5193583 was isolated and ligated into the EcoRI-XbaI-restricted polylinker region of pcDNA3 v. 1.1 (Invitrogen) generating pcDNA3-hCygb. Rat hepatocytes were transfected with pcDNA3-hCygb or pcDNA3 DNA using FuGENETM 6 transfection reagent (Roche Applied Science), and stable transfectants were selected with 0.3 mg/ml G418. The clones were isolated and screened for Cygb expression by Western blot analysis.
NO Consumption Assays
NO consumption was measured with ISO-NOP and microchip NO electrodes (WPI Instruments, Inc.) as previously described (34). NO scavenging activities of globins were routinely measured at 37 °C in 2 ml of 100 mm sodium phosphate buffer containing 0.3 mm EDTA and 1.0 mg of MnSOD (11 μm of the dimer). MnSOD was routinely added to NO consumption assays to competitively scavenge interfering O2˙̄ generated by reductants and reductases (34). Copper- and zinc-containing SOD reacted with NO and was thus unsuitable. Cell NO metabolism was measured at 37 °C in Dulbecco's phosphate-buffered saline containing 5 mm glucose and 100 μg/ml cycloheximide (27, 36). NO consumption measurements were corrected for background rates. NO-saturated water (1.94 mm) was prepared over AG 1-X8 resin as previously described (34). CO-saturated water (1.0 mm) was prepared under 99.5% CO (34). O2-saturated buffer (1.14 mm) was prepared, and O2 concentrations were varied in reactions as previously described (27, 34, 36). O2 was depleted from buffers by bubbling with 99.99% argon and by preincubating for 5 min with 4 units of glucose oxidase and 260 units of catalase. O2 concentration was measured with an O2 electrode (YSI Co., Yellow Springs, OH). [O2] was 200 and 260 μm in air-saturated buffers at 37 and 20 °C, respectively. Buffer conductivity was measured at 37 °C with an inductive conductivity monitor (GE Healthcare).
Nitrate and Nitrite Assays
Nitrate was reduced to nitrite with NADPH and nitrate reductase. Nitrite was assayed using the Griess reagent (34).
Globin Reduction Assays
Reduction of ferric Cygb, Ngb, and Mb was measured by following the formation of the ferrous-CO complex at 422, 416, and 422 nm, respectively, in a 1-cm thermostatted quartz cuvette at 37 °C. The reactions were initiated by adding 10 μm of globin (heme) to an anaerobic 1-ml reaction mix containing buffer, 20 μm CO, and the indicated concentrations of ascorbate. O2 was depleted from reactions by scrubbing the reaction mix with N2 and reacting 5 mm glucose with 2 units of glucose oxidase and 260 units of catalase for 10 min prior to adding 4–8 μl of the concentrated globin.
Cell NO Exposures and Aconitase Assays
Freshly harvested K9neo and K9Cygb hepatocytes were resuspended at a density of 3.2 × 106 cells/ml in 3-ml of serum-free F12K medium buffered to pH 7.4 with 50 mm sodium HEPES and containing 100 μg/ml cycloheximide. O2, N2, and NO gas mixtures were delivered at 30 ml/min using three-way gas proportioners (27). The cells were harvested, and the extracts were assayed for aconitase activity and protein as previously described (27).
Data Analysis
The Tukey-Kramer (Honestly Significant Difference) statistical analysis method in the program JMP (SAS Institutes Inc.) was used for the analysis of significance (p < 0.05). The data presented are representative of the results of two or more trials.
RESULTS
Ascorbate-driven NO Metabolic Activity of Cygb, Ngb, and Mb
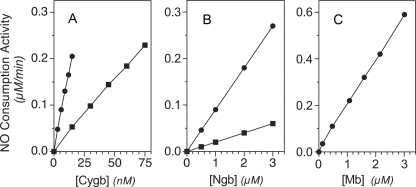
Ascorbate, a potential electron donor for plant hemoglobins (37, 38) and Mb (39), was investigated for its ability to support enzymic NO scavenging by Cygb, Ngb, and Mb. With 10 mm ascorbate, the enzymic NO scavenging activity is linearly dependent upon globin concentration. Human Cygb shows a turnover rate of 0.25 NO/heme/s with 100 nm NO at 37 °C (Fig. 1A, ●). An ∼4-fold slower turnover is seen at 20 °C (Fig. 1A, ■). Mouse Ngb (Fig. 1B, ●) and sperm whale Mb (Fig. 1C, ●) show respectively 150- and 70-fold lower activities than Cygb.
FIGURE 1.
Catalytic NO metabolism by globins in vitro. Human Cygb (A), mouse Ngb (B), and sperm whale Mb (C) were measured for NO metabolic activity at 37 (●) or 20 °C (■) in a 2-ml reaction volume containing 10 mm l-ascorbate, 100 mm sodium phosphate, pH 7.0, 0.3 mm EDTA, and 11 μm MnSOD. The reactions were initiated with 400 nm NO, and the activities were determined for 100 nm NO. The concentrations of globins are relative to heme content.
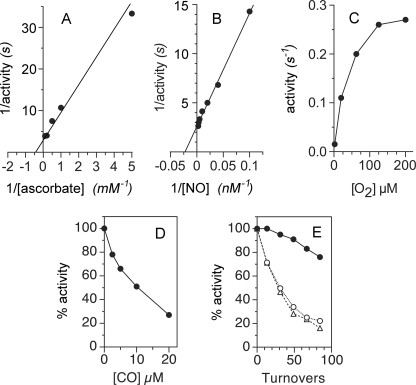
The Cygb activity shows an apparent Km(ascorbate) value of 2 mm (Fig. 2A), an apparent Km for NO of 40 nm, and kcat of 0.5 s−1 with saturating ascorbate and O2 in neutral phosphate buffer (Fig. 2B). With 100 nm NO, half-maximal activity is seen at ∼20 μm O2 (Fig. 2C). Inhibition of the activity occurs at [NO]/[O2] ratios >1:500 (see below), thus complicating the determination of a Km(O2) value. Similar to other NOD activities (3, 23, 27, 40), the activity is inhibited by CO and inactivated cyanide. With 20 μm O2, 10 μm CO inhibits the Cygb activity by ∼50% (Fig. 2D). NO progressively inhibits the activity during turnover with 400 nm NO and 200 μm O2 (Fig. 2E, ●), and the activity is slowly inactivated by 250 μm cyanide (Fig. 2E, ○). Preincubation of Cygb with cyanide for 7 min does not cause a greater loss of activity (Fig. 2E, compare ○ and ▵), thus demonstrating the importance of turnover for cyanide-mediated inactivation.
FIGURE 2.
Cygb-catalyzed NO metabolism with ascorbate. A, Cygb activity was measured by varying concentrations of ascorbate with 200 μm O2 and 200 nm NO. B, activity was measured with 200 μm O2 for different [NO]. C, activity was measured at different [O2] with 100 nm NO. D, activity was measured with 20 μm O2, 150 nm NO, and varying [CO]. E, activity was measured for 100 nm NO with no addition (●), the addition of 250 μm sodium cyanide (○), or following a 7-min preincubation with 250 μm sodium cyanide (▵). All of the measurements were made with 22.5 nm Cygb (heme) at 37 °C in 100 mm sodium phosphate buffer, pH 7.0, containing 0.3 mm EDTA, 11 μm MnSOD, and 10 mm ascorbate, unless otherwise specified. The reactions were initiated with 400 nm NO.
The results demonstrate Km values for NO and O2 within physiologically relevant concentration ranges. However, the Km(ascorbate) value of 2 mm is greater than the Km values of 0.4–0.9 mm reported for ascorbate-utilizing enzymes (41, 42), and the ascorbate concentrations measured in some tissues (43).
Anions Competitively Inhibit Activity
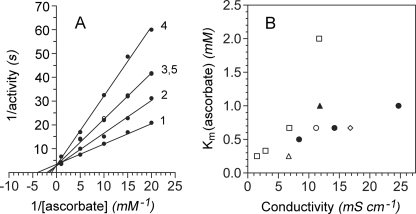
Salts were tested for effects on the Km(ascorbate) value and steady-state behavior of Cygb. Lowering the buffer sodium phosphate concentration from 100 to 10 mm decreases the Km(ascorbate) value to 0.25 mm (Fig. 3A, line 1). Salts, including potassium chloride (lines 2–4) or sodium chloride (line 5), competitively inhibit the activity with respect to [ascorbate]. Increasing the sodium phosphate concentration or including sodium phosphate or NaCl in a 25 mm sodium citrate buffer also competitively inhibits the activity and increases the Km(ascorbate) value (Fig. 3B). Furthermore, the competitive effect of buffer salt depends more on the anion than the ionic strength. Salts do not significantly affect the kcat achieved with saturating ascorbate (Fig. 3A) or the Km(NO) (data not shown). The results suggest electrostatic interactions in the binding of the negatively charged ascorbate anion.
FIGURE 3.
Anions competitively inhibit ascorbate-dependent NO metabolism. A, NO metabolism was measured with 7.5–30 nm Cygb (heme) at 37 °C in 10 mm sodium phosphate buffer with 0, 50, 100, and 200 mm KCl (lines 1–4, respectively) or 100 mm NaCl (line 5). B, Km(ascorbate) values are plotted versus buffer conductivity. The values are for 10, 20, 50, or 100 mm sodium phosphate buffer (□), for 10 mm sodium phosphate buffer containing either 50, 100, or 200 mm KCl (●) or 100 mm NaCl (○), and for 25 mm sodium citrate buffer alone (▵) or containing 100 mm NaCl (◇) or 50 mm sodium phosphate (▴). All of the buffers were pH 7.0 and contained 0.3 mm EDTA and 11 μm MnSOD.
Cygb Reduction by Ascorbate
The rates of ferric globin reduction were measured under conditions similar to those for ascorbate-driven NO metabolism (Fig. 2A). Globin reduction was measured in the presence of excess CO and the absence of O2 to stabilize the reduced heme from reoxidation. The rate of Fe2+(CO) complex formation was measured by following the absorbance increase at 422 nm as described under “Materials and Methods.” Under these conditions, the Cygb reduction rate shows a linear dependence upon [ascorbate] up to 8 mm (data not shown). The apparent second order rate constant for ascorbate-mediated reduction of ferric Cygb is 1.3 ± 0.2 m−1 s−1. The corresponding rate constants for Ngb and Mb are 0.10 and 0.13 m−1 s−1, respectively. The results demonstrate an ∼10-fold faster reduction rate for Cygb than for Ngb and Mb. However, these differences do not account for the larger differences in turnover rates measured in Fig. 1.
Cygb NO Metabolic Activity Produces Nitrate
Slow and gradual addition of 32 nmol of NO-saturated water (16-μl) with a gas tight syringe to a 2-ml reaction mix containing 50 mm Tris-Cl, pH 7.5, 2 mm ascorbate, 200 μm O2, 11 μm MnSOD, and 1.0 μm Cygb at 37 °C yields 28.7 ± 1.0 nmol of NO3− and 2.8 ± 0.5 nmol of NO2− or 91 ± 3% NO3− and 9 ± 2% NO2−. The results demonstrate a nitrate-generating NOD mechanism for Cygb (4, 5). However, the data also indicate secondary NO oxidation reactions, possibly representing residual reactions of O2˙̄ with NO to form peroxynitrite and NO2− (44).
Cygb-NOD Activities with NADH, NADPH, cytochrome b5, and Cellular Reductases
Several cellular reductants and reductases with globin reducing capacity (5, 13, 45, 46) were tested for their ability to support the NOD activity of Cygb in the presence of a high [SOD].
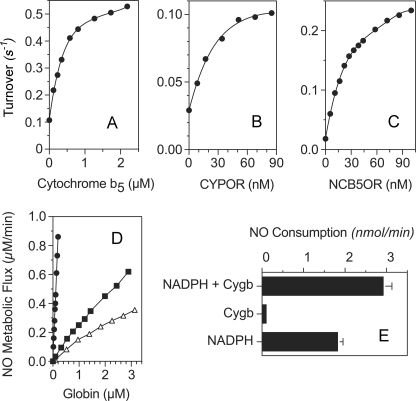
NADH and NADPH support a low Cygb-NOD turnover that is >15-fold slower than with ascorbate under otherwise comparable conditions (Table 1), demonstrating a preference for ascorbate. Reduced cytochrome b5 also supports the NOD activity of Cygb and shows saturation with half-maximal activity at 0.3 μm (Fig. 4A). Ferredoxin reductase and NADPH produce a low background activity in the cytochrome b5 reducing system. CYPOR can also support a low activity of Cygb-NOD (Fig. 4B). NCB5OR, an enzyme with a flavin-containing reductase domain and a cytochrome b5 domain that is found in the lumen of the endoplasmic reticulum (29), also supports the NO metabolic activities of Cygb (Fig. 4C), Ngb (Fig. 4D, ■), and Mb (▵). Cygb shows the highest activity of the three (Fig. 4D, compare ● with ■ and ▵). Microsomes containing membrane-bound CYPOR (14 milliunits) and incubated with 100 μm NADPH can supply electrons for the NO metabolic activity of Cygb. With 300 nm Cygb, microsomes show greater activity than that observed with NADPH alone and catalyzed by the microsomal NOD activity (Fig. 4E) (23).
TABLE 1.
Cygb-NOD activity with pyridine nucleotides and ascorbate
NO consumption activity of 30 nm Cygb (heme) in 2 ml of 100 mm sodium phosphate buffer, pH 7.0, containing 0.3 mm EDTA, 11 μm MnSOD, and 200 μm O2 at 37 °C. The turnover rates were determined for 100 nm NO. The results are the averages of three measurements ± S.D.
| Reductant | Activity |
|---|---|
| min−1 | |
| NADH, 1.0 mm | 0.32 ± 0.03 |
| NADPH, 1.0 mm | 0.40 ± 0.04 |
| Ascorbate, 1.0 mm | 6.1 ± 0.3 |
FIGURE 4.
Cygb-catalyzed NO metabolism with cellular reductants and reductases. A, activity of 15 nm Cygb in the presence of varying concentrations of rat cytochrome b5, 100 μm NADPH, and 20 milliunits of ferredoxin-NADP+ oxidoreductase. B, activity of 15 nm Cygb with varying concentrations of soluble human CYPOR (2 milliunits/nmol) and 20 μm NADPH. C, activity of 30 nm Cygb with varying concentrations of NCB5OR and 100 μm NADH. D, activities of various concentrations of Cygb (●), Ngb (■), and Mb (▵) with 28 nm NCB5OR and 100 μm NADH. Globin concentrations represent heme content. The reactions in A–D were in 2-ml of 100 mm sodium phosphate buffer, pH 7.0, containing 0.3 mm EDTA, 11 μm MnSOD, and 200 μm O2. The reactions were initiated with 400 nm NO, and all of the activities were measured at 150 nm NO. E, human microsomes (20 μl) were measured for NO metabolic activity at 1 μm NO with or without 100 μm NADPH and with or without 300 nm Cygb in 100 mm sodium HEPES buffer, pH 7.8, containing 0.25 m sucrose, 11 μm MnSOD, and 200 μm O2 at 37 °C. The error bars represent the S.D. of three measurements.
The results demonstrate that, in addition to ascorbate, cytochrome b5 and NCB5OR effectively support Cygb-NOD activity. We can estimate apparent second order rate constants for cytochrome b5 and NCB5OR-mediated reduction of Cygb of 3 × 106 and 6 × 106 m−1 s−1, respectively. None of the electron donors is as effective in supporting the NO scavenging activity of either Ngb or Mb, thus suggesting electron donor specificity for Cygb.
NO Consumption and Cytoglobin Expression by Cultured Hepatocytes and A549 and Caco-2 Cells
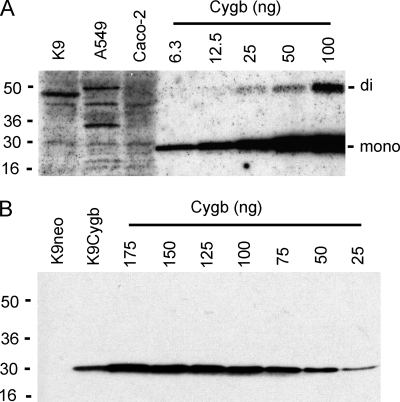
Cultured rat hepatocytes (K9) and human A549 lung and intestinal Caco-2 cells catalytically consume NO at 1.2 ± 0.1, 5.7 ± 0.4, and 22.6 ± 1.2 nmol of NO/min/107 cells (± S.D., n = 3), respectively, with 1 μm NO and 200 μm O2. None of the cells express Cygb as measured by Western blot analysis (Fig. 5A). Human Cygb migrates as a monomer (20.9 kDa) on reducing SDS-PAGE gels. A weak signal from the disulfide-linked dimeric Cygb is detected with >25 ng of purified Cygb/lane after long film exposures. The results are consistent with immunohistochemical assays of Cygb in rat hepatocytes (15, 16).
FIGURE 5.
Cygb expression in cultured rat hepatocytes (K9), A549, and Caco-2 cells. Human Cygb was measured in K9, A549, and Caco-2 cell extracts (A) and in K9neo and K9Cygb cell extracts (B) by Western blot analysis as under “Materials and Methods.” The gel lanes were loaded with 200 (A) or 100 μg (B) of cell-free extract protein or the indicated amounts of human Cygb. The Cygb monomer (mono) and dimer (di) signals are labeled.
Rat hepatocytes offered a cell model for measuring the capacity of Cygb to function as a NO scavenger. Rat hepatocytes express negligible Cygb and relatively low levels of NO metabolic activity, synthesize ascorbate (47, 48), and contain microsomal detoxification enzymes including CYPOR and cytochrome b5. Transfection of hepatocytes with pcDNA3-hCygb expressing the human Cygb gene under control of the cytomegalovirus promoter produced several stable cell lines with elevated Cygb expression (data not shown). A representative clone, K9Cygb, was selected for further investigation. K9Cygb expressed 50 ± 10 ng of Cygb/100 μg of soluble protein, whereas the control cell line, K9neo, which was stably transfected with the pcDNA3 vector, showed no detectable Cygb protein (Fig. 5B). If we assume that ∼90% of the cell weight is water, we can estimate that K9Cygb hepatocytes express 1.3 ± 0.2 μm 20.9-kDa Cygb monomer, because the wet weight of 107 K9 cells is 21 ± 3 mg (± S.D., n = 3), and 107 cells release 1.10 ± 0.21 mg of soluble protein (± S.D., n = 3) with detergent lysis.
NO Metabolism in K9Cygb and K9neo Hepatocytes
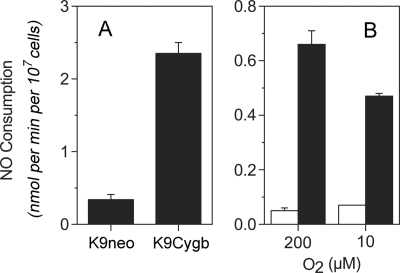
K9Cygb cells show ∼7-fold greater NO metabolic activity than K9neo cells with 200 nm NO and 200 μm O2 (Fig. 6A). With 10 nm NO, the activity difference is larger. The Cygb-NOD activity is ∼13-fold greater for 200 μm O2 and ∼9-fold higher at a more physiological O2 concentration of 10 μm (Fig. 6B).
FIGURE 6.
NO consumption by K9neo and K9Cygb cells. Cellular NO consumption rates were measured for 200 nm NO in the presence of 200 μm O2 (A). K9neo (white bars) and K9Cygb (black bars) NO consumption activities were measured at 10 nm NO with the indicated O2 concentrations (B). NO metabolism was measured at 37 °C in Dulbecco's phosphate-buffered saline containing 5 mm glucose and 100 μg/ml cycloheximide. The error bars represent the S.D. of three measurements.
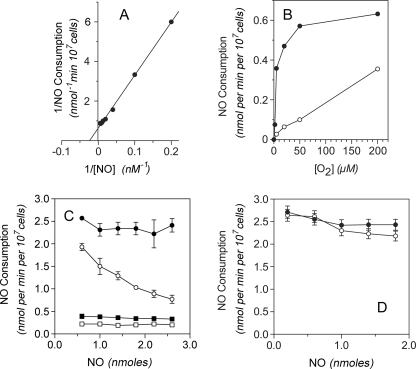
The NO metabolic activity shows an apparent Km(NO) value of 40 nm with 200 μm O2 (Fig. 7A). Half-maximal activity is observed with ≤5 μm O2 with 20 nm NO (Fig. 7B, ●). NO inhibits the activity at higher NO:O2 concentration ratios precluding the determination of a true Km(O2) value. CO also inhibits the activity, and inhibition is also less pronounced with higher O2 concentrations (Fig. 7B, ○). Similar to in vitro reactions (Fig. 2E), the NO metabolic activity is weakly yet progressively inactivated by 250 μm cyanide during turnover (Fig. 7C, compare ○ with the control ●). In contrast, the basal NO metabolic activity in K9neo cells is rapidly inactivated by 250 μm cyanide (Fig. 7C, compare □ with the control ■). NO metabolism by K9Cygb cells is also insensitive to the CYPOR and flavoenzyme inhibitor, DPI. DPI (50 μm) inactivates <10% of the total activity (Fig. 7D, compare ○ with the control ●). The results demonstrate the capacity of Cygb to function as a NOD. Further, the data show that the activity is distinct from the cyanide and DPI-sensitive microsomal NOD (23, 27). The results also suggest that the cyanide-sensitive NO metabolic activity previously attributed to Cygb in NIH3T3 fibroblasts (25) is due to the ubiquitous microsomal NOD.
FIGURE 7.
Cygb-catalyzed NO consumption in rat hepatocytes. A, activity was measured at different NO concentrations with 200 μm O2. B, activity was measured for 20 nm NO with varying O2 concentrations in the absence (●) or presence of 20 μm CO (○). C, activities of K9Cygb and K9neo cells were measured in the absence (● and ■, respectively) and presence of 250 μm NaCN (○ and □, respectively). The activities were measured for 100 nm NO following repetitive additions of 200 nm NO in the presence of 200 μm O2. D, activity of K9Cygb cells was measured in the presence of either Me2SO solvent (0.1% v/v) (●) or 50 μm DPI in Me2SO (0.1% v/v) (○). The activities were measured for 100 nm NO following repetitive additions of 200 nm NO with 200 μm O2. All of the measurements were at 37 °C. The error bars represent the S.D. of three independent measurements and in some cases are within data points.
Sensitivity of Aconitase to NO-mediated Inactivation in Cygb-expressing Hepatocytes
Exposure of K9neo and K9Cygb hepatocytes to an atmosphere containing 480 ppm of NO balanced with 21% O2 and N2 for 60 min inactivates the NO-sensitive aconitase (27, 32) to 61 ± 9% (± S.D., n = 4) and 75 ± 5% (± S.D., n = 4) of its control activity levels, respectively. The 14% difference between the values is significant (p < 0.05). In the absence of cells and under similar exposure conditions, NO rapidly achieves a concentration of 400 ± 50 nm. Exposure of either cell line to 240 ppm NO did not result in significant inactivation of aconitase, suggesting effective protection by the basal NO metabolic activity under these conditions. Aconitase activities in control K9neo and K9Cygb cell extracts were 11.5 ± 1.0 and 11.3 ± 1.5 milliunits/mg (± S.D.; n = 5), respectively. The results demonstrate Cygb-dependent protection of aconitase in rat hepatocytes.
DISCUSSION
Like other globins (4, 5), Cygb, Ngb, and Mb can function as enzymic NODs when coupled to suitable electron donors. With ascorbate and reduced cytochrome b5, the human Cygb-NOD shows a maximal turnover rate of ∼0.5 s−1. With a net NOD activity of 2 nmol NO/min/107 cells at 200 nm NO and 200 μm O2 (Fig. 6A) and expression of 550 ng of Cygb/1.10 ± 0.21 mg of soluble protein (± S.D., n = 3) or 107 cells (Fig. 5B), we can estimate a similar turnover rate of 1.2 s−1 within engineered K9Cygb hepatocytes. We can also calculate a kcat/Km(NO) (k′NOD) value of 1.3–3 × 107 m−1 s−1 for the Cygb-catalyzed NO dioxygenation reaction, which is in range of the 3.4 to ∼7 × 107 m−1 s−1 measured for the structurally similar Mb and Ngb using stopped flow analysis (12, 30) and the ∼2.2 × 107 m−1 s−1 estimated from NO electrode measurements of the reaction of NO with oxy-Cygb (13). Hence, electron transfer to Cygb does not significantly limit enzymatic activity in vitro or in rat hepatocytes.
With saturating ascorbate or reduced cytochrome b5, the Cygb-NOD activity is limited by its k′NOD and susceptibility to NO inhibition. These are determined, respectively, by NO diffusion and reactivity with the active site oxy-complex (k′NOD) and the competition between O2 and NO for binding to the ferrous heme (Kd(O2)/Kd(NO)) (5). NOD activity is ultimately determined by globin structure. By comparison, flavohemoglobin-NOD is >5-fold less sensitive to NO inhibition with a relatively small Kd(O2)/Kd(NO), reacts with NO up to 100-fold faster with a relatively large k′NOD ≤ 2.9 × 109 m−1 s−1, and shows >100-fold faster turnover rates (5, 49, 50).
By analogy to the peroxidases and peroxiredoxins and the catalases that scavenge H2O2 in cells (51), individual NODs may be suited for scavenging NO under different conditions. Cygb-NOD is clearly distinct from the microsomal CYPOR-dependent NOD activity. Caco-2, A549 cells, and hepatocytes express negligible Cygb (Fig. 5A) but express a DPI and cyanide-sensitive microsomal activity that shows only weak NO inhibition at an [NO]/[O2] ratio of 1:100 (23, 27). In contrast, the NO metabolic activity of Cygb in hepatocytes is DPI-resistant, slowly inactivated by cyanide (Fig. 7C), and inhibited by NO at [NO]/[O2] ratios of >1:500 (Fig. 2E).
Our results demonstrate that ascorbate and cytochrome b5 both act as efficient electron donors for the Cygb-NOD activity and, moreover, suggest that the preference for electron donors in cells will be dependent upon their relative abundance. The low Km(ascorbate) value of 0.25–0.67 mm measured at physiological salt concentrations (Fig. 3B) argues for a role for ascorbate in ascorbate-utilizing collagen-synthesizing hepatic stellate cells and fibroblasts and ascorbate-rich neurons (15, 16). We have not measured ascorbate or cytochrome b5 levels in cultured rat (K9) hepatocytes. However, hepatocytes synthesize ascorbate and are a major source of ascorbate in rats (43, 48). Ascorbate concentrations of 6 nmol/mg of protein (∼1.2 mm) and 2 mm have been measured in cultured primary rat hepatocytes (47, 48).
The Km(ascorbate) and kcat values for the Cygb-NOD activity (Figs. 2 and 3) allow us to estimate apparent second order rate constants for Cygb reduction ranging from 125 to 1000 m−1 s−1 and dependent upon anion concentrations, whereas the ferric Cygb was reduced by ascorbate with an ∼100-fold smaller rate constant under similar buffer conditions. The disparate rate constants indicate that ferric Cygb, per se, is not an obligate intermediate in the catalytic cycle. The rate of Cygb reduction may be greater for the high potential Fe3+(−OONO) intermediate (4, 5) like the fast ascorbate-mediated reduction of the ferryl form of leghemoglobin (52) and Compound I or II in ascorbate peroxidase (53–55). The high reduction rate constants estimated for cytochrome b5 and NCB5OR may be similarly explained. The steady-state kinetic results suggest caution when evaluating roles of potential electron donors based solely on transient measurements of ferric globin reduction (12, 13, 45, 56, 57).
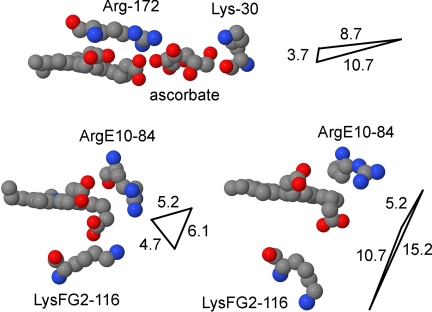
The Michaelis-Menten behavior of the Cygb-NOD activity with ascorbate and the competitive inhibition by anions (Figs. 2A and 3) strongly suggest a positively charged binding site for ascorbate. Cygb has unique invariant ArgE10–84 and LysFG2–116 residues near a solvent-exposed heme proprionate carboxylate (58, 59) that may form a binding site similar to that in ascorbate peroxidase (60). In the peroxidase-ascorbate complex (Protein Data Bank code 1OAF) (Fig. 8, top panel), Arg-172 facilitates ferric heme reduction by hydrogen bonding and stabilizing ascorbate ene-diol(ate) (53, 55, 60), and a Lys-30 amine hydrogen bonds the ascorbate 6-hydroxyl. In the dimeric Cygb structure (Protein Data Bank code 1UMO) (58), the heme proprionate, ArgE10 and LysFG2 residues are located in different positions in the A and B subunits (Fig. 8, compare bottom left and bottom right panels), indicating a large conformational shift, and a possible inducible ascorbate-binding site. Moreover, the A and B subunit conformations are associated with changes between hexa- and pentacoordinate iron (58), implying communication between the iron and the putative ascorbate-binding site during catalysis. It is noteworthy that the B subunit carboxylate O-atom and LysFG2 N-atom are separated by 10.7 Å (Fig. 8, bottom right panel) as seen in the ascorbate peroxidase-ascorbate complex (Fig. 8, top panel). In the monomeric Cygb structure (Protein Data Bank code 1V5H) (59), the ArgE10 guanidinium nitrogen is 6.6 Å from the carboxylate O-atom and 8.5 Å from the LysFG2 N-atom, the latter two being only 2.9 Å from each other, further illustrating the large structural flexibility of the site.
FIGURE 8.
Putative ascorbate-binding site in Cygb. Ascorbate hydrogen bonds Arg-172 and Lys-30 in ascorbate peroxidase (Protein Data Bank code 1OAF) (55, 60). Hydrogen bonding stabilizes the ene-diol(ate) intermediate and facilitates the transfer of an electron via the heme proprionate (top). The A subunit of human Cygb dimer (Protein Data Bank code 1UMO) (58) shows conserved ArgE10–84 and LysFG2–116 residues for potential binding and stabilization of ascorbate (bottom left). The B subunit (bottom right) shows an alternate orientation of ArgE10–84, LysFG2–116, and the heme proprionate. Distances are given in Å by the triangles where the vertices represent the proprionate carboxylate O-atom and the arginine or lysine side chain N-atoms.
Elevated Cygb expression in activated hepatic stellate cells, chondroblasts, osteoblasts, and hypoxic neurons (15, 16, 21, 61) suggests an important role for Cygb in the protection of these cells against NO toxicity; however, quantitative knowledge of Cygb expression levels and the capacity for NO detoxification has been lacking. We have determined that ∼1.3 μm Cygb confers a NOD activity equal to ∼1.6 μm NO s−1 (kcat [Cygb]) to cells at saturating [O2] and [NO]. This NOD activity protected the NO-sensitive aconitase by a modest ∼14% during a continuous exposure of cells to ≤0.45 μm NO at 200 μm O2. It should be noted, however, that sustained [NO]/[O2] ratios of >1:500 progressively inhibit the NOD activity. Cells would require a higher [Cygb] to achieve a lower steady-state [NO] and afford greater protection. By scavenging NO, Cygb would also decrease peroxynitrite formation (44) and protect respiration (25) and enzymes important to fibrogenesis such as the prolyl 4-hydroxylase (61, 62). Cygb could also regulate the nanomolar [NO] found in tissues (63) and feedback regulate O2 delivery to tissues (8, 64). The large effects of [O2] on activity (Fig. 7B) may also explain the benefit of hypoxic induction of Cygb (15, 65). Experiments can now be directed toward determining the roles of an ascorbate and cytochrome b5-driven Cygb-NOD activity during fibrosis (16) and other conditions and elucidating the electron transfer mechanisms for ascorbate and cytochrome b5.
Acknowledgments
We are indebted to Drs. Thorsten Burmester and Thomas Hankeln for generously providing the expression plasmid for mouse Ngb and the rabbit antisera to Cygb. E. coli expressing soluble rat cytochrome b5 were kindly provided by Dr. Stephen Sligar. We thank Drs. Hao Zhu and Franklin Bunn for providing NCB5OR. We are also grateful to Dr. John Olson for supplying sperm whale Mb.
This work was supported in part by National Institutes of Health Grant GM65090. This work was also supported by American Heart Association Scientist Development Grant 9730193N. Part of this work was presented at the 12th Annual Meeting of the Society for Free Radical Biology and Medicine (Gardner, P. R. and Gardner, A. M., Free Radic. Biol. Med. 39, (Suppl. 1) 100 (abstr.)).
- NOD
- nitric-oxide dioxygenase
- Mb
- myoglobin
- Cygb
- cytoglobin
- Ngb
- neuroglobin
- SOD
- superoxide dismutase
- MnSOD
- manganese-containing superoxide dismutase
- NCB5OR
- novel cytochrome b5 oxidoreductase
- CYPOR
- NADPH-cytochrome P450 oxidoreductase
- DPI
- diphenyleneiodonium chloride.
REFERENCES
- 1.Weber R. E., Vinogradov S. N. (2001) Physiol. Rev. 81, 569–628 [DOI] [PubMed] [Google Scholar]
- 2.Wittenberg J. B., Wittenberg B. A. (1990) Annu. Rev. Biophys. Biophys. Chem. 19, 217–241 [DOI] [PubMed] [Google Scholar]
- 3.Gardner P. R., Gardner A. M., Martin L. A., Salzman A. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner P. R., Gardner A. M., Brashear W. T., Suzuki T., Hvitved A. N., Setchell K. D., Olson J. S. (2006) J. Inorg. Biochem. 100, 542–550 [DOI] [PubMed] [Google Scholar]
- 5.Gardner P. R. (2005) J. Inorg. Biochem. 99, 247–266 [DOI] [PubMed] [Google Scholar]
- 6.Dordas C., Hasinoff B. B., Igamberdiev A. U., Manac'h N., Rivoal J., Hill R. D. (2003) Plant J. 35, 763–770 [DOI] [PubMed] [Google Scholar]
- 7.Flögel U., Merx M. W., Godecke A., Decking U. K., Schrader J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittenberg J. B., Wittenberg B. A. (2003) J. Exp. Biol. 206, 2011–2020 [DOI] [PubMed] [Google Scholar]
- 9.Brunori M., Vallone B. (2007) Cell. Mol. Life Sci. 64, 1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giuffrè A., Moschetti T., Vallone B., Brunori M. (2008) IUBMB Life 60, 410–413 [DOI] [PubMed] [Google Scholar]
- 11.Burmester T., Hankeln T. (2009) J. Exp. Biol. 212, 1423–1428 [DOI] [PubMed] [Google Scholar]
- 12.Brunori M., Giuffrè A., Nienhaus K., Nienhaus G. U., Scandurra F. M., Vallone B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8483–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smagghe B. J., Trent J. T., 3rd, Hargrove M. S. (2008) PLoS One 3, e2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentmann A., Schmidt M., Reuss S., Wolfrum U., Hankeln T., Burmester T. (2005) J. Biol. Chem. 280, 20660–20665 [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M., Gerlach F., Avivi A., Laufs T., Wystub S., Simpson J. C., Nevo E., Saaler-Reinhardt S., Reuss S., Hankeln T., Burmester T. (2004) J. Biol. Chem. 279, 8063–8069 [DOI] [PubMed] [Google Scholar]
- 16.Kawada N., Kristensen D. B., Asahina K., Nakatani K., Minamiyama Y., Seki S., Yoshizato K. (2001) J. Biol. Chem. 276, 25318–25323 [DOI] [PubMed] [Google Scholar]
- 17.Fordel E., Thijs L., Martinet W., Schrijvers D., Moens L., Dewilde S. (2007) Gene 398, 114–122 [DOI] [PubMed] [Google Scholar]
- 18.Pesce A., Bolognesi M., Bocedi A., Ascenzi P., Dewilde S., Moens L., Hankeln T., Burmester T. (2002) EMBO Rep. 3, 1146–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakasugi K., Nakano T., Morishima I. (2003) J. Biol. Chem. 278, 36505–36512 [DOI] [PubMed] [Google Scholar]
- 20.Herold S., Fago A., Weber R. E., Dewilde S., Moens L. (2004) J. Biol. Chem. 279, 22841–22847 [DOI] [PubMed] [Google Scholar]
- 21.Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M., Laufs T. L., Roesner A., Schmidt M., Weich B., Wystub S., Saaler-Reinhardt S., Reuss S., Bolognesi M., De Sanctis D., Marden M. C., Kiger L., Moens L., Dewilde S., Nevo E., Avivi A., Weber R. E., Fago A., Burmester T. (2005) J. Inorg. Biochem. 99, 110–119 [DOI] [PubMed] [Google Scholar]
- 22.Coffey M. J., Natarajan R., Chumley P. H., Coles B., Thimmalapura P. R., Nowell M., Kühn H., Lewis M. J., Freeman B. A., O'Donnell V. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8006–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallstrom C. K., Gardner A. M., Gardner P. R. (2004) Free Radic. Biol. Med. 37, 216–228 [DOI] [PubMed] [Google Scholar]
- 24.Palacios-Callender M., Hollis V., Mitchison M., Frakich N., Unitt D., Moncada S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18508–18513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halligan K. E., Jourd'heuil F. L., Jourd'heuil D. (2009) J. Biol. Chem. 284, 8539–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall C. N., Keynes R. G., Garthwaite J. (2009) Biochem. J. 419, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner P. R., Martin L. A., Hall D., Gardner A. M. (2001) Free Radic. Biol. Med. 31, 191–204 [DOI] [PubMed] [Google Scholar]
- 28.Beck von Bodman S., Schuler M. A., Jollie D. R., Sligar S. G. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 9443–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H., Larade K., Jackson T. A., Xie J., Ladoux A., Acker H., Berchner-Pfannschmidt U., Fandrey J., Cross A. R., Lukat-Rodgers G. S., Rodgers K. R., Bunn H. F. (2004) J. Biol. Chem. 279, 30316–30325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou Y., Maillett D. H., Eich R. F., Olson J. S. (2002) Biophys. Chem. 98, 127–148 [DOI] [PubMed] [Google Scholar]
- 31.Burmester T., Weich B., Reinhardt S., Hankeln T. (2000) Nature 407, 520–523 [DOI] [PubMed] [Google Scholar]
- 32.Gardner P. R., Costantino G., Szabó C., Salzman A. L. (1997) J. Biol. Chem. 272, 25071–25076 [DOI] [PubMed] [Google Scholar]
- 33.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 34.Gardner P. R. (2008) Methods Enzymol. 436, 217–237 [DOI] [PubMed] [Google Scholar]
- 35.Burmester T., Ebner B., Weich B., Hankeln T. (2002) Mol. Biol. Evol. 19, 416–421 [DOI] [PubMed] [Google Scholar]
- 36.Gardner P. R., Gardner A. M., Hallstrom C. K. (2004) Methods Mol. Biol. 279, 133–150 [DOI] [PubMed] [Google Scholar]
- 37.Becana M., Klucas R. V. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 7295–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igamberdiev A. U., Bykova N. V., Hill R. D. (2006) Planta 223, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 39.Galaris D., Cadenas E., Hochstein P. (1989) Arch. Biochem. Biophys. 273, 497–504 [DOI] [PubMed] [Google Scholar]
- 40.Gardner P. R., Costantino G., Salzman A. L. (1998) J. Biol. Chem. 273, 26528–26533 [DOI] [PubMed] [Google Scholar]
- 41.Tschank G., Sanders J., Baringhaus K. H., Dallacker F., Kivirikko K. I., Günzler V. (1994) Biochem. J. 300, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner M. C., Murray C. J., Klinman J. P. (1989) Biochemistry 28, 4656–4664 [DOI] [PubMed] [Google Scholar]
- 43.Corpe C. P., Tu H., Eck P., Wang J., Faulhaber-Walter R., Schnermann J., Margolis S., Padayatty S., Sun H., Wang Y., Nussbaum R. L., Espey M. G., Levine M. (2010) J. Clin. Invest. 120, 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckman J. S., Koppenol W. H. (1996) Am. J. Physiol. 271, C1424–C1437 [DOI] [PubMed] [Google Scholar]
- 45.Fago A., Mathews A. J., Moens L., Dewilde S., Brittain T. (2006) FEBS Lett. 580, 4884–4888 [DOI] [PubMed] [Google Scholar]
- 46.Zhu H., Qiu H., Yoon H. W., Huang S., Bunn H. F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H. S., Loughran P. A., Rao J., Billiar T. R., Zuckerbraun B. S. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 295, G146–G152 [DOI] [PubMed] [Google Scholar]
- 48.Chan T. S., Shangari N., Wilson J. X., Chan H., Butterworth R. F., O'Brien P. J. (2005) Free Radic. Biol. Med. 38, 867–873 [DOI] [PubMed] [Google Scholar]
- 49.Gardner A. M., Martin L. A., Gardner P. R., Dou Y., Olson J. S. (2000) J. Biol. Chem. 275, 12581–12589 [DOI] [PubMed] [Google Scholar]
- 50.Gardner P. R., Gardner A. M., Martin L. A., Dou Y., Li T., Olson J. S., Zhu H., Riggs A. F. (2000) J. Biol. Chem. 275, 31581–31587 [DOI] [PubMed] [Google Scholar]
- 51.Rhee S. G., Chae H. Z., Kim K. (2005) Free Radic. Biol. Med. 38, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 52.Moreau S., Puppo A., Davies M. J. (1995) Phytochemistry 39, 1281–1286 [DOI] [PubMed] [Google Scholar]
- 53.Bursey E. H., Poulos T. L. (2000) Biochemistry 39, 7374–7379 [DOI] [PubMed] [Google Scholar]
- 54.Lad L., Mewies M., Raven E. L. (2002) Biochemistry 41, 13774–13781 [DOI] [PubMed] [Google Scholar]
- 55.Macdonald I. K., Badyal S. K., Ghamsari L., Moody P. C., Raven E. L. (2006) Biochemistry 45, 7808–7817 [DOI] [PubMed] [Google Scholar]
- 56.Weiland T. R., Kundu S., Trent J. T., 3rd, Hoy J. A., Hargrove M. S. (2004) J. Am. Chem. Soc. 126, 11930–11935 [DOI] [PubMed] [Google Scholar]
- 57.Trandafir F., Hoogewijs D., Altieri F., Rivetti di Val Cervo P., Ramser K., Van Doorslaer S., Vanfleteren J. R., Moens L., Dewilde S. (2007) Gene 398, 103–113 [DOI] [PubMed] [Google Scholar]
- 58.de Sanctis D., Dewilde S., Pesce A., Moens L., Ascenzi P., Hankeln T., Burmester T., Bolognesi M. (2004) J. Mol. Biol. 336, 917–927 [DOI] [PubMed] [Google Scholar]
- 59.Sugimoto H., Makino M., Sawai H., Kawada N., Yoshizato K., Shiro Y. (2004) J. Mol. Biol. 339, 873–885 [DOI] [PubMed] [Google Scholar]
- 60.Sharp K. H., Mewies M., Moody P. C., Raven E. L. (2003) Nat. Struct. Biol. 10, 303–307 [DOI] [PubMed] [Google Scholar]
- 61.Nakatani K., Okuyama H., Shimahara Y., Saeki S., Kim D. H., Nakajima Y., Seki S., Kawada N., Yoshizato K. (2004) Lab. Invest. 84, 91–101 [DOI] [PubMed] [Google Scholar]
- 62.Cao M., Westerhausen-Larson A., Niyibizi C., Kavalkovich K., Georgescu H. I., Rizzo C. F., Hebda P. A., Stefanovic-Racic M., Evans C. H. (1997) Biochem. J. 324, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hall C. N., Garthwaite J. (2009) Nitric Oxide 21, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gödecke A. (2006) Cardiovasc Res. 69, 309–317 [DOI] [PubMed] [Google Scholar]
- 65.Fordel E., Geuens E., Dewilde S., De Coen W., Moens L. (2004) IUBMB Life 56, 681–687 [DOI] [PubMed] [Google Scholar]